Abstract
Quiescent hair follicle (HF) bulge stem cells (SCs) differentiate to early progenitor (EP) hair germ (HG) cells, which divide to produce transit-amplifying (TA) matrix cells. EPs can revert to SCs upon injury, but whether this de-differentiation occurs in normal HF homeostasis (hair cycle), and the mechanisms regulating both differentiation and de-differentiation are unclear. Here we use lineage tracing, gain of function, transcriptional profiling, and functional assays to examine the role of observed endogenous Runx1 level changes in the hair cycle. We find that forced Runx1 expression induces hair degeneration (catagen) and simultaneously promotes changes in the quiescent bulge SC transcriptome towards a cell-state resembling the EP HG fate. This cell-state transition is functionally reversible. We propose that SC differentiation and de-differentiation are likely to occur during normal HF degeneration and niche restructuring in response to changes in endogenous Runx1 levels associated with SC location with respect to the niche.
Keywords: hair follicle stem cells, Runx1, skin, reversible fate, catagen, target genes
Introduction
Mammalian development and adult homeostasis are generally modeled as irreversible transitions between different cell states (Waddington, 1957). De-differentiation can be achieved by nuclear transfer or forced expression of master transcription factors (Pournasr et al., 2011). Germ line transit-amplifying (TA) cells revert to stem cells (SCs) in the adult mouse and fly testis (Simons and Clevers, 2011; Spradling et al., 2011). In mammals, somatic TA cells, and sometimes even terminally differentiated lineages (TDL), can de-differentiate to SCs in injury or cancer (Porrello et al., 2011; Schwitalla et al., 2013; Yanger et al., 2013). However, within normal un-injured somatic mammalian tissues, it is unclear to what extent distinct molecular and functional cell-states may be reversible.
The adult HF is composed largely of epithelial cells that form: (1) a permanent region (bulge) housing the HF SCs; (2) the temporary region (bulb) containing TA cells (matrix) and the TDL (inner root sheath (IRS) and hair core/shaft) (Blanpain, 2010). The outer-most root sheath (ORS) is contiguous with the bulge SC layer. The dermal papillae (DP), is a mesenchymal signaling center at the base of the bulb important for SC activation. HFs undergo cyclic phases of morphological remodeling known as the hair cycle (Blanpain, 2010). The hair cycle phases are: growth (anagen) when the bulge generates a new bulb, regression (catagen) when bulb cells die by apoptosis, and rest (telogen) when the bulge is quiescent (Muller-Rover et al., 2001).
In telogen the bulb is replaced by the HG, which arises from quiescent bulge cells (Ito et al., 2004; Zhang et al., 2009). The HG fate is distinct from matrix and bulge fates, as shown by gene expression (Greco et al., 2009). Moreover, HG cells proliferate rapidly and then are lost from the dish (Greco et al., 2009), and at least the late-stage HG cells arising directly from bulge cells that migrate at telogen do not self-renew (Zhang et al, 2009). Thus, the HG acts as an “early progenitor” (EP), defined here as the first step of a bulge SC embarked on the path of differentiation towards a TA matrix cell. Bulge SCs self-renew at anagen by rare, symmetric divisions with respect to the basement membrane (Zhang et al., 2009; Zhang et al., 2010).
Some of the ORS cells that migrated from the bulge at late anagen/early catagen remain below the bulge to eventually make a “new” HG and some move up to form a “new” bulge (Hsu et al, 2011). Importantly, it is not known whether the bulge SCs displaced into the ORS simply change location, or actually differentiate into HG cells and then de-differentiate upon returning to the new bulge. It is known that in response to injury, such as hair plucking or LASER ablation, hair germ (HG) cells can de-differentiate to bulge SCs (Ito et al., 2004; Rompolas et al., 2013). Whether this plasticity of fate is employed during normal hair homeostasis, the significance of a potential flexibility in cell fate in the absence of injury, and a potential molecular mechanism remain a mystery.
Previously we showed that Runx1, a transcription factor from the Runt family (Blyth et al., 2005) is highly expressed in HG cells and is essential for their activation/proliferation and subsequent anagen onset (Hoi et al., 2010; Lee et al., 2013; Osorio et al., 2008; Scheitz et al., 2012). Runx1 is even more highly expressed in the epithelial strand at late catagen (Fig. 1A and (Hsu et al., 2011)), suggesting a possible role at this stage of the hair cycle.
Figure 1. Runx1+ cells in the ORS at catagen generate new bulge and hair germ cells.
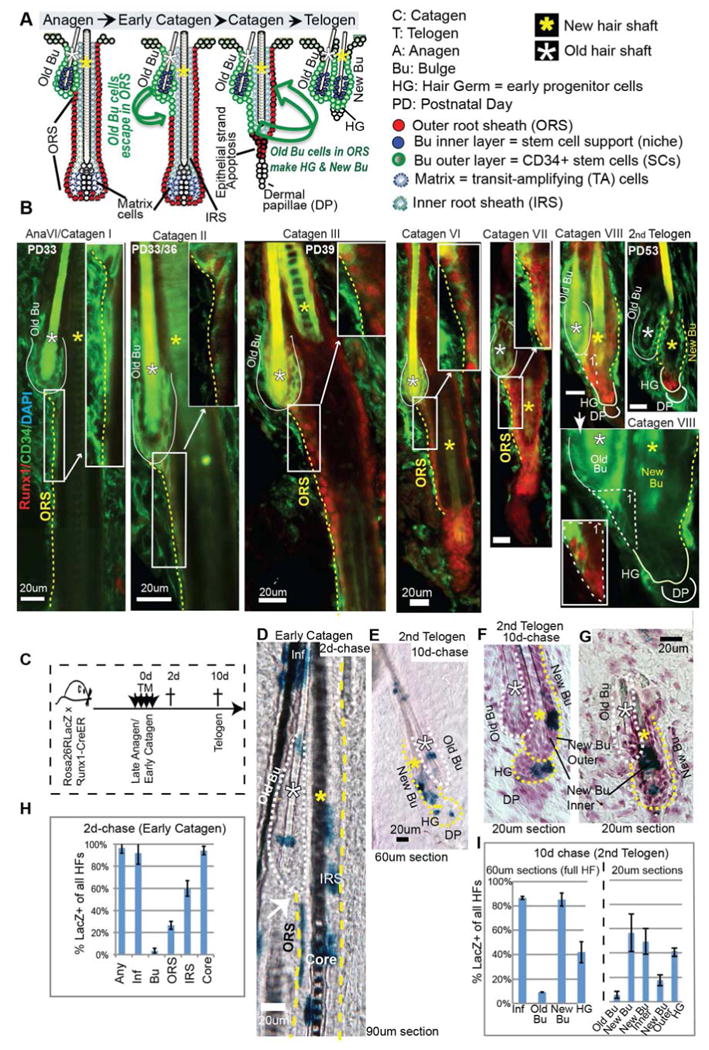
(A) Cartoon representation of cellular dynamics in the hair follicle, show the bulge SCs displaced into the outer root sheath (ORS) at late anagen/early catagen, which survive apoptosis and return upward to form the new bulge and the HG. (B) Skin sections stained for Runx1 and bulge SC marker CD34 show cells in the ORS with low CD34 and high Runx1 expression starting at Catagen II-III. Regions of the ORS with Runx1/CD34 overlap are enlarged in the upper right corner (white box). Note that Runx1 is up-regulated in the most bulge-proximal CD34+ ORS cells that lost contact with the inner layer. The ORS on the bulge side is made of cells placed immediately right to the dotted yellow lines. (C) Scheme of tamoxifen (TM) injection to genetically label and trace Runx1 expressing cells from early catagen to telogen. (D, E) Thick sections encompassing full hair follicles (HFs) stained with X-Gal to detect LacZ+ cells. (F, G) Same as D but in thin sections counterstained with hematoxylin to delineate the inner and the outer bulge layers. (H, I) Counts of LacZ+ HFs with distribution of label in each compartment. For abbreviations see panel 1A.
Here we provide experimental evidence suggesting that increase in endogenous Runx1 levels during normal catagen has a dual function: (1) to induce degeneration of TA and TDL lineages; and (2) to promote a reversible differentiation of bulge SCs to HG EPs. This reversibility may ensure a proper balance between SC and EP populations during niche restructuring in normal tissue homeostasis.
Results
ORS cells that once expressed Runx1 contribute to both “new” bulge and hair germ formation
Skin sections stained for endogenous Runx1 and CD34 (a bulge SC marker), showed that Runx1 became detectable in CD34+ ORS cells just below the bulge by catagen II (Fig. 1B). Based on location and CD34 expression, this ORS region must contain the migrated “old” bulge cells previously traced via H2B-GFP and Lgr5-CreER (Hsu et al., 2011). CD34 levels were generally higher in the old bulge than the ORS, while Runx1 showed the opposite trend (Fig. 1B). At catagen VIII, the forming new bulge initially showed high Runx1/low CD34 levels, but switched to low Runx1/high CD34 by the 2nd telogen. These data suggest that quiescent bulge cells that have migrated into the ORS up-regulate Runx1 at early catagen. By the 2nd telogen, if they become “new” bulge cells they down-regulate Runx1 and if they become HG they keep Runx1.
