Abstract
The human colon plays host to a diverse and metabolically complex community of microorganisms. While the colonic microbiome has been suggested to contribute to the development of colorectal cancer (CRC), a definitive link has not been made. The role in which the colon microflora could contribute to the initiation and/or progression of CRC is explored in this review. Potential mechanisms of bacterial oncogenesis are presented, along with lines of evidence derived from animal models of microbially induced CRC. Particular focus is given to the oncogenic capabilities of enterotoxigenic Bacteroides fragilis. Recent progress in defining the microbiome of CRC in the human population is evaluated, and the future challenges of linking specific etiologic agents to CRC are emphasized.
Keywords: bacterial toxin, chronic inflammation, colonic microbiome, colorectal cancer, genotoxins, oncogenesis
Each year, approximately 1.2 million individuals are diagnosed with colon cancer worldwide [1]. As the second leading cancer affecting both men and women, colorectal cancer (CRC) claims the lives of over 600,000 individuals annually [1]. Greater than 90% of CRC cases are spontaneous, occurring in people with little or no family history of the disease. Once thought to be a cancer predominantly afflicting the western world, incidence rates of CRC are rapidly increasing in areas that have historically been considered low-risk, including South America, eastern Asia and eastern Europe [1]. This trend has been attributed to changes in dietary patterns, along with decreased physical activity, leading to a rise in obesity within these populations [2,3]. As a prominent public health threat, potential contributions to the development of CRC have been the focus of intense study. Colorectal carcinomas usually begin as benign tumors, called polyps or adenomas, which can develop anywhere along the colon from the epithelial cells lining the mucosa. Typically over a period of 10 or more years, some polyps become cancers. Importantly, however, colon cancer can be fully prevented by the early detection and removal of polyps. This progression from normal epithelium to adenoma to adenocarcinoma has been well characterized by Fearon and Vogelstein to involve the cumulative accumulation of genetic mutations [4]. The proposed classes of optimal target genes include tumor suppressors and oncogenes, along with mismatch repair genes. Common examples include APC (a tumor-supressor gene), KRAS (an oncogene), and MLH1 and MLH2 (mismatch repair genes) [4,5]. While there is general consensus about the stepwise transition to colorectal carcinoma, the initiating mechanism(s) remain unclear.
The notion that the endogenous enteric microbiome contributes to the etiopathogenesis of colon cancer has been proposed for decades. The human gastrointestinal (GI) tract is colonized by a vast and complex community of microorganisms totaling approximately 1013 bacteria composed of over 500 microbial species [6]. The commensal intestinal microbiota outnumbers human cells nine to one, and perhaps more impressively, their collective genes outnumber that of their human host 100 to one. The microbial community are immense. The colon is colonized soon after birth, facilitating the essential roles played by the colon microbiota in host physiology, including mucosal immune development, regulation of cell proliferation and modulation of gene expression in host epithelial cells [7,8]. Other beneficial functions of the metabolically complex microbiome include providing usable forms of nutrients as a byproduct of metabolism and protection against exogenous pathogens. In the healthy colon, the microbiota interactions with the host are at homeostasis; however, intrinsic or extrinsic factors can cause perturbations, leading to abnormalities in microbiome composition or function that have been associated with several diseases, including inflammatory bowel disease (IBD) and colon cancer [9,10].
The entirety of the healthy human colon is covered by a mucus layer that consists of an inner gel-like layer and a loose outer layer, both primarily composed of a secreted network of highly glycosylated MUC2 mucins. Among the family of mucin genes expressed in the human colon, the gene product of MUCB has also been detected in minor quantities at the base of the crypt [11]. In addition, MUC5AC and MUC6 have been associated with colorectal adenomas and ulcerative colitis [12]. The outer mucus layer serves as a semipermeable network providing a habitat for commensal bacteria to reside in, while the inner gel-like mucus layer acts as a physical barrier excluding bacteria from direct contact with the epithelium [13]. It is likely that bacteria transiently penetrate the inner mucus barrier in a healthy state; however, they are thought to be cleared quickly through host immune responses [13]. The inner mucus layer ranges in width from 30 to 170 μm in the human colon, increasing in depth from the ascending to the descending colon [14]. Bacteria mainly colonize two major niches within the human colon: the lumen and the outer mucus layer. Characterization of these distinct microbial communities has been the focus of a series of recent studies [6,15–17]. The distinction between these communities is important, as the microbial milieu in these two ecological niches may contribute differently to the etiology of disease. It is well accepted that microbial dysbiosis (an imbalance of the microbiota) with bacterial invasion and persistence in the inner mucus layer (biofilm formation), contributes to the development or progression of IBD [9,18,19]. Massive bacterial biofilms within the empty mucus layer, constituting invasions of greater than 109 bacteria/ml, were identified in 94% of ulcerative colitis patients, 98% of Crohn’s disease patients and 78% of self-limiting colitis patients, compared with just 11% in controls [19]. The phylum-level 16S profiles were observed to involve a shift in major populations, most notably an increase in Proteobacteria and a decrease in Firmicutes and Bacteroidetes [18]. IBD is associated with an increased risk for the development of GI malignancies. While the development of CRC in the setting of IBD involves many of the same genetic mutations as the stepwise transition to sporadic CRC, the timing and frequency of these mutations often differ [20,21]. Furthermore, chronic colitis-associated CRC tends to be macroscopically heterogeneous compared with sporadic CRC and arises from flat dysplastic tissue rather than distinct polyps [21]. This stresses the importance of characterizing the microbial–epithelial interactions in various CRC disease states, analyses that have been less detailed to date [17,22,23].
Mechanisms of bacteria-induced oncogenesis
A significant amount of effort has been employed to determine the mechanisms of microbially induced oncogenesis. Proposed mechanisms include the inhibition, alteration or exacerbation of normal host responses such as apoptosis, inflammation and cellular proliferation. Alternatively, bacteria may also promote cancer through production of secondary metabolites, such as reactive oxygen intermediates, or direct effects on cell transformation through the production of oncogenic toxins. An overview of the mechanisms of bacterial initiation or progression of oncogenesis is shown in Figure 1. It is also important to mention that it has been proposed that the microbiome may also serve a protective role against the development of CRC through its effects on host physiology [24].
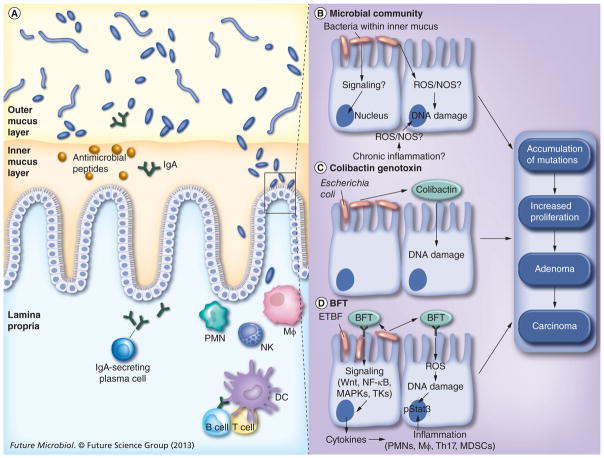
Figure 1. Overview of tissue- and cell-level mechanisms of bacterial oncogenesis.
(A) In the healthy human colon, the inner mucus layer serves as a physical barrier separating the mucosal epithelium from luminal contents. The mucus layer is further protected through epithelial cell secretion of antimicrobial peptides and plasma cell secretion of IgA. This spatial segregation largely maintains the host–microbe homeostasis; nevertheless, bacterial invasion of the inner mucus layer does occur. (B) It is this perturbation that facilitates direct interactions between microbes and host cells, resulting in pathology. The precise mechanisms by which the bacterial community may induce oncogenesis when invading the inner mucus layer are, as yet, uncertain. (C & D) By contrast, for select bacteria for which preliminary epidemiologic data suggest an association with some human colorectal cancer, linkages between the mechanism of action of secreted toxins and colorectal cancer are shown. (C) Genotoxin colibactin secreted by Escherichia coli harboring the PKS island damages DNA. DNA damage by colibactin can be direct and/or through as yet unidentified colonic epithelial and/or other mechanisms. (D) Steps supported by experimental data regarding the action of BFT secreted by ETBF. See text for details.
DC: Dendritic cell; ETBF: Enterotoxigenic Bacteroides fragilis; Mφ: Macrophage; MDSC: Myeloid-derived suppressor cell; NOS: Nitric oxide synthase; PMN: Polymorphonuclear cell; ROS: Reactive oxygen species.
Chronic inflammation
The association between inflammation and tumorigenesis has been appreciated since 1863, when Rudolph Virchow hypothesized that cancer developed from sites of chronic inflammation, termed ‘the chronic irritation hypothesis’ [25]. Today, the connection between inflammation and cancer is well established; however, the mechanisms and pathways are not fully characterized. Infection triggers inflammation as a means to effectively combat an invading pathogen. Polymorphonuclear phagocytes are typically normally the first cells recruited to the site of infection and serve as potent producers of proinflammatory cytokines and chemokines that amplify the response by recruiting more immune cells [26,27]. These cells produce an abundance of reactive oxygen species that can damage lipids, proteins and DNA, leading to increased mutations in proliferating cells and ultimately alterations in cell turnover and death [26–29].
