Abstract
High power densities have been obtained from MFC reactors having a purple color characteristic of Rhodopseudomonas. We investigated the microbial community structure and population in developed purple MFC medium (DPMM) and MFC effluent (DPME) using 16S rRNA pyrosequencing. In DPMM, dominant bacteria were Comamonas (44.6%), Rhodopseudomonas (19.5%) and Pseudomonas (17.2%). The bacterial community of DPME mainly consisted of bacteria related to Rhodopseudomonas (72.2%). Hydrogen oxidizing bacteria were identified in both purple-colored samples: Hydrogenophaga and Sphaerochaeta in the DPMM, and Arcobacter, unclassified Ignavibacteriaceae, Acinetobacter, Desulfovibrio and Wolinella in the DPME. The methanogenic community of both purple-colored samples was dominated by hydrogenotrophic methanogens including Methanobacterium, Methanobrevibacter and Methanocorpusculum with significantly lower numbers of Methanosarcina. These results suggeste that hydrogen is actively produced by Rhodopseudomonas that leads to the dominance of hydrogen consuming microorganisms in both purple-colored samples. The syntrophic relationship between Rhodopseudomonas and hydrogenotrophic microbes might be important for producing high power density in the acetate-fed MFC under light conditions.
Keywords: Purple color characteristic of Rhodopseudomonas, Microbial community, Hydrogen oxidizing bacteria, Hydrogenotrophic methanogens, Syntrophic relationship
Introduction
Microbial fuel cell (MFC) is a new technology in renewable energy. It generates electrical power while accomplishing waste water treatment by utilizing microorganisms (Pant et al. [2012]). Although MFCs have been comprehensively investigated, this technology is still at an early stage and under extensive laboratory research. Over the past 10 years, considerable effort has been made to improving power generation efficiency, focusing mainly on different MFC setups and new materials (Kim et al. [2008]; Lovley [2006]; Sleutels et al. [2012]). Microbial ecology studies in MFC systems are important for understanding the mechanism of microbial electricity generation (Rabaey et al. [2007]; Rabaey and Rozendal [2010]). In addition, MFC systems also provide insight into the physiological roles of microbes and better understanding of interactions in complex microbial communities within natural environments (Bretschger et al. [2010]).
The bacterial population and predominant species vary depending on operational conditions such as inocula, substrate nature and electrode materials (Sun et al. [2012]; Logan and Regan [2006]; Logan [2009]). In general, MFCs using mixed culture produce more power than ones with pure culture (Watson and Logan [2010]). However, complex syntrophic interactions existing in MFC systems relating to high power densities have not been well studied. Previous studies have also shown that MFC performances are affected by light, which can cause the solution medium of respective MFCs to enrich in the phototrophic purple nonsulfur (PNS) bacterium, Rhodopseudomonas palustris DX-1. This bacterium has been shown to produce higher power densities in pure culture than in mixed culture and increase power production along with light intensity. In contrast to, the R. palustris ATCC 17001 does not generate power (Xing et al. [2008]). The genus Rhodopseudomonas liberates hydrogen when illuminated anaerobically in the presence of a carbon source such as acetate and malate (Barbosa et al. [2001]). The hydrogen production is mediated by a nitrogenase enzyme and dependent on light (Basak and Das [2007]; Rey et al. [2007]).
Previous MFC microbial ecology studies have used Sanger-based 16S rRNA sequencing for microbial community characterization. Recent advances in next generation sequencing such as pyrosequencing offer a better alternative for comprehensively characterizing microbial communities, especially in terms of less abundant members (Dowd et al. [2008]; Huse et al. [2007]; Lee et al. [2010]). 454 pyrosequencing of 16S rRNA gene has been revealed highly diverse microbial communities in MFCs and MECs (Lee et al. [2010]; Jia et al. [2013]; Lu et al. [2012a]). The present study aimed at better understanding the high power density achieved in MFC reactors having purple color characteristic of Rhodopseudomonas. To do this, the microbial communities in the purple-colored samples collected from a media bottle and a MFC reactor respectively were investigated using 16S rRNA amplicon pyrosequencing.
Materials and methods
Sample source
The MFC medium was kept in a common media bottle (320 mL) sealed with its cap and placed under natural sunlight and fluorescent lamp lighting (around 400 lx) without any inoculum. Four weeks later the developed purple medium was collected. It was observed that coloring of the medium serendipitously became purple over time. The naturally developed purple MFC effluent was collected from a single-chamber air-cathode MFC (4.5 L) having four homemade brush anodes (60 mm in diameter and 150 mm in length) inoculated with the primary clarifier overflow of a local wastewater treatment plant. The setup has been operated in batch mode for approximately two years using acetate (1.0 g/L) as a substrate at 30°C. Both media had the following composition: 1.0 g/L sodium acetate in 50 mM phosphate buffer solution (PBS, pH 7.0) containing (per liter deionized water): KCl, 0.13 g/L; NaH2PO4 · 2H2O, 2.75 g/L; Na2HPO4 · 12H2O, 11.466 g/L; NH 4Cl, 0.31 g/ L, metal (12.5 mL/L) and vitamins (5 mL/L) (Lovley and Phillips [1988]). For simplicity, the MFC media bottle medium will now be referred to as “developed purple MFC medium (DPMM)” and the MFC reactor effluent as “developed purple MFC effluent (DPME)”.
