Abstract
Cellulosic materials constitute most of the biomass on earth, and can be converted into biofuel or bio-based materials if fermentable sugars can be released using cellulose-related enzymes. Acremonium cellulolyticus is a mesophilic fungus which produces a high amount of cellulose-related enzymes. In the genome sequence data of A. cellulolyticus, ORFs showing homology to GH10 and GH11 xylanases were found. The xylanases of A. cellulolyticus play an important role in cellulolytic biomass degradation. Search of a draft genome sequence of A. cellulolyticus for xylanase coding regions identified seven ORFs showing homology to GH 11 xylanase genes (xylA, xylB, xylC, xylD, xylE, xylF and xylG). These genes were cloned and their enzymes were prepared with a homologous expression system under the control of a glucoamylase promoter. Six of the seven recombinant enzymes were successfully expressed, prepared, and characterized. These enzymes exhibited optimal xylanase activity at pH 4.0 – 4.5. But this time, we found that only XylC had enormously higher relative activity (2947 U•mg −1) than the other xylanases at optimum pH. This result is surprising because XylC does not retain a carbohydrate-binding module 1 (CBM-1) that is necessary to bind tightly own substrate such as xylan. In this study, we discuss the relationship between activity, pH and sequence of seven xylanases in A. cellulolyticus.
Keywords: Acremonium cellulolyticus, Xylanase, Hemicellulose, Homologous expression, Biomass, Xylan
Introduction
Lignocellulosic biomass can be converted into biofuel or bio-based materials (Deutschmann and Dekker [2012]; Kumar et al. [2008]) but must be converted into fermentable sugars by saccharification. Xylan is one of the major structural components of plant cell walls and is the second most abundant renewable biomass resource (York and O’Neill [2008]). Xylan consists of a backbone of β-1,4-D-xylan with short side chains of O-acetyl, β-L-arabinofuranosyl, D-α-glucuronic acid and phenolic acid (Coughlan and Hazlewood [1993]). Xylanases (endo-1,4-β-xylanases; EC 3.2.1.8) catalyze the hydrolysis of the β-1,4 bonds of xylan and thus are important enzymes for the degradation of hemicellulosic polysaccharides (Collins et al. [2005]; Prade [1996]). Based on their amino acid sequence similarities, xylanases are mainly classified into families 10 and 11 of the glycoside hydrolases (GH; http://www.cazy.org/Glycoside-Hydrolases.html; (Coutinho and Henrissat [1999]). GH10 xylanases generally have a molecular weight ≥30 kDa and a low pI, while GH11 xylanases are generally smaller (approximately 20 kDa) and have a high pI (Beaugrand et al. [2004]). The crystal structures of xylanases show that GH10 enzymes fold into a (β/α)8-barrel (Dimarogona et al. [2012]; Harris et al. [1994]; Lo Leggio et al. [1999]), whereas family 11 enzymes have a β-jellyroll structure (Paës et al. [2012]; Sidhu et al. [1999]). Cellulose and xylan are closely linked together in plant cell walls (Carpita and Gibeaut [1993]). Thus, cellulases and hemicellulases work coordinately in the enzymatic degradation of these polysaccharides.
Filamentous fungi produce a wide spectrum of degradation enzymes for cellulose and xylan (van den Brink and de Vries [2011]). Acremonium cellulolyticus, isolated by Yamanobe et al., is a high cellulolytic enzyme-producing fungus (Yamanobe et al. [1987]). Fujii et al. reported that the culture supernatant from A. cellulolyticus has a higher cellulase specific activity and yields more glucose from lignocellulosic materials than the culture supernatant from Trichoderma reesei (Fujii et al. [2009]). Thermostable xylanase from A. cellulolyticus was found by Mitsuishi et al. (Mitsuishi et al. [1987]), but this enzyme has not been identified. Furthermore, there is no report of the expression and characterization of the xylanases (GH11) from A. cellulolyticus. The cellulose-induced xylanases of A. cellulolyticus play an important role in the cellulolytic biomass degradation process. In this study, we searched the xylanase coding regions from the draft genome sequence of A. cellulolyticus (unpublished data) and attempted to clarify the characteristics of the all xylanases (GH11).
Materials and Methods
Materials
Birch-wood xylan was purchased from Sigma-Aldrich (St. Louis, MO, USA). All other chemicals were of the highest grade commercially available.
Strain and culture conditions for Acremonium cellulolyticus
Acremonium cellulolyticus CF-2612 and Y-94 (CBS136886) were maintained on potato dextrose agar plates (Fang et al. [2009]). The A. cellulolyticus YP-4 uracil autotroph was maintained on potato dextrose agar plates containing uracil and uridine at final concentrations of 1 g/l each (Inoue et al. [2013]). Transformants of A. cellulolyticus YP-4 were maintained on MM agar plates (Fujii et al. [2012]). For measurement of gene expression, the strains were cultivated in 50 ml of basic medium (24 g/l of KH2PO4, 1 g/l of Tween 80, 5 g/l of (NH4)2SO4, 1.2 g/l of MgSO4•7H2O, 0.01 g/l of ZnSO4•7H2O, 0.01 g/l of MnSO4•6H2O, 0.01 g/l of CuSO4•7H2O; pH 4.0) supplemented with 20 g/l soluble starch (Wako Pure Chemical Industries, Osaka, Japan) as a carbon source in 500-ml Erlenmeyer flasks at 30°C for 96 h on a rotary shaker operated at 230 rpm. The cells were centrifuged for 15 min at 3,500 rpm, then the supernatants were centrifuged for 10 min at 13,500 rpm. The pooled enzyme solution was filtered through a 0.45 μM membrane and adjusted with acetate buffer (pH 5.0) containing 0.01% NaN3. The samples were stored at 4°C until use.