To test this, we marked Runx1+ ORS cells at early 2nd catagen and traced their lineage. Runx1-CreER;Rosa26RLacZ adult mice were injected with tamoxifen (TM) for 4 days starting at the 6th day after their skin turned from pink to black, indicative of anagen (∼PD32) (Fig. 1C). We used thick frozen skin sections that encompass full HFs (90μm for catagen and 60μm for telogen) (Fig. 1C-I). We scored the compartments of each HF as positive or negative for LacZ+ cells, irrespective of the total number of LacZ+ cells in the compartment, to rule out proliferation effects.
Two days after the last TM injection (catagen, 2d-chase), we quantified the initial labeling of Runx1-expressing cells and found LacZ+ cells in >95% of HFs, in a pattern expected from endogenous Runx1 expression (Fig. 1D, H). In this stage, endogenous Runx1 is expressed weakly in the bulge, stronger in the ORS, and strongest in bulb cells including the differentiated lineages (Fig. 1B and S1A), and also in the infundibulum (Inf) cells (Scheitz et al., 2012). About 60% of HFs showed LacZ+ cells in the IRS (a terminally differentiated lineage –TDL), and >90% of HFs showed LacZ+ cells in the hair shaft/core (another TDL) and Inf (Fig. 1D). None of these lineages are expected to contribute normally to bulge SCs (Blanpain, 2010; Jaks et al., 2010) and HG regeneration (Greco et al., 2009; Hsu et al., 2011).
The lineages expected to contribute to the “new” bulge and HG are the “old” bulge and ORS (Hsu et al., 2011), which showed LacZ+ cells in ∼3% and ∼30% of all HFs, respectively, at 2d-chase (Fig, 1D, H and S1B). Therefore, if Runx1-expressing cells of the ORS at early first catagen contribute to the “new” bulge and HG we expected that after 10d-chase (2nd telogen) a maximum of ∼33% of HFs would contain LacZ+ cells in the “new” bulge and HG. This appears to be the case for HG, since we found ∼30% of HFs with LacZ+ cells in the HG at 10d-chase (Fig. 1E, F, I)
Surprisingly, ∼ 80% of HFs were labeled in the “new” bulge at 10d-chase, suggesting additional contribution from other HF compartments besides the ORS and “old” bulge (Fig. 1I, left). The bulge has two layers, the outer layer containing SCs and the inner layer serving as the SC niche (Hsu et al., 2011). To distinguish these we analyzed thin (20μm) sections and indeed we found that only ∼20% of HFs in thin sections (corresponding to ∼30% of full HFs in thick sections) had LacZ+ cells in the outer bulge SC layer (Fig. 1F, I right), as expected from their known origin in the ORS. These data confirm that in addition to contribution to HG, Runx1+ ORS cells contribute to the new bulge. LacZ+ cells were present in the inner bulge in ∼50% of HF in thin sections, which corresponds to ∼80% of full HFs in thick (60μm) sections (Fig. 1F, G and I, right panel), which were most likely originating in HF TDL. Moreover, the data suggest that Runx1+ORS cells also contribute to new bulge and germ in the 1st telogen (Fig. S1C, D), suggesting that the same reorganization of the SC niche takes place repeatedly during homoestasis.
Runx1 expression is down-regulated in the hair germ as it begins proliferation to make the matrix at anagen and up-regulated in apoptotic matrix cells
These new data raised the possibility that Runx1 up-regulation in the migrated bulge cells in early catagen ORS that act as precursors of the HG may promote a bulge fate transition towards HG cells. Conversely, Runx1 down-regulation in the ORS cells that returned to the “new” bulge may revert them to a bulge fate. To further investigate this, we extended our previously reported analysis of Runx1 expression (Osorio et al., 2008) in bulge SC, HG EP, and matrix TA cell compartments throughout the hair cycle. Quantification of Runx1 protein by immuno-staining revealed striking changes in levels in different compartments (Fig. 2A and S1A, A′).
Figure 2. Transgenic mouse to model Runx1 dynamic expression levels throughout the hair cycle.
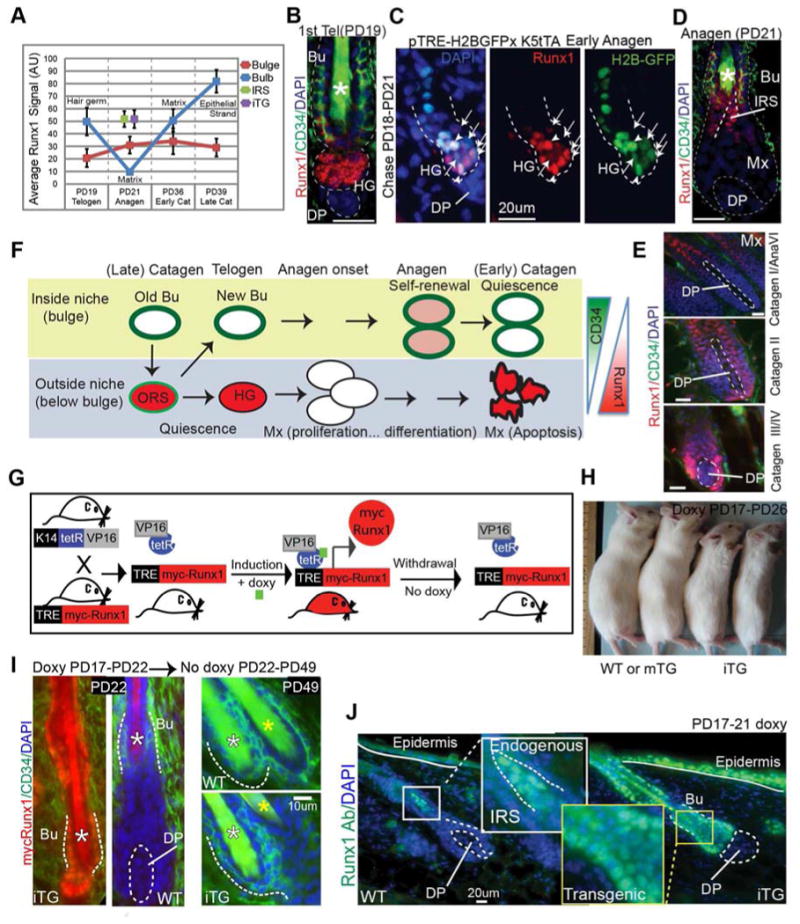
(A) Quantification of endogenous Runx1 levels throughout the hair cycle using immunofluorescence microscopy. See also Fig. S1A, A′ and 1B. Data in purple represents the Runx1 levels quantified in the transgenic mouse model described in Fig. 2G-J. (B, D, E) Skin sections stained for endogenous Runx1 and CD34 show abrupt down-regulation of Runx1 in the transition from the hair germ (HG) to the matrix (Mx) at anagen followed by up-regulation in matrix and lower bulb cells at catagen. (C) Skin sections from TRE-H2BGFP;K5-tTA mice doxy chased (PD18-PD21) show cells with an apparent correlation between H2BGFP and Runx1 levels (arrowheads indicate undivided cells, while arrows point to cells that divided and diluted H2BGFP during the 3-day chase). See also Fig. S2A for mRNA levels in divided versus un-divided cells isolated by FACS. (F) Cartoon summarizing Runx1 expression levels in different compartments throughout the hair cycle. (G) Scheme of transient mycRunx1 induction and withdrawal in transgenic mice responsive to doxy. (H) Mice induced for 9-days with doxy are small and die within a few days. mTG mice are normal size and survive. (I) Skin sections from WT and Runx1iTG at stages indicated stained for mycRunx1 and CD34. (J) Skin sections stained side-by-side for Runx1 and photographed at the same exposure. Note comparable transgenic Runx1 levels in the bulge (Bu) and HG to endogenous levels in the forming IRS quantified in Fig. 2A and S1A′. See Fig. 1A for abbreviations.
In the bulge, Runx1 protein levels are generally low (Fig. 2A), although necessary for normal bulge proliferation at anagen (Hoi et al., 2010). The mRNA level is higher in dividing bulge-sorted cells compared to non-dividing bulge-sorted cells (Fig. S2A, two asterisks). Bulge cells migrated in the upper ORS at catagen (Fig. 1B and 2A)), and at the bulge/HG junction at telogen express high Runx1 protein levels ((Osorio et al., 2008) and Fig. 2B).