Total DNA extraction and 16S rRNA gene pyrosequencing
Each purple-colored sample (5 ml) was centrifuged at 5630× g for 10 minutes to collect microbial biomass. Total genomic DNA was extracted using a PowerSoil DNA isolation kit (MO-BIO) according to the manufacturer’s protocol. Amplicon libraries were constructed for 454 sequencing using different sets of primers: 27 F (5′GAGTTTGATCMTGGCTCAG-3′) and 518R (5′-WTTACCGCGGCTGCTGG-3′) targeting the bacterial domain, 519 F (5′-CAGCMGCCGCGGTAA −3′) and 915R (5′-GTGCTCCCCCGCCAATTCCT −3′) targeting the archaeal domain (Baker et al. [2003]). The forward primers contained a 10 base barcode sequence positioned between the adapter and the primer sequence. The barcode sequences were unique for each sample.
The amplification of 16S rDNA was performed in a final volume of 50 μl containing 5 μl of 10X buffer, 1 μl of dNTP mixture (200 mM), 0.5 μl of (20XU) FastStart Taq DNA Polymerase (Roche), 1 μl of each primer (10 μM), 1 μL of DNA template (50 ng/μL) and water. The PCR conditions were as follows: 94°C for 3 min; 30 cycles of 94°C for 45 S, 55°C for 45S, followed by 72°C for 1 min; and a final extension at 72°C for 10 min. The PCR products were gel purified on 1% agarose gels using a Quick Gel Extraction Kit (Invitrogen). The purified 16 s amplicons were quantified with Picogreen and pooled in equal concentrations prior to emulsion PCR. Amplicon pyrosequencing was performed using a 454/Roche GS Junior instrument (454 Life Sciences).
Sequence analysis
The sffinfo command was used to extract fasta format (sequence data) and quality (quality scores) files from a raw.sff file with –s and –q options, respectively. The following steps were carried out by the QIIME (Quantitative Insights into Microbial Ecology) software package (Caporaso et al. [2011]). Output fasta and quality files were filtered to exclude sequences with lengths (<150 bp) and quality score (<50). Raw sequences with uncorrectable barcodes were removed. The remaining sequences were sorted by a mapping file to their relevant samples according to the barcode. The denoised dataset was clustered into operational taxonomic units (OTUs) with 97% similarity threshold, using the UCLUST algorithm. The representative sequences were selected and aligned with greengenes core set of aligned sequences using Pynast. Taxonomy was assigned using the greengenes database. For OTUs that represented more than 5% of the sequences for any sample, taxonomic identification at the genus level was determined using basic local alignment search tool (BLAST) to compare the representative sequence for that OTU against the National Center for Biotechnology Information (NCBI) nucleotide non-redundant database. Phylogenetic trees were constructed using the software MEGA v5 with the neighbor joining criterion, and 1,000 times of bootstrap resampling was performed to assess the confidence of tree topologies (Kumar et al. [2008]).
Results
Bacterial community revealed on the V1-V3 region of the 16S rRNA gene
The bacterial communities from DPMM and DPME were revealed using the 16 s rRNA primer set (V1- V3). The number of total reads was 8253 reads in DPMM and 4008 reads in DPME. We obtained 77 and 151 operational taxonomic units (OTUs) from DPMM and DPME at 97% similarity respectively (Figure 1). The rarefaction curve of the DPMM tends to reach a plateau, but the DPME showed that new bacterial phylotypes continued to emerge even after 4,000 reads. Betaproteobacteria (49.3%), Alphaproteobacteria (19.8%) and Gammaproteobacteria (17.8%) were major bacterial phyla in DPMM. On the other hand the DPME primarily had Alphaproteobacteria (75.4%), Epsilonproteobacteria (6.5%) and Bacteroidetes (5.7%) phyla (Figure 2).
Figure 1.
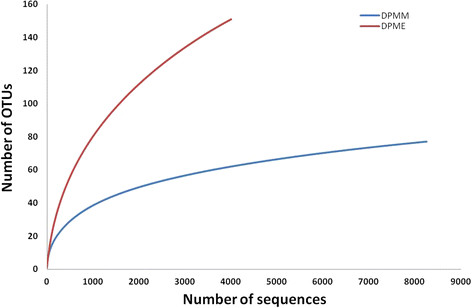
Rarefaction curves for bacterial 16S rRNA gene sequences with cut-off threshold of 97% similarity in DPMM and DPME.
Figure 2.
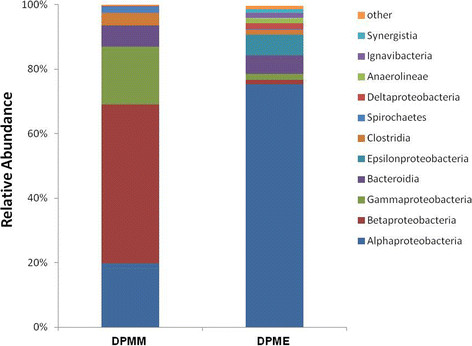
Relative abundances of bacterial phyla and proteobacterial classes in DPMM and DPME.