While bacterial infection was once thought of as an acute condition, it is clear that many bacteria are able to persist in the host and lead to chronic infections accompanied by inflammation. Study of the molecular mechanisms that link chronic infections to inflammation and cancer is an area of intense investigation. Persistent generation of microbially induced inflammation mediators such as TNF- α, IL-1 or even lipopolysaccharide on its own can lead to the induction of the NF-κB family of transcription factors, which have been shown to play a role in inflammation-driven carcino-genesis [28]. In an unstimulated cell, the family of NF-κB transcription factors are bound to the IκB inhibitory family of molecules, which effectively prevents translocation to the nucleus. Activation of this signaling pathway leads to the stimulation of the IKK complex, which phosphorylates the inhibitory IκB proteins, targeting them for ubiquitin-dependent degradation. This releases NF-κB proteins, allowing for translocation to the nucleus, in turn leading to the transcription of target genes [28]. Some of the genes targeted encode inflammatory cytokines, including IL-1β, IL-6 and VEGF, which leads to a positive feedback loop of continuing inflammation [26–28]. In addition, antiapoptotic genes, such as those of the Bcl2 family, are upregulated by NF-κB, preventing routine cell turnover. Furthermore, expression of genes involved in cell cycle regulation is altered (e.g., cyclins are upregulated and cell cycle inhibitors are down-regulated). Ultimately, NF-κB plays a key role in inflammation-driven tumor development by generating an environment that promotes mutations and simultaneously prevents damaged cells from undergoing apoptosis, both key features of cancerous cells [26–29].
Reactive oxygen species and nitric oxide are also generated by inflamed epithelial cells under the stress of bacterial toxin exposure or chronic bacterial infection [29,30]. These molecules play important roles in the initiation and progression of carcinogenesis by directly altering DNA, leading to mutations, deletions and chromosomal instability; if left unrepaired, these can lead to carcinogenesis [26,31]. In addition to direct effects on DNA, reactive species canduction pathways. For example, reactive oxygen species can direct cell proliferation and inhibit apoptosis through activation of the transcription factors MAPK, AP-1 and NF-κB [31]. Persistent asymptomatic bacterial infection of the colon, in which the inner mucus layer is penetrated, is proposed as being capable of inducing chronic inflammation, resulting in a cascade of diverse and complex events that combine to generate a procarcinogenic microenvironment.
Oncogenic bacterial metabolites & toxins
In addition to the indirect bacterial infection, there are also direct bacterial mechanisms of oncogenesis. Through their metabolically complex processes, bacteria also produce reactive species, such as the derivatives of molecular oxygen, including superoxide, hydrogen peroxide and hydroxyl radicals [31,32]. These free radicals contribute to genomic instability by the mechanisms discussed above. Alternatively, several bacterial toxins have been identified that are predicted to be carcinogenic These toxins have the capacity to modify host physiology, leading either to direct DNA damage, augmentation of cellular proliferation and/or disruption of cellular differentiation and apoptosis [32]. One thoroughly studied example is CagA of Helicobacter pylori, which is considered to be the most important risk factor that links H. pylori infection to the development of gastric cancer [33,34]. CagA binds the cellular tyrosine phosphatase, SHP2, leading to modulation of cell structure [28]. It has also been shown to target multiple host proteins that regulate inflammation, and several studies suggest that it has the ability to activate NF-κB and β-catenin signaling [35–38]. Recently, CagA was associated directly with a tumor-suppressor pathway when it was shown to usurp the tumor suppressor ASPP2 and modify its activity, thus promoting cell survival [39]. Strains of H. pylori expressing active VacA are associated with an increased risk of gastric cancer [40]. VacA has been shown to stimulate VEGF, increase cell proliferation and inhibit and induce apoptosis, as well as suppress the host immune response to favor long-term colonization, along with a heightened risk for transformation [41–43]. The Pasteurella multocida toxin is known to act as a highly potent mitogen and inhibitor of apoptosis [44]. P. multocida toxin is an AB toxin that activates G proteins through its deamidase function, influencing downstream signaling pathways, including MAPK cytoplasmic and JAK–STAT [45,46].
Another example of a bacterial toxin is CDT. This genotoxin is produced by several bacteria, including selected strains of Escherichia coli, Actinobacillus actinomycetem-comitans, Campylobacter jejuni, Shigella spp., Salmonella spp., Helicobacter hepaticus and Helicobacter cinaedi, as well as other entero-hepatic Helicobacter spp. [47]. CDT is composed of three subunits, one of which, CdtB, functions similarly to mammalian DNase by directly damaging host DNA [48]. Another CRC genotoxin family of interest can be found within the PKS genotoxic island of E. coli. Recent studies have shown that this island encodes a hybrid peptide–polyketide, colibactin, which is capable of directly inducing DNA double-strand breaks both in vitro and in vivo [49,50]. Furthermore, deletion of the genotoxic PKS island from an E. coli strain diminished its oncogenic potential [51]. In addition to the toxins mentioned above, the microbial community contains a repertoire of toxins, listed. in Table 1, proposed to have oncogenic abilities: CNF-1 and CIF in E. coli, and BFT discussed, in detail below [52–55]. Of this group, toxins produced by E. coli, Salmonella spp., Shigella spp. or Bacteroides fragilis are potential contributors to CRC pathogenesis within the microbiota. One consistent feature of these bacterial carcinogenic mechanisms is that, through either direct or indirect methods, they interfere with key eukaryotic processes.
Table 1.
Oncogenic bacterial toxins and linkage to human colorectal cancer.
| Toxin | Bacterial species | Potential oncogenic mechanisms | Human CRC studies† | Ref. |
|---|---|---|---|---|
| CagA | Helicobacter pylori | Binds cellular SHP2, leading to cytoskeletal rearrangements; dysregulates NF-κB and β-catenin signaling; hijacks the tumor suppressor ASPP2 of p53 to inhibit cell cycle progression | Meta-analysis of 16 case–control studies revealed that infection with H. pylori strains containing cagA increased the risk for gastric cancer 1.64-fold over H. pylori strains lacking cagA | [33–39] |
| VacA | H. pylori | Induces the formation of vacuolar compartments and increases proliferation through stimulation of VEGF; has been shown to suppress immune responses and inhibit apoptosis; has also been reported to induce apoptosis, an antitumor mechanism | Several epidemiological studies have reported that H. pylori strains that express active toxin have a higher association with gastric cancer than strains expressing inactive toxin (as measured in vitro) | [40–43] |
| PMT | Pasteurella multocida | Activates Rho GTPases, influencing cell signaling and proliferation | Not available | [44–46] |
| CDT | Escherichia coli, Actinobacillus actinomycetemcomitans, Helicobacter spp., Shigella spp., Salmonella spp., Haemophilus spp., Campylobacter spp. | A subunit of CDT called Cdtb (for binding subunit) acts as a DNase directly damaging host DNA | Not available | [47,48] |
| Colibactin | E. coli | Directly induces DNA double-strand breaks in vivo and in vitro | 67 and 21% of CRC (n = 21) and control (n = 24) patients, respectively, were positive for PKS+ E. coli (see text) | [49–51] |
| MAP | Citrobacter rodentium | Targets host mitochondria, causes epithelial barrier disruption and cytoskeletal rearrangement | Not relevant. C. rodentium is a murine pathogen | [47] |
| CNF | E. coli, Yersinia pseudotuberculosis | Modifies Rho signaling, inhibits cell cycle progression | Not available | [52] |
| CIF | E. coli, Burkholderia pseudomallei, Y. pseudotuberculosis, Photorhabdus luminescens and Photorhabdus asymbiotica | Inhibits apoptosis | Not available | [53] |
| BFT | Bacteroides fragilis | Cleaves E-cadherin, leads to Wnt/β-catenin nuclear signaling, leading to increased colon epithelial cell proliferation; direct DNA damage in colonic epithelial cells; induces oncogenic Stat3/ Th17 mucosal inflammation and cell signaling; induces spermine oxidase and NF-κB cell signaling | 38 and 12% of CRC (n = 73) and control (n = 59) patients, respectively, were positive for ETBF | [54,55,58, 65–67] |
Except for Helicobacter pylori, where the associations of select virulence determinants to gastric cancer are provided.
CRC: Colorectal cancer; ETBF: Enterotoxigenic Bacteroides fragilis
APCMin model of B. fragilis-induced CRC
One of the more compelling pieces of evidence displaying a direct link between an infectious bacterial agent in the induction of CRC lies with the murine models of enterotoxigenic B. fragilis (ETBF) infection. The genus Bacteroides is one of the most numerically prominent members of the intestinal microbial flora. One species in particular, B. fragilis, is a Gram-negative obligate anaerobe and common symbiote colonizing nearly all humans. However, B. fragilis is also an important opportunistic pathogen, as it is the most common anaerobe isolated from clinical infections despite comprising only a small portion (<1–2%) of the total micro-biota [56,57]. Long recognized for roles in intestinal infections and more recently viewed as a molecular subtype, ETBF was revealed to induce colitis in wild-type C57BL/6 mice and promote oncogenic transformation in APCMin mice (a murine intestinal cancer model) [54,58]. B. fragilis consists of two molecular subtypes, termed nontoxigenic B. fragilis (NTBF) and ETBF. NTBF is proposed to be a probiotic organism, serving a crucial role in immune development and providing the host with usable forms of dietary products [7]. By contrast, ETBF has been identified as a cause of inflammatory diarrheal disease in animals and humans, and has also been suggested to be associated with active IBD and CRC [55,59–61]. Interestingly, a recent study by Zitomersky et al. found that ETBF carriage is potentially quite common in the US population, as they detected ETBF in 40% (six out of 15) of healthy asymptomatic individuals between 31 and 66 years of age in Boston (MA, USA) [62].