In the HG, Runx1 is necessary for proliferation presumably by down-regulating cyclin dependent kinase inhibitors (CKIs) (Hoi et al., 2010; Lee et al., 2013; Osorio et al., 2008). Thus, we expected that Runx1 would remain high in dividing HG cells at anagen onset. Surprisingly, some HG cells with reduced H2B-GFP retention after 3-d doxycline chase on TRE-H2BGFP;K5-tTA mice at anagen onset, indicative of 1-2 divisions (Zhang et al., 2009), showed reduced Runx1 protein and mRNA levels (Fig. 2C, arrows, Fig. S2A, one asterisk). The expression profiles of these divided HG cells at anagen onset resemble the matrix signature (Zhang et al., 2009), while the profiles of quiescent HG cells at telogen (prior to divisions) resemble the bulge (Greco et al., 2009). These observations suggest that as HG EPs begin to divide they rapidly transit to matrix TA cells and that Runx1 levels quickly decrease during this transition.
In the matrix, Runx1 protein remained undetectable throughout anagen but became suddenly high in early catagen, when these cells are known to undergo apoptosis (Fig. 2A, D, E and S1A, A′). Runx1 levels in matrix cells increased even more as catagen progressed, and reached its highest level in the regressing apoptotic epithelial strand in late catagen (Fig. 2A, 1B and S1A, A′).
The dynamics of Runx1 expression are summarized in Fig. 2F).
Generation of tet-inducible transgenic mice to modulate Runx1 expression levels
To directly test a potential role of observed endogenous Runx1 level changes in the hair cycle, we generated tetracycline inducible transgenic mice. The K14-rtTA construct (“on” in skin and other epithelia) will drive a newly generated TRE-mycRunx1 allele (see Experimental Procedures and Supplementary Information (SI)) upon doxycycline (doxy) induction. The K14-rtTA mice previously drove physiologically meaningful levels of another transcription factor in the skin (Nguyen et al., 2006). Within 4-6 hrs of doxy induction mycRunx1 expression would turn on, while withdrawing doxy would revert expression to undetectable levels (Fig. 2G).
As expected, the doxy-induced TRE-mycRunx1;K14-rTA double transgenic mice (Fig. 2H and S2B) turned on mycRunx1 nuclear expression, as shown by anti-myc staining (Fig. 2I and S2C). Withdrawing doxy-food shut-down mycRunx1 expression by PD49 (Fig. 2I, right). Two TRE-mycRunx1 mouse lines proved fully inducible by staining of different body regions (not shown), and we refer to them here as Runx1iTG. Two mouse lines showed mycRunx1 expression in skin patches throughout the mouse back skin, and we refer to them here as mosaic Runx1mTG lines. Double transgenic (TRE-MycRunx1;K14-rtTA) littermates without doxy and all single transgenic littermates showed no detectable staining, and are designated as wild type (WT) (Fig. 2I and S2C). Runx1iTG mice began losing weight 6-days after doxy induction (Fig. 2H), and died within 9-12 days, but they recovered if we withdrew doxy food after 5 days of induction (Fig. 6B). Runx1mTG mice were viable and we used them for long-term studies along with skin transplants from Runx1iTG (Fig. 2H and 4B, C). All the experiments shown here were performed at least twice with nmouse≥2-3 of each genotype per experiment.
Figure 6. Bulge stem cell fate transition induced by Runx1 is functionally reversible.
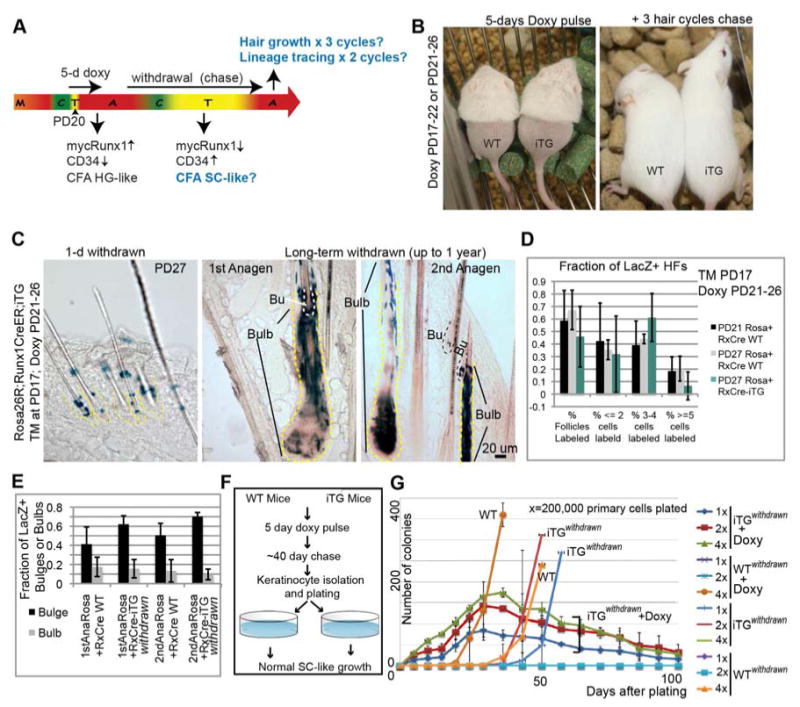
(A) Scheme of doxy induction and withdrawal to assess reversibility of Runx1-induced effects on bulge-cell function. (B) Mice on doxy food (left) were gently removed hair on their lower back to monitor hair growth without causing activation of growth by injury. They regrew hair after doxy withdrawal (right) for 3 consecutive cycles (compare with Fig. 4B). (C) Skin sections from Runx1-CreER;Rosa26R;Runx1iTG mice injected with TM at PD17, doxy treated from PD21-PD26, and sacrificed at PD27 (left panel) show localization of LacZ+ cells to the bulge. Middle and right panels show LacZ+ cells in the differentiated HF compartment (bulb) and in the SC compartment (Bu) at the 2 subsequent anagen stages after TM injection. (D, E) LacZ+ HF counts of images collected from all stages in (C), show similar patterns of LacZ contribution to HF regeneration in iTG and WT skin. See also Fig. S7. (F) Scheme of doxy treatment and withdrawal for colony formation assays (CFA) to examine the growth potential (self-renewal ability) of cells that transiently experienced mycRunx1 expression. (G) CFA of cells from (F) show recovery of SC-like behavior in cells from mice in which doxy was withdrawn for several weeks after the initial 5-days of doxy treatment (note iTGwithdrawn vs. WT plots). Keratinocytes from these mice, cultured with doxy upon plating, displayed once again the behavior described previously for the HG (rapid proliferation followed by death; see plots marked iTGwithdrawn +doxy). See also Fig. S7.
Figure 4. Long-term Runx1 induction in bulge SCs confer a short-lived progenitor cell behavior both in vivo and in vitro.
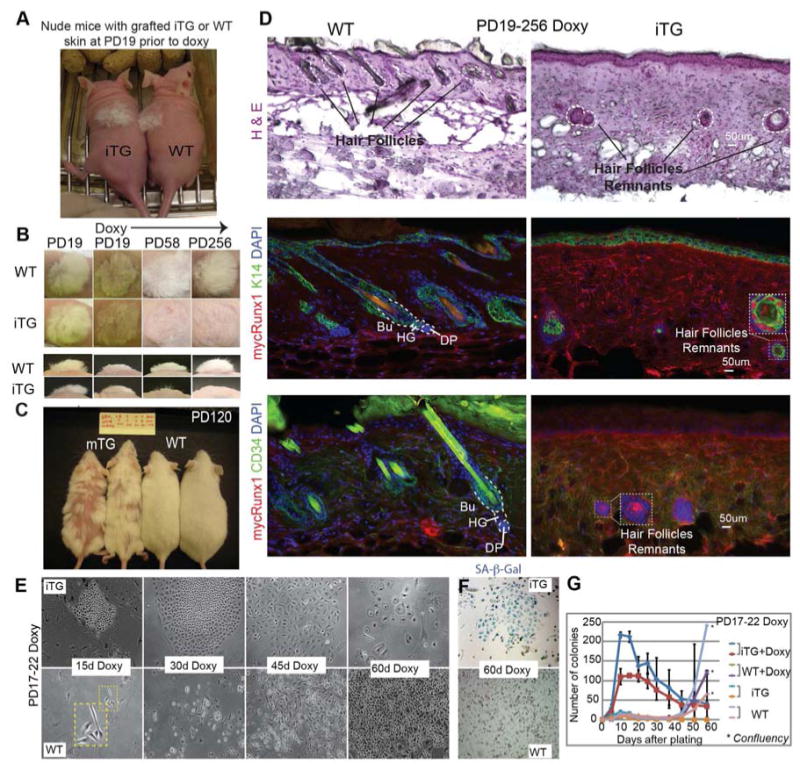
(A) Grafted newborn Runx1iTG skin onto nude mice, to bypass lethality of Runx1 induction, prior to doxy feeding at the equivalent PD19. (B) Grafted areas on nude mice at times post-induction as indicated. Note permanent hair loss in Runx1iTG skin. (C) Runx1mTG mice induced with doxy for >3 months show permanent hair loss in a patchy manner, as expected from mosaic expression of Runx1. (D) Skin sections from (B) sectioned and stained as indicated show lack of HFs in Runx1iTG skin. Circles show rare remnants of HF cells, which are stained positive for mycRunx1 and K14, but are negative for the SC marker CD34. MycRunx1 staining shows higher dermal background in Runx1iTG than WT, possibly due to increased cellularity in the former. (E-G) Keratinocytes isolated from PD17-PD22 doxy treated mice were plated on irradiated mouse embryonic fibroblast in low Ca2+ E medium. Colonies were counted every week; representative phase contrast images are shown in (E), and the overall counts are shown in (G). See also S6A-B. Note diminished colony size and flat cell morphology indicative of senescence at 60 days in Runx1iTG cells. (F) Endogenous SA-β-Gal staining indicates senescence of Runx1iTG cells by 60 days. See also Fig. S6C.
