Abstract
Chromatin immunoprecipitation (ChIP) allows specific protein-DNA interactions to be isolated. Combining ChIP with high-throughput sequencing reveals the DNA sequence involved in these interactions. Here, we describe how to perform ChIP-seq starting with whole tissues or cell lines, and ending with millions of short sequencing tags that can be aligned to the reference genome of the species under investigation. We also outline additional procedures to recover ChIP-chip libraries for ChIP-seq and discuss contemporary issues in data analysis.
1. INTRODUCTION
Protein-DNA interactions play vital roles in the regulation of gene expression, genome integrity and chromatin organization. The in vivo mapping of transcription factor binding and modified histones has greatly broadened our understanding of how the genome can be deployed to achieve tissue and developmental stage-specific gene regulation. Computational methods have provided substantial insight into our understanding of transcriptional regulation [1], and yet recent experimental discoveries have underscored the need for a simple and reproducible method for mapping protein-DNA interactions on a global basis. These include recent discoveries that: (i) sequence-specific transcription factors (TFs) do not occupy all positions in the genome that would be predicted by their corresponding binding matrices [2,3], (ii) sequence specific transcription factors often bind regions that do not show similarity to their canonical binding matrices [2-5] and (iii) the binding patterns of TFs between species are poorly conserved [6-8].
Chromatin Immunoprecipitation (ChIP) [9,10] is a commonly used technique to detect interactions between proteins and DNA, which is based on the enrichment of DNA associated with a protein of interest. The development of ChIP coupled with high-throughput sequencing analysis (ChIP-seq) allows the unbiased identification of binding sites of a given transcription factor and has overcome several limitations inherent to microarray analysis of ChIP (ChIP-chip) [11,12].
Due to their size and more repetitive nature, higher eukaryotic genomes are a challenge for tiling microarray design. Most of the repetitive sequence cannot be interrogated with high confidence, whereas direct sequencing can reveal binding events located in repetitive regions in the mammalian genome [13-15]. Every model organism requires species-specific microarray designs before ChIP-chip can be performed, while ChIP-seq can be done without prior knowledge of the underlying sequence and relies only on the subsequent DNA sequence alignment to the reference genome of interest. Furthermore, the nature of the microarray hybridization signal makes detection and rigorous quantification of low abundance signals problematic. Taken together, ChIP-seq can provide greater resolution, sensitivity and specificity compared to ChIP-chip [11,14,16].
A number of high-throughput sequencing technology platforms have been developed that are suitable for ChIP-seq, including the Genome Analyzer (Illumina, formerly Solexa), SOLiD (Applied Biosystems), 454-FLX (Roche) and HeliScope (Helicos) [17]. The Illumina Genome Analyzer and the ABI SOLiD sequencers produce shorter reads but give a higher number of sequencing reads per run, whereas the 454-FLX sequencer gives longer yet fewer sequencing reads per run [18]. Sequencing depth is a critical factor in identifying weaker binding positions and it has been shown that millions of mapped sequencing tags are needed to detect enrichments significantly higher than twofold [19].
Here, we outline detailed methodologies for ChIP-seq using the Illumina Genome Analyzer to produce tens of millions of aligned sequencing tags. Our protocol adapts methods described previously [14,20] with additional modifications and technical improvements to the chromatin immunoprecipitation (ChIP) and library generation steps.
2. Description of method
2.1. Overview
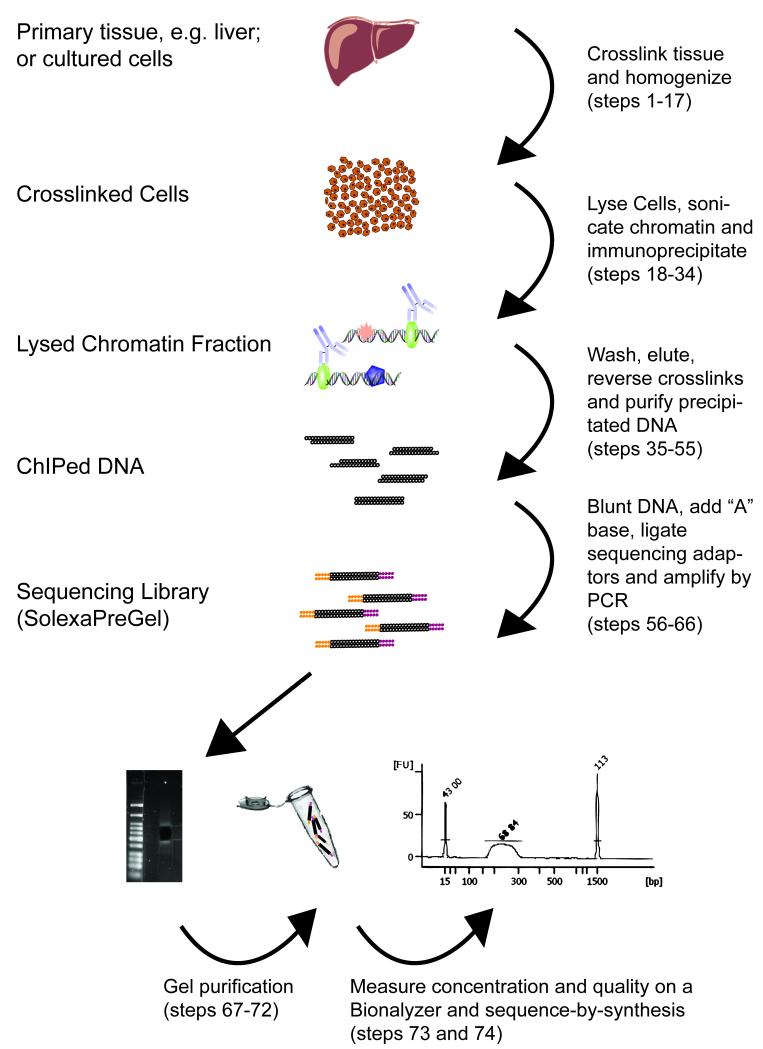
A successful ChIP experiment begins with the crosslinking of protein-DNA interactions using formaldehyde (Fig 1). Histone modifications can also be successfully identified using non-crosslinked native chromatin in the ChIP protocol [21], but the ability to capture weaker and transient protein-DNA interactions has made formaldehyde fixation of starting materials a standard practice. After crosslinking, the tissue is homogenized, and the cells are lysed. Subsequently, the chromatin is sheared using sonication and incubated with magnetic beads coupled to an antibody specific for the target protein. The success of the ChIP is dependent on the antibody being used; indeed, we have found that a large fraction of highly specific, IP-proven antisera do not perform well against cross-linked chromatin. We therefore strongly recommend the use of a positive control antibody as described below when testing new antibodies, tissues or performing ChIP-seq for the first time. In principle, the generation of a sequencing library from DNA is relatively straightforward. However, as opposed to ChIP analyzed by real-time PCR, ChIP-seq requires a larger quantity of precipitated DNA to minimize the generation of adapter dimer artefacts and to preserve the complexity of the DNA sample. This protocol is routinely used in our laboratory and has been successful with a variety of antibodies, tissues and cells from a wide range of vertebrate species.
