Abstract
Drug resistance is a major issue in the treatment of glioblastoma. Almost all glioblastomas are intrinsically resistant to chemotherapeutic temozolomide (TMZ) or develop resistance during treatment. The interaction networks of microRNAs (miRNAs) and mRNAs likely regulate most biological processes and can be employed to better understand complex processes including drug resistance in cancer. In this study, we examined if integrative miRNA/mRNA network analysis using the web-service tool mirConnX could be used to identify drug resistance factors in glioblastoma. We used TMZ-resistant glioblastoma cells and their integrated miRNA/mRNA networks to identify TMZ-sensitizing factors. TMZ resistance was previously induced in glioblastoma cell lines U87, Hs683, and LNZ308. miRNA/mRNA expression profiling of these cells and integration of the profiles using mirConnX resulted in the identification of plant homeodomain (PHD)-like finger 6 (PHF6) as a potential TMZ-sensitizing factor in resistant glioblastoma cells. Analysis of PHF6 expression showed significant upregulation in glioblastoma as compared to normal tissue. Interference with PHF6 expression in three TMZ-resistant subclones significantly enhanced TMZ-induced cell kill in two of these cell lines. Altogether, these results demonstrate that mirConnX is a feasible and useful tool to investigate miRNA/mRNA interactions in TMZ-resistant cells and has potential to identify drug resistance factors in glioblastoma.
Glioblastoma is a significant problem in both adults and children. It is the most common primary brain tumour in adults and despite the standard treatment, consisting of surgery, chemoradiation, and adjuvant temozolomide (TMZ), the median survival for glioblastoma patients is only 14 months1,2. Drug resistance is a major issue in the management of glioblastoma. DNA repair mechanisms hamper the cytotoxic effect of the DNA damaging chemotherapeutic agent temozolomide in glioblastoma3,4,5,6,7,8,9,10. Canonical DNA repair systems shown to be involved in TMZ resistance are the O6-methylguanine DNA methyltransferase (MGMT) enzyme5,6,7 and the mismatch-repair (MMR) machinery3,4,8,9,10. Besides these DNA repair mechanisms, post-transcriptional regulation of gene expression by microRNAs (miRNAs) has also been reported to play a role in TMZ resistance11,12,13,14,15,16,17,18,19,20,21,22.
Most, if not all, biological processes are driven by the interactive networks of mRNAs and miRNAs23. Several tools are currently available for integrative miRNA/mRNA regulatory network analysis, including mirConnX24, MAGIA25, MMIA26, and GenMir++27. The algorithms of these programs combine sequence information with gene expression data to create a condition-specific regulatory network. Such integrated miRNA/mRNA networks can be potentially related to aberrant cellular processes and specific diseases, including drug resistance and cancer24,28,29.
miRNAs are short non-coding single stranded molecules, consisting of ~22 nucleotides which regulate cellular processes by complementary binding to target messenger RNA (mRNA). Binding of the miRNA with its ‘seed' region to the 3′ untranslated region (3′UTR) of the target mRNA results in translational inhibition of the mRNA30,31,32. Importantly, multiple miRNAs can regulate the same mRNA and single miRNAs can regulate multiple mRNAs, indicating a complex system of translational regulation30,31,32. Over the past decade multiple studies have demonstrated the importance of miRNAs in cancer in general, and their involvement in TMZ resistance in glioblastoma in particular11,12,13,14,15,16,17,18,19,20,21,22,31. miRNAs reported to influence TMZ resistance in glioblastoma include miR-195, miR-455, miR-1011, miR-181b12,18, miR-2113,16, miR-125b14, miR-14515, miR-21117, miR-1719, miR-920, the miR-183/96/182 cluster21, and miR-221/22222. However, so far no comprehensive network analysis has been performed on integrated miRNA/mRNA expression profiles of drug resistance in glioblastoma cells.
In this study, we examined whether mirConnX can be used to identify drug resistance factors in glioblastoma. As proof of concept, we employed three glioblastoma cell lines and their TMZ-resistant subclones and performed comprehensive integrative analysis of their mRNA/miRNA expression profiles in order to identify potential TMZ-sensitizing factors. Integration of these profiles using mirConnX resulted in the identification of plant homeodomain (PHD)-like finger 6 (PHF6) as being a potential TMZ-sensitizing factor. Altogether, these results demonstrate that mirConnX is a feasible and useful tool to examine miRNA/mRNA interactions in TMZ-resistant cells and is potentially useful for the identification of new drug targets.