The community of the DPMM consisted mainly of bacteria related to the genera Comamonas (42.8%), Rhodopseudomonas (19.6%) and Pseudomonas (17.1%). The genus Rhodopseudomonas (73.3%) was the most dominant followed by Arcobacter (5.25%) in the DPME (Table 1). The phylogenetic analysis based on sequences of partial 16S rRNA genes indicated that Rhodopseudomonas was closely related to Rhodopseudomonas faecalis (AF123085) with 99% similarity (Figure 3). However, the predominant bacteria in DPMM, Comamonas and Pseudomonas, were found with low abundance in DPME accounting for 0.47% and 0.05% respectively. Comamonas and Arcobacter had a high similarity of 99% to Comamonas testosteroni (NR102841) and Arcobacter butzleri ED-1 (NR074567), respectively. Both are known exoelectrogenic bacteria in an acetate-fed MFC (Fedorovich et al. [2009]; Juang et al. [2011]). Hydrogenophaga (3.99%) and Sphaerochaeta (1.88%) were found at higher abundance in the DPMM compared to the DPME (Table 1). It has been reported that the genus Hydrogenophaga were highly abundant in acetate-fed MFC and some Hydrogenophaga species showed hydrogen-dependent power generation (Kimura and Okabe [2013b]). The genus Sphaerochaeta was detected in the hydrogen-producing MECs (Lu et al. [2012b]). Higher abundance of the unclassified Ignavibacteriaceae (1.66%), Acinetobacter (0.68%), Desulfovibrio (0.8%), Wolinella (1.01%) and Azospirillum (1.14%) were present in DPMM than in DPME; these genera were commonly found in electrode biofilm community for MFCs and MECs (Pisciotta et al. [2012]; Liu et al. [2012]; Yu et al. [2012]). These bacteria are known to be able to use hydrogen as an electron donor (Wong et al. [1986]; Gross and Simon [2003]; Yong et al. [2002]; Iino et al. [2010]; Yuan et al. [2012]). Geobacter sp., a known exoelectrogenic bacteria, was found to account for 0.4% abundance in DPME, but was negligible in the DPMM.
Table 1.
Relative abundances of bacterial OTUs based on the V1-V3 primer set (>0.1% of total population) in DPMM and DPME
| Classification | DPMM (%) | DPME (%) | OTUs | Genbank closest match (accession no. and similarity) |
|---|---|---|---|---|
|
Rhodopseudomonas(Alphaproteobacteria) |
19.63 |
73.28 |
100 |
Rhodopseudomonas faecalis (JX282402, 99%) |
|
Comamonas |
42.82 |
0.47 |
27 |
Comamonas testosteroni (NR102841, 99%) |
|
Pseudomonas |
17.12 |
0.05 |
148 |
Pseudomonas stutzeri (KF171338, 99%) |
| Unclassfied Bacteroidales |
3.27 |
0.23 |
25 |
Uncultured bacterium (JQ724350, 99%) |
|
Arcobacter |
0 |
5.25 |
66 |
Arcobacter butzleri ED-1 (NR074567 99%) |
| Unclassfied Porphyromonadaceae |
1.48 |
0.29 |
7 |
Uncultured bacterium (JF568463, 99%) |
|
Hydrogenophaga |
3.99 |
0.05 |
153 |
Hydrogenophaga sp. (AB746948, 98%) |
| Unclassfied Clostridiales |
1.46 |
0.23 |
137 |
Uncultured Anaerovorax sp. (JQ087107, 98%) |
|
Sphaerochaeta |
1.88 |
0.13 |
127 |
Uncultured bacterium (JQ245575, 99%) |
|
Azospirillum(Alphaproteobacteria) |
0 |
1.14 |
73 |
Uncultured bacterium (GQ152964, 99%) |
| Unclassfied Methylocystaceae(Alphaproteobacteria) |
0 |
1.38 |
135 |
Uncultured Rhizobiales (JQ723649, 99%) |
|
Acinetobacter(Gammaproteobacteria) |
0.3 |
0.68 |
136 |
Uncultured bacterium (KC001216, 99%) |
|
Geobacter(Deltaproteobacteria) |
0.01 |
0.23 |
190 |
Uncultured Geobacter (JQ724335, 99%) |
|
Desulfovibrio(Deltaproteobacteria) |
0.1 |
0.8 |
122 |
Bacterium enrichment (GU196246, 98%) |
| Unclassfied Desulfobacteraceae(Deltaproteobacteria) |
0 |
0.31 |
140 |
Uncultured bacterium (GQ152952, 98%) |
|
Wolinella(Epsilonproteobacteria) |
0 |
1.01 |
29 |
Uncultured bacterium (JQ987976, 100%) |
|
Clostridium(Firmicutes) |
0.33 |
0 |
79 |
Iron-reducing bacterium (FJ802349, 97%) |
|
Bacteroides(Firmicutes) |
0.51 |
0 |
46 |
Uncultured bacterium (JQ983412, 97%) |
|
Oscillospira(Firmicutes) |
0.59 |
0 |
168 |
Uncultured Oscillibacter (JX462523, 99%) |
| Unclassfied Catabacteriaceae(Firmicutes) |
0 |
0.36 |
9 |
Uncultured bacterium (KC408589, 99%) |
| Unclassfied Ruminococcaceae(Firmicutes) |
0.22 |
0.1 |
125 |
Uncultured bacterium (JQ983937, 99%) |
|
Dysgonomonas(Bacteroidetes) |
0.4 |
0.1 |
156 |
Uncultured Dysgonomonas (JX462548, 99%) |
| Unclassfied Porphyromonadaceae(Bacteroidetes) |
0 |
1.4 |
113 |
Uncultured bacterium (JN792255, 99%) |
| WCHB1-05 (Chloroflexi) |
0 |
0.26 |
141 |
Uncultured bacterium (JX224526, 99%) |
| WCHB1-50 (Chloroflexi) |
0 |
1.01 |
191 |
Uncultured bacterium (AJ387902, 96%) |
| Unclassfied Ignavibacteriaceae (Chlorobi) |
0 |
1.66 |
54 |
Uncultured Ignavibacterium (JQ724356, 100%) |
| Aminiphilus (Synergistetes) | 0 | 1.01 | 75 | Uncultured bacterium (KC736302, 99%) |
Figure 3.