Figure 1.
Outline of ChIP-seq procedure.
2.2. Step-by-step protocol
2.2.1. Crosslinking of cells or primary tissues
Covalent fixation of the protein-DNA complexes is achieved by brief formaldehyde fixation. Ideally the starting material for one ChIP uses 5×107 cells from culture or the equivalent of one-quarter of an adult mouse liver. While it is possible to start with limited material [22,23], we have found that higher amounts of starting material yield more consistent and reproducible protein-DNA enrichments. To crosslink material for ChIP, follow steps 1 to 6 for cultured cells and steps 7 to 17 for whole tissue.
Cells
Add 1/10 volume of fresh 11% formaldehyde solution (50mM Hepes-KOH, 100 mM NaCl, 1mM EDTA, 0.5 mM EGTA, 11% Formaldehyde) to plates or flasks. Alternatively, pour off cell culture media and cover cells in a solution of 1% formaldehyde (final concentration) in 50mM Hepes-KOH, 100 mM NaCl, 1mM EDTA, 0.5 mM EGTA.
Swirl briefly and let sit at room temperature for 10 min.
Add 1/20 volume of 2.5 M glycine to quench formaldehyde.
Rinse cells twice with ice cold PBS.
Transfer cells to 15ml conical tubes and spin 4 min at 2000 × rcf.
Proceed with cell lysis or freeze cells in liquid nitrogen and store pellets at −80 °C. Continue with step 18.
Primary tissue
7. Whenever possible, perfuse tissue with PBS to remove blood.
8. On a kimwipe wetted with PBS, mince tissue quickly with a razorblade into small pieces. The pieces should be not bigger than 0.5 cm3.
9. Add tissue to at least five volumes of freshly prepared solution A (1% formaldehyde, 50mM Hepes-KOH, 100 mM NaCl, 1mM EDTA, 0.5 mM EGTA).
10. Mix and leave at room temperature for 20 min.
11. Add 1/20 volume of 2.5 M glycine to quench formaldehyde.
12. Rinse tissue with PBS and flash freeze or proceed directly to step 13.
13. Dounce tissue in ice-cold PBS first with the loose and later with the tight pestle (Dounce Tissue Grinder from Wheaton Science, Catalog #357544). We do not add protease inhibitors during this step.
14. The equivalent of one dounced mouse liver is filtered into a 50 ml conical tube through a 100 μm cell strainer to remove connective tissue. Fill tube with ice-cold PBS to 40 ml and centrifuge at 4 °C at 2500 × rcf for 3 min.
15. Discard supernatant and repeat wash.
16. Resuspend pellet in 10 ml ice cold PBS and transfer to 15 ml conical tube, centrifuge as above. Distribute into several 15 ml tubes if there would be more than 2ml of tissue per tube.
17. Proceed with lysis, or freeze cells in liquid nitrogen and store pellets at −80 °C
2.2.2. Preblock and binding of antibody to magnetic beads
Like all immunoprecipitation experiments, successful ChIP requires a suitable antibody. With ambitious antibody generation efforts led by both academic and industrial labs, many candidate antibodies corresponding to DNA binding proteins are available. Numerous antibodies have been shown to work in ChIP; nevertheless, it is often the case that a series of antibodies must be tested against a protein of interest. Often the creation of new antisera targeted to different epitopes is required to create ChIP-grade antibodies. When testing new antibodies or performing ChIP (and especially ChIP-seq) for the first time we recommend using a positive control such as anti-H3K4me3 (ab8580, Abcam) which detects the trimethylated lysine 4 form of histone H3 in a wide range of species, provides robust reliable enrichments, and highlights potential transcription start sites in the genome.
Magnetic beads are less porous than traditional agarose beads [24][21] and easier to handle, and hence highly recommended for chromatin immunoprecipitation to reduce background precipitation of nonspecific DNA. The exact type of magnetic beads depends of the species and subclass of the antibody being used. Protein G coated beads have high affinity to most rabbit and goat antibodies. Antibodies are incubated with the magnetic beads prior to the addition of the nuclear extracts, and excess, unbound antibodies are then washed away. This ensures that unbound antibodies cannot compete with the antibodies attached to the magnetic beads for target epitopes during ChIP. All antibody incubations and washes are performed at 4 °C.