Results
RNA profiling of TMZ-sensitive glioblastoma cells and their resistant subclones
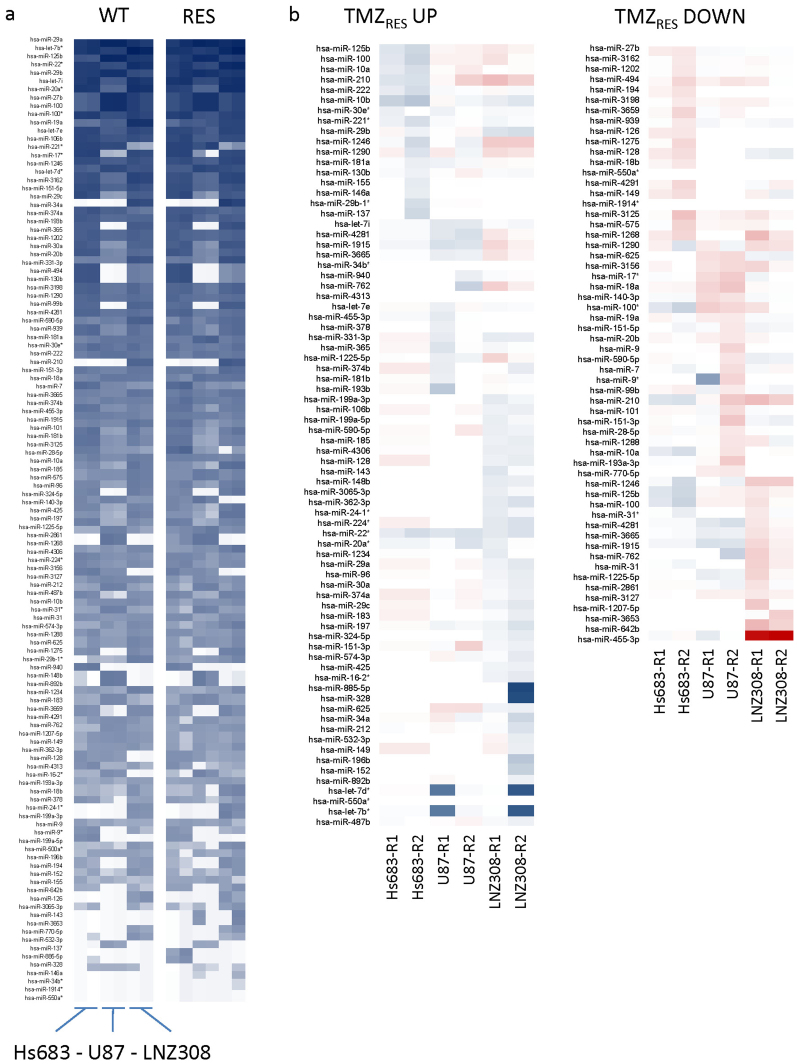
TMZ resistance was induced in duplicate in the glioblastoma cell lines Hs683, U87, and LNZ308, creating two independent resistant subclones of each wild type cell line as described previously33. TMZ IC50 values of the resistant subclones were increased >2-fold compared to the wild type cells33. RNA from both the wild type and the resistant cells was isolated and used for mRNA microarray expression profiling as described previously33. The same RNA was also used to perform miRNA microarray expression analysis. miRNA expression profiles of Hs683, U87, and LNZ308 wild type and their resistant subclones are depicted in heatmap format (Fig. 1a). The miRNAs listed were sorted based on overall expression levels in all cell lines, both TMZ-sensitive and TMZ-resistant, with highly expressed miRNAs at the top and lower expressed miRNAs at the bottom. In order to quickly scan if specific miRNAs were differentially expressed in the resistant subclones compared to the wild type cell lines, we used the simple ad hoc method fold change to calculate the resistant/wild type expression ratio for each miRNA in each wild type and resistant subclone pair separately. miRNAs with an expression ratio of ≥1.5 were considered upregulated and miRNAs with an expression ratio of ≤0.67 were considered downregulated. We identified differentially expressed miRNAs in individual resistant subclones when comparing them to their wild type counterparts. However, no common up- or downregulated miRNAs were identified that were differentially expressed in all resistant subclones (Fig. 1b). Since multiple miRNAs can regulate the same mRNA in a complex regulatory system of translational repression, we decided to use both mRNA and miRNA expression profiles of the wild type and TMZ-resistant cell lines for integrative network analysis using mirConnX to identify potential drug targets that would likely be missed when considering only mRNAs targeted by individual miRNAs.