Cloning of xylanases and construction of the expression vector
A draft genome sequence of A. cellulolyticus (unpublished data) was searched for the xylanase (GH11) genes using In silico Molecular Cloning gene analysis software (In Silico Biology, Inc., Yokohama, Japan) based on the internal amino acid sequence. Construction of the xylanase expression vector for A. cellulolyticus basically followed a procedure described previously (Inoue et al. [2013]). The genomic regions encoding xylanases were amplified by PCR from CF-2612 chromosomal DNA using primers as indicated in Table 1. The seven amplified xylanase genes were constructed by introducing the appropriate fragment digested with HpaI/SbfI (xylA, xylB, xylC, xylD, xylE and xylF) into the EcoRV/SbfI (xylG) site of a series of pANC expression vectors containing a glucoamylase (glaA) promoter and terminator (Inoue et al. [2013]): pANC209 (xylA), pANC210 (xylB), pANC223 (xylC), pANC230 (xylD), pANC231 (xylE), pANC232 (xylF) and pANC233 (xylG) (Table 2).
Table 1.
Summary of nucleotide primers used in this study
| Primer | Nucleotide sequence (5’ → 3’) |
|---|---|
|
For plasmids construction |
|
| AC-F12 (xylA) |
ATTGTTAACAAGATGAAGATCACATCAGTGTTCG |
| AC-R12 (xylA) |
AATCCTGCAGGTTAAGATACAGTAACAGTGGCACTTC |
| AC-F13 (xylB) |
ATTGTTAACATCATGGGCATCTCATCTATTCTTC |
| AC-R13 (xylB) |
AATCCTGCAGGCTATTGGCACTGACTGTAGTAAGCGT |
| AC-F25 (xylC) |
ATTGTTAACAAGATGAAGCTCTCTCTGGCTGCAA |
| AC-R25 (xylC) |
AATCCTGCAGGCTAGGACACGGTGATGGTACTAGAAC |
| AC-F26 (xylD) |
ATTGTTAACAAGATGCGGTCATTTGCTCGCCTTGTC |
| AC-R26 (xylD) |
AATCCTGCAGGTCAGCTAACAGTAAAATCCAGGTAAC |
| AC-F27 (xylE) |
ATTGTTAACAAGATGATTTATTTCCCTCAGCTCATG |
| AC-R27 (xylE) |
AATCCTGCAGGCTATTGAGTGGCAGTCTGCTGGGCA |
| AC-F28 (xylF) |
ATTGTTAACAAGATGTTCTCTTTCAGTACTGCCTT |
| AC-R28 (xylF) |
AATCCTGCAGGCTACAAGCATTGATAGTAGTACGGGT |
| AC-F29 (xylG) |
ATTGATATCAAGATGGTTGCTTTCTCGAGCTTATTTAC |
| AC-R29 (xylG) |
AATCCTGCAGGGTCCAACATCAATGCTACTTACAGC |
|
For quantitative PCR |
|
|
xylA-f |
GAGGACGGAGTCAATGGAGA |
|
xylA-r |
CCGAGAGGTAGGAGCCAGAA |
|
xylB-f |
TGCTCTCGGTGTTGATGTTG |
|
xylB-r |
GTGGTCTGGTAGTCGGTGGA |
|
xylC-f |
GTGTCGACCAACCCTCCATC |
|
xylC-r |
GTCCAAGTGCCTTCCATGCT |
|
xylD-f |
TTTAGCGATAGCGGCAGGTT |
|
xylD-r |
CCCATGTAAGGGAGCGAGTT |
|
xylE-f |
CGGGCCACAAACTATATCCA |
|
xylE-r |
TTCCAGCCAACACCAACAAC |
|
xylF-f |
ACTAGCAAGGACGGCGTAGA |
|
xylF-r |
CACCGAGGAACTCAGACGAA |
|
xylG-f |
CAGCACGGGGTTTGAGGTT |
|
xylG-r |
CCGAAGTTGATGGGAGTGGT |
|
gpdA-f |
AACATCATTCCCAGCAGCAC |
| gpdA-r | CGGCAGGTCAAGTCAACAAC |
Table 2.