18. Add 100 μl magnetic beads (Invitrogen, Dynabeads) to a 1.5 ml microfuge tube. Add 1 ml block solution (0.5% BSA (w/v) in PBS). Set up 1 tube per IP.
19. Collect the beads using magnetic stand. Remove supernatant by aspiration.
20. Wash beads in 1.0 ml block solution two more times.
21. Resuspend beads in block solution and add 2 - 15 μg of antibody in a final volume of 250 μl.
22. Incubate overnight or a minimum of 4h on a rotating platform at 4°C.
23. Wash magnetic beads as described above (3 times in 1 ml block solution).
24. Resuspend in 100 μl block solution.
2.2.3 Cell Lysis and Sonication
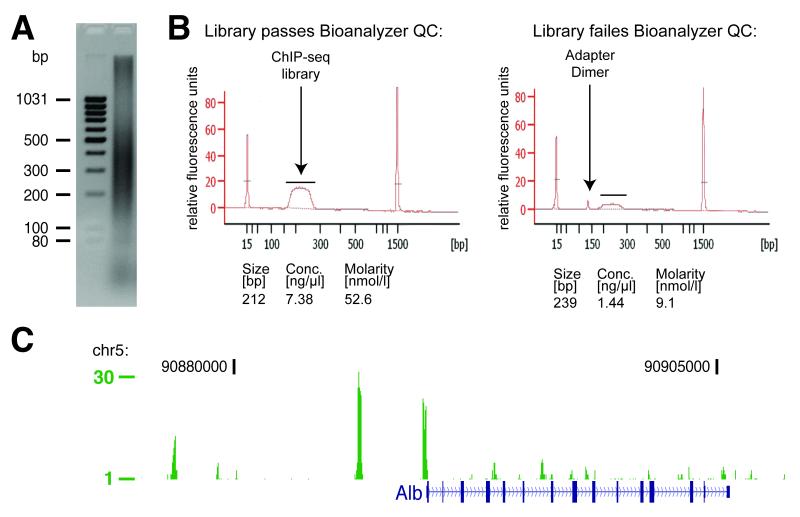
The cells are lysed to remove the bulk of cytosolic proteins, leaving only the contents of the nucleus for ChIP. This lysis step can improve ChIP results in cases where the protein of interest is not only bound to chromatin but also abundant in the cytosol. The successful isolation of nuclei can be confirmed after step 2 using standard Trypan blue staining. All lysis buffers should be supplemented with protease inhibitors (Complete, EDTA-free, Roche, #11873580001). Settings for the sonication of chromatin must be pre-determined based on equipment and material. The equipment and settings described here work well with most cell lines and primary tissues. After sonication, the opaque lysate should become clear as a first indicator of a successful sonication. If the lysate does not clear after additional cycles of sonication, the material may be over cross-linked and the cross-linking time in step 2 (for cells) or step 10 (for tissues) should be reduced. Ideally, most chromatin fragments resulting from sonication occur between 200-400 bp. This size range can be confirmed by running the whole-cell extract (WCE) on an agarose gel or an Agilent Bioanalyzer after reversing formaldehyde crosslinking and the DNA purification subsequent to step 54. (Fig 2A)
Figure 2.
(A) Example whole-cell extract (WCE) sonication result. (B) Agilent Bioanalyzer 2100 traces for two ChIP-seq libraries. The left panel shows a successful library preparation. The right panel shows a library with significant amounts of adapter dimers. The quantification of the libraries is shown underneath each panel. (C) C/EBPα ChIP-seq genome track (absolute fragment count) at the albumin locus in mouse hepatocytes showing several strong and weaker binding events.
25. Resuspend each pellet of cross-linked tissue in 10 ml of LB1 (50 mM Hepes-KOH, pH 7.5; 140 mM NaCl; 1mM EDTA; 10% Glycerol; 0.5% NP-40 or Igepal CA-630; 0.25% Triton X-100). Rock at 4°C for 10 min. Spin at 2,000 × rcf for 4 minutes at 4°C in a tabletop centrifuge.
26. Resuspend each pellet in 10 ml of LB2 (10 mM Tris-HCL, pH8.0; 200 mM NaCl; 1 mM EDTA; 0.5 mM EGTA) . Rock gently at 4°C for 5 min. Pellet nuclei in tabletop centrifuge by spinning at 2,000 × rcf for 5 minutes at 4°C.
27. Resuspend each pellet in each tube in 3 ml LB3 (10 mM Tris-HCl, pH 8; 100 mM NaCl; 1 mM EDTA; 0.5 mM EGTA; 0.1% Na-Deoxycholate; 0.5% N-lauroylsarcosine).
28. Transfer cells to a homemade “sonication tube” (cut a polypropylene, 15ml conical tube into two pieces at the 7 ml mark).
29. Sonicate suspension with a microtip attached to a Misonix Sonicator 3000 Homogenizer sonicator. Samples should be kept in an ice-water bath during sonication. Sonicate 8 - 12 cycles of 30 sec ON and 60 sec OFF. Power-output should be between 27 and 33 Watt.
30. Add 300 μl of 10% Triton X-100 to sonicated lysate. Split into 2 ml centrifuge tubes. Spin at 20,000 × rcf for 10 minutes at 4°C to pellet debris.