To achieve near physiological Runx1 protein levels, and avoid artifacts of excessively high expression, we chose the doxy concentration of several tested (Experimental Procedures) that gave us consistently detectable mycRunx1 expression in the bulge, which appeared comparable to endogenous Runx1 expression in the IRS, HG and early catagen matrix (Fig. 2J, quantified in 2A, S1A′).
Runx1 is insufficient for anagen onset but sufficient to induce catagen
Runx1 is necessary for HG activation and proliferation and for timely anagen onset (Osorio et al., 2008), so we tested whether forced expression of Runx1 was sufficient to induce anagen. We induced Runx1iTG during the 2nd telogen, a period of prolonged quiescence in most mouse strains but found no evidence of anagen onset, as shown by normal telogen morphology (Fig. 3A, F-blue) and lack of BrdU staining in Runx1iTG skin (Fig. S2D). These results may suggest that the presence of inductive anagen signals, not present in the skin at 2nd telogen are required in addition to Runx1 for normal anagen onset. Forced Runx1 expression in 2nd telogen did, however, result in CD34 down-regulation (Fig. S4C).
Figure 3. Runx1 expression is sufficient for catagen implementation in bulb cells but requires additional anagen inductive signals to promote stem cell proliferation.
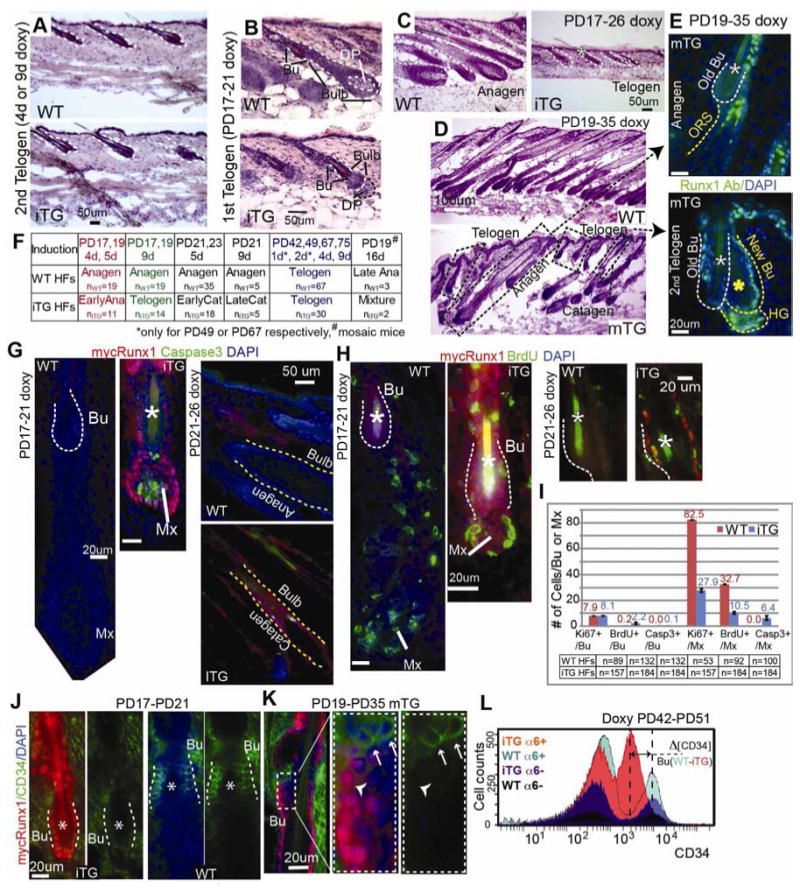
(A-D) Hematoxylin and Eosin (H&E) stained skin sections from doxy-induced mice at stages indicated. WT were littermate controls. Note normal telogen morphology in Runx1iTG and WT in (A). Note Runx1iTG HFs with smaller bulb and lack of hair shaft indicative of earlier anagen, with a DP unenclosed by the epithelial cells in (B) followed by return to telogen with a single hair shaft (*) in (C). (D) HFs in the 16-d induced mosaic Runx1mTG skin showed variable hair cycle stages (dotted rectangles). Corresponding stages are shown in (E), with Runx1 Ab staining (green). Note endogenous Runx1 expression pattern in anagen HF (top) and transgenic pattern (with a positive Bu and interfollicular epidermis) in late catagen/telogen HF (bottom). Note the presence of two hair shafts (*) indicating this HF had been through at least a portion of the 1st adult anagen. White asterisk marks the old shaft (club hair) in the old bulge and yellow asterisk marks the new shaft. (F) Summary of short-term doxy induction and results. (G, H) Staining for markers indicated and quantified in (I) reveal increased BrdU incorporation (2-hr pulse) in Runx1iTG bulge (Bu) and massive apoptosis in the matrix (Mx) and bulb. (J, K) Skin sections from Runx1iTG and Runx1mTG mice show down-regulation of CD34 by immunostaining. Note mosaic expression of mycRunx1 in bulge (Bu) in (K), correlating with lack of CD34 expression. (L) FACS histogram shows overlay of 4 populations isolated from Runx1iTG and WT littermate mouse skin when bulge SCs were at catagen/telogen. Note several fold decrease in CD34 level in CD34+/α6+ Runx1iTG bulge cells compared to WT bulge. See also Fig. S3 and S4.
We next induced mycRunx1 elevation in the short and synchronous 1st quiescence period (PD17∼PD20) where the proliferative signals arise shortly after (PD20∼PD21). We doxy-fed Runx1iTG mice for 4-5 days starting in quiescence at PD17, 19 and examined the HF morphology in skin sections stained with Hematoxylin and Eosin (H&E) (Fig. 3B, F-red). In this Fvb mouse strain, WT skin was already at almost full anagen by PD21, showing a developed bulb and a newly forming hair shaft (Fig. 3B, top). Contrary to our expectation that anagen would be accelerated, Runx1iTG HFs were behind in anagen progression by 4 days of induction relative to controls (Fig. 3B, bottom), and prematurely returned to telogen without producing a new hair shaft by 9-days (Fig. 3C, F-green). Mosaic Runx1mTG induced for 16-days at PD19 showed an abnormal mixture of different HF cycle stages (Fig. 3D, F#). HFs at morphological stages comparable with WT littermates (anagen) showed the endogenous Runx1 staining pattern (Fig 3E, top). In contrast, HFs that expressed Runx1 at elevated levels in a transgenic pattern were found prematurely at catagen or the 2nd telogen, as indicated by HF morphology and the presence of two hair shafts (Fig. 3E, bottom). We attribute this phenotype to the mis-expression of Runx1 not only in the quiescent bulge, but also in the matrix and the lower ORS, where Runx1 is normally not expressed during anagen until these cells begin apoptosis at catagen onset.
Runx1iTG induction for 5- and 9- days in early- and mid-anagen (PD21, PD23), resulted in catagen morphology (Fig. 3F, black, G right panel and S3C). This suggested that Runx1 is sufficient to induce catagen at any stage of anagen, and may explain why endogenous Runx1 is suddenly turned on at its highest levels in the lower bulb in catagen.
Terminal differentiation markers analysis (GATA3 for the IRS, and AE13 for the hair shaft), revealed complete lack of TDL in Runx1iTG HFs induced at PD17/19 (Fig. S3A). Apoptosis (Caspase-3) and proliferation (BrdU injection 2-hr prior to sacrifice) analysis revealed massive apoptosis in the matrix (Fig. 3G, and S3C).
In the bulge, mycRunx1 down-regulated CD34 (Fig. 3J-L, S4), and also increased proliferation (Fig. 3H, S3B), as we expected would occur in conjunction with inductive proliferative signals. CD34 reduction was cell autonomous as suggested by mosaic analysis (Fig. 3K), which ruled out a hair cycle stage effect of Runx1 on bulge-marker CD34 expression. Quantification of BrdU, Ki67 and Caspase-3 for the PD17-21 induction is provided in Fig. 3I. Note that the difference in hair cycle stage observed here cannot account for the differences in BrdU incorporation. This result implied that Runx1 elevation might promote the bulge cell transition to a HG-like cell state that is CD34- and more responsive to proliferative anagen inducing signals present in the skin at the 1st telogen.