To date, no ETBF strains have been fully sequenced. However, through identification and sequencing of a transposon-flanked pathogenicity island, ETBF was determined to encode a 20-kDa zinc-dependent metalloprotease termed BFT [63,64]. BFT is the only known virulence factor of ETBF, and all strains harbor one of three highly related bft isoforms (bft-1, bft-2 or bft-3) present on the B. fragilis chromosome. All molecular isoforms are capable of exhibiting biological activity; however, the relationship between isoform and disease severity is not yet known. BFT binds to a currently unknown colonic epithelial cell receptor, triggering rapid cleavage of the tumor-suppressor protein E-cadherin, which, in turn, frees its associated β-catenin, allowing its nuclear localization [65]. The subsequent expression of the β-catenin/Wnt signaling pathway leads to an increase in colonic epithelial cell proliferation. It also stimulates additional signaling pathways through NF-κB. While the precise contribution of the plethora of colonic epithelial cell signaling triggered by BFT to ETBF pathogenesis remains unknown, one clear biologic outcome is the recruitment and activation of inflammatory cells, as well as epithelial cell secretion of pro-inflammatory cytokines and the production of reactive oxygen species [30,66,67]. A recent study by our group demonstrated that ETBF induces a rapid-onset acute symptomatic colitis, followed by chronic subclinical colonic inflammation and hyperplasia in specific pathogen-free wild-type C57BL/6 mice. Unique to this model is the ability of ETBF to persistently colonize the mice for an extended period of time after a single oral exposure to ETBF; in this study, mice infected for 16 months exhibited low-level colitis [58]. The acute ETBF murine colitis mimics the inflammatory diarrhea detected in humans with ETBF infection, whereas the long-term murine colonization is analogous to what is observed in ETBF colonization in the human population, suggesting that ETBF carriers may be susceptible to asymptomatic ETBF-induced colitis.
The pro-oncogenic cellular signaling induced by BFT in concert with the persistent chronic inflammation induced by ETBF in wild-type mice suggest that ETBF is an oncogenic bacterium. This was recently tested by our group using the APCMin mouse strain, a well-established cancer model in which loss of a single copy of the Apc gene predisposes mice to the development of numerous tumors in the small intestine when the second allele spontaneously mutates [68]. However, importantly for this animal model of bacterial-induced carcinogenesis, adenomas are primarily observed in the small intestine and not in the colon [69]. APC is a multidomain tumor-suppressor protein that binds to and promotes proteosomal degradation of β-catenin to regulate downstream Wnt signaling [69]. Loss or mutation of the Apc gene is the cause of the inherited disease familial adenomatous polyposis, and occurs in virtually all sporadic colon cancers [70]. By 4 months of age, sham APCMin mice developed an average of one to three tumors in the colon. By contrast, APCMin mice colonized with ETBF developed chronic asymptomatic colitis, with colon tumor foci detected as early as 1 week postinoculation. At 1 month of age, a marked increase in colon tumor formation (~12 tumors/mouse on average) occurs predominantly in the distal colon of ETBF-colonized mice. By contrast, APCMin mice colonized with NTBF did not exhibit colon tumors in excess of the sham mice. ETBF induces rapid activation of Stat3 both in the colonic epithelial cells, which are the targets of transformation in the colon, and in a subset of mucosal immune cells. Stat3 activation is required for Th17 cell development and, consistent with this, ETBF induces a rapid mucosal Th17 inflammatory response within 1 week of colonization. Colon tumors induced by ETBF also have a marked increase in Stat3 activation. Furthermore, excess tumor formation is significantly inhibited by administration of IL-17-blocking antibody, indicating that IL-17 is necessary for tumorigenesis in this model [54]. These studies suggest that persistent long-term colonization with ETBF may induce chronic colonic inflammation, with the potential for oncogenic transformation [54,58]. Furthermore, while Th17 inflammatory responses typically help the host control bacterial and fungal infection, the ETBF murine model demonstrates that endogenous Th17 responses can yield oncogenesis in the colon, a result that is supported by additional murine and human data [71–75].
Other animal models of bacterial influences on CRC
Animal models of bacteria-driven oncogenesis have proven to be valuable tools in elucidating the link between microbes and CRC. Genetic knockout, germ-free and chemical mouse models have been developed and extensively used in studies connecting bacteria and CRC. The APCMin model previously mentioned was the first mutant murine model for colon cancer and is an important tool, given the importance of inactivation of the Apc gene in the initiation of sporadic CRC. This initial APCMin mutant, carrying a truncation at codon 850 of the Apc gene, was identified among a colony of mice following random ethylnitrosourea mutagenesis [76]. Utilizing gene-knockout technology, alternative Apc mutants have subsequently been constructed, including mouse strains with truncations at codon 716 and 1638 that also develop polyps [69,77]. There are several additional genetically engineered models of intestinal neoplasia, which are extensively covered in a review by Taketo and Edelmann, including single knockouts of Muc2, IL10, Smad3 and Gαi2, and double knockouts of APC with Smad4, TCRβ with p53, Gpx1 with Gpx2 and Tgfb1 with Rag2 [78–86]. In the absence of intestinal microbiota, under germ-free conditions, IL10-knockout mice and TCRβ with p53, Gpx1 with Gpx2 and Tgfb1 with Rag2 double-knockout mice all showed decreased or completely inhibited tumor formation [84–90]. Together, these studies indicate a role for the intestinal microflora in the development of inflammation and neoplasia. Mixed results have been reported in germ-free APCMin mice. While Dove et al. noted a twofold decreased tumor load in the medial small intestine, they did not see a significant overall decrease in tumors[88]. By contrast, a recent study by Li et al. found a significant decrease of tumor load in both the small intestine and the colon. Furthermore, they identified two pathways triggered by microbiota, c-Jun/JNK and STAT3, which act to enhance tumor formation [89].
Mouse models of chemically induced colitis have also been used in studies to address bacterial involvement in colitis and tumorigenesis. The most commonly used agents are azoxymethane (AOM) and dextran sulfate sodium (DSS). A recent study by Uronis et al. found that IL10-knockout mice that were colonized with complex microbiota and exposed to AOM developed tumors; however, germ-free conditions abolished tumor formation [91]. It was further shown that conventional IL10/MyD88 double-knockout mice showed no signs of tumor development upon treatment with AOM, suggesting that microbially induced tumorigenesis in this system was dependent on the TLR/MyD88 pathway [91]. Johansson et al. showed that bacteria penetrate the inner mucus layer before inflammation is observed in the DSS colitis model, suggesting that invasion of the protective inner mucus layer and subsequent bacterial contact with the epithelium triggers the host immune response and inflammation [92]. Further studies have shown that DSS-induced colitis can be ameliorated under gnotobiotic conditions, indicating that the presence of microflora facilitates DSS colitis [93]. A more recent study by Elinav et al. further supports the role of intestinal flora in DSS-induced inflammation [94]. Using mice deficient in the NLRP6 inflammasome, they showed that the resulting altered microbiota, characterized by increased levels of Bacteroidetes, led to an increased recruitment of inflammatory cells and worsened colitis upon DSS exposure when compared with wild-type mice. This report, among others, emphasizes that the microbiota composition is shaped not only by diet, but also by the host immune make-up, suggesting that human host polymorphisms modulating the inflammatory response may be important contributors to the influence of the microbiota on CRC pathogenesis [51,91,95–97]. Furthermore, several studies have shown that antibiotic treatment is capable of blocking colitis in the murine DSS colitis model [98–100]. Under germ-free conditions, DSS treatment alone, however, is able to induce slight inflammatory cell infiltration and edema, but without tumor induction [101].
While abundant data implicate the aggregate microbiome as a cofactor in colon tumor development, individual pathogens thought to promote colonic tumorigenesis have also been investigated using animal models. A study by Ellmerich and colleagues showed that Streptococcus bovis, long associated with colon cancer through epidemiological studies, is capable of markedly increasing the production of inflammatory cytokines and aberrant crypt foci in the colonic mucosa of rats through exposure to S. bovis cell wall antigens [102]. This study, however, lacked controls to demonstrate that the response was specific to S. bovis cell wall antigens. Furthermore, it is important to mention that the strain utilized in this study was later classified as S. bovis biotypeII/1 (Streptococcus infantarius subsp. infantarius), which shows a less convincing link to human CRC compared with bio-type I, a topic thoroughly covered in a recent review by Boleij and Tjalsma [103]. Another suspect, Helicobacter hepaticus, colonizes the liver and colon of several mouse strains and has been linked to hepatitis, chronic colitis and CRC, and is discussed in a recent review by Fox et al. [104]. It was recently shown that H. hepaticus triggers nitric oxide and TNF-α production, leading to inflammation and carcinogenesis in Rag2-deficient mice, implicating the innate immune response induced by H. hepaticus as carcinogenic [105]. A subsequent study utilized transcriptional profiling ofH. hepaticus-infected Rag2-knockout mice to reveal that colon and liver tissues exhibited different stress responses to infection. The colon was found to have a significant upregulation of genes involved in the generation of reactive species, while genes involved in DNA repair showed lower expression; this was directly contrasted with the liver, which showed upregulation of all major DNA repair pathways during infection [106]. These findings support the role of H. hepaticus inflammation-induced carcinogenesis, and also leads to interesting insights into the complexity of tissue-specific microbial pathophysiology. Similarly, the colon microbiota has been thought to play a role in the progression of certain diseases, such as HIV and HCV, both of which are conditions associated with an increased risk of cancer [107,108]. Other studies show that certain strains of Enterococcus faecalis produce extracellular superoxide and hydrogen peroxide, which induce aneuploidy and tetraploidy in colonic epithelial cells [109,110]. E. faecalis also encodes a metalloprotease, GelE, which contributes to the development of colitis, dysplasia and adenocarcinoma in monocolonized IL-10-deficient mice[87,111]. However, to date, a link between E. faecalis and human CRC has not been identified [112].