31. Combine supernatants from the 2 ml centrifuge tubes in a new 15 ml conical tube. The amount of LB3 and Triton X-100 is adjusted to the number of ChIPs to be performed. For example to prepare 3 ChIPs from one 3 ml sonication you would top up the centrifuged sonication to 9 ml with LB3 and 600 μl of 10% Triton X-100 (1% final concentration). Mix well and split into three 15 ml conical tubes, so that each contains 3 ml of cell lysate with 300 μl Triton per ChIP.
32. Save 50 μl of cell lysate from each sonication as whole-cell extract (WCE) DNA. Store at −20°C.
2.2.4. Chromatin immunoprecipitation
33. Add 100 μl antibody/magnetic bead mix from step 24 to cell lysates.
34. Gently mix overnight on rotator or rocker at 4°C.
2.2.5. Wash, elution, and cross-link reversal
Steps 35 through 40 should be done in a 4°C cold room.
35. Pre-chill one 1.5 ml microfuge tube for each IP.
36. Transfer half the volume of an IP to a pre-chilled tube.
37. Let tubes sit in magnetic stand to collect the beads. Remove supernatant and add remaining IP. Let tubes sit again in magnetic stand to collect the beads and remove supernatant.
38. Add 1 ml RIPA Buffer (50 mM Hepes-KOH, pH 7.5; 500 mM LiCl; 1mM EDTA; 1% NP-40 or Igepal CA-630; 0.7% Na-Deoxycholate) to each tube. Remove tubes from magnetic stand and shake or agitate tube gently to resuspend beads. Replace tubes in magnetic stand to collect beads. Remove supernatant. Repeat this wash 4-6 more times.
39. Wash once with 1 ml TBS (20 mM Tris-HCl, pH 7.6; 150 mM NaCl).
40. Spin at 960 × rcf for 3 minutes at 4°C and remove any residual TBS buffer using the magnetic stand.
41. Add 200 μl of elution buffer (50 mM Tris-HCl, pH 8; 10 mM EDTA; 1% SDS).
42. Elute and perform reverse crosslinking at 65°C for 6-18 h. Resuspend beads in the first 15 min with brief vortexing every five minutes.
43. Thaw 50 μl of the WCE from step 32, add 150 μl of elution buffer and mix. Reverse the formaldehyde crosslinking as in step 42 simultaneously with the ChIP samples.
2.2.6. Digestion of cellular protein and RNA
The proteins and RNA in the samples are enzymatically digested and the DNA is further purified by phenol-chloroform extraction and ethanol precipitation. GlycoBlue (Ambion, AM9516) can be used instead of glycogen as carrier for the ethanol precipitation, which substantially improves visualization of the DNA pellet.
44. Remove 200 μl of supernatant and transfer to new tube.
45. Add 200 μl of TE to each tube of IP and WCE DNA to dilute SDS in elution buffer.
46. Add 8 μl of 1 mg/ml RNaseA (Ambion Cat # 2271).
47. Mix and incubate at 37°C for 30 min.
48. Add 4 μl of 20 mg/ml proteinase K (Invitrogen, 25530-049).
49. Mix and incubate at 55°C for 1-2 hours.
50. Add 400ul phenol:chloroform:isoamyl alcohol (P:C:IA) and separate phases with 2 ml Phase Lock Gel Light tubes FPR5101 Flowgen Bioscience and follow the instructions provided.
51. Transfer aqueous layer to new centrifuge tube containing 16 μl of 5M NaCl (200 mM final concentration) and 1 μl of 20 μg/μl GlycoBlue (Ambion, AM9516).
52. Add 800 μl 100% EtOH. Incubate for 30 min at −80°C.
53. Spin at 20,000 × rcf for 10 minutes at 4°C to pellet DNA. Wash pellets with 500 μl of 80% EtOH and spin at 20,000 × rcf for 5 minutes.
54. Dry pellets 10 to 20 minutes in a speedvac at 45 °C and resuspend each in 30 μl of 10 mM Tris-HCl, pH 8.0.
55. Measure DNA concentration of WCE with NanoDrop 1000 (Thermo Fisher Scientific). Note that ChIP samples are too low in DNA concentration to give reliable results using a NanoDrop.
2.2.7. Perform End-Repair of the DNA
The final steps of this protocol convert the ChIP-enriched DNA into a library suitable for high-throughput sequencing using an Illumina Genome Analyzer. Historically, the Illumina Genomic Sample Preparation Kit has been used for ChIP reactions, since no dedicated ChIP-seq kit was available. Indeed, the recently-released ChIP-seq kit is very similar to the Genomic Sample Preparation Kit. Here, we have replaced all the enzymes from the Illumina Genomic Sample Prep Kit using standard, commercially available products. Only the Adapter Oligonucleotide Mix and the PCR primers 1.1 and 2.1 should be directly ordered from Illumina, as they contain proprietary modifications that, based on our experience, greatly improve library synthesis.
Using commercial reagents as opposed to pre-assembled kits greatly reduces the price per library generation, and allows the preparation of a master mix for the following reactions. For new users, or if only a small number of samples are to be processed at a time, it may be simpler to use the Illumina Genomic Sample Preparation Kit. ChIP-seq libraries can also be prepared using paired-end adapters and PCR primers, as they are compatible with both single- and paired-end flowcells. However, we have not optimized the protocol for the paired-end adapters. Based on our prior experiences, we predict that concentration of the paired-end adapter in the ligation reaction will need to be optimized carefully.
-
56. Pipette the following mix into PCR tubes and keep on ice, or make a master mix on ice containing water, buffer and enzymes, and add to the samples. Incubate 30 min at 20 °C in a thermal cycler.