To distinguish direct versus systemic effects, we induced mycRunx1 expression in TRE-mycRunx1;K14-rtTA keratinocyte cell lines and in transiently transfected CD34+/a6+ bulge and CD34-/α6+ non-bulge sorted cultured cells (Fig. S3D-F). Runx1 elevation resulted in a brief burst of BrdU incorporation, followed by a plunge below WT control levels, accompanied by increased apoptosis (Fig. S3E-F). These data demonstrate a cell-autonomous effect on apoptosis and proliferation of keratinocytes by high Runx1 levels.
Long-term functional assays show that stem cells with continuous high Runx1 expression switch to a short-lived progenitor behavior
Next we performed long-term functional assays to ask if mycRunx1-expressing bulge cells continue to behave in vivo and in vitro like long-lived SCs or switch to a shortlived EP cell behavior.
We employed both skin grafts of the Runx1iTG mice onto nude mice to avoid lethality after 9-day induction and the two Runx1mTG mouse lines, which appeared healthy in long-term induction. Transplanted nude mice were fed doxy chow at the equivalent PD19, after shaving the original hair coat, which permanently arrested new hair coat growth (Fig. 4A, B). Both Runx1mTG mice and grafted Runx1iTG skin began to lose the hair coat (in patches for Runx1mTG) within a few weeks of induction, and remained bald for the entire ∼1-year duration of the experiment (Fig. 4A-C). Upon inspection of sections from the long-term Runx1iTG skin, rare degenerated HF remnants were detected in Runx1iTG dermis, which showed epithelial K14 expression but lacked the SC marker CD34 (Fig. 4D). Thus, long-term expression of mycRunx1 in the HF in vivo inhibited the long-lived regenerative potential of bulge SCs and resulted in complete loss of the hair follicle, thereby causing a behavior expected of short-lived progenitors.
Next we turned to colony formation assays (CFA), an in vitro functional measure of self-renewal ability (Blanpain, 2010). Bulge SC cells are slow in seeding colonies but then they expand continuously whereas HG cells are fast in seeding colonies but shortly exhaust their growth potential (Greco et al., 2009). Primary keratinocytes isolated from Runx1iTG skin of 5-day doxy-fed adult mice (PD17-PD22) displayed rapid initial proliferation with increased colony number and size, followed in ∼40 days by exhaustion and cell senescence, being mostly lost from the dish by ∼60 days (Fig. 4E-G, S6A-C), therefore behaving as short-lived HG EPs. As expected, WT cells showed the typical growth pattern of adult skin keratinocytes exhibited by long-lived SCs (Fig. 4E-G and S6A-C).
High Runx1 expression in bulge cells promotes a massive change in gene expression signature that largely resembles the early progenitor hair germ
Next we analyzed the gene expression changes that occur in bulge cells upon mycRunx1 induction, to further test if the bulge cells are converting to a HG expression signature. Supporting this the mRNA of CD34 and several other known HF SC markers was down-regulated in the Runx1iTG CD34+/α6-integrin+ bulge cells, sorted within 1-5 days of doxy induction (Fig. 5A, A′).
Figure 5. Runx1 initiates a hair germ early progenitor fate in bulge cells.
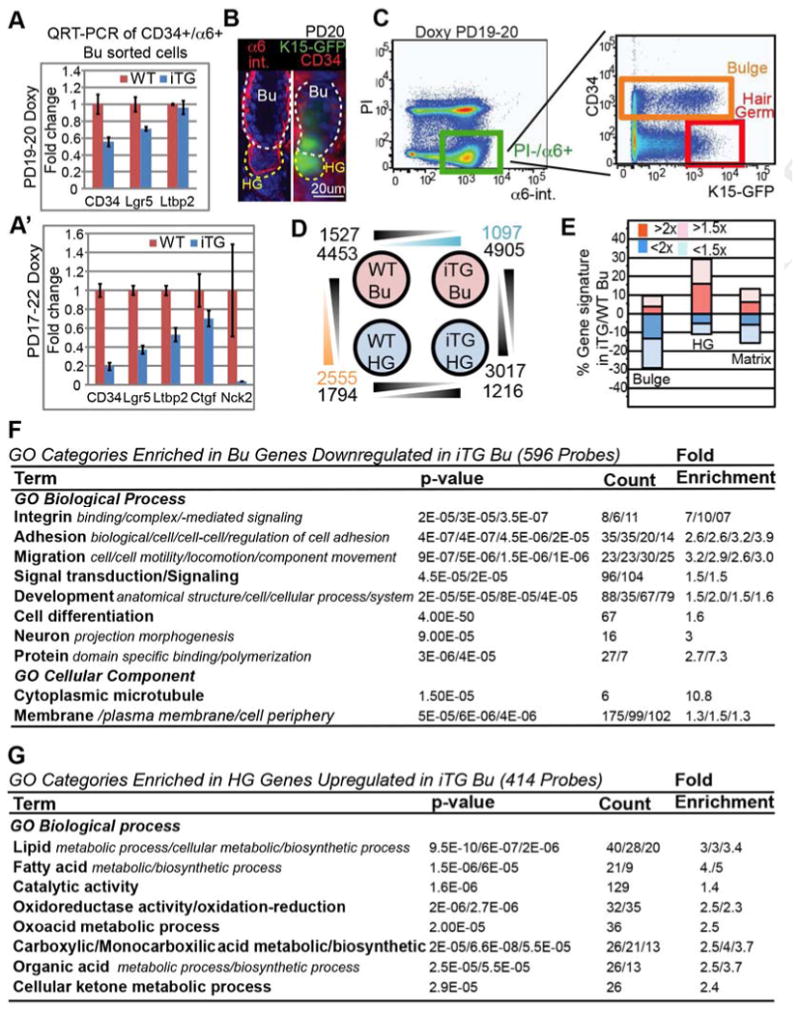
(A, A′) QRT-PCR of sorted bulge cells show robust down-regulation of several bulge stem cell markers tested. (B) Skin sections from K15-EGFP;Runx1iTG mice at PD20 show telogen morphology and GFP expression in the HG and in some bulge (Bu) cells. (C) FACS dot plot show sorting gates to isolate bulge (as CD34+/α6+/GFP+/- cells) and HG cells as (CD34−/α6+/GFP+). The cut-off for HG GFP was for the brightest 1/3 of the cells to avoid potential contamination from dimmer cells occasionally found in epidermis and sebaceous gland (not shown). (D) Microarray analysis of wild type (WT) and iTG bulge (Bu) and hair germ (HG) sorted populations; shown are number and probes which decrease or increase among each paired comparison. (E) Comparison of bulge iTG/WT by microarrays shows changes in bulge (Bu), hair germ (HG) and matrix gene signature. Note down-regulation of bulge signature and up-regulation of HG signature in iTG bulge. The effect on matrix signature was neutral. See also Fig. S5C. (F, G) GO categories enriched among Runx1 targets in two selected gene subsets, as indicated.
To examine global gene expression changes we turned to Affymetrix microarrays. K15-EGFP mice (Morris et al., 2004) showed bright GFP expression in the HG and in some bulge cells at telogen (Fig. 5B, C). Runx1iTG;K15-EGFP mice and WT;K15-EGFP control littermates were doxy-induced at ∼PD19 for 1-day, and their skin cells were isolated by FACS as CD34+/α6+ bulge cells irrespective of GFP expression and as CD34-/α6+/GFP+ HG cells (Fig. 5B, C). We examined skin from 6 different regions of the body to ensure telogen morphology and quiescence of bulge cells, in all skin samples used for microarrays (Fig. S5A, B). Affymetrix microarrays and GeneSpring software analysis revealed that over 2500 probe sets (∼2500 genes) changed expression in Runx1iTG bulge relative to WT bulge cells, suggesting profound and rapid changes in the molecular signature of quiescent bulge cells upon Runx1 elevation after only 1 day (Tables S1, S2 and GEO database GSE53077 [NCBI #16928442]).
Next we compared the WT databases, and extracted a bulge (4453 probe sets) and a HG (2556 probe sets) signature, as genes increased by >2x in one population vs. the other (Tables S1, S2). We used previously published databases to define the matrix signature, which was different from both the HG and bulge (Greco et al., 2009; Lien et al., 2011). Consistent with the known role of Runx1 in both activating and repressing transcription, 1527 probe sets were down-regulated in CD34+/a6+ Runx1iTG bulge compared to WT bulge cells, and 1097 probe sets were up-regulated (Fig. 5D). Importantly, the “bulge signature genes” were largely down-regulated while the “HG signature genes” were largely up-regulated in Runx1iTG bulge compared to WT bulge (Fig. 5E and S5C). A comparison with the matrix signature did not show a meaningful correlation with changing Runx1 levels (Fig. 5E, right). These data strongly suggest that short-term induction of high Runx1 levels in quiescent bulge cells turns the molecular signature of bulge cells into that of a cell-state resembling HG cells.