Summary of seven xylanases (GH11) in A. cellulolyticus
| Strain | Plasmid | Xyl11 | AA | MW (Da) | CBM-1 | N-terminal sequence (5 aa) |
|---|---|---|---|---|---|---|
| Y209 |
pANC209 |
A |
209 |
22377 |
- |
AGGIN |
| Y210 |
pANC210 |
B |
282 |
29510 |
CBM |
AEAIN |
| Y223 |
pANC223 |
C |
223 |
23955 |
- |
QSITT |
| Y230 |
pANC230 |
D |
190 |
21029 |
- |
- |
| Y231 |
pANC231 |
E |
234 |
25788 |
- |
ATNYI |
| Y232 |
pANC232 |
F |
276 |
29210 |
CBM |
NTPNS |
| Y233 | pANC233 | G | 233 | 24906 | - | SAINY |
Homologous expression of the recombinant xylanases
Protoplasts of A. cellulolyticus YP-4 were transformed with pANC209, 210, 223, 230, 231, 232 and 233 by nonhomologous integration into the host chromosomal DNA (Fujii et al. [2012]). Gene integration into prototrophic transformants was verified by genomic PCR. The expression of each recombinant xylanase was carried out using the following cultures: Y209 (YP-4 transformed with pANC209; XylA), Y210 (YP-4 transformed with pANC210; XylB), Y223 (YP-4 transformed with pANC223; XylC), Y230 (YP-4 transformed with pANC230; XylD), Y231 (YP-4 transformed with pANC231; XylE), Y232 (YP-4 transformed with pANC232; XylF), Y233 (YP-4 transformed with pANC233; XylG) (Table 2). All cultures were grown in medium containing 20 g/l soluble starch and 5 g/l urea using the method described previously (Inoue et al. [2013]). The recombinant xylanases expressed were purified from the culture supernatant.
Purification of the recombinant xylanases
A. cellulolyticus Y209, 210, 223, 231, 232 and 233 were grown at 30°C on a starch-induced medium. The purification procedures were basically as previously reported (Lee et al. [2009]). The purified enzyme was concentrated, dialyzed against 20 mM sodium acetate buffer (pH 5.0) using a Vivaspin 20 concentrator (10,000 MWCO, Sartorius AG, Goettingen, Germany) and stored at 4°C until use. The purity and size of the protein was analyzed by SDS-PAGE using precast NuPAGE 4 - 12% polyacrylamide Bis-Tris gels (Life Technologies, Carlsbad, CA, USA) (Hachmann and Amshey [2005]). All proteins were identified by N-terminus sequence analysis.
Protein assay
Protein concentration was determined by a Pierce BCA Protein Assay Kit (Pierce, Rockford, IL, USA) using bovine serum albumin as the standard (Shibuya et al. [1989]; Smith et al. [1985]). Protein solution (25 μl) was mixed with 200 μl of BCA reagent and then incubated at 37°C for 30 min. Protein concentration was determined by measuring the absorption at 570 nm.
N-terminus sequence analysis
The N-terminal sequences of six xylanases (XylA, XylB, XylC, XylE, XylF and XylG) were commercially determined by Edman degradation using Procise 494 HT (ABI, Foster City, California, USA). Each enzyme was adjusted to 2.0 mg/ml (600 μg) in 20 mM sodium acetate buffer (pH 5.5) prior to degradation.
Enzyme assay
The xylanase activity assay was based on the method described by Bailey et al. ([1992]), using 1% (w/v) birch-wood xylan (Sigma-Aldrich) as the substrate. Activity was examined under a variety of buffer conditions between pH 3.0 and 8.0 (McIlvaine [1921]). The appropriately diluted protein solution was mixed with 0.45 ml of substrate, followed by incubation for 10 min at 50°C. The reducing sugars produced in the reaction mixture were measured by a 3,5-dinitrosalicylic acid (DNS) assay. One unit of xylanase activity was defined as the quantity of enzyme required to liberate 1 μmol of xylose equivalent per minute at 50°C.
Differential scanning fluorimetry (DSF)
A series of DSF experiments generally followed the protocol published by Niesen and others (Niesen et al. [2007]; Lo et al. [2004]). The experiments were carried out with a CFX96 Real-Time PCR System™ (Bio-Rad) using 450/490 excitation and 560/580 emission filters in a 96-well plate format. Each enzyme was prepared at a final concentration of 0.1 mg/ml (2 μg) in 50 mM sodium acetate pH 4.0 buffer. SYPRO orange dye [5,000-fold stock solution in dimethyl sulfoxide (DMSO); Invitrogen, Carlsbad, CA, USA] was added in twentyfold dilution to the enzyme (Senisterra and Finerty [2009]). The negative control comprised buffer and the SYPRO mixture, without xylanase. Each sample (20 μl per well) was measured between 25 and 90°C using a stepwise gradient of 0.5°C per 5 s. Following curve fitting of the data to the Boltzmann equation, the melting temperature (Tm) of each xylanase was calculated using Bio-Rad CFX Manager software (Bio-Rad). The results are shown in Table 3.
Table 3.