ChIP sample, or 5-50 ng of WCE 30.0 μl Water 45.0 ul T4 DNA ligase buffer(NEB, B0202S) 10.0 μl dNTP mix,each 10 mM (NEB, N0447L) 4.0 μl T4 DNA polymerase (NEB, M0203L) 5.0 μl Klenow DNA polymerase (NEB, M0210L) 1.0 μl T4 PNK (NEB, M0201L) 5.0 μl Total 100.0 μl 57. Cleanup samples using the DNA Clean&Concentrator-5 kit, (Zymo Research, USA), following the manufacturer’s protocol.
58. Elute with 33 μl EB preheated to 50 °C. Chill on ice.
2.2.8. Add “A” Bases to the DNA
Pipette the following mix in 1.5 ml tubes and keep on ice, or make a master mix on ice containing buffer and enzyme and add to the samples.
| DNA sample | 32.0 μl |
| Klenow buffer (NEB, B7002S) | 5.0 ul |
| dATP (1mM) | 10.0 μl |
| Klenow 3′-5′ exo minus (NEB, M0212L) | 3.0 μl |
|
| |
| Total | 50.0 μl |
59. Incubate 30 min at 37 °C in a water bath.
60. Cleanup samples using the DNA Clean&Concentrator-5 kit, following the manufacturer’s protocol.
61. Elute with 9 μl EB preheated to 50 °C. Chill on ice.
2.2.9. Ligate Sequencing Adapters to DNA Fragments
One of the most persistent problems we have observed is the formation of adapter dimers generated during adapter-target DNA ligations. Dimers form clusters on the flowcell of the Illumina Genome Analyzer and thus compete with the desired sample for sequencing. This can reduce the sequencing reads from the actual ChIP experiment. A number of steps can be taken to significantly reduce adapter dimers: (i) the amount of Adapter Oligonucleotide mix can be titrated by diluting the Adapter Oligonucleotide mix 40-fold. This gives robust results with as little as 5 ng of DNA; (ii) pooling of multiple ChIPs can be used to increase the relative amount of sample DNA versus Adapter Oligonucleotides; (iii) ultrapure ligases can be used, such as those from Enzymatics [25]; (iiii) after PCR amplification the library can be purified by solid-phase reversible immobilization technology as described in [25]; and (iv) Illumina recommends a gel purification step following the ligation reaction which will likely minimize these adaptor dimers but may result in loss of sample complexity in the case of ChIP-seq.
Pipette the following mix in 1.5 ml tubes on ice. Alternatively, add a master mix containing buffer and Adapter Oligo to the samples followed by the ligase.
| DNA sample | 8.0 μl |
| Quick Ligation Reaction Buffer (NEB, M2200L) | 12.5 ul |
| 40-fold diluted Genomic Adapter Oligo mix (Illumina) | 2.0 μl |
| Quick T4 DNA Ligase (NEB, M2200L) | 2.5 μl |
|
| |
| Total | 25.0 μl |
62. Incubate 15 min at RT.
63. Cleanup samples using the DNA Clean&Concentrator-5 kit, following the manufacturer’s protocol.
64. Elute with 24 μl EB preheated to 50 °C. Chill on ice.
2.2.10. Amplify Adapter-Modified DNA by PCR
ChIP-seq libraries at this stage have by nature only a small mass. To reduce the risk of complexity loss, we perform the PCR amplification before the size-selection step in agarose gel.
The library is amplified using a DNA polymerase that: (i) is high fidelity and (ii) produces blunt ends. The recommended 2X master mix (NEB, F-531L) containing Phusion DNA polymerase should be distributed into convenient aliquots to avoid multiple freeze-and-thaw cycles. Alternatively, PCR yield can also be improved using Platinum Pfx polymerase (Invitrogen) see [25] for details.
Pipette the following mix directly into PCR tubes and keep on ice.
| DNA sample | 23.0 μl |
| Phusion Master Mix with HF Buffer (NEB, F-531L) | 25.0 ul |
| Genomic PCR primer 1.1 (Illumina) | 1.0 μl |
| Genomic PCR primer 2.1 (Illumina) | 1.0 μl |
|
| |
| Total | 50.0 μl |
-
65. Run program:
Step1: 98°C 30 sec
Step2: 98°C 10 sec
Step3: 65°C 30 sec
Step4: 72°C 30 sec
Step5 GOTO Step2 17 times
Step6: 72°C 5 min
Step7: 4°C HOLD
66. Purify with QIAquick and elute with 32 μl preheated EB. This sample is called SolexaPreGel. If validation of ChIP-seq library is desired, follow the optional protocol for reamplification (steps 75-80). Alternatively purify using solid-phase reversible immobilization technology as described in [25].
2.2.11. Gel purification of SolexaPreGel for ChIPseq
The amplified library is purified on an agarose gel to select a specific size-range for cluster generation, as well as to remove potential adapter dimers. One sample per gel can be used to avoid cross contamination of different libraries.
Using Xylene cyanol (Sigma, X4126) as loading dye has the advantage that it runs above the actual library, and does not interfere with the visualisation of the critical size range (150 bp – 700 bp) on a transilluminator.
67. Cast the appropriate number of 50 ml 2% agarose (BIO-RAD,161-3106) TAE gels with 5 μl SybrSafe (S33102).
68. Add 3 μl of loading buffer (50% glycerol supplemented with 0.25% Xylene cyanol) to 8 μl of DNA ladder (NEB, N3233L).
69. Add 10 μl of loading buffer to each sample.
70. Load the entire ladder into the first well of the gel, leave one lane empty and load the sample into the next well. Load only one sample per gel to eliminate any possibility of cross-contamination.