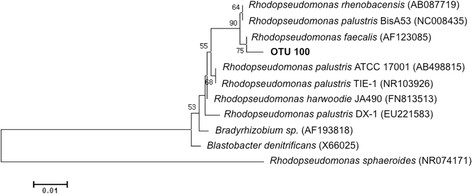
Phylogenetic tree based on 16S rRNA gene sequences showing the relationships of OUT 100 with members of the genusRhodopseudomonas. Bootstrap values (>50%) are shown on the nodes from 1000 bootstrap replicates. The scale bar represents 5% difference in nucleotide sequences.
Archaeal and bacterial communities revealed by the V3-V5 region of the 16S rRNA gene
The number of total high quality sequences obtained from raw pyrosequencing data was 4586 reads. We obtained 59 OTUs for DPMM and 38 OTUs for DPME at 97% similarity. The rarefaction curves were far from the plateauing due to the limitation in sequencing depth for covering a much higher bacterial diversity than archaeal diversity (Additional file 1: Figure S1). The dominance of Euryarchaeota was found in both purple-colored samples, accounting for 57.2% (DPMM) and 93.2% (DPME) of total sequence reads. The remaining bacterial 16S rRNA gene sequences were affiliated with Bacteroidetes, Proteobacteria and Firmicutes phyla and their relative abundance in DPMM was higher than in DPME (Additional file 2: Figure S2). The archaeal community in DPMM was mainly dominated by 16 s rRNA gene sequences closely related to hydrogenotrophic methanogens comprising of Methanobacterium (20.7%), Methanobrevibacter (18.8%) and Methanocorpusculum (15.3%). Methanobrevibacter was found to be most dominant in DPME (85.86%) (Figure 4). While Methanosarcina was found to account for 1.1% of DPMM, there was no detection of Methanosarcina in DPME. The archaeal communities in both purple-colored samples mainly comprised of hydrogenotrophic methanogens.
Figure 4.

Relative abundances of archaeal genera in DPMM and DPME.
Discussion
Light conditions significantly enhanced power densities (around 8–16%) in the MFCs using mixed and pure cultures of R. palustris strains DX-1 and RE-2 respectively (Xing et al. [2009]). In order to better understand electricity generation in MFCs containing purple photosynthetic Rhodopseudomonas palustris in mixed culture, microbial communities in DPMM and DPME were investigated using 16S rRNA gene pyrosequencing. As expected, Rhodopseudomonas was found to be dominant in both samples, but it was present in much higher abundance in DPME than DPMM; this might be due to the fact that DPME has developed for a longer period of time (2 years) than DPMM (4 weeks). The Rhodopseudomonas sp. detected in our study does not provide insight on its capability for electricity generation. The Rhodopseudomonas sp. was found to be more closely related with R. faecalis and R. rhenobacensis which are unknown for their ability to generate electricity, and R. palustris ATCC 17001 which does not produce an electric current, but it is less closely related to known electrogenic bacteria R. palustris strains DX-1 (Xing et al. [2008]).
Previous studies have shown that Geobacter, Comamonas and Pseudomonas dominated the anode communities of acetate-fed MFCs under light conditions, revealed by PCR/DGGE and 16S rRNA gene library analyses (Xing et al. [2009]). The capacity of electricity generation in acetate-fed MFCs by those genera has also been revealed (Xing et al. [2010]; Pham et al. [2008]; Xing et al. [2008]). However, these genera were found with low abundance in the DPME. This contrast in abundance might indicate that the species structure of the bacterial community differ between the MFC anode biofilm and effluent, whilst also being dependent on inoculum source.