Specific activity and thermal shift assay of six xylanases (GH11)
| Enzyme (GH11) | Specific activity at pH 5.5 (U/mg) | Specific activity at optimum pH (U/mg) | Tm (°C) at pH 5.5 | Tm (°C) at optimum pH | Δ°C |
|---|---|---|---|---|---|
| XylA |
320 |
999 (pH 4.0) |
55 |
59 |
4 |
| XylB |
252 |
667 (pH 4.0) |
62.5 |
66 |
3.5 |
| XylC |
1434 |
2947 (pH 4.0) |
58 |
61.5 |
3.5 |
| XylE |
43 |
304 (pH 4.0) |
59 |
65.5 |
6.5 |
| XylF |
16 |
34 (pH 4.0) |
54 |
60 |
6 |
| XylG | 35 | 285 (pH 4.0) | 55 | 64.5 | 9.5 |
Real-time quantitative PCR
Total RNA was extracted from disrupted fungal cells. Single-stranded cDNA was synthesized and then real-time quantitative PCR was conducted as described previously (Fujii et al. [2010]). The expression of each gene was normalized against that of the glyceraldehyde-3-phosphate dehydrogenase gene (gpdA accession number AB847425). Results are shown as relative expressions. The gene-specific primers used were shown in Table 1. The nucleotide sequences of each gene from CF-2612 will appear in the GenBank/EMBL/DDBJ nucleotide database under accession nos. AB847990 (xylA), AB847991 (xylB), AB847992 (xylC), AB847993 (xylD), AB847994 (xylE), AB847995 (xylF), and AB847996 (xylG).
Results
Cloning of xylanase genes from A. cellulolyticus
A search of the genome sequence database of A. cellulolyticus identified seven ORFs exhibiting homology to xylanase (GH11) genes: xylA (689 bp), xylB (915 bp), xylC (735 bp), xylD (845 bp), xylE (760 bp), xylF (952 bp) and xylG (761 bp). The PCR-amplified DNA fragments were ligated into the expression vectors containing a glucoamylase (glaA) promoter and terminator, as described in Materials and Methods. All ligated gene fragments and their ligation sites were verified by sequencing. The xylD gene was found to be smaller than the other xylanase genes (Figure 1) and the speculative conserved active site residue (Glu) near the C-terminus of xylanase was absent.
Figure 1.
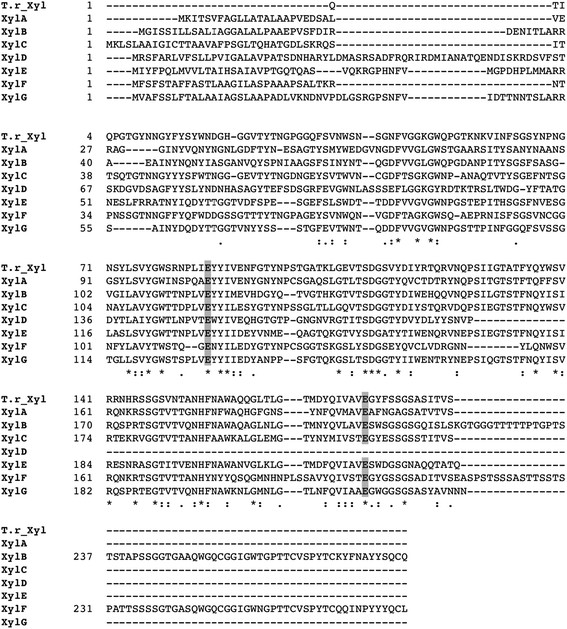
Sequence alignment betweenA. cellulolyticusGH11s andT. reeseiGH11. All seven xylanases (XylA-G) were detected from A. cellulolyticus. Strictly conserved residues are highlighted with an asterisk and similarly conserved residues are marked as a colon. The two glutamate catalytic residues are strictly conserved (highlighted with gray background). All xylanases have a signal peptide at the N-terminus whereas only XylB and XylF have a CBM at their C-terminal end. The figure was made using ClustalW Thompson et al. ([1994]).
Homologous expression and purification of the recombinant xylanases
For xylA, xylB, xylC, xylD, xylE, xylF and xylG genes, the homologous expression vector (pANC209, 210, 223, 230, 231, 232 and 233, respectively) was transformed into A. cellulolyticus YP-4 to provide Y209, Y210, Y223, Y230, Y231, Y232 and Y233 (Table 2). SDS-PAGE analysis showed that the molecular size of XylA (22.3 kDa), XylB (29.5 kDa), XylC (23.9 kDa), XylE (25.7 kDa), XylF (29.2 kDa) and XylG (24.9 kDa) was in accordance with the molecular size calculated from the individual sequences (Figure 2). XylD activity was not detetcted in this expression system because of its incomplete length (Figure 1). Presumably the higher molecular masses of XylB and XylF are largely due to the presence of a CBM-1 with each catalytic domain (Figure 2).
Figure 2.
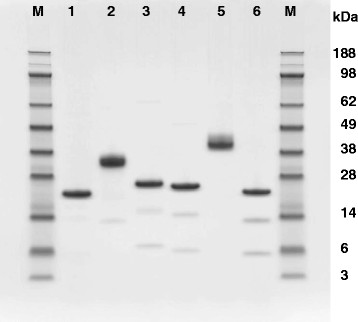
SDS-PAGE analysis of six kinds of xylanase (GH11). Lanes: M. protein marker, 1. XylA, 2. XylB, 3. XylC, 4. XylE, 5. XylF, 6. XylG. All of these proteins are final products after purification on a Resource ISO column.