71. Run gel at 120 V for 40 min.
72. Excise the 200 – 300 bp fragments on a Dark Reader (Claire Chemical Research) and purify the DNA with a Qiagen MinElute Gel Extraction Kit (Qiagen, 28606). When the DNA is extracted it might be advantageous not to heat the gel slice to 50 °C but to dissolve the gel slice at room temperature as discussed in [25]. Elute with 15 μl EB preheated to 50 °C. You can excise and store the larger fragments (300 – 800 bp) as a backup.
73. Run the library on a Bioanalyzer DNA 1000 assay (Agilent) to estimate the concentration and to check that no adapter dimers are present (Figure 2B). If there are adapter dimers visible, the library could be rescued by solid-phase reversible immobilization technology as described in [25].
74. The sample is now ready for sequencing on an Illumina Genome Analyzer (section 2.3.).
2.2.12. Optional Protocol for Reamplification
We have described and validated an additional procedure that begins by diluting 1 microlitre of the amplified ChIP-seq library (SolexaPreGel) in 9 μl of EB buffer for subsequent analysis using real-time PCR or ChIP-chip [14]. This approach allows direct testing of libraries or to confirm sequencing results with readily available technologies, such as DNA microarrays or real-time PCR. This portion of material should be set aside routinely. For this method, it is necessary to process a WCE sample at the same time as a reference for real-time PCR and/or ChIP-chip.
75. Use 2 μl of the diluted SolexaPreGel sample (1 μl SolexaPreGel in 9 μl EB) per PCR reaction. One should also amplify the WCE sample that will be used as an input control for subsequent analyses.
-
76. Make PCR mix:
-
77. Add 48 μl of PCR Mix to each sample and run program:
Step1: 95°C 2 minutes
Step2: 95°C 30 sec
Step3: 65°C 30 sec
Step4: 72°C 1 minute
Step5: GOTO Step2 24 times
Step6: 72°C 5 minutes
Step7: 4°C HOLD
78. After PCR is completed, cleanup samples with QIAquick minelute PCR Purification Kit. Elute with 25 μl EB.
79. Measure DNA concentration of all samples with NanoDrop.
80. Samples are ready for further processing towards microarray or real-time PCR.
2.2.13. Optional rescue of traditional ChIP-chip libraries for ChIP-seq
Ligation mediated PCR (LMPCR) [26] has been extensively used to amplify ChIP enriched DNA fragments [3,5,7,27]. Like the procedure presented here for building ChIP-seq libraries, LMPCR involves ligating annealed linkers to the DNA of interest, followed by a PCR amplification and (often) microarray analysis (ChIP-chip). While it is generally preferable to repeat ChIP-seq with new experiments, there are cases where the original material used for ChIP-chip was valuable and difficult to obtain; for instance, our laboratory uses primary human islets and hepatocytes samples that have limited supply. While the original ChIP-chip libraries could be processed into a ChIP-seq library by addition of new linkers, the short nature of the reads currently obtained by high-throughput sequencing technology necessitates the removal of the original ChIP-chip linkers prior to library generation. Otherwise, the first 25 bases would be the original ChIP-chip linker sequence. Here, we present an optional procedure to remove a ChIP-chip linker from a previously made LMPCR library, so it can be re-ligated to Illumina linkers, and used for ChIP-seq. While it is possible that additional amplifications during the Solexa library preparation could introduce bias into the results, we have obtained ChIP-seq results from ChIP-chip libraries that are fully consistent with the original ChIP-chip experiment. In order to control for any such bias, we recommend performing the same procedure on the LMPCR amplified input material that was used in the original ChIP-chip experiment.
81. Clean up ChIP-chip library using the DNA Clean&Concentrator-5 kit, following the manufacturer’s protocol. Elute in 42 μl EB preheated to 50 °C.
82. Add 5 ul of 10X PNK buffer (NEB, B0201S) to cleaned ChIP-chip library and heat to 70 °C for 10 min and chill on ice immediately (this may increase the efficiency of the subsequent phosphorylation step but can be omitted). Add 5 μl (50U) PNK (NEB, M0201S) and 5 μl 10 mM ATP and incubate at 37 °C for 1-2 hours.
83. Cleanup samples using the DNA Clean&Concentrator-5 kit, following the manufacturer’s protocol. Elute in 9 μl pre-warmed 50°C EB.
84. Exonuclease digestion of linkers on phosphorylated LMPCR library: Mix directly on ice in a PCR tube: 8.5 μl sample; 1 μl 10 × reaction buffer (NEB, B0262S) and 0.5 μl Lambda exonuclease (NEB, M0262S). Digest for 10 min at 37 °C in a PCR block. Heat inactivate for 10 min at 75 °C.
85. Clean up samples using the DNA Clean&Concentrator-5 kit and elute in 30 μl EB preheated to 50 °C.
86. Continue with ChIP-seq library generation (step 56 to 74).
2.3. Sequencing process
The Illumina Genome Analyser sequencing processes and biochemistry have been well described [28]. The sequencing capacity of next generation high-throughput sequencing machines is increasing at an almost exponential rate; for instance, the Illumina Genome Analyzer was able to produce 1Gb of sequence per flow cell in January 2008, yet by December 2009, predicted yields are estimated to be 100Gb. The amount of DNA sequence, the length of DNA reads and quality of the data produced by the next-generation sequencing technologies are only likely to increase.