Another well-studied bacterial agent of interest is Citrobacter rodentium, which is known to induce self-limiting colitis, epithelial cell proliferation and tumorigenesis in the murine colon. C. rodentium is not a human pathogen, but is considered to be the mouse homolog of human attaching and effacing E. coli strains, which are yet another proposed procarcinogenic species. Early studies found that C. rodentium infection increases the carcinogenic effect of 1,2-dimethylhydrazine treatment in NIH Swiss mice [113]. Later, a study revealed that C. rodentium infection leads to cytokinetic alterations and is sufficient to promote colon tumor development in APCMin mice [114]. Maddocks et al. reported that human attaching and effacing enteropathogenic E. coli strains downregu-late DNA mismatch repair genes and provided preliminary data identifying these bacteria in human CRC [115]. A recent publication by Arthur et al. showed that tightly adherent E. coli strains harboring the PKS genotoxic island were able to induce tumor formation in AOM-treated IL-10-deficient mice under germfree conditions [51]. The authors further showed that conventionally housed IL-10-deficient mice developed an altered microbiome in association with colitis in 100% of the mice. Importantly, they showed that inflammation, not the carcinogen AOM, modified the microbiota structure, with the emergence of potential procarcinogenic phyla. Furthermore, a specific microbial virulence factor (the PKS island), not inflammation alone, was required for microbially induced carcinogenesis in this model. This study stresses the interplay between specific carcinogenic species, the microbial community and the host. Consideration of these multifactorial influences is important when transitioning to studies concerning microbial involvement in human CRC.
Human studies
Despite a long quest, direct links between the bacterial microbiome and CRC in humans have not yet been established. Culture-based, observational or case–control studies largely focusing on fecal analyses from patients with CRC and healthy control patients have suggested that Bacteroides, Streptococcus gallolyticus subsp. gallolyticus (previously known as S. bovis biotypeI), E. coli and Enterococcus spp., among others, may be associated with the development of CRC. Particularly notable over time has been the association of S. gallolyticus endocarditis and/or bacteremia with a high likelihood of having an underlying GI tract malignancy, most commonly CRC [116–122]. Clostridium septicum aortitis and/or bacteremia have also been suggested to be indicators of GI malignancy [123]. Culture-based human studies combined with recent experimental mechanistic studies have provided the greatest support for potential roles for ETBF, S. gallolyticus, enteroadherent E. coli and E. coli possessing the PKS island in human CRC [51,61,115,120].
Molecular approaches, in particular the advent of next-generation sequencing techniques, have facilitated studies to examine more comprehensively the microbial associations with CRC (Table 2). These approaches enhance culture-based methods because they allow the detection of ‘noncultivable’ microbes. Overall, the available data suggest that the tumor-associated microbiome differs from that detected on matched normal tissue in the same patient. Furthermore, the fecal microbiome of CRC patients appears to differ from that associated with their tumor and also from the fecal microbiome of healthy volunteers [124,125]. A wide range of bacteria have been reported to be enriched in tumor tissue samples, including E. coli, Proteobacteria (especially Enterobacteriaceae), Bacteroides- spp., Prevotella spp., Streptococcus spp., Peptostreptococcus spp., Enterococcus spp. and Fusobacterium spp. [22,23,126,127]. However, the differences detected between sample groups vary among studies, without clear patterns having yet been identified that might be useful, for example, to identify an individual at risk for or suffering from CRC. The methodologic differences, varying sample types analyzed, varying populations studied and limited patient data provided make differences among the studies difficult to interpret. Two recent studies, representing the largest set of CRC and matched normal tissue samples analyzed to date, identified a predominance of Fusobacterium spp. (F. nucleatum and other Fusobacterium spp.) to be associated with CRC as compared with adjacent normal tissue [23,126]. No healthy control populations were included in either study, and experimental models of F. nucleatum for testing its oncogenicity have not yet been reported. Most studies have focused on patients with CRC; however, to begin to implicate bacteria in the pathogenesis of CRC, it is important to determine bacterial associations with colonic adenomas, precursors of CRC. Similar studies considering the unique pathogenic associations for IBD (discussed earlier) would also be helpful. In the one molecular study evaluating adenomas available to date, the bacterial population distributions also differed between adenoma and control patients when rectal biopsies of normal tissue were compared using 16S rRNA sequence analysis [17]. It is clear that additional studies are needed not only to delineate the microbial populations associated with CRC compared with diverse control populations, but also to understand how the microbial populations may relate to disease outcome and contribute to the pathogenesis of CRC.
Table 2.
Molecular analyses of microbiota in human colorectal cancer.
| Population studied | Method | Sites analyzed | Key findings | Comments | Ref. |
|---|---|---|---|---|---|
| Tubular-villous adenoma (29 patients/83 biopsies†), CRC (31 patients/83 biopsies‡), Screening colonoscopy (31 patients/97 biopsies), Colonoscopy for symptoms (34 patients/113 biopsies§); Germany | 16S rDNA PCR (5′, 600-bp product) on colon biopsies with 40–100 cloned 16S sequences analyzed/ biopsied; gentamicin protection assay | Not stated | Number of 16S PCR-positive patients (number with Eschericheria coli): 1 (1) asymptomatic control, 10 (4) symptomatic controls, 27 (18) adenoma, 28 (24) carcinoma Gentamicin protection assay detected intracellular E. coli in biopsies of 81% of adenoma and CRC patients (n = 16) and 0% of controls (n = 25) | No antibiotics in prior 8 weeks; sites of biopsies and pathology of biopsies (i.e., tumor or normal mucosa) not stated; bowel preparation not stated | [127] |
| Adenoma patients (21), nonadenoma (control) patients (23); participants from DHS study; USA | 16S rRNA PCR analyzed by T-RFLP; 16S rRNA clone libraries (=30 clones/ patient) for four adenoma and four control patients; 16S rRNA FISH | Colonoscopy-obtained biopsy samples, 10–12 cm from anal verge | T-RFLP data: bacterial community distribution differed between adenoma and control patients Clone data: adenoma patients had >Proteobacteria, <Bacteroidetes (phyla), >Faecalibacterium, >Dorea, <Bacteroides and <Coprococcus (genera) FISH: bacteria present in mucus layer in adenoma and controls without difference noted | No antibiotics in prior 8 weeks; only rectal biopsies analyzed (normal tissue); excluded patients with prior CRC or adenoma; all received polyethylene glycol bowel preparation | [17] |
| Fecal population: CRC patients (60) and normal colonoscopy patients (119) Mucosal population: normal colon (22) and CRC (22; 16 colon, 6 rectal); France | Replicate 16S rRNA (V3–V4) pyrosequencing of 6 controls and 6 CRC fecal samples (n = 24 samples); qPCR for select genera and species¶ on all stools and 44 mucosal DNA samples | Feces collected between 3 days and 2 weeks prior to colonoscopy; mucosal samples from surgery | PCA indicated the phylogenetic core of CRC and control stools differ qPCR revealed higher Bacteroides/ Prevotella in CRC versus control patients | No prior history of cancer or colitis; specific exposure to antibiotics not stated; fecal and mucosal results not discussed separately; site of mucosal tissue harvesting in colon not stated | [74] |
| 6 CRC patients; The Netherlands | 16S rRNA (V1–V3) pyrosequencing | Tumor and adjacent normal mucosa 5–10 cm from tumor in surgical resection specimens | On-and off-tumor bacterial populations differed significantly, although consistent patient-to-patient changes were not shown Overall, tumor tissue associated with Bacteroidetes and putative commensals, whereas Proteobacteria, especially Enterobacteriaceae, favored off-tumor tissue | No patient details provided; site of tissue acquisition in colon not stated; no healthy control population | [22] |
| CRC patients (46), healthy volunteers (56); China | 16S rRNA (V3) pyrosequencing; PCR to detect Bacteroides species and butyryl-CoA transferase genes | Fecal samples collected before surgery for CRC patients and in association with routine clinic visits for controls | PCA differentiated CRC from control patients Bacteroides species, except Bacteroides fragilis and butyrate-producing bacteria, were enriched in control samples, whereas the family Enterobacteriaceae and genera Streptococcus, Peptostreptococcus, Enterococcus and Fusobacterium, among others, were enriched in CRC samples | Antibiotic exposure not defined for CRC group; no antibiotic exposure for 3 months in control group; other patient data not stated | [124] |
| CRC patients (46) including stool (21), rectal swab (32), tumor tissue with matched normal tissue (2–5 and 10–20 cm from tumor) (27); healthy volunteers (56) including stool (22) and rectal swab (34); China | 16S rDNA (V1–V3) bacterial pyrosequencing | Feces and rectal swab prior to bowel cleansing; tissue samples from surgical resection | Microbiota of CRC and lumen differ By PCA analysis, tumor and matched normal tissues were similar; however, in additional analyses, genus-level#, but not phylum-level, differences were detected in tumor versus matched normal tissues Both mucosally adherent bacteria (rectal swab comparison) and luminal microbial composition (stool) differed between CRC and healthy volunteers†† | No antibiotics within 1 month of sampling; no patient data provided; ‘within patient’ versus ‘between patient’ analyses not clearly defined | [125] |
| 11 CRC and matched normal tissue samples‡‡ for metagenomic sequencing; 88 CRC and matched normal tissue samples for qPCR; Canada | Paired-end, PCR-amplified RNA libraries sequenced on Illumina® GAIIx platform; qPCR for Fusobacterium nucleatum rRNA | CRC and matched normal colon mucosa from tumor tissue repository of surgical specimens | Only F. nucleatum subsp. nucleatum was markedly disproportionate between CRC and matched control tissues (nine of 11 subjects =twofold higher for F. nucleatum Illumina reads in CRC tissue and ~70% of 88 patients evaluated by qPCR) | Only paper to date to evaluate transcriptionally active bacteria; colon sites of tissue samples not defined; no patient data provided; no healthy control population | [126] |
| Nine CRC and adjacent normal tissue samples for whole-genome sequencing; 95 CRC and adjacent normal tissue samples for PCR and 16S rRNA pyrosequencing; Spain; USA; Vietnam | Whole-genome Illumina sequencing; 16S rDNA (V3–V5) pyrosequencing; FISH using all bacteria and Fusobacterium-targeted probes | CRC and adjacent normal colon mucosa from surgical specimens | Overall Fusobacterium species enriched in CRC (range: <1 to >20% relative abundance in nine Illumina-sequenced samples; 16S rDNA sequencing, qPCR and FISH suggested increased Fusobacterium in CRC compared with normal colon tissues) | Colon sites of tissue samples not defined; no patient data provided; OTU with greatest similarity to F. nucleatum was the most dominant phylotype within CRC, but some tumors contained multiple Fusobacterium species; no healthy control population | [23] |
Nineteen with new adenoma and ten with colonoscopy within 1 year of polypectomy (six with recurrent adenoma).