Comparison of sequence identity and N-terminal sequence analysis
The deduced amino acid sequence of the ORFs indicated that XylA, XylB, XylC, XylD, XylE, XylF and XylG are encoded by 209, 282, 223, 190, 234, 276 and 233 amino acids, respectively, including the individual signal peptides at the N-terminus. Table 2 shows the five N-terminal amino acids of each expressed xylanase. In this homologous expression system, these signal peptides were cleaved off by a signal peptidase on the N-terminal side of the arginine residue. Both XylB and XylF include a carbohydrate-binding module (CBM-1) at their C-terminus. Figure 1 shows a sequence alignment of the active domain in the xylanases. Six xylanases (XylA, XylB, XylC, XylE, XylF and XylG) exhibit amino acid sequence homology of around 11 ~ 52%, with the two catalytic glutamates being strictly conserved.
Specific activity and the effect of pH on xylanase activity
Xylanase activity assays were carried out using 1% (w/v) birch-wood xylan under a variety of buffer conditions between pH 3.0 and 8.0. Calculation of the specific activity of each xylanase showed that XylC has much higher specific activity, ranging from about 4.5 to 90-fold higher than the other xylanases at pH 5.5 (Table 3). The optimum pH of all the xylanases is between pH 4.0 – 4.5, although XylA and XylC retain activity between pH 4.0 – 6.0 and pH 4.0 – 7.0, respectively (Figure 3). XylC exhibited twofold higher activity at its optimum pH (pH 4.0) than at pH 5.5.
Figure 3.
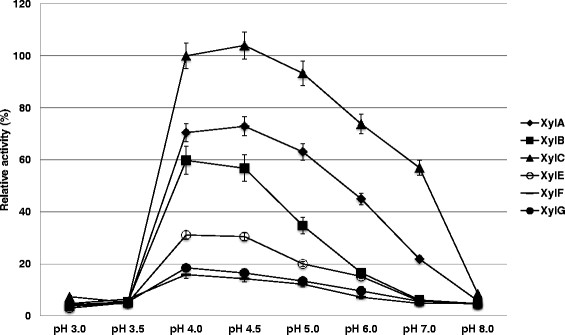
Optimum pH of six kinds of xylanase (GH11). The optimal pH of each xylanase was determined using the DNS method with 1% (w/v) birch-wood xylan. The buffers were: 0.1 M citric acid buffer pH 3.0 – 3.5; 0.1 M sodium acetate pH 4.0 − 4.5; 0.1 M sodium citrate pH 5.0 – 6.0; 0.1 M Hepes-NaOH pH 7.0; 0.1 M Tris–HCl pH 8.0. The enzymes are: XylA (black diamond), XylB (black square), XylC (black triangle), XylE (white circle), XylF (black dash) and XylG (black circle). The percent relative activity is based on the activity of XylC at pH 4.0. The data plotted are averages of triplicate experiments.
Thermostability of the xylanases
Fluorescence-based thermal shift assays of the xylanases were performed in a 96-well plate format using a real-time PCR detection system and a volume of 20 μl per well. The samples were heated at 0.1°C/s from 25 to 90°C. All the xylanases were found to be thermostable between 52 and 61°C at pH 5.5, and their thermostability increased by 4 to 10°C at pH 4.0 (Table 3). These results show that all xylanases are in a more active and thermostable form at pH 4.0.
Gene expression in cultures grown on cellulose and xylan
A. cellulolyticus Y-94 was cultured on Solka Floc or xylan as the carbon source for 24, 72, and 120 h, then the levels of expression of their xylanase gene were measured by real-time quantitative PCR (Figure 4). No expression of any xylanase gene was observed after 24 h under both growth conditions (undetectable). A higher expression level of xylA and xylB was observed in 72 h, indicating that these genes contribute to xylan degradation by A. cellulolyticus. xylC, which showed the highest xylanase activity, was expressed slightly compared with xylA and xylB; the expression of xylD, xylE, xylF and xylG was not detected under any of the conditions tested, with the exception of xylG at 120 h, xylan cultivation. Figure 4 shows the expression levels relative to that of gpdA used as an internal control.
Figure 4.
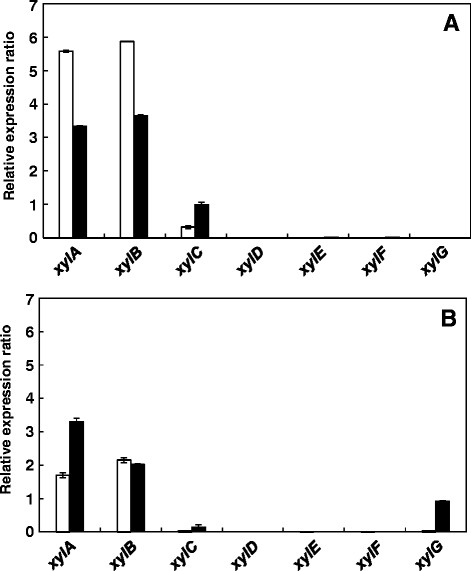
Expression of xylanase genes.A. cellulolyticus Y-94 was cultured in medium containing 50 g/l of cellulose (A) or 50 g/l of xylan (B). Culture time was 24 h (data not shown), 72 h (white bars), and 120 h (black bars). Expression levels are shown relative to that of gpdA as an internal control.