Many groups have published improvements to the different technologies [25,29], most of which focus on the upfront sample preparation rather than the particular sequencing biochemistry. There are also ongoing developments in data analysis, primarily in genome alignment tools and peak calling, but also in image processing (see below). The most significant hurdle for efficient operation of the Genome Analysers involves quantification of the DNA library before cluster generation. The original methodologies of standard UV spectroscopy followed by titration of libraries to achieve optimal cluster density often afforded variable densities, and were laborious. The use of the Agilent Bioanalyser allows much better quality control of libraries prior to sequencing. In our hands, implementing the use of the Bioanalyser has increased the quality of library submissions to our sequencing service and improved the output of high quality sequencing data. The new high-sensitivity DNA1000 kit from Agilent improves detection of samples 20-fold. This allows quantification and QC of libraries from smaller amounts of starting material or fewer cycles of PCR amplification, both desirable to most users. The next generation technologies all require generation of a “sample prep spike” where minute quantities of DNA are prepared into libraries for sequencing, massively amplified for quantitation, and then massively diluted for sequencing. It would be preferable to bypass this amplification entirely and directly quantitate adapter ligated nucleic acid molecules, opening the way to improved analysis of limited clinical or biological resources.
Quantitative real-time PCR [25] allows very robust quantitation and uniform cluster densities from Illumina ready libraries. The protocols are slightly complicated by the need for multiple primer-probe combinations as the adapter molecules are different for single end and paired end, or DNA and RNA libraries. This is being addressed by Illumina and standard adapter sequences are scheduled for release in 2009/10.
2.4. Quality Control and Data Analysis
2.4.1. Basic Data Pipeline
The raw data format of the Illumina sequencer is images files. After each completed sequencing run these images are computationally processed to obtain nucleotide-base calls. Besides the standard analysis pipeline provided by Illumina, alternative base-calling algorithms exist, including Alta-Cyclic [30] and Rolexa [31], which are reported to reduce error rates and thus produce a higher number of alignable reads. Alternative programs tend to be more CPU intensive than the standard Illumina pipeline, a cost that somewhat counterbalances the sequence gains. However, improved base-calling will allow for longer and more reliable sequence reads and should in principle help map reads that cross into repetitive regions by anchoring them in the surrounding non-repetitive sequence. This procedure could also improve the reliability of the identification of single nucleotide polymorphisms (SNPs), and thus allele-specific protein-DNA contacts.
Subsequent to base-calling, the sequencing reads have to be aligned to a reference genome. Several applications are available to align the sequencing reads to a reference. Among many others there are ELAND [32], MAQ (maq.sourceforge.net) and Bowtie (bowtie-bio.sourceforge.net). The main differences among these algorithms are the use of quality values and the treatment of reads that map to multiple locations. MAQ uses the quality values provided by the base caller (which indicate the probability that the base is called correctly) to resolve mismatches in alignments. With MAQ, a mismatch at a low quality base is penalized less than a mismatch at a high quality base, since it is more likely that the difference is a sequencing error in that case. Bowtie and ELAND do not use quality values. If a read aligns to multiple positions in the reference genome equally well, MAQ and Bowtie choose one of those positions uniformly at random. In this case ELAND assigns these reads to an arbitrary, but not necessarily random, locations. Note that since MAQ uses quality values in scoring alignments and Bowtie does not, it is more likely that Bowtie will assign the same score to two alignments than will MAQ.
It should be noted that for the identification of binding events in some repetitive areas of the genome, the precise treatment of sequencing reads that map multiple times to the reference genome can be critical. However, these cases seem to represent a minority compared to the bulk of binding events.
2.4.2. Examination of Aligned Data as First Quality Control
In order to inspect if a ChIP-seq experiment was successful, it is convenient to view the alignment results as continuous-valued data in track formats, such as wiggle (WIG), GFF (General Feature Format), or bedGraph using for example the UCSC or Ensembl genome browser. Figure 2C shows a wiggle track for a ChIP-seq experiment against the liver master regulator C/EBPα at the albumin locus performed in primary mouse liver. The height of the track represents the number of overlapping sequencing reads at bp resolution. This visualization allows a quick evaluation of the enrichments present in the data.
2.4.3. Automated Identification of Binding Events
Following confirmation of successful genomic enrichment in a ChIP experiment, the next task is to identify the regions across the whole genome that are enriched in sequencing reads and thus harbor the DNA-protein interaction in vivo. Several algorithms have been developed to analyze chip-seq data and identify the locations of transcription factor binding sites and histone marks along the genome.
ChipSeq Peak Finder
ChipSeq Peak Finder [11] clusters the reads and uses the ratio of the counts in the immunoprecipitated and the control sample to call peaks. An updated version of the method, eRange [33], also allows the use of reads which map to multiple locations in the genome which results in a significant increase in the amount of data utilized.
XSET
The extended set method XSET [16] uses the full estimated length of the DNA fragments to call the regions with highest numbers of overlapping fragments.
Mikkelsen methodology
The method in Mikkelsen et.al. [34] takes into account the ‘mappability’ of the underlying sequence, a measure of how many reads could be uniquely mapped at each location, and computes p-values to find significant differences between the observed and expected number of fragments.
PeakSeq
PeakSeq [35] allows for this mappability effect, which starts with a normalization step comparing the control with the background component of the ChIP sample and then detects significantly high concentrations of reads using the Binomial distribution.
MACS
Model- based Analysis for Chip-Seq (MACS) [36] shifts the tags on the forward and reverse strand together and uses the Poisson distribution to detect enrichment. In addition, the method ignores multiple identical reads to avoid biases during amplification and sequencing library preparation.
QuEST
Quantitative enrichment of sequence tags (QuEST) [37] shifts the peaks from opposite strands together and produces a kernel density estimation-derived score to call the enriched regions.
FindPeaks
FindPeaks [38] calls peaks according to some minimum height criteria without including a control sample in the analysis.