Ten new diagnoses of CRC: 15 with colonoscopy within 1 year of partial colectomy (seven adenoma, five adenoma and CRC); three with colonoscopy 5–10 years after CRC resection.
Normal colonoscopy.
Clostridium/Leptum group, Clostridium/Coccoides group, Bacteroides/Prevotella group, Escherichia coli, Bifidobacterium genus, Lactobacillus/Leuconostoc/ Pediococcus group and Faecalibacterium prausnitzii species.
Eleven genera were reported as differing between tumor and matched normal tissues by unweighted UniFrac PCA.
Sixteen and ten genera were reported as differing between rectal swab and stool, respectively, of CRC versus healthy volunteers by unweighted UniFrac PCA.
Distance of normal tissue from tumor not defined.
CRC: Colorectal cancer; OTU: Operational taxonomic unit; PCA: Principal component analysis; qPCR: Real-time PCR; T-RFLP: Terminal restriction fragment length polymorphism.
Conclusion & future perspective
Sporadic CRC is ultimately a genetic disease, where gene alterations and chromosomal instability are central to the stepwise progression towards neoplasia [4,5]. This complex process is undoubtedly the result of numerous influences ranging from age, gender, nutritional intake, physical activity and host genetic background to the diverse and variable intestinal micro-biome. The epidemiological and experimental evidence discussed here strongly suggest a role for several bacterial agents in CRC. However, traditional bacteriological approaches are built on the assumption that an etiologic pathogen can be isolated, cultured and identified, and that pathogenesis can be explained through confirmation of disease. Throughout the 19th century and beyond, these concepts, grounded in Koch’s postulates, have proven to be crucial in the identification of countless infectious pathogens, including the etiologic agent of gastric cancer, H. pylori [128]. Yet unlike the archetypal infectious disease consisting of a single causative agent, the colon plays host to a variety of commensal organisms, many of which have been implicated, both alone and in consort, to contribute to the genesis of colon cancer. The challenge for traditional epidemiological approaches to identify links between bacterial agents and CRC is further hampered by the long length of time between initiation and detectable carcinogenesis. Searching for the responsible agent(s) among the multiple constituents of the intestinal flora presents a challenging prospect, since it is possible that the critical inciting microbiotic agent or composition is no longer present at the time of disease discovery. As such, we are then potentially reliant on the detection of an immune signature of the microbe or microbiota to provide the epidemiologic link to CRC.
Two recent reviews draw attention to the potential for bacterial ‘alpha’-bugs or drivers in the context of the aggregate flora to shape the microbial community in order to create a procarcinogenic environment [129,130]. This emphasizes the need for detailed knowledge about specific microbes, as well as alterations of whole microbial communities under diseased and healthy states, to better understand the etiology of CRC. The advent of next-generation sequencing technologies has facilitated these types of studies, which can take into account the community of a specimen, many of which were discussed here. However, limitations in the experimental evidence to date include small sampling numbers and absent or inadequate control populations for comparison. Furthermore, no information regarding host genetics has been analyzed. As revealed by the numerous mouse studies, commensal bacteria have pathogenic capabilities in the context of genetic abnormalities in the host.
While advances have been made in the early stages of characterizing what species are present on a tumor and its have been no attempts to determine the spatial organization of those microbes with respect to the host epithelium. The spatial arrangement of the bacterial community is likely to dictate both microbe–microbe interactions and microbe–host interactions. Proximity to the host epithelium facilitates the way in which microbes are recognized and responded to by the host innate and adaptive immune system [131]. A systematic study of the distribution of microbes along both the length of the colon, as well as a cross-sectional characterization of the lumen and mucus layer members, is essential to further elucidate the role of specific bacterial community members in the cancerous disease state.
As the field moves forward, several types of evidence will be needed to link the microbiota to human CRC [132]. Prospectively conducted studies, initiated at a time point before the onset of disease, and with relevant samples (ideally blood, tissue and stool) for analysis, would be ideal. Capturing information about the microbiome structure and composition in the early stages of disease initiation and throughout disease development would be invaluable. Ideally, the detection of microbiome dysbiosis or specific putative etiologic agents before disease development would help to address the ‘cause or consequence’ conundrum. However, population-based microbiome studies are both cost-prohibitive and impractical for evaluating long-term (20–40 years in the case of colon cancer) disease development. Attention to designing control groups and using varied controls is also important in order to help determine whether a microbe or a microbiota composition exhibits a strong, consistent association with human CRC. We should seek to detect an immunologic response to the purported microbial etiologies of CRC. It was the combined criteria of either detection of H. pylori or an immune response to H. pylori that provided crucial data that defined H. pylori as the cause of most gastric cancers [128]. Murine models of colon oncogenesis will likely provide key insights into molecules and mediators with translational importance, enabling us to understand how the microbiota contribute to human CRC. Ultimately, elimination of the inciting microbe or restructuring of the microbiome, whether by diet, probiotics, antibiotics or vaccination, with subsequent prevention of CRC, is required for definitive declaration of disease association. While these criteria are stringent and create a necessarily high bar for investigators to reach, there has never been more interest in understanding the microbial inciters of human CRC. The emerging data are exciting and capture our imagination, making the future for discovery in this field bright.
Executive summary.
Background
Colorectal cancer (CRC) is a public health issue both within the USA and globally, leading to the deaths of over 600,000 individuals annually.
The majority of CRC is sporadic in nature. Accumulation of mutations in normal colonic epithelial cells characterizes the progression from adenoma to carcinoma.
The microbiota have been implicated in contributing to the initiation or progression of CRC for years.
In the human colon, bacteria are kept spatially segregated from the colonic epithelium by a mucus layer; perturbation of this mucus layer has been associated with states of pathology, including CRC.
Mechanisms of bacteria-induced oncogenesis
Chronic bacterial infection associated with invasion of the inner mucus layer can lead to chronic inflammation through the persistent generation of inflammatory mediators, affecting cell turnover, apoptosis and increasing the likelihood of mutations.
Reactive oxygen and nitrogen species may arise from several sources, including bacteria and epithelial and immune cells. Through direct DNA damage or modification of cellular signaling, these molecules can generate a procarcinogenic environment.
Bacteria are capable of producing a variety of oncogenic toxins that can directly damage DNA, influence cellular signaling and/or induce mucosal inflammation in order to initiate or promote colon tumorigenesis.
APCMin model of Bacteroides fragilis-induced CRC
Bacteroides fragilis is a symbiotic organism with the capacity to become an opportunistic pathogen.
Enterotoxigenic B. fragilis (ETBF) express the oncogenic BFT and has been associated with diarrheal disease accompanied by colitis in both humans and animals.
ETBF induces acute colitis followed by chronic subclinical colitis in specific pathogen-free wild-type C57BL/6 mice, which is associated with long-term ETBF colonization.
ETBF promotes tumor formation under specific pathogen-free conditions in the APCMin model of murine colon cancer through, in part, a Stat3-mediated Th17 mucosal immune response.
Other animal models of bacterial influences on CRC
Under germ-free conditions, several murine models of genetically engineered neoplasia show decreased colon tumor formation.
Several mouse models of chemically induced colitis can be ameliorated with antibiotic treatment or through a gnotobiotic environment.
Multiple pathogenic bacteria are capable of inducing tumorigenesis in genetically engineered mice.
Human studies
Culture-based and observational studies have associated multiple bacterial species with CRC; however, the most convincing combined clinical and experimental evidence exists for ETBF, Streptococcus gallolyticus and Escherichia coli species, especially those harboring the PKS genotoxic island.