Chemolithotrophic bacterium Hydrogenophaga, which consumes hydrogen and carbon dioxide as energy and carbon sources respectively (Willems et al. [1989]). They were found in DPMM but not in DPME. Geobacter sulfurreducens has been found in syntrophic cooperation with the Hydrogenophaga sp. strain AR20 in acetate-fed MFC (Kimura and Okabe [2013a]). In DPMM, Hydrogenophaga might use the hydrogen produced by Rhodopseudomonas but not the acetate. Members of chemolithotrophic bacteria were also found in DPME, including unclassified Ignavibacteriaceae, Acinetobacter, Desulfovibrio and Azospirillum which are known to be capable of using hydrogen as an electron donor (Wong et al. [1986]; Gross and Simon [2003]; Yong et al. [2002]; Iino et al. [2010]). It has been reported that the growth of G. sulfurreducens was more efficient when co-cultured with Wolinella and Desulfovibrio, which act as hydrogen-consuming partners; with nitrate as the electron acceptor, acetate oxidation was more rapid, resulting in faster growth of Geobacter sulfurreducens under low hydrogen partial pressure (Cord-Ruwisch et al. [1998]). The removal of hydrogen by the hydrogen-consuming bacteria in the DPME might have stimulated the growth of Rhodopseudomonas. Our results suggested that the growth of Rhodopseudomonas was due to the syntrophic cooperation with hydrogen-consuming bacteria in both purple-colored samples.
Anodic hydrogen oxidation by hydrogenotrophic exoelectrogens produces electrical current after complete oxidation of acetate, and a rapid current increase occurs when hydrogen gas is supplied to the reactor (Lee et al. [2009]). However, a previous study showed that electricity generation by hydrogenotrophic exoelectrogens was excluded due to inhibition of nitrogenase-dependent hydrogen production in Rhodopseudomonas by NH4Cl in the medium (Hillmer and Gest [1977]; Rey et al. [2007]). The presence of hydrogen-oxidizing bacteria in the purple-colored samples suggested that hydrogen production by Rhodopseudomonas could not be completely inhibited by NH4Cl in the medium (Rey et al. [2007]). Potential hydrogenotrophic exoelectrogens might coexist with Rhodopseudomonas in the anode biofilm due to their involvement in electricity generation via hydrogen oxidation. This potential syntrophic interaction between Rhodopseudomonas and hydrogen-oxidizing bacteria might be one of the explanations for the high power density obtained from illuminated MFCs.
Bacterial 16S rRNA gene sequences were also detected by the V3-V5 primer set and inconsistent results in the bacterial diversity and the abundance patterns of bacterial community were observed between V1-V3 and V3-V5 data sets. In our previous study (unpublished data), the V3-V5 primer set was also applied for methane-producing biocathodes in MECs, which accounted for 99% of the total sequences belonged to the archaea domain. The PCR amplification of the V3–V5 region of 16S rRNA can be biased depending on the activity of the archaeal population present in the samples. The differences in portions of archaeal sequences between two purple-colored samples suggest that the DPME could have a more active archaeal population than DPMM.
Archaeal community in MFC reactors enriched with Rhodopseudomonas sp. has not been reported yet. The predominant hydrogenotrophic methanogens were found in both purple-colored samples, but not acetoclastic methanogens despite the MFC medium containing high concentration of acetate. Under anaerobic conditions, methanogenic archaea often partner with heterotrophic H2-producing bacteria which catalyze oxidation of a variety of organic compounds (fatty acids, alcohols, and aromatic compounds). The methanogens utilize the H2 produced by these heterotrophic bacteria during methanogenesis while the heterotrophic bacteria benefit from the methanogens, which play a role in the removal of excess hydrogen that would inhibit their growth (McInerney et al. [2008]; Sakai et al. [2009]). Hydrogenotrophic methanogenesis by Methanobrevibacter in DPME might be relying on the predominance of Rhodopseudomonas for interspecies H2 transfer. However, it seems likely that CO2 derived from Rhodopseudomonas is limited as a major source of carbon for methane generation due to its known role in being recycled into biomass (McKinlay and Harwood [2010]). Hydrogen generation by Rhodopseudomonas in both purple-colored samples containing acetate could share similar environmental condition with hydrogen-producing MEC reactors fed with acetate, where hydrogenotrophic methanogens have emerged as the most active methanogens (Lu et al. [2012a]).
In general, few acetoclastic methanogens are present in acetate fed-MFCs due to their inhibition in growth as a result of air exposure and outcompetition for organic substrates by facultative anaerobes and exoelectrogens (Chae et al. [2010]; Dar et al. [2008]). In addition, hydrogen inhibition of growth and acetate metabolism in Methanosarcina species has been reported (Ahring et al. [1991]; Ferguson and Mah [1983]). Minor acetoclastic methanogen (Methanosarcina) present in the DPMM and the non-detection of Methanosarcina in the DPME might be indication of the robust inhibition by hydrogen produced by Rhodopseudomonas and its acetate competition.
In conclusion, although the electric generation ability of Rhodopseudomonas detected in our study is unknown, putative hydrogenotrophic exoelectrogens existing in MFC reactors having purple-colored effluents might contribute to increasing power density. For future studies the above observations are needed to investigate how known-exoelectrogenic phototrophic bacteria can be applied to efficiently generate electricity via syntrophic relationships with the hydrogenotrophic exoelectrogens and methanogens in the acetate-fed MFC under light conditions.