SISSR
Site Identification from Short Sequence Reads (SISSR) [39] estimates high read counts using Poisson probabilities and calls regions where the peaks shift from the forward to the reverse strand.
Other methods
In Kharchenko et.al. [19] three similar peak calling methods are proposed, scoring read counts upstream and downstream of the each region to match tag patterns in the forward and reverse strands. In addition, Nix et. al. [40] have simulated spike-in data, combined them with input reads from real experiments and used different metrics to score the peaks controlling for false discoveries. Another method that has been developed is BayesPeak which uses hidden Markov models and Bayesian techniques to identify the enriched regions based on posterior probabilities [41].
As with any new technology, it will take some time until the analysis of ChIP-seq experiments is a more standardized process. The growing number of tailored web-based tools and the advances made in sequencing throughput and quality will facilitate and improve routine analysis in the future, and make this technology available to a broader group of researchers.
3. Concluding remarks
Using ChIP-seq, it is possible to ask, at a genome wide level, where and when proteins interact with DNA. As more high-throughput sequencers become available, the amount of information obtained through ChIP-seq is limited only by the available antibodies, sufficient starting material, and an accurate reference genome sequence on which to align results. The maps of transcription factor binding and modified histones generated by ChIP-seq are important resources for further functional investigation of the processes and mechanisms involved in gene regulation.
ACKNOWLEDGEMENTS
We thank J. S. Carroll for discussions.
Footnotes
Order the following primer: Primer 1.2 for reamplification: 5′AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT
Order the following primer: Primer 2.2 for reamplification: 5′CAAGCAGAAGACGGCATACGAGCTCTTCCGATCT Oligonucleotide sequences reference: http://intron.ccam.uchc.edu/groups/tgcore/wiki/013c0/Solexa_Library_Primer_Sequences.html
References
- [1].Elnitski L, Jin VX, Farnham PJ, Jones SJ. Locating mammalian transcription factor binding sites: a survey of computational and experimental techniques. Genome Res. 2006;16:1455–1464. doi: 10.1101/gr.4140006. [DOI] [PubMed] [Google Scholar]
- [2].Rabinovich A, Jin VX, Rabinovich R, Xu X, Farnham PJ. E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res. 2008;18:1763–1777. doi: 10.1101/gr.080622.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- [4].Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- [6].Wilson MD, Barbosa-Morais NL, Schmidt D, Conboy CM, Vanes L, et al. Species-specific transcription in mice carrying human chromosome 21. Science. 2008;322:434–438. doi: 10.1126/science.1160930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat. Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, et al. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- [9].Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 2000;25:99–104. doi: 10.1016/s0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- [10].Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- [11].Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- [12].Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- [13].Wold B, Myers RM. Sequence census methods for functional genomics. Nat. Methods. 2008;5:19–21. doi: 10.1038/nmeth1157. [DOI] [PubMed] [Google Scholar]
- [14].Schmidt D, Stark R, Wilson MD, Brown GD, Odom DT. Genome-scale validation of deep-sequencing libraries. PLoS ONE. 2008;3:e3713. doi: 10.1371/journal.pone.0003713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bourque G, Leong B, Vega VB, Chen X, Lee YL, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- [17].Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- [18].Smith DR, Quinlan AR, Peckham HE, Makowsky K, Tao W, et al. Rapid whole-genome mutational profiling using next-generation sequencing technologies. Genome Res. 2008;18:1638–1642. doi: 10.1101/gr.077776.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat. Biotechnol. 2008 doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].O’Neill LP, Turner BM. Immunoprecipitation of native chromatin: NChIP. Methods. 2003;31:76–82. doi: 10.1016/s1046-2023(03)00090-2. [DOI] [PubMed] [Google Scholar]
- [22].Acevedo LG, Iniguez AL, Holster HL, Zhang X, Green R, et al. Genome-scale ChIP-chip analysis using 10,000 human cells. Biotechniques. 2007;43:791–797. doi: 10.2144/000112625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].O’Neill LP, VerMilyea MD, Turner BM. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat. Genet. 2006;38:835–841. doi: 10.1038/ng1820. [DOI] [PubMed] [Google Scholar]
- [24].Kim TH, Ren B. Genome-wide analysis of protein-DNA interactions. Annu Rev Genomics Hum. Genet. 2006;7:81–102. doi: 10.1146/annurev.genom.7.080505.115634. [DOI] [PubMed] [Google Scholar]
- [25].Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, et al. A large genome center’s improvements to the Illumina sequencing system. Nat. Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mueller PR, Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- [27].Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- [28].Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Meyer M, Briggs AW, Maricic T, Hober B, Hoffner B, et al. From micrograms to picograms: quantitative PCR reduces the material demands of high-throughput sequencing. Nucleic Acids Res. 2008;36:e5. doi: 10.1093/nar/gkm1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Erlich Y, Mitra PP, delaBastide M, McCombie WR, Hannon GJ. Alta-Cyclic: a self-optimizing base caller for next-generation sequencing. Nat. Methods. 2008;5:679–682. doi: 10.1038/nmeth.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Rougemont J, Amzallag A, Iseli C, Farinelli L, Xenarios I, et al. Probabilistic base calling of Solexa sequencing data. BMC Bioinformatics. 2008;9:431. doi: 10.1186/1471-2105-9-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cox AJ. Ultra high throughput alignment of short sequence tags. in preparation.
- [33].Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- [34].Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Fejes AP, Robertson G, Bilenky M, Varhol R, Bainbridge M, et al. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nix DA, Courdy SJ, Boucher KM. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics. 2008;9:523. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Spyrou C. personal communication.