To understand how Runx1 might act in quiescent bulge cells to promote a HG fate, we examined known pathways that regulate hair growth and homeostasis (Lee and Tumbar, 2012). However, these pathways were represented broadly among Runx1 target genes, and did not suggest a single mode of action in this fate conversion (Table S2). Consistent with a role of Runx1 as master regulator of many pathways in a fate switch, a number of chromatin-remodeling factors, which are generally known to promote and stabilize cell fate transitions, changed their mRNA expression (Fig. S5D). The over-represented GO categories among down-regulated bulge-signature genes were signal transduction, development, differentiation, cell motility, and cell adhesion (Fig. 5F, and Table S3, SI). Conversely, the GO categories over-represented among the up-regulated HG-signature genes were all implicated in metabolism (Fig. 5G and Table S3, SI (see Discussion).
The HG-like cell-state induced by high Runx1 levels in bulge SCs is reversible
All the data so far suggest that endogenous Runx1 expressed in bulge SCs residing in the ORS at catagen may promote the early steps of SC differentiation towards early progenitor HG fate. Since some of these Runx1+ ORS cells return to the “new” bulge, lose Runx1 expression and regain CD34, the HG-like state induced by Runx1 in ORS cells might be functionally reversible. To test this, we doxy-fed Runx1iTG mice for 5-days starting at PD17 (or PD21) to express mycRunx1 and induce the HG-like cell-state in bulge cells, followed by doxy withdrawal (Fig. 6A). These mice are now designated as Runx1iTG, withdrawn. After >23 days of doxy food withdrawal, mycRunx1 expression was no longer detectable (Fig. 2I). While bulge CD34 expression is lost upon 1-5 days of mycRunx1 expression (Fig. 3J-L, S4A, B), it was fully restored in Runx1iTG, withdrawn mice by PD49 (Fig. 2I).
We then monitored hair growth, lineage contribution, and cell-culture behavior to examine if bulge cells that experienced transient high Runx1 levels can return to a SC phenotype (Fig. 6A). In marked contrast to Runx1mTG mice or skin grafts from Runx1iTG mice, in which long-term mycRunx1 expression led to HF and SC loss, hair growth in Runx1iTG, withdrawn mice showed a slight delay in the first anagen onset, but then cycled normally for 2 additional hair cycles (Fig. 6B).
Next we examined the behavior of the WT and Runx1iTG, withdrawn bulge SCs to verify that they self-renew and differentiate in a similar manner, despite the transient Runx1 expression in the latter. Thus, we performed lineage tracing experiment using quadruple Runx1-CreER;Rosa26R-LacZ;Runx1iTG (TRE-mycRunx1;K14-rtTA) transgenic mice. We first marked bulge cells by TM injection at PD17 (catagen) and then induced mycRunx1 expression by feeding mice doxy food at PD21. Subsequent tracking of marked Runx1iTG, withdrawn bulge SCs and their progeny showed no difference in LacZ+ cell distribution and frequency, within the bulge (indicating self-renewal) or bulb (indicating differentiation) compared to WT bulge cells (Fig. 6C-E and S7). Thus, the bulge SCs that had functionally behaved as HG cells after Runx1 level elevation reverted to a normal SC behavior, self-renewing and differentiating over long-term at similar rates with the WT bulge cells upon doxy withdrawal.
To further test the reversibility of the cell-state induced by transient Runx1 expression in bulge SCs, we also performed in vitro functional assays (Fig. 6F). Keratinocytes isolated from PD79 Runx1iTG, withdrawn mice (but not earlier) showed normal SC-like patterns of colony formation, like the WT controls (Fig. 6G). In contrast, if we added doxy to Runx1iTG, withdrawn cells in culture, they behaved again as early progenitor HG-like cells (Fig. 6G – iTG, withdrawn+doxy). All these data were consistent with reversion of HG-like fate induced by Runx1 to a bulge SC fate upon Runx1 withdrawal.
Discussion
With this work we examined the role of changing endogenous Runx1 levels throughout the hair cycle, which appears to be specialized in different HF compartments. We propose that Runx1 acts as a determining factor in the transition from bulge SCs to the early progenitor HG cell state. This state induced by Runx1 in quiescence is reversible, and represents a metastable intermediate in the SC to TA cell fate transition (Fig. 7 and Graphical abstract). Moreover, we found that Runx1 is sufficient for catagen induction in full anagen HF.
Figure 7. Proposed role of changing levels of Runx1 in adult hair follicle homeostasis.
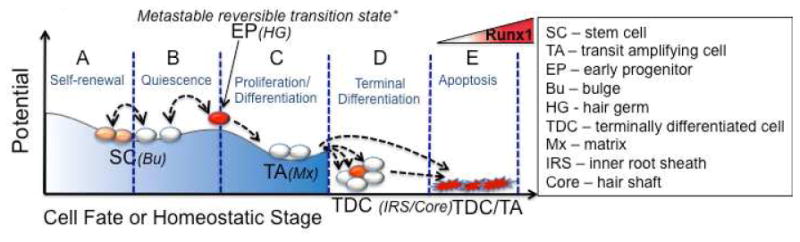
(A) Low levels of Runx1 expression in bulge cells enhance self-renewal (Hoi et al., 2010). (B) Runx1 up-regulation in quiescent bulge cells found outside the niche results in early progenitor fate, a metastable and reversible state, that functions as an intermediate between stem cells (bulge) and transit-amplifying cells (matrix). (C) Inductive proliferation and differentiation signals cause the early progenitor to proliferate, down-regulate Runx1, and fall down the hill of biological potential to a more stable (irreversible) matrix TA cell fate. (D) TA cells terminally differentiate to specific HF lineages. Relatively high Runx1 expression in one (IRS) of these lineages during anagen is shown in red. (E) TDC/TA cells with high Runx1 expression undergo apoptosis and therefore are plotted at zero potential. * The reversible fate change induced by Runx1 is based on transgenic transient expression experiments.
Runx1 expression in bulge cells promotes the transition towards the hair germ fate
An important outcome of this study is that mycRunx1 elevation in bulge SCs induces significant changes in gene expression and cell behavior that closely resemble the HG EP fate. Previously, our candidate approach revealed Wnt, Stat3 and p21 as regulated downstream of Runx1 in the HF (Lee et al., 2013; Osorio et al., 2011; Scheitz et al., 2012). Here we show that Runx1 broadly regulates bulge expression of ∼2500 genes that fit multiple pathways, including global chromatin remodeling factors. This is consistent with a probable role of Runx1 as one of the master regulators that switch off the bulge and on the HG gene signature. Moreover, while WT keratinocytes behave as SCs in culture, the mycRunx1-expressing keratinocytes adopt rapid proliferation and short-lived behavior typical of early progenitors, previously described for HG cells (Greco et al., 2009). In vivo, bulge cells with mycRunx1 expression initially proliferate faster in response to anagen signals, but are lost from the skin in long-term, therefore also behaving essentially as shortlived progenitors.
Previously, we have implicated Runx1 as a proliferation factor in HG and bulge cell activation at anagen onset by loss of function studies (Osorio et al., 2008). Here we carefully describe endogenous Runx1 levels, which appear tightly coupled with cell location relative to the bulge SC niche. Within the bulge, low Runx1 levels are apparently needed for normal bulge SC proliferation rates during anagen ((Fig. 7A) and (Hoi et al., 2010)). Quiescent SCs outside the niche in the ORS at catagen exhibit elevated Runx1 levels that remain high as these cells form the HG at telogen, and then decrease again right at proliferation onset as these HG cells become matrix TA cells (Fig. 7B). Although Runx1 expression is necessary for HG and bulge cell activation/proliferation and for anagen onset (Osorio et al., 2008), we find here by transgenic expression that it is not sufficient. We propose that Runx1 endogenous expression in the ORS at quiescence (catagen/telogen) in bulge SCs transitioning towards HG may prepare these cells for overcoming their “slow-cycling” SC character, thus becoming fit for rapid response to subsequent proliferation signals, as expected of HG cells.