Figure 1. miRNA expression profiles of wild type and TMZ-resistant subclones.
(a), overall miRNA expression profiles of Hs683, U87, and LNZ308 wild type and resistant subclones, sorted based on overall expression levels in all cell lines, both TMZ-sensitive and TMZ-resistant, with highly expressed miRNAs at the top and lowly expressed miRNAs at the bottom. (b), heatmap of upregulated (left) and down regulated (right) miRNAs in resistant subclones. TMZRES UP heatmap: blue = strong upregulation, light blue = upregulation, red = modest upregulation. TMZRES DOWN heatmap: red = strong downregulation, light red = downregulation, blue = modest downregulation.
Integrative miRNA/mRNA network analysis using mirConnX identifies PHF6 as a potential target of multiple miRNAs in TMZ-resistant glioblastoma cells
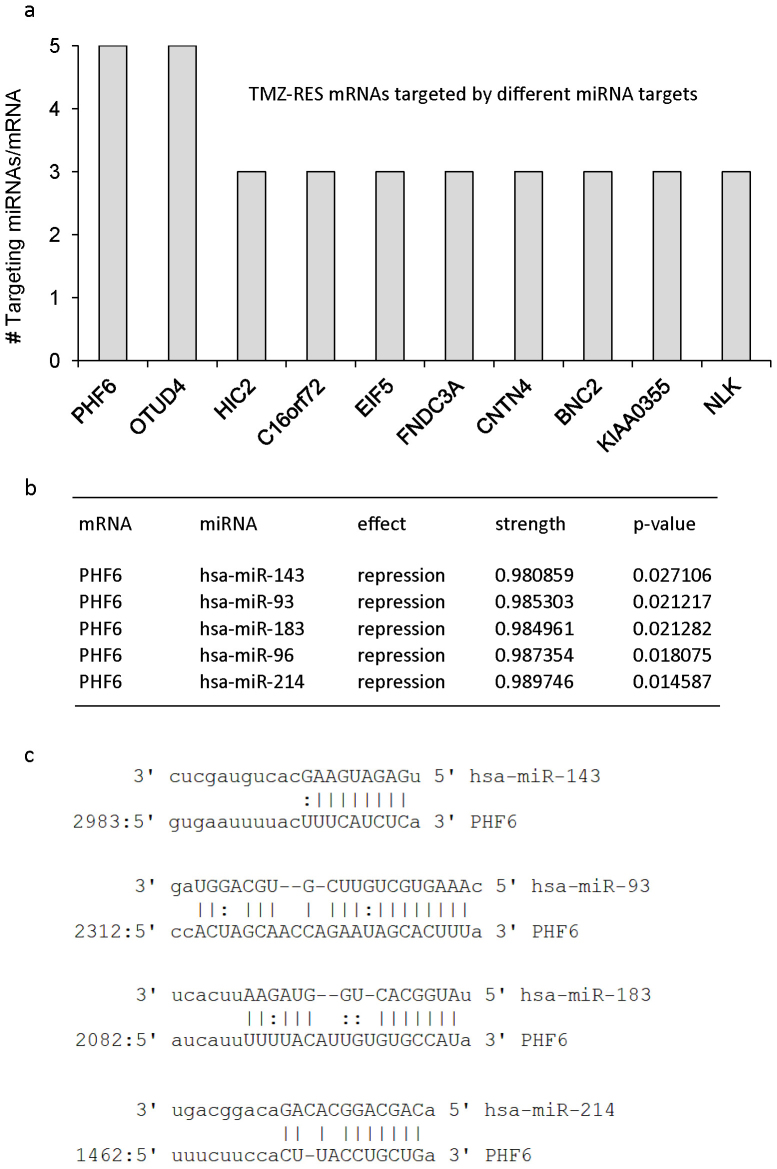
The input requirements for the mirConnX web interface include at least six matched columns of mRNA and miRNA expression data24. Furthermore, type of species and statistical analysis method need to be specified. We used Pearson's correlation as an association measure of choice to examine correlations between miRNAs and mRNAs in our datasets, generating a dynamic network of miRNA/mRNA interactions extracted from the provided expression data. Subsequently, mirConnX combined this dynamic network with a prior constructed static network consisting of computationally-based miRNA target predictions supplemented with interactions found in literature. mirConnX used an integration function with user-specified weights to combine these networks24. We analysed the expression profiles of the wild type cells and the resistant cells separately. Integration of the miRNA and mRNA expression profiles of the TMZ-sensitive or TMZ-resistant subclones using mirConnX resulted in the regulatory networks as depicted in Fig. 2. The miRNAs are depicted as blue squares and mRNAs as orange circles. The effect of the interaction (inactivation or activation) is represented by arrows in green (activation) and bar-headed lines in red (inactivation). Because we were interested in the regulatory miRNA/mRNA processes involved in TMZ resistance, we focussed on the interaction network obtained from the expression profiles of the TMZ-resistant subclones. We determined which mRNA depicted in the network is potentially targeted by the largest set of either upregulated or downregulated miRNAs in these cells. The plant homeodomain (PHD)-like finger 6, PHF6 and OTU domain containing 4, OTUD4 were potential targets of each of five miRNAs in the network of the TMZ-resistant subclones (Fig. 3a). Importantly, PHF6 and OTUD4 were not present in the interaction network obtained