Competing interests
The authors declare that they have no competing interests.
Additional files
Supplementary Material
Rarefaction curves based on the V3-V5 primer set with cut-off threshold of 97% similarity in DPMM and DPME.
Microbial community distribution based on the V3-V5 primer set in DPMM and DPME.
Contributor Information
Tae-Jin Park, Email: parktaejin1@gmail.com.
Weijun Ding, Email: superding@zju.edu.cn.
Shaoan Cheng, Email: shaoancheng@zju.edu.cn.
Manreetpal Singh Brar, Email: manreet@hku.hk.
Angel Po Yee Ma, Email: angelma.hku@gmail.com.
Hein Min Tun, Email: heinmin@gmail.com.
Frederick C Leung, Email: fcleung@hkucc.hku.hk.
Acknowledgements
This study was funded by grant from Initiative on Clean Energy and Environment, The University of Hong Kong and grants funded by the National Natural Science Foundation of China (No. 21073163, No. 51278448).
References
- Ahring BK, Westermann P, Mah RA. Hydrogen inhibition of acetate metabolism and kinetics of hydrogen consumption by methanosarcina-thermophila Tm-1. Arch Microbiol. 1991;4(1):38–42. [Google Scholar]
- Baker GC, Smith JJ, Cowan DA. Review and re-analysis of domain-specific 16S primers. J Microbiol Methods. 2003;4(3):541–555. doi: 10.1016/j.mimet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Barbosa MJ, Rocha JM, Tramper J, Wijffels RH. Acetate as a carbon source for hydrogen production by photosynthetic bacteria. J Biotechnol. 2001;4(1):25–33. doi: 10.1016/s0168-1656(00)00368-0. [DOI] [PubMed] [Google Scholar]
- Basak N, Das D. The prospect of purple non-sulfur (PNS) photosynthetic bacteria for hydrogen production: the present state of the art. World J Microb Biot. 2007;4(1):31–42. [Google Scholar]
- Bretschger O, Osterstock JB, Pinchak WE, Ishii S, Nelson KE. Microbial fuel cells and microbial ecology: applications in ruminant health and production research. Microb Ecol. 2010;4(3):415–427. doi: 10.1007/s00248-009-9623-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. P Natl Acad Sci USA. 2011;4:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae KJ, Choi MJ, Kim KY, Ajayi FF, Chang IS, Kim IS. Selective inhibition of methanogens for the improvement of biohydrogen production in microbial electrolysis cells. Int J Hydrogen Energ. 2010;4(24):13379–13386. [Google Scholar]
- Cord-Ruwisch R, Lovley DR, Schink B. Growth of geobacter sulfurreducens with acetate in syntrophic cooperation with hydrogen-oxidizing anaerobic partners. Appl Environ Microbiol. 1998;4(6):2232–2236. doi: 10.1128/aem.64.6.2232-2236.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar SA, Kleerebezem R, Stams AJM, Kuenen JG, Muyzer G. Competition and coexistence of sulfate-reducing bacteria, acetogens and methanogens in a lab-scale anaerobic bioreactor as affected by changing substrate to sulfate ratio. Appl Microbiol Biot. 2008;4(6):1045–1055. doi: 10.1007/s00253-008-1391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM, James GA, Wolcott RD. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;4:43. doi: 10.1186/1471-2180-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorovich V, Knighton MC, Pagaling E, Ward FB, Free A, Goryanin I. Novel electrochemically active bacterium phylogenetically related to arcobacter butzleri, isolated from a microbial fuel cell. Appl Environ Microbiol. 2009;4(23):7326–7334. doi: 10.1128/AEM.01345-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson TJ, Mah RA. Effect of H2-Co2 on methanogenesis from acetate or methanol in methanosarcina Spp. Appl Environ Microbiol. 1983;4(2):348–355. doi: 10.1128/aem.46.2.348-355.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross R, Simon J. The hydE gene is essential for the formation of wolinella succinogenes NiFe-hydrogenase. Fems Microbiol Lett. 2003;4(2):197–202. doi: 10.1016/S0378-1097(03)00681-5. [DOI] [PubMed] [Google Scholar]
- Hillmer P, Gest H. H2 metabolism in the photosynthetic bacterium Rhodopseudomonas capsulata: production and utilization of H2 by resting cells. J Bacteriol. 1977;4(2):732–739. doi: 10.1128/jb.129.2.732-739.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML, Mark Welch D. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 2007;4(7):R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino T, Mori K, Uchino Y, Nakagawa T, Harayama S, Suzuki K. Ignavibacterium album gen. nov., sp nov., a moderately thermophilic anaerobic bacterium isolated from microbial mats at a terrestrial hot spring and proposal of Ignavibacteria classis nov., for a novel lineage at the periphery of green sulfur bacteria. Int Syst Evol Micr. 2010;4:1376–1382. doi: 10.1099/ijs.0.012484-0. [DOI] [PubMed] [Google Scholar]