Our gene expression studies begin to reveal some of the potential specific functions Runx1 may be performing during the bulge SC to the early progenitor HG transition. In particular, cell adhesion genes are down-regulated, suggesting that Runx1 might promote the bulge cell release from the niche and migration down the basement membrane into the ORS. Consistent with this interpretation, Runx1-deficient keratinocytes display defects in cell migration (Osorio et al., 2011). Conversely, HG signature genes preferentially up-regulated by Runx1 in the quiescent bulge cells are metabolic enzymes. We previously showed that Runx1 promotes the G1/S transitions of the cell cycle in skin keratinocytes (Hoi et al., 2010). Therefore, a potential involvement of Runx1 in cell growth during the G1 phase in vivo, prior to divisions at anagen onset, may explain at least in part how Runx1 might act in G0/G1 quiescence during catagen to prepare HG cells for their proliferation at anagen onset. An interesting twist is that even though ORS cells down-regulate Runx1 upon entering the “new” bulge, their transient endogenous Runx1 expression in the ORS may also prepare them for the subsequent proliferation at anagen for proper SC self-renewal. These aspects of Runx1 regulation will be subject of a future study.
In summary, all our data suggest that the role of endogenous high Runx1 levels in ORS cells that migrated from the bulge at catagen is to promote the early steps of their fate transition towards the early progenitor HG.
Runx1 expression in the lower hair follicle bulb is sufficient to implement catagen
Despite the highest endogenous Runx1 expression in the apoptotic bulb and epithelial strand at catagen (Fig 7E), our previous knockout studies did not reveal a role of Runx1 at this stage. Here, however, we show that early elevation of Runx1 can initiate apoptosis exclusively in those cells that would normally express high levels of Runx1 just as they begin to undergo apoptosis. Because the affected HFs progress through catagen to teleogen, we propose that expression of Runx1 is a normal trigger of apoptosis. However, there must be a redundant system(s) to trigger apoptosis in the absence of Runx1. Interestingly, endogenous high Runx1 expression at catagen below the bulge appears to conveniently couple the removal of the “used” matrix TA and TDL cells with the differentiation of bulge SCs towards supplying new HG early progenitors.
Reversible hair germ like cell-state induced by Runx1 in quiescent hair follicle stem cells during normal homeostasis
While previous work demonstrated that “old” bulge cells that migrated into the upper ORS returned to the “new” bulge (Hsu et al., 2011), experimental evidence supporting significant functional and/or molecular changes during this journey outside the niche was lacking. Here we provide expression and lineage tracing data demonstrating that the migrated bulge cells in the upper ORS up-regulate endogenous Runx1 and down-regulate it again when they return to the “new” bulge. Furthermore, we find that Runx1 up-regulation in bulge cells rapidly promotes a cell-state resembling the early progenitor HG (see the first section of Discussion), and that this effect is functionally reversed when Runx1 is down-regulated.
De-differentiation of the HG to bulge cells was previously proposed as a response to injury (hair plucking), although lineage-tracing data are still missing (Ito et al., 2004), and was also shown to occur in response to injury by LASER ablation (Rompolas et al., 2013). Our data suggest that differentiation and de-differentiation of bulge SCs are likely to occur naturally at catagen, during the bulge SC journey outside the niche (old bulge) in the ORS followed by a return (new bulge), when they experience changing endogenous Runx1 levels. Moreover, these SC fate changes are likely to occur in the absence of cell division, since bulge, upper- and mid-ORS cells are known to withdraw from the cell cycle during this catagen/telogen period (Hsu et al., 2011). This flexibility in fate may provide robustness to produce the correct ratio of “new” bulge and “new” HG cells from a fixed pool of quiescent “old” bulge cells, to re-create the proper telogen HF structure for the next regeneration cycle.
Lessons for tissue stem cell early differentiation: quiescent reversible transition state
Our data fit a model for HF SC fate determination that follows Ferrell's theory proposed for early development fate transitions (Ferrell, 2012), as an alternative to the irreversible fates described by Waddington (Waddington, 1957). Specifically, the first step in stem cell differentiation towards a progenitor cell may occur in quiescence (catagen/telogen), when some SCs exit the niche and become early progenitors (HG cells). The differentiation of SCs to TA or progenitor cells would occur through a reversible transition state, the “early progenitor”, conveniently represented in the HF by an identifiable and sufficiently long-lived (although still short-term) cell state, the HG. SC differentiation into early progenitor cells is likely driven by up-regulation of certain fate determinants, of which Runx1 appears to be an early one in the HF. The early progenitor transition state is metastable as it would either fall backwards from the hill of biological potential to remake SCs (when HG-like cells in ORS de-differentiate to the “new” bulge), or fall forward to divide and make the TA cells (differentiation of HG cells to matrix cells), which are more stable cell states (Graphical Abstract). Additional inductive proliferative signals (anagen promoting) are also needed for the early progenitors to engage in cell division and become TA cells (Fig. 7F). The fate reversibility is likely to cease upon cell division, as suggested in the HF by the fact that dividing HG cells rapidly change to acquire matrix TA characteristics upon 1-2 divisions (Zhang et al., 2009), and matrix cells cannot make SCs as shown by lineage-tracing (Greco et al., 2009).
Our proposed model in the HF, which implicates a reversible transition cell-state in quiescence mediating the SC to TA cell fate commitment, may provide new insight especially to the SC systems that are more active or less synchronous than the HF, where the steps of SC to TA cell fate transition are difficult to delineate and the transition state is very short-lived.
Experimental procedures
Mice
Mouse work followed Cornell IACUC guidelines. The c-Myc tag was inserted to the Runx1 N-terminus in the pcDNA3.1Runx1b (a gift from Nancy Speck) and the mycRunx1 fragment was transferred to pTRE2 (Clontech) vector. Cornell Mouse Transgenic Core Facility generated 10 transgenic TRE-mycRunx1+ mice, which were tested for inducibility by crossing with K14-rtTA mice (Nguyen et al., 2006), and feeding mice with 20- 60- 200-and 1000mg/kg mouse chow. The last concentration was used in all subsequent experiments. We used TRE-H2BGFP; K5-tTA for in vivo cell division counting (Tumbar et al., 2004), Runx1-LacZ mice for Runx1 expression studies (North et al., 2002) and Runx1-CreER mice (Samokhvalov et al., 2007) crossed with Rosa26RLacZ mice (JAX) for lineage tracing. Sequential crosses were performed to obtain quadruple transgenic mice: Runx1-CreER;Rosa26LacZ;TRE-mycRunx1 (iTG, fully inducible);K14-rtTA. For hair germ and bulge cell sorting after 1-day doxy induction at ∼PD19 we crossed K15-EGFP mice (Morris et al., 2004) with TRE-mycRunx1(iTG);K14-rtTA mice.
Microarrays
Sorted CD34+/α6+ (bulge) and CD34-/α6+/GFP+ (hair germ) cells were used to prepare mRNA from 2 different mice of each genotype, then subjected to Affymetrix microarray assay (Cornell Core Facilities) followed by GeneSpring analysis. After excluding the probes with a signal value <100 or called absent, genes designated as on average ≥2-fold up in WT bulge vs. WT HG (4453 probes) and ≥2-fold up in WT HG vs. WT bulge (2555 probes) were defined as bulge and HG signature genes, respectively. The matrix signature genes were defined as ≥2-fold up in anagen matrix vs. anagen bulge and telogen bulge, utilizing previously published databases (Lien et al., 2011).
Immunofluoresce staining
The frozen-cut skin sections were incubated with primary antibodies in the blocking solution for 1-4 hrs at room temperature or overnight at 4°C with following dilution: rabbit anti-Runx1 (1:4000, a gift from Thomas Jessell), mouse anti-Myc tag (1:200, Zymed), rat anti-CD34 (1:50, BD Biosciences), rabbit anti-K5 or K14 (1:1000, Covance), rabbit anti-activated Caspase-3 (1:500, R&D Systems), mouse anti-AE13 (1:50, Immunoquest), mouse anti-AE15 (1:10, a gift from Tung-Tien Sun), mouse anti-GATA3 (1:100, Santa Cruz Biotechnologies), rat anti-BrdU (1:300, Abcam), rabbit anti-Ki67 (1:1000, NovoCastra Laboratories). This was followed by incubation in secondary antibody fluorescently coupled with FITC or TxRed.
Microscopy and image processing
We examined all stained samples under a fluorescent upright light microscope (Nikon). Images were obtained with a charge-coupled-device 12-bit camera (Retiga EXi; QImaging) using the IP-Lab software (MVI), color combined and assembled in montages. All the images were processed with Photoshop and Illustrator (Adobe) to enhance the contrast, brightness, and levels to the same extent among samples within the same experiment.
Cell culture and colony formation assay
200,000 live skin isolated in sterile conditions from adult mice were plated onto the embryonic feeder cell layers, in low-Ca2+ keratinocyte E medium (Barrandon and Green, 1987). 0.5 μg of doxy/ml of media was added to cultures to maintain Runx1 expression. The number of colonies was counted by phase-contrast microscopy every 5 days during the first 30 days and every 7 days during the last 30 days.
See also SI for additional details on the procedures.
Supplementary Material
Table S1. Probe sets up-regulated in WT Bu vs WT HG = “Bu signature” list (4453) and iTG signals
Table S2. Probe sets up-regulated in WT HG vs WT Bu = “HG signature” list (2555) and iTG signals
Table S3. GO categories and pathways found over-represented among Runx1 target genes split in down-regulated Bu and up-regulated HG signatures, respectively.