Sequencing data suggest that the microbiome of CRC patients differs from that of healthy individuals, and that the tumor microbiome differs from that detected on flanking normal tissue from the same patient.
Future perspective
Further experimental evidence is necessary to link members of the microbiome to CRC.
Thorough characterization of the spatial arrangement of microbes along the length and cross-section of the colon in the disease and healthy state is needed.
Future studies of the human population will benefit from increased sampling sizes, the inclusion of multiple and varied controls, consideration of host genetics and prospectively collected human samples, including colon tissues, to allow analysis before the onset of disease.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
This work was supported by T32AI007417 to C Dejea, K08DK087856 to E Wick, R01CA098454 to R Casero and R01DK080817, R01CA151393 and R01 CA151325 to CL Sears, all from the NIH. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• ▪ of interest
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Center MM, Jemal A, Ward E. International trends in colorectal cancer incidence rates. Cancer Epidemiol Biomarkers Prev. 2009;18(6):1688–1694. doi: 10.1158/1055-9965.EPI-09-0090. [DOI] [PubMed] [Google Scholar]
- 3.Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59(6):366–378. doi: 10.3322/caac.20038. [DOI] [PubMed] [Google Scholar]
- 4.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 5.Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9(4):138–141. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- 6▪.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. A 16S RNA analysis of mucosal-associated and fecal bacteria from three people over time that demonstrated significant variation between bacteria identified in stool versus mucosa within individuals, as well as in bacteria identified between individuals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 8.Reikvam DH, Erofeev A, Sandvik A, et al. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression inflammatory bowel diseases. PLoS ONE. 2011;6(3):e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleessen B, Kroesen AJ, Buhr HJ, Blaut M. Mucosal and invading bacteria in patients with inflammatory bowel disease compared with controls. Scand J Gastroenterol. 2002;37(9):1034–1041. doi: 10.1080/003655202320378220. [DOI] [PubMed] [Google Scholar]
- 10.Rowland IR. The role of the gastrointestinal microbiota in colorectal cancer. Curr Pharm Des. 2009;15(13):1524–1527. doi: 10.2174/138161209788168191. [DOI] [PubMed] [Google Scholar]
- 11.Sheng YH, Hasnain SZ, Florin TH, McGuckin MA. Mucins in inflammatory bowel diseases and colorectal cancer. J Gastroenterol Hepatol. 2012;27(1):28–38. doi: 10.1111/j.1440-1746.2011.06909.x. [DOI] [PubMed] [Google Scholar]
- 12.McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9(4):265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 13.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host–microbial interactions. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuo K, Ota H, Akamatsu T, Sugiyama A, Katsuyama T. Histochemistry of the surface mucous gel layer of the human colon. Gut. 1997;40(6):782–789. doi: 10.1136/gut.40.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmed S, Macfarlane GT, Fite A, McBain AJ, Gilbert P, Macfarlane S. Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol. 2007;73(22):7435–7442. doi: 10.1128/AEM.01143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen XJ, Rawls JF, Randall T, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010;1(3):138–147. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular–phylogenetic characterization of microbial community imbalances in human. Proc Natl Acad Sci USA. 2007;104(34):13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swidsinski A, Loening-Baucke V, Herber A. Mucosal flora in Crohn’s disease and ulcerative colitis – an overview. J Physiol Pharmacol. 2009;60(Suppl 6):61–71. [PubMed] [Google Scholar]
- 20.Tarmin L, Yin J, Harpaz N, et al. Adenomatous polyposis coli gene mutations in ulcerative colitis-associated dysplasias and cancers versus sporadic colon neoplasms. Cancer Res. 1995;55(10):2035–2038. [PubMed] [Google Scholar]
- 21.Xie J, Itzkowitz SH. Cancer in inflammatory bowel disease. World J Gastroenterol. 2008;14(3):378–389. doi: 10.3748/wjg.14.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchesi JR, Dutilh BE, Hall N, et al. Towards the human colorectal cancer microbiome. PLoS ONE. 2011;6(5):e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23▪.Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–298. doi: 10.1101/gr.126573.111. This human colorectal cancer microbiome study utilized 454 sequencing, real-time PCR and FISH to identify an increased association between Fusobacterium and colon tumors when compared with flanking tissues (published in the same issue as [126]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pitari GM, Zingman LV, Hodgson DM, et al. Bacterial enterotoxins are associated with resistance to colon cancer. Proc Natl Acad Sci USA. 2003;100(5):2695–2699. doi: 10.1073/pnas.0434905100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357(9255):539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 26.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karin M, Greten FR. NFκB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5(10):749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 29.Nath G, Gulati AK, Shukla VK. Role of bacteria in carcinogenesis, with special reference to carcinoma of the gallbladder. World J Gastroenterol. 2010;16(43):5395–5404. doi: 10.3748/wjg.v16.i43.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodwin AC, Destefano Shields CE, Wu S, et al. Polyamine catabolism contributes to enterotoxigenic. Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci USA. 2011;108(37):15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waris G, Ahsan H. Reactive oxygen species. role in the development of cancer and various chronic conditions. J Carcinog. 2006;5:14. doi: 10.1186/1477-3163-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lax AJ. Opinion Bacterial toxins and cancer – a case to answer? Nat Rev Microbiol. 2005;3(4):343–349. doi: 10.1038/nrmicro1130. [DOI] [PubMed] [Google Scholar]
- 33.Huang JQ, Zheng GF, Sumanac K, Irvine EJ, Hunt RH. Meta-analysis of the relationship between cagA seropositivity and gastric cancer. Gastroenterology. 2003;125(6):1636–1644. doi: 10.1053/j.gastro.2003.08.033. [DOI] [PubMed] [Google Scholar]
- 34.Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2(1):28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 35.Hatakeyama M, Higashi H. Helicobacter pylori CagA: a new paradigm for bacterial carcinogenesis. Cancer Sci. 2005;96(12):835–843. doi: 10.1111/j.1349-7006.2005.00130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brandt S, Kwok T, Hartig R, Konig W, Backert S. NFκB activation and potentiation by the of Helicobacter pylori CagA protein. Proc Natl Acad Sci USA. 2005;102(26):9300–9305. doi: 10.1073/pnas.0409873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamb A, Yang XD, Tsang YH, et al. Helicobacter pylori CagA activates NF-kappaB by targeting TAK1 for TRAF6-mediated Lys 63 ubiquitination. EMBO Rep. 2009;10(11):1242–1249. doi: 10.1038/embor.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franco AT, Israel DA, Washington MK, et al. Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc Natl Acad Sci USA. 2005;102(30):10646–10651. doi: 10.1073/pnas.0504927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buti L, Spooner E, Van der Veen AG, Rappuoli R, Covacci A, Ploegh HL. Helicobacterpylori cytotoxin-associated geneA (CagA) subverts the apoptosis-stimulating protein of p53 (ASPP2) tumor suppressor pathway of the host. Proc Natl Acad Sci USA. 2011;108(22):9238–9243. doi: 10.1073/pnas.1106200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miehlke S, Kirsch C, Agha-Amiri K, et al. The Helicobacter pylori vacA s1, m1 genotype and cagA is associated with gastric carcinoma in Germany. Int J Cancer. 2000;87(3):322–327. [PubMed] [Google Scholar]
- 41.Strowski MZ, Cramer T, Schafer G, et al. Helicobacter pylori stimulates host vascular endothelial growth factor-A (vegf-A) gene expression via MEK/ERK-dependent activation of Sp1 and Sp3. FASEB J. 2004;18(1):218–220. doi: 10.1096/fj.03-0055fje. [DOI] [PubMed] [Google Scholar]
- 42.Gebert B, Fischer W, Weiss E, Hoffmann R, Haas R. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science. 2003;301(5636):1099–1102. doi: 10.1126/science.1086871. [DOI] [PubMed] [Google Scholar]
- 43.Cover TL, Krishna US, Israel DA, Peek RM., Jr Induction of gastric epithelial cell apoptosis by Helicobacter pylori vacuolating cytotoxin. Cancer Res. 2003;63(5):951–957. [PubMed] [Google Scholar]
- 44.Preuss I, Hildebrand D, Orth JH, Aktories K, Kubatzky KF. Pasteurella multocida toxin is a potent activator of anti-apoptotic signalling pathways. Cell Microbiol. 2010;12(8):1174–1185. doi: 10.1111/j.1462-5822.2010.01462.x. [DOI] [PubMed] [Google Scholar]
- 45.Orth JH, Aktories K. Molecular biology of Pasteurella multocida toxin. Curr Top Microbiol Immunol. 2012;361:73–92. doi: 10.1007/82_2012_201. [DOI] [PubMed] [Google Scholar]