Discussion
Analysis of the A. cellulolyticus genome identified seven ORFs exhibiting homology to the xylanase (GH11) genes: xylA, xylB, xylC, xylD, xylE, xylE and xylG, which encode XylA (209 aa), XylB (282 aa), XylC (223 aa), XylD (190 aa), XylE (234 aa), XylF (276 aa) and XylG (233 aa), respectively. Each gene also encodes the signal peptide at the N-terminus. N-terminal sequence analysis showed that each signal peptide is cleaved by an aminopeptidase N-terminal to the arginine residue in this homologous expression system (Inoue et al. [2013]). In addition, Blast alignments indicated that only XylB and XylF have a carbohydrate-binding module (CBM-1) at their C-terminus, suggesting that XylB and XylF bind more strongly to their substrate than the other xylanases. Figure 1 shows the sequence alignment of the active domain in the xylanases. Six xylanases (XylA, XylB, XylC, XylE, XylF and XylG) exhibit amino acid sequence homology of around 11 ~ 52% and strictly conserve the two catalytic glutamate residues, and also strongly conserve other resides located in the speculative catalytic cleft (Paës et al. [2012]). Although the structure of XylC or any of the other xylanases has not been determined, it is expected to have a β-jellyroll structure based on multiple sequence alignment and molecular weight analyses. This protein folding motif is extremely common in small molecular weight (<30 kDa) proteins like the GH11 family (Paës et al. [2012]). Analysis of the A. cellulolyticus genome showed the xylD gene to be smaller than the other family members (Figure 1), and that it lacks some of the regions conserved in the other xylanases, including one of the catalytic glutamic acids. It seems that the xylD gene cannot translate normal xylanase protein (XylD) in A. cellulolyticus. Amino acid sequence alignments suggest that the active domains of six xylanases (Xyl11A, B, C, E, F, and G) in A. cellulolyticus have common activity and structure, and share high homology with the GH11 family of Trichoderma reesei, which is well known for its hyper-productivity (Zou et al. [2012]). There is over 50% sequence identity between XylC and T. reesei GH11.
Six of the seven xylanases (Xyl11A, B, C, E, F, and G) were expressed and purified successfully in A. cellulolyticus transformant strains. SDS-PAGE analysis shows that the purified XylB, XylE, XylF and XylG are accompanied by one or two smaller, faint bands (Figure 2). However, these proteins exhibited the significant xylanase activity toward the birch-wood xylan (Bailey et al. [1992]). Therefore, their faint bands suggest the nick regions of the recombinant proteins expressed. The nick regions seem to be located somewhere in these folded proteins (unclear). The N-terminus of the expressed protein giving rise to each major band was also determined (Table 2). These data indicate that A. cellulolyticus efficiently translates and expresses these xylanase genes using this homologous expression system (Inoue et al. [2013]).
Activity assays conducted at various pH values showed that all the xylanases have their optimum pH between pH 4.0 – 4.5, although XylA and XylC retain activity between pH 4.0 – 6.0 and pH 4.0 – 7.0, respectively (Figure 3). In addition, we examined the specific activity of all the xylanases against xylan and found that XylC has much higher specific activity than the other xylanases (Table 3). XylC exhibited between 4.5 to 90-fold higher activity at pH 5.5 than the other xylanases, and twofold higher activity at its optimum pH (pH 4.0) than at pH 5.5. These results clearly show that XylC plays the important role as a glycoside hydrolase 11 (GH11) of all the xylanases in A. cellulolyticus. It is possible that XylC uses a different mechanism for substrate recognition and catalysis compared to other xylanases. Detailed structural studies will be required to clarify this issue.
The levels of expression of the seven xylanase genes in Y-94 cultured for 24, 72, and 120 h were measured by real-time quantitative PCR (Figure 4). No expression of any xylanase gene was observed after 24 h (data not shown). A higher expression level of xylA and xylB was observed compared to the other xylanases, indicating that these genes contribute to xylan degradation by A. cellulolyticus after 72 h. XylC, which showed the highest xylanase activity, was expressed slightly less than xylA and xylB. This result suggests that expression of xylA and xylB is important for the saccharification of biomass, and that the detailed substrate specificity of xylA and xylB may be different from that of xylC. The expression of xylD, xylE, xylF and xylG was not detected under any of the conditions tested, with the exception of xylG at 120 h, xylan cultivation (Figure 4).
Thermal shift assays conducted using differential scanning fluorimetry showed that the xylanases have a Tm of around 60°C at pH 5.5, and that their thermostability increases by up to 10°C at pH 4.0 (Table 3). These results show that all GH11s in A. cellulolyticus are most stable and active at pH 4.0. Industrial use of this enzyme will require thermostability at least up to 70°C. XylC plays an important role in degrading xylan and is a good candidate for industrial use in food, paper and chemical production (Deutschmann and Dekker [2012]; Polizeli et al. [2005]). We are currently attempting to crystallize XylC to determine its atomic-level structure by X-ray crystallography.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Masahiro Watanabe, Email: masa-watanabe@aist.go.jp.
Hiroyuki Inoue, Email: inoue-h@aist.go.jp.
Benchaporn Inoue, Email: inoue-b@aist.go.jp.
Miho Yoshimi, Email: m-yoshimi@aist.go.jp.
Tatsuya Fujii, Email: tatsuya.fujii@aist.go.jp.
Kazuhiko Ishikawa, Email: kazu-ishikawa@aist.go.jp.