- Jia J, Tang Y, Liu B, Wu D, Ren N, Xing D. Electricity generation from food wastes and microbial community structure in microbial fuel cells. Bioresour Technol. 2013;4:94–99. doi: 10.1016/j.biortech.2013.06.072. [DOI] [PubMed] [Google Scholar]
- Juang DF, Yang PC, Chou HY, Chiu LJ. Effects of microbial species, organic loading and substrate degradation rate on the power generation capability of microbial fuel cells. Biotechnol Lett. 2011;4(11):2147–2160. doi: 10.1007/s10529-011-0690-9. [DOI] [PubMed] [Google Scholar]
- Kim BS, Kim BK, Lee JH, Kim M, Lim YW, Chun J. Rapid phylogenetic dissection of prokaryotic community structure in tidal flat using pyrosequencing. J Microbiol. 2008;4(4):357–363. doi: 10.1007/s12275-008-0071-9. [DOI] [PubMed] [Google Scholar]
- Kimura Z, Okabe S. Acetate oxidation by syntrophic association between geobacter sulfurreducens and a hydrogen-utilizing exoelectrogen. Isme J. 2013a;4(8):1472–1482. doi: 10.1038/ismej.2013.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Z, Okabe S. Hydrogenophaga electricum sp nov., isolated from anodic biofilms of an acetate-fed microbial fuel cell. J Gen Appl Microbiol. 2013;4(4):261–266. doi: 10.2323/jgam.59.261. [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;4(4):299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Torres CI, Parameswaran P, Rittmann BE. Fate of H-2 in an upflow single-chamber microbial electrolysis cell using a metal-catalyst-free cathode. Environ Sci Technol. 2009;4(20):7971–7976. doi: 10.1021/es900204j. [DOI] [PubMed] [Google Scholar]
- Lee TK, Doan TV, Yoo K, Choi S, Kim C, Park J. Discovery of commonly existing anode biofilm microbes in two different wastewater treatment MFCs using FLX Titanium pyrosequencing. Appl Microbiol Biot. 2010;4(6):2335–2343. doi: 10.1007/s00253-010-2680-6. [DOI] [PubMed] [Google Scholar]
- Liu W, Wang A, Sun D, Ren N, Zhang Y, Zhou J. Characterization of microbial communities during anode biofilm reformation in a two-chambered microbial electrolysis cell (MEC) J Biotechnol. 2012;4(4):628–632. doi: 10.1016/j.jbiotec.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Logan BE. Exoelectrogenic bacteria that power microbial fuel cells. Nat Rev Microbiol. 2009;4(5):375–381. doi: 10.1038/nrmicro2113. [DOI] [PubMed] [Google Scholar]
- Logan BE, Regan JM. Electricity-producing bacterial communities in microbial fuel cells. Trends Microbiol. 2006;4(12):512–518. doi: 10.1016/j.tim.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Lovley DR. Bug juice: harvesting electricity with microorganisms. Nat Rev Microbiol. 2006;4(7):497–508. doi: 10.1038/nrmicro1442. [DOI] [PubMed] [Google Scholar]
- Lovley DR, Phillips EJP. Novel mode of microbial energy-metabolism - organic-carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microb. 1988;4(6):1472–1480. doi: 10.1128/aem.54.6.1472-1480.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Xing D, Ren N. Pyrosequencing reveals highly diverse microbial communities in microbial electrolysis cells involved in enhanced H2 production from waste activated sludge. Water Res. 2012;4(7):2425–2434. doi: 10.1016/j.watres.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Lu L, Xing DF, Ren NQ, Logan BE. Syntrophic interactions drive the hydrogen production from glucose at low temperature in microbial electrolysis cells. Bioresour Technol. 2012b;4:68–76. doi: 10.1016/j.biortech.2012.08.040. [DOI] [PubMed] [Google Scholar]
- McInerney MJ, Struchtemeyer CG, Sieber J, Mouttaki H, Stams AJM, Schink B, Rohlin L, Gunsalus RP. Physiology, ecology, phylogeny, and genomics of microorganisms capable of syntrophic metabolism. Ann N Y Acad Sci. 2008;4:58–72. doi: 10.1196/annals.1419.005. [DOI] [PubMed] [Google Scholar]
- McKinlay JB, Harwood CS. Carbon dioxide fixation as a central redox cofactor recycling mechanism in bacteria. P Natl Acad Sci USA. 2010;4(26):11669–11675. doi: 10.1073/pnas.1006175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant D, Singh A, Van Bogaert G, Olsen SI, Nigam PS, Diels L, Vanbroekhoven K. Bioelectrochemical systems (BES) for sustainable energy production and product recovery from organic wastes and industrial wastewaters. Rsc Adv. 2012;4(4):1248–1263. [Google Scholar]
- Pham TH, Boon N, Aelterman P, Clauwaert P, De Schamphelaire L, Vanhaecke L, De Maeyer K, Hofte M, Verstraete W, Rabaey K. Metabolites produced by pseudomonas sp enable a gram-positive bacterium to achieve extracellular electron transfer. Appl Microbiol Biot. 2008;4(5):1119–1129. doi: 10.1007/s00253-007-1248-6. [DOI] [PubMed] [Google Scholar]