Figure S1. Runx1 is expressed at different levels throughout the hair cycle and is found in early catagen ORS cells, which serve as precursors of the “new” bulge and HG.
Figure S2. Increasing Runx1 levels is insufficient for activating bulge and hair germ proliferation during quiescence.
Figure S3. High levels of Runx1 induce differentiation block and apoptosis in matrix and bulb cells and increase proliferation rates in dividing bulge cells
Figure S4. High Runx1 expression down-regulates protein levels of bulge stem cell marker CD34 regardless of the hair cycle stages.
Figure S5. Affymetrix microarray analysis reveals change in bulge signature towards a hair germ-like phenotype upon transient expression of Runx1
Figure S6. Cell culture functional analysis reveals loss of long-term self-renewal ability of Runx1iTG stem cells.
Figure S7. Lineage tracing in Runx1iTG, withdrawn and WT full hair follicles reveals normal behavior of Runx1iTG, withdrawn bulge SCs after 5 days doxy induction and withdrawal.
Highlights.
Runx1 promotes hair follicle stem cell differentiation to early progenitors
The cell state induced by Runx1 expression in stem cells is reversible
De-differentiation of adult tissue stem cells can occur without injury
Runx1 expression triggers catagen
Acknowledgments
We thank Y. V. Zhang and C. M. Piskun for experiments in Fig. 2C and S2A and many colleagues for reagents (see Experimental Procedures and SI). The Cornell Cytometry core is supported in part by the ESSCF, NYS-DOH, Contract C026718. Opinions do not necessarily reflect the view of ESSCF, the NYS-DOH, or NYS. Funding was from NYSTEM C024354 and NIH R01AR053201 to T.T. and HSF to A. S. We thank colleagues, in particular M. Rendl (Mount Sinai) for constructive criticism and V. Vogt (Cornell) for moral support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Blanpain C. Stem cells: Skin regeneration and repair. Nature. 2010;464:686–687. doi: 10.1038/464686a. [DOI] [PubMed] [Google Scholar]
- Blyth K, Cameron ER, Neil JC. The RUNX genes: gain or loss of function in cancer. Nat Rev Cancer. 2005;5:376–387. doi: 10.1038/nrc1607. [DOI] [PubMed] [Google Scholar]
- Ferrell JE., Jr Bistability, bifurcations, and Waddington's epigenetic landscape. Current biology : CB. 2012;22:R458–466. doi: 10.1016/j.cub.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco V, Chen T, Rendl M, Schober M, Pasolli HA, Stokes N, Dela Cruz-Racelis J, Fuchs E. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell. 2009;4:155–169. doi: 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoi CS, Lee SE, Lu SY, McDermitt DJ, Osorio KM, Piskun CM, Peters RM, Paus R, Tumbar T. Runx1 directly promotes proliferation of hair follicle stem cells and epithelial tumor formation in mouse skin. Mol Cell Biol. 2010;30:2518–2536. doi: 10.1128/MCB.01308-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YC, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Kizawa K, Hamada K, Cotsarelis G. Hair follicle stem cells in the lower bulge form the secondary germ, a biochemically distinct but functionally equivalent progenitor cell population, at the termination of catagen. Differentiation. 2004;72:548–557. doi: 10.1111/j.1432-0436.2004.07209008.x. [DOI] [PubMed] [Google Scholar]
- Jaks V, Kasper M, Toftgard R. The hair follicle-a stem cell zoo. Exp Cell Res. 2010;316:1422–1428. doi: 10.1016/j.yexcr.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Lee J, Hoi CS, Lilja KC, White BS, Lee SE, Shalloway D, Tumbar T. Runx1 and p21 synergistically limit the extent of hair follicle stem cell quiescence in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4634–4639. doi: 10.1073/pnas.1213015110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Tumbar T. Hairy tale of signaling in hair follicle development and cycling. Semin Cell Dev Biol. 2012;23:906–916. doi: 10.1016/j.semcdb.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien WH, Guo X, Polak L, Lawton LN, Young RA, Zheng D, Fuchs E. Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell. 2011;9:219–232. doi: 10.1016/j.stem.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G. Capturing and profiling adult hair follicle stem cells. Nature biotechnology. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- Muller-Rover S, Handjiski B, van der Veen C, Eichmuller S, Foitzik K, McKay IA, Stenn KS, Paus R. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J Invest Dermatol. 2001;117:3–15. doi: 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- Nguyen H, Rendl M, Fuchs E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 2006;127:171–183. doi: 10.1016/j.cell.2006.07.036. [DOI] [PubMed] [Google Scholar]
- North TE, de Bruijn MF, Stacy T, Talebian L, Lind E, Robin C, Binder M, Dzierzak E, Speck NA. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 2002;16:661–672. doi: 10.1016/s1074-7613(02)00296-0. [DOI] [PubMed] [Google Scholar]
- Osorio KM, Lee SE, McDermitt DJ, Waghmare SK, Zhang YV, Woo HN, Tumbar T. Runx1 modulates developmental, but not injury-driven, hair follicle stem cell activation. Development. 2008;135:1059–1068. doi: 10.1242/dev.012799. [DOI] [PubMed] [Google Scholar]
- Osorio KM, Lilja KC, Tumbar T. Runx1 modulates adult hair follicle stem cell emergence and maintenance from distinct embryonic skin compartments. J Cell Biol. 2011;193:235–250. doi: 10.1083/jcb.201006068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pournasr B, Khaloughi K, Salekdeh GH, Totonchi M, Shahbazi E, Baharvand H. Concise review: alchemy of biology: generating desired cell types from abundant and accessible cells. Stem cells. 2011;29:1933–1941. doi: 10.1002/stem.760. [DOI] [PubMed] [Google Scholar]
- Rompolas P, Mesa KR, Greco V. Spatial organization within a niche as a determinant of stem-cell fate. Nature. 2013;502:513–518. doi: 10.1038/nature12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samokhvalov IM, Samokhvalova NI, Nishikawa S. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature. 2007;446:1056–1061. doi: 10.1038/nature05725. [DOI] [PubMed] [Google Scholar]
- Scheitz CJ, Lee TS, McDermitt DJ, Tumbar T. Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J. 2012;31:4124–4139. doi: 10.1038/emboj.2012.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Simons BD, Clevers H. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 2011;145:851–862. doi: 10.1016/j.cell.2011.05.033. [DOI] [PubMed] [Google Scholar]
- Spradling A, Fuller MT, Braun RE, Yoshida S. Germline stem cells. Cold Spring Harbor perspectives in biology. 2011;3:a002642. doi: 10.1101/cshperspect.a002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, Fuchs E. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. The Strategy of the Gene. London: George Allen and Unwin; 1957. [Google Scholar]
- Yanger K, Zong Y, Maggs LR, Shapira SN, Maddipati R, Aiello NM, Thung SN, Wells RG, Greenbaum LE, Stanger BZ. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes & development. 2013;27:719–724. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YV, Cheong J, Ciapurin N, McDermitt DJ, Tumbar T. Distinct self-renewal and differentiation phases in the niche of infrequently dividing hair follicle stem cells. Cell Stem Cell. 2009;5:267–278. doi: 10.1016/j.stem.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YV, White BS, Shalloway DI, Tumbar T. Stem cell dynamics in mouse hair follicles: a story from cell division counting and single cell lineage tracing. Cell Cycle. 2010;9:1504–1510. doi: 10.4161/cc.9.8.11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Probe sets up-regulated in WT Bu vs WT HG = “Bu signature” list (4453) and iTG signals
Table S2. Probe sets up-regulated in WT HG vs WT Bu = “HG signature” list (2555) and iTG signals
Table S3. GO categories and pathways found over-represented among Runx1 target genes split in down-regulated Bu and up-regulated HG signatures, respectively.
Figure S1. Runx1 is expressed at different levels throughout the hair cycle and is found in early catagen ORS cells, which serve as precursors of the “new” bulge and HG.
Figure S2. Increasing Runx1 levels is insufficient for activating bulge and hair germ proliferation during quiescence.
Figure S3. High levels of Runx1 induce differentiation block and apoptosis in matrix and bulb cells and increase proliferation rates in dividing bulge cells
Figure S4. High Runx1 expression down-regulates protein levels of bulge stem cell marker CD34 regardless of the hair cycle stages.
Figure S5. Affymetrix microarray analysis reveals change in bulge signature towards a hair germ-like phenotype upon transient expression of Runx1
Figure S6. Cell culture functional analysis reveals loss of long-term self-renewal ability of Runx1iTG stem cells.
Figure S7. Lineage tracing in Runx1iTG, withdrawn and WT full hair follicles reveals normal behavior of Runx1iTG, withdrawn bulge SCs after 5 days doxy induction and withdrawal.