Acknowledgements
This work was supported by the Japan-U.S. Cooperation Project for Research and Standardization of Clean Energy Technologies.
References
- Bailey MJ, Biely P, Poutanen K. Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol. 1992;4:257–270. doi: 10.1016/0168-1656(92)90074-J. doi:10.1016/0168-1656(92)90074-J. [DOI] [Google Scholar]
- Beaugrand J, Chambat G, Wong V, Goubet F, Rémond C, Paës G, Benamrouche S, Debeire P, O'Donohue M, Chabbert B. Impact and efficiency of GH10 and GH11 thermostable endoxylanases on wheat bran and alkali-extractable arabinoxylans. Carbohydr Res. 2004;4:2529–2540. doi: 10.1016/j.carres.2004.08.012. doi:10.1016/j.carres.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary-cell walls in flowering plants - consistency of molecular-structure with the physical-properties of the walls during growth. Plant J. 1993;4:1–30. doi: 10.1111/j.1365-313X.1993.tb00007.x. doi:10.1111/j.1365-313X.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Collins T, Gerday C, Feller G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol Rev. 2005;4:3–23. doi: 10.1016/j.femsre.2004.06.005. doi:10.1016/j.femsre.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Coughlan MP, Hazlewood GP. Beta-1,4-D-xylan-degrading enzyme-systems - biochemistry, molecular-biology and applications. Biotechnol Appl Biochem. 1993;4:259–289. doi:10.1111/j.1470-8744.1993.tb00244.x. [PubMed] [Google Scholar]
- Coutinho PM, Henrissat B. Carbohydrate-active enzymes: an integrated database approach. Roy Soc Ch. 1999;4:3–12. [Google Scholar]
- Deutschmann R, Dekker RF. From plant biomass to bio-based chemicals: latest developments in xylan research. Biotechnol Adv. 2012;4:1627–1640. doi: 10.1016/j.biotechadv.2012.07.001. doi:10.1016/j.biotechadv.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Dimarogona M, Topakas E, Christakopoulos P, Chrysina ED. The structure of a GH10 xylanase from Fusarium oxysporum reveals the presence of an extended loop on top of the catalytic cleft. Acta Crystallogr D. 2012;4:735–742. doi: 10.1107/S0907444912007044. doi:10.1107/S0907444912007044. [DOI] [PubMed] [Google Scholar]
- Fang X, Yano S, Inoue H, Sawayama S. Strain improvement of Acremonium cellulolyticus for cellulase production by mutation. J Biosci Bioeng. 2009;4:256–261. doi: 10.1016/j.jbiosc.2008.11.022. doi:10.1016/j.jbiosc.2008.11.022. [DOI] [PubMed] [Google Scholar]
- Fujii T, Fang X, Inoue H, Murakami K, Sawayama S. Enzymatic hydrolyzing performance of Acremonium cellulolyticus and Trichoderma reesei against three lignocellulosic materials. Biotechnol Biofuels. 2009;4:24. doi: 10.1186/1754-6834-2-24. doi:10.1186/1754-6834-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Murakami K, Sawayama S. Cellulase hyperproducing mutants derived from the fungus Trichoderma reesei QM9414 produced large amounts of cellulase at the enzymatic and transcriptional levels. Biosci Biotechnol Biochem. 2010;4:419–422. doi: 10.1271/bbb.90655. http://dx.doi.org/10.1271/bbb.90655 http://dx.doi.org/10.1271/bbb.90655. [DOI] [PubMed] [Google Scholar]
- Fujii T, Iwata K, Murakami K, Yano S, Sawayama S. Isolation of uracil auxotrophs of the fungus Acremonium cellulolyticus and the development of a transformation system with the pyrF gene. Biosci Biotechnol Biochem. 2012;4:245–249. doi: 10.1271/bbb.110498. http://dx.doi.org/10.1271/bbb.110498 http://dx.doi.org/10.1271/bbb.110498. [DOI] [PubMed] [Google Scholar]
- Hachmann JP, Amshey JW. Models of protein modification in Tris-glycine and neutral pH Bis-Tris gels during electrophoresis: effect of gel pH. Anal Biochem. 2005;4:237–245. doi: 10.1016/j.ab.2005.04.015. http://dx.doi.org/10.1016/j.ab.2005.04.015 http://dx.doi.org/10.1016/j.ab.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Harris GW, Jenkins JA, Connerton I, Cummings N, Loleggio L, Scott M, Hazlewood GP, Laurie JI, Gilbert HJ, Pickersgill RW. Structure of the catalytic core of the family F xylanase from Pseudomonas fluorescens and identification of the xylopentaose-binding sites. Structure. 1994;4:1107–1116. doi: 10.1016/S0969-2126(94)00112-X. [DOI] [PubMed] [Google Scholar]
- Inoue H, Fujii T, Yoshimi M, Taylor LE 2nd, Decker SR, Kishishita S, Nakabayashi M, Ishikawa K. Construction of a starch-inducible homologous expression system to produce cellulolytic enzymes from Acremonium cellulolyticus. J Ind Microbiol Biotechnol. 2013;4:823–830. doi: 10.1007/s10295-013-1286-2. doi:10.1007/s10295-013-1286-2. [DOI] [PubMed] [Google Scholar]