- Pisciotta JM, Zaybak Z, Call DF, Nam JY, Logan BE. Enrichment of microbial electrolysis cell biocathodes from sediment microbial fuel cell bioanodes. Appl Environ Microb. 2012;4(15):5212–5219. doi: 10.1128/AEM.00480-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabaey K, Rozendal RA. Microbial electrosynthesis - revisiting the electrical route for microbial production. Nat Rev Microbiol. 2010;4(10):706–716. doi: 10.1038/nrmicro2422. [DOI] [PubMed] [Google Scholar]
- Rabaey K, Rodriguez J, Blackall LL, Keller J, Gross P, Batstone D, Verstraete W, Nealson KH. Microbial ecology meets electrochemistry: electricity-driven and driving communities. Isme J. 2007;4(1):9–18. doi: 10.1038/ismej.2007.4. [DOI] [PubMed] [Google Scholar]
- Rey FE, Heiniger EK, Harwood CS. Redirection of metabolism for biological hydrogen production. Appl Environ Microb. 2007;4(5):1665–1671. doi: 10.1128/AEM.02565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai S, Imachi H, Sekiguchi Y, Tseng IC, Ohashi A, Harada H, Kamagata Y. Cultivation of methanogens under low-hydrogen conditions by using the coculture method. Appl Environ Microb. 2009;4(14):4892–4896. doi: 10.1128/AEM.02835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels THJA, Ter Heijne A, Buisman CJN, Hamelers HVM. Bioelectrochemical systems: an outlook for practical applications. Chemsuschem. 2012;4(6):1012–1019. doi: 10.1002/cssc.201100732. [DOI] [PubMed] [Google Scholar]
- Sun Y, Wei J, Liang P, Huang X. Microbial community analysis in biocathode microbial fuel cells packed with different materials. AMB Express. 2012;4(1):21. doi: 10.1186/2191-0855-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson VJ, Logan BE. Power production in MFCs inoculated with shewanella oneidensis MR-1 or mixed cultures. Biotechnol Bioeng. 2010;4(3):489–498. doi: 10.1002/bit.22556. [DOI] [PubMed] [Google Scholar]
- Willems A, Busse J, Goor M, Pot B, Falsen E, Jantzen E, Hoste B, Gillis M, Kersters K, Auling G, Deley J. Hydrogenophaga, a New genus of hydrogen-oxidizing bacteria that includes hydrogenophaga-flava comb-Nov (formerly pseudomonas-flava), hydrogenophaga-palleronii (formerly pseudomonas-palleronii), hydrogenophaga-pseudoflava (formerly pseudomonas-pseudoflava and pseudomonas-carboxydoflava), and hydrogenophaga-taeniospiralis (formerly pseudomonas-taeniospiralis)N. Int J Syst Bacteriol. 1989;4(3):319–333. [Google Scholar]
- Wong TY, Graham L, Ohara E, Maier RJ. Enrichment for hydrogen-oxidizing acinetobacter spp in the rhizosphere of hydrogen-evolving soybean root-nodules. Appl Environ Microb. 1986;4(5):1008–1013. doi: 10.1128/aem.52.5.1008-1013.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D, Zuo Y, Cheng S, Regan JM, Logan BE. Electricity generation by rhodopseudomonas palustris DX-1. Environ Sci Technol. 2008;4(11):4146–4151. doi: 10.1021/es800312v. [DOI] [PubMed] [Google Scholar]
- Xing D, Cheng S, Regan JM, Logan BE. Change in microbial communities in acetate- and glucose-fed microbial fuel cells in the presence of light. Biosens Bioelectron. 2009;4(1):105–111. doi: 10.1016/j.bios.2009.06.013. http://dx.doi.org/10.1016/j.bios.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Xing D, Cheng SA, Logan BE, Regan JM. Isolation of the exoelectrogenic denitrifying bacterium comamonas denitrificans based on dilution to extinction. Appl Microbiol Biot. 2010;4(5):1575–1587. doi: 10.1007/s00253-009-2240-0. [DOI] [PubMed] [Google Scholar]
- Yong P, Farr JPG, Harris IR, Macaskie LE. Palladium recovery by immobilized cells of desulfovibrio desulfuricans using hydrogen as the electron donor in a novel electrobioreactor. Biotechnol Lett. 2002;4(3):205–212. [Google Scholar]
- Yu J, Park Y, Cho H, Chun J, Seon J, Cho S, Lee T. Variations of electron flux and microbial community in air-cathode microbial fuel cells fed with different substrates. Water Sci Technol: J Int Assoc Water Pollut Res. 2012;4(4):748–753. doi: 10.2166/wst.2012.240. [DOI] [PubMed] [Google Scholar]
- Yuan H, Ge T, Chen C, O’Donnell AG, Wu J. Significant role for microbial autotrophy in the sequestration of soil carbon. Appl Environ Microbiol. 2012;4(7):2328–2336. doi: 10.1128/AEM.06881-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rarefaction curves based on the V3-V5 primer set with cut-off threshold of 97% similarity in DPMM and DPME.
Microbial community distribution based on the V3-V5 primer set in DPMM and DPME.