- 46.Orth JH, Preuss I, Fester I, Schlosser A, Wilson BA, Aktories K. Pasteurella multocida toxin activation of heterotrimeric G proteins by deamidation. Proc Natl Acad Sci USA. 2009;106(17):7179–7184. doi: 10.1073/pnas.0900160106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jinadasa RN, Bloom SE, Weiss RS, Duhamel GE. Cytolethal distending toxin: a conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology. 2011;157(Pt 7):1851–1875. doi: 10.1099/mic.0.049536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elwell C, Chao K, Patel K, Dreyfus L. Escherichia coli CdtB mediates cytolethal distending toxin cell cycle arrest. Infect Immun. 2001;69(5):3418–3422. doi: 10.1128/IAI.69.5.3418-3422.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nougayrede JP, Homburg S, Taieb F, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313(5788):848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 50.Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrede JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci USA. 2010;107(25):11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51▪.Arthur JC, Perez-Chanona E, Muhlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338(6103):120–123. doi: 10.1126/science.1224820. This murine study demonstrated that IL10-deficient mice colonized with Escherichia coli NC101 (pks+) develop invasive colon cancer independent of inflammation, and also provided evidence correlating this strain with colorectal cancer in humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miraglia AG, Travaglione S, Meschini S, et al. Cytotoxic necrotizing factor 1 prevents apoptosis via the Akt/IκB kinase pathway role of nuclear factor-kappaB and Bcl-2. Mol Biol Cell. 2007;18(7):2735–2744. doi: 10.1091/mbc.E06-10-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Samba-Louaka A, Nougayrede JP, Watrin C, Oswald E, Taieb F. The enteropathogenic Escherichia coli effector Cif induces delayed apoptosis in epithelial cells. Infect Immun. 2009;77(12):5471–5477. doi: 10.1128/IAI.00860-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54▪.Wu S, Rhee KJ, Albesiano E, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15(9):1016–1022. doi: 10.1038/nm.2015. Identifies enterotoxigenic Bacteroides fragilis as being capable of inducing tumorigenesis in Apc+/− multiple intestinal neoplasia mice through a Th17-dependent mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toprak NU, Yagci A, Gulluoglu BM, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12(8):782–786. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 56.Moore WE, Holdeman LV. Human fecal flora: the normal flora of 20 Japanese Hawaiians. Appl Microbiol. 1974;27(5):961–979. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Polk BF, Kasper DL. Bacteroides fragilis subspecies in clinical isolates. Ann Intern Med. 1977;86(5):569–571. doi: 10.7326/0003-4819-86-5-569. [DOI] [PubMed] [Google Scholar]
- 58.Rhee KJ, Wu S, Wu X, et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect Immun. 2009;77(4):1708–1718. doi: 10.1128/IAI.00814-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Basset C, Holton J, Bazeos A, Vaira D, Bloom S. Are Helicobacter species and enterotoxigenic Bacteroides fragilis involved bowel disease? Dig Dis Sci. 2004;49(9):1425–1432. doi: 10.1023/b:ddas.0000042241.13489.88. [DOI] [PubMed] [Google Scholar]
- 60.Prindiville TP, Sheikh RA, Cohen SH, Tang YJ, Cantrell MC, Silva J., Jr Bacteroides fragilis enterotoxin gene sequences in patients with inflammatory bowel disease. Emerg Infect Dis. 2000;6(2):171–174. doi: 10.3201/eid0602.000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sears CL. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev. 2009;22(2):349–369. doi: 10.1128/CMR.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zitomersky NL, Coyne MJ, Comstock LE. Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut. Infect Immun. 2011;79(5):2012–2020. doi: 10.1128/IAI.01348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Franco AA, Mundy LM, Trucksis M, Wu S, Kaper JB, Sears CL. Cloning and characterization of the Bacteroides fragilis metalloprotease toxin gene. Infect Immun. 1997;65(3):1007–1013. doi: 10.1128/iai.65.3.1007-1013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Franco AA. The Bacteroides fragilis pathogenicity island is contained in a putative novel conjugative transposon. J Bacteriol. 2004;186(18):6077–6092. doi: 10.1128/JB.186.18.6077-6092.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu S, Lim KC, Huang J, Saidi RF, Sears CL. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc Natl Acad Sci USA. 1998;95(25):14979–14984. doi: 10.1073/pnas.95.25.14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu S, Morin PJ, Maouyo D, Sears CL. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology. 2003;124(2):392–400. doi: 10.1053/gast.2003.50047. [DOI] [PubMed] [Google Scholar]
- 67.Wu S, Powell J, Mathioudakis N, Kane S, Fernandez E, Sears CL. Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor-kappaB pathway. Infect Immun. 2004;72(10):5832–5839. doi: 10.1128/IAI.72.10.5832-5839.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Su LK, Kinzler KW, Vogelstein B, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256(5057):668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 69.Fodde R, Edelmann W, Yang K, et al. A targeted chain-termination mutation in the mouse Apc gene results in multiple intestinal tumors. Proc Natl Acad Sci USA. 1994;91(19):8969–8973. doi: 10.1073/pnas.91.19.8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70▪.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. First report of the Cancer Genome Atlas project profiling genetic alterations in 276 colorectal carcinoma samples, demonstrating significant similarity in patterns of genetic alterations from patient to patient. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu J, Duan Y, Cheng X, et al. IL-17 is associated with poor prognosis and promotes angiogenesis via stimulating VEGF production of cancer cells in colorectal carcinoma. Biochem Biophys Res Commun. 2011;407(2):348–354. doi: 10.1016/j.bbrc.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 72.Chae WJ, Gibson TF, Zelterman D, Hao L, Henegariu O, Bothwell AL. Ablation of IL-17A abrogates progression of spontaneous intestinal tumorigenesis. Proc Natl Acad Sci USA. 2010;107(12):5540–5544. doi: 10.1073/pnas.0912675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Su X, Ye J, Hsueh EC, Zhang Y, Hoft DF, Peng G. Tumor micro environments direct the recruitment and expansion of human Th17 cells. J Immunol. 2010;184(3):1630–1641. doi: 10.4049/jimmunol.0902813. [DOI] [PubMed] [Google Scholar]
- 74.Sobhani I, Tap J, Roudot-Thoraval F, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE. 2011;6(1):e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morikawa T, Baba Y, Yamauchi M, et al. STAT3 expression, molecular features, inflammation patterns, and prognosis in a database of 724 colorectal cancers. Clin Cancer Res. 2011;17(6):1452–1462. doi: 10.1158/1078-0432.CCR-10-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247(4940):322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 77.Oshima M, Oshima H, Kitagawa K, Kobayashi M, Itakura C, Taketo M. Loss of Apc heterozygosity and abnormal tissue building in nascent intestinal polyps in mice carrying a truncated Apc gene. Proc Natl Acad Sci USA. 1995;92(10):4482–4486. doi: 10.1073/pnas.92.10.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Velcich A, Yang W, Heyer J, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science. 2002;295(5560):1726–1729. doi: 10.1126/science.1069094. [DOI] [PubMed] [Google Scholar]
- 79.Berg DJ, Davidson N, Kuhn R, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4+ Th1-like responses. J Clin Invest. 1996;98(4):1010–1020. doi: 10.1172/JCI118861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu Y, Richardson JA, Parada LF, Graff JM. Smad3 mutant mice develop metastatic colorectal cancer. Cell. 1998;94(6):703–714. doi: 10.1016/s0092-8674(00)81730-4. [DOI] [PubMed] [Google Scholar]
- 81.Rudolph U, Finegold MJ, Rich SS, et al. Ulcerative colitis and adenocarcinoma of the colon in G alpha i2-deficient mice. Nat Genet. 1995;10(2):143–150. doi: 10.1038/ng0695-143. [DOI] [PubMed] [Google Scholar]
- 82.Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin MF, Taketo MM. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell. 1998;92(5):645–656. doi: 10.1016/s0092-8674(00)81132-0. [DOI] [PubMed] [Google Scholar]
- 83.Funabashi H, Uchida K, Kado S, Matsuoka Y, Ohwaki M. Establishment of a Tcrb and Trp53 genes deficient mouse strain as an animal model for spontaneous colorectal cancer. Exp Anim. 2001;50(1):41–47. doi: 10.1538/expanim.50.41. [DOI] [PubMed] [Google Scholar]
- 84.Chu FF, Esworthy RS, Chu PG, et al. Bacteria-induced intestinal cancer in mice with disrupted Gpx1 and Gpx2 genes. Cancer Res. 2004;64(3):962–968. doi: 10.1158/0008-5472.can-03-2272. [DOI] [PubMed] [Google Scholar]
- 85.Engle SJ, Ormsby I, Pawlowski S, et al. Elimination of colon cancer in germ-free transforming growth factor β 1-deficient mice. Cancer Res. 2002;62(22):6362–6366. [PubMed] [Google Scholar]
- 86.Taketo MM, Edelmann W. Mouse models of colon cancer. Gastroenterology. 2009;136(3):780–798. doi: 10.1053/j.gastro.2008.12.049. [DOI] [PubMed] [Google Scholar]
- 87.Balish E, Warner T. Enterococcus faecalis induces inflammatory bowel disease in IL-10 knockout mice. Am J Pathol. 2002;160(6):2253–2257. doi: 10.1016/S0002-9440(10)61172-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dove WF, Clipson L, Gould KA, et al. Intestinal neoplasia in the ApcMin mouse independence from the microbial and natural killer (beige locus) status. Cancer Res. 1997;57(5):812–814. [PubMed] [Google Scholar]