- Kumar R, Singh S, Singh OV. Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives. J Ind Microbiol Biotechnol. 2008;4:377–391. doi: 10.1007/s10295-008-0327-8. doi:10.1007/s10295-008-0327-8. [DOI] [PubMed] [Google Scholar]
- Lee JW, JY P k, Kwon M, Choi IG. Purification and characterization of a thermostable xylanase from the brown-rot fungus Laetiporus sulphureus. J Biosci Bioeng. 2009;4:33–37. doi: 10.1016/j.jbiosc.2008.09.006. doi:10.1016/j.jbiosc.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Lo Leggio L, Kalogiannis S, Bhat MK, Pickersgill RW. High resolution structure and sequence of T. aurantiacus xylanase I: implications for the evolution of thermostability in family 10 xylanases and enzymes with (beta)alpha-barrel architecture. Proteins. 1999;4:295–306. doi: 10.1002/(SICI)1097-0134(19990815)36:3<295::AID-PROT4>3.0.CO;2-6. doi:10.1002/(SICI)1097-0134(19990815)36:3. [DOI] [PubMed] [Google Scholar]
- Lo MC, Aulabaugh A, Jin GX, Cowling R, Bard J, Malamas M, Ellestad G. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem. 2004;4:153–159. doi: 10.1016/j.ab.2004.04.031. http://dx.doi.org/10.1016/j.ab.2004.04.031 http://dx.doi.org/10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- McIlvaine TC. A buffer solution for colorimetric comparison. J Biol Chem. 1921;4:183–186. [Google Scholar]
- Mitsuishi Y, Yamanobe T, Yagisawa M, Takasaki Y. Purification and properties of thermostable xylanases from mesophilic fungus strain Y-94. Agric Biol Chem. 1987;4:3207–3213. doi: 10.1271/bbb1961.51.3207. http://dx.doi.org/10.1271/bbb1961.51.3207 http://dx.doi.org/10.1271/bbb1961.51.3207. [DOI] [Google Scholar]
- Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;4:2212–2221. doi: 10.1038/nprot.2007.321. doi:10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- Paës G, Berrin JG, Beaugrand J. GH11 xylanases: Structure/function/properties relationships and applications. Biotechnol Adv. 2012;4:564–592. doi: 10.1016/j.biotechadv.2011.10.003. doi:10.1016/j.biotechadv.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Polizeli ML, Rizzatti AC, Monti R, Terenzi HF, Jorge JA, Amorim DS. Xylanases from fungi: properties and industrial applications. Appl Microbiol Biotechnol. 2005;4:577–591. doi: 10.1007/s00253-005-1904-7. doi:10.1007/s00253-005-1904-7. [DOI] [PubMed] [Google Scholar]
- Prade RA. Xylanases: from biology to biotechnology. Biotechnol Genet Eng Rev. 1996;4:101–131. doi: 10.1080/02648725.1996.10647925. [DOI] [PubMed] [Google Scholar]
- Senisterra GA, Finerty PJ Jr. High throughput methods of assessing protein stability and aggregation. Mol Biosyst. 2009;4:217–223. doi: 10.1039/b814377c. doi:10.1039/b814377c. [DOI] [PubMed] [Google Scholar]
- Shibuya T, Watanabe Y, Nalley KA, Fusco A, Salafsky B. The BCA protein determination system. An analysis of several buffers, incubation temperature and protein standards. Tokyo Ika Daigaku Zasshi. 1989;4:677–682. [Google Scholar]
- Sidhu G, Withers SG, Nguyen NT, McIntosh LP, Ziser L, Brayer GD. Sugar ring distortion in the glycosyl-enzyme intermediate of a family G/11 xylanase. Biochemistry. 1999;4:5346–5354. doi: 10.1021/bi982946f. doi:10.1021/bi982946f. [DOI] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;4:76–85. doi: 10.1016/0003-2697(85)90442-7. http://dx.doi.org/10.1016/0003-2697(85)90442-7 http://dx.doi.org/10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;4:4673–4680. doi: 10.1093/nar/22.22.4673. doi:10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink J, de Vries RP. Fungal enzyme sets for plant polysaccharide degradation. Appl Microbiol Biotechnol. 2011;4:1477–1492. doi: 10.1007/s00253-011-3473-2. doi:10.1007/s00253-011-3473-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanobe T, Mitsuishi Y, Takasaki Y. Isolation of a cellulolytic enzyme producing microorganism, culture conditions and some properties of the enzymes. Agric Biol Chem. 1987;4:65–74. doi: 10.1271/bbb1961.51.65. [DOI] [Google Scholar]
- York WS, O’Neill MA. Biochemical control of xylan biosynthesis – which end is up? Curr Opin Plant Biol. 2008;4:258–265. doi: 10.1016/j.pbi.2008.02.007. doi:10.1016/j.pbi.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Zou G, Shi S, Jiang Y, van den Brink J, de Vries RP, Chen L, Zhang J, Ma L, Wang C, Zhou Z. Construction of a cellulase hyper-expression system in Trichoderma reesei by promoter and enzyme engineering. Microb Cell Fact. 2012;4:21. doi: 10.1186/1475-2859-11-21. doi:10.1186/1475-2859-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]