- 89.Li Y, Kundu P, Seow SW, et al. Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APCMin/+ mice. Carcinogenesis. 2012;33(6):1231–1238. doi: 10.1093/carcin/bgs137. [DOI] [PubMed] [Google Scholar]
- 90.Kado S, Uchida K, Funabashi H, et al. Intestinal microflora are necessary for development of spontaneous adenocarcinoma of the large intestine in T-cell receptor beta chain and p53 double-knockout mice. Cancer Res. 2001;61(6):2395–2398. [PubMed] [Google Scholar]
- 91.Uronis JM, Muhlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS ONE. 2009;4(6):e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johansson ME, Gustafsson JK, Sjoberg KE, et al. Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PLoS ONE. 2010;5(8):e12238. doi: 10.1371/journal.pone.0012238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Perse M, Cerar A. Dextran sodium sulphate colitis mouse model: traps and tricks. J Biomed Biotechnol. 2012;2012:718617. doi: 10.1155/2012/718617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Elinav E, Strowig T, Kau AL, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145(5):745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Garrett WS, Lord GM, Punit S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131(1):33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vijay-Kumar M, Aitken JD, Carvalho FA, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328(5975):228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hans W, Scholmerich J, Gross V, Falk W. The role of the resident intestinal flora in acute and chronic dextran sulfate sodium-induced colitis in mice. Eur J Gastroenterol Hepatol. 2000;12(3):267–273. doi: 10.1097/00042737-200012030-00002. [DOI] [PubMed] [Google Scholar]
- 99.Rath HC, Schultz M, Freitag R, et al. Different subsets of enteric bacteria induce and perpetuate experimental colitis in rats and mice. Infect Immun. 2001;69(4):2277–2285. doi: 10.1128/IAI.69.4.2277-2285.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fukuda M, Kanauchi O, Araki Y, et al. Prebiotic treatment of experimental colitis with germinated barley foodstuff: a comparison with probiotic or antibiotic treatment. Int J Mol Med. 2002;9(1):65–70. [PubMed] [Google Scholar]
- 101.Kitajima S, Morimoto M, Sagara E, Shimizu C, Ikeda Y. Dextran sodium sulfate-induced colitis in germ-free IQI/Jic mice. Exp Anim. 2001;50(5):387–395. doi: 10.1538/expanim.50.387. [DOI] [PubMed] [Google Scholar]
- 102.Ellmerich S, Scholler M, Duranton B, et al. Promotion of intestinal carcinogenesis by Streptococcus bovis. Carcinogenesis. 2000;21(4):753–756. doi: 10.1093/carcin/21.4.753. [DOI] [PubMed] [Google Scholar]
- 103.Boleij A, Tjalsma H. Gut bacteria in health and disease: a survey on the interface between intestinal microbiology and colorectal cancer. Biol Rev Camb Philos Soc. 2012;87(3):701–730. doi: 10.1111/j.1469-185X.2012.00218.x. [DOI] [PubMed] [Google Scholar]
- 104.Fox JG, Ge Z, Whary MT, Erdman SE, Horwitz BH. Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol. 2011;4(1):22–30. doi: 10.1038/mi.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Erdman SE, Rao VP, Poutahidis T, et al. Nitric oxide and TNF-alpha trigger colonic inflammation and carcinogenesis in Helicobacter hepaticus-infected, Rag2-deficient mice. Proc Natl Acad Sci USA. 2009;106(4):1027–1032. doi: 10.1073/pnas.0812347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mangerich A, Knutson CG, Parry NM, et al. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci USA. 2012;109(27):E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Balagopal A, Philp FH, Astemborski J, et al. Human immunodeficiency virus-related microbial translocation and progression of hepatitis C. Gastroenterology. 2008;135(1):226–233. doi: 10.1053/j.gastro.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Klatt NR, Funderburg NT, Brenchley JM. Microbial translocation, immune activation, and HIV disease. Trends Microbiol. 2013;21(1):6–13. doi: 10.1016/j.tim.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huycke MM, Moore D, Joyce W, et al. Extracellular superoxide production by Enterococcus faecalis requires demethylmenaquinone and is attenuated by functional terminal quinol oxidases. Mol Microbiol. 2001;42(3):729–740. doi: 10.1046/j.1365-2958.2001.02638.x. [DOI] [PubMed] [Google Scholar]
- 110.Wang X, Allen TD, May RJ, Lightfoot S, Houchen CW, Huycke MM. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008;68(23):9909–917. doi: 10.1158/0008-5472.CAN-08-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ruiz PA, Shkoda A, Kim SC, Sartor RB, Haller D. IL-10 gene-deficient mice lack TGF-beta/Smad signaling and fail to inhibit proinflammatory gene expression in intestinal epithelial cells after the colonization with colitogenic Enterococcus faecalis. J Immunol. 2005;174(5):2990–2999. doi: 10.4049/jimmunol.174.5.2990. [DOI] [PubMed] [Google Scholar]
- 112.Winters MD, Schlinke TL, Joyce WA, Glore SR, Huycke MM. Prospective case-cohort study of intestinal colonization with enterococci that produce extracellular superoxide and the risk for colorectal adenomas or cancer. Am J Gastroenterol. 1998;93(12):2491–2500. doi: 10.1111/j.1572-0241.1998.00710.x. [DOI] [PubMed] [Google Scholar]
- 113.Barthold SW, Osbaldiston GW, Jonas AM. Dietary, bacterial, and host genetic interactions in the pathogenesis of transmissible murine colonic hyperplasia. Lab Anim Sci. 1977;27(6):938–945. [PubMed] [Google Scholar]
- 114.Newman JV, Kosaka T, Sheppard BJ, Fox JG, Schauer DB. Bacterial infection promotes colon tumorigenesis in ApcMin/+ mice. J Infect Dis. 2001;184(2):227–230. doi: 10.1086/321998. [DOI] [PubMed] [Google Scholar]
- 115.Maddocks OD, Short AJ, Donnenberg MS, Bader S, Harrison DJ. Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans. PLoS ONE. 2009;4(5):e5517. doi: 10.1371/journal.pone.0005517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Keusch GT. Opportunistic infections in colon carcinoma. Am J Clin Nutr. 1974;27(12):1481–1485. doi: 10.1093/ajcn/27.12.1481. [DOI] [PubMed] [Google Scholar]
- 117.Kok H, Jureen R, Soon CY, Tey BH. Colon cancer presenting as Streptococcus gallolyticus infective endocarditis. Singapore Med J. 2007;48(2):e43–e45. [PubMed] [Google Scholar]
- 118.Klein RS, Recco RA, Catalano MT, Edberg SC, Casey JI, Steigbigel NH. Association of Streptococcus bovis with carcinoma of the colon. N Engl J Med. 1977;297(15):800–802. doi: 10.1056/NEJM197710132971503. [DOI] [PubMed] [Google Scholar]
- 119.Boleij A, van Gelder MM, Swinkels DW, Tjalsma H. Clinical importance of Streptococcus gallolyticus infection among colorectal cancer patients: systematic review and meta-analysis. Clin Infect Dis. 2011;53(9):870–878. doi: 10.1093/cid/cir609. [DOI] [PubMed] [Google Scholar]
- 120.Boleij A, Muytjens CM, Bukhari SI, et al. Novel clues on the specific association of Streptococcus gallolyticus subsp gallolyticus with colorectal cancer. J Infect Dis. 2011;203(8):1101–1109. doi: 10.1093/infdis/jiq169. [DOI] [PubMed] [Google Scholar]
- 121.Corredoira-Sanchez J, Garcia-Garrote F, Rabunal R, et al. Association between bacteremia due to Streptococcus gallolyticus subsp gallolyticus (Streptococcus bovis I) and colorectal neoplasia: a case–control study. Clin Infect Dis. 2012;55(4):491–496. doi: 10.1093/cid/cis434. [DOI] [PubMed] [Google Scholar]
- 122.Schlegel L, Grimont F, Ageron E, Grimont PA, Bouvet A. Reappraisal of the taxonomy of the Streptococcus bovis/Streptococcus equinus complex and related species: description of Streptococcus gallolyticus subsp gallolyticus subsp nov., S gallolyticus subsp macedonicus subsp nov and S gallolyticus subsp pasteurianus subsp nov. Int J Syst Evol Microbiol. 2003;53(Pt 3):631–645. doi: 10.1099/ijs.0.02361-0. [DOI] [PubMed] [Google Scholar]
- 123.Seder CW, Kramer M, Long G, Uzieblo MR, Shanley CJ, Bove P. Clostridium septicum aortitis: report of two cases and review of the literature. J Vasc Surg. 2009;49(5):1304–1309. doi: 10.1016/j.jvs.2008.11.058. [DOI] [PubMed] [Google Scholar]
- 124.Wang T, Cai G, Qiu Y, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6(2):320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE. 2012;7(6):e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126▪.Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. This human colorectal cancer microbiome study utilized 454 sequencing, real-time PCR and FISH to identify an increased association between Fusobacterium and colon tumors when compared with flanking tissues (published in the same issue as [23]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Swidsinski A, Khilkin M, Kerjaschki D, et al. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology. 1998;115(2):281–286. doi: 10.1016/s0016-5085(98)70194-5. [DOI] [PubMed] [Google Scholar]
- 128.Uemura N, Okamoto S, Yamamoto S, et al. Helicobacter pylori infection and the development of gastric cancer. N Engl J Med. 2001;345(11):784–789. doi: 10.1056/NEJMoa001999. [DOI] [PubMed] [Google Scholar]
- 129.Sears CL, Pardoll DM. Perspective: alpha-bugs, their microbial partners, and the link to colon cancer. J Infect Dis. 2011;203(3):306–311. doi: 10.1093/jinfdis/jiq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver–passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10(8):575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 131.Hand TW, Dos Santos LM, Bouladoux N, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337(6101):1553–1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Evans AS. Causation and disease: the Henle–Koch postulates revisited. Yale J Biol Med. 1976;49(2):175–195. [PMC free article] [PubMed] [Google Scholar]