from the miRNA/mRNA expression profiles of the wild type TMZ-sensitive cell lines. Interestingly, both PHF6 and OTUD4 were not identified in our previous mRNA expression analysis of these cells33, and are thus good candidates to demonstrate the added value of using integrative analysis of miRNA and mRNA expression profiles. We continued our analysis with PHF6, based on its predicted target interactions as shown in Fig. 3b. miR-143, miR-93, miR-183, miR-96, and miR-214 show a predicted repressive effect on PHF6, with the strength of the interactions, based on a regulation score calculated by mirConnX24, between the five miRNAs and the 3′UTR of PHF6 close to 1 (p<0.05), indicating strong interactions (range, 0–1). In Fig. 3c, we confirm the alignment between the different miRNAs and the 3′UTR region of PHF6 in independent databases. miR-143, miR-93, miR-183, and miR-214 show efficient base-pairing with PHF6. However, we were not able to confirm PHF6 as a potential target of miR-96 in these databases. Altogether, the mirConnX interaction network analysis demonstrates a potential role for PHF6 in the integrative mRNA/miRNA network of TMZ-resistant glioblastoma cells.
Figure 2. miRNA/mRNA interaction networks of wild type glioblastoma cells (left) and TMZ-resistant subclones (right), generated by mirConnX.
The miRNAs are depicted as blue squares, mRNAs as orange circles, and transcription factors (TFs) as green triangles. The effect of the interaction (activation or inactivation) is represented by arrows in green (activation) and bar-headed lines in red (inactivation). Magnification of network of TMZ-resistant cells is shown in boxed area.
Figure 3. Integrative miRNA/mRNA network analysis using mirConnX identifies PHF6 as a target of multiple miRNAs in TMZ-resistant glioblastoma cells.
(a), mRNAs targeted by at least three miRNAs in the interaction network of TMZ-resistant subclones. (b), miRNAs miR-143, -93, -183, -96, and -214 potentially target PHF6 mRNA in our TMZ-resistant subclones. Shown are the effect of the interaction (repression) between miRNA and mRNA, the strength of the regulation (regulation score), and its respective p-value. The regulation score calculated by mirConnX is based on the weight of the prior static network constructed by mirConnX and the weight of the association measure (Pearson's correlation) extracted from the user-supplied expression data24. (c), alignment between the different miRNAs and the 3′UTR region of PHF6 in independent databases. miR-96 is not identified as targeting PHF6 in these databases. miR-96 is encoded within <10 kb of miR-183.
PHF6 expression is increased in TMZ-resistant cells and primary glioblastoma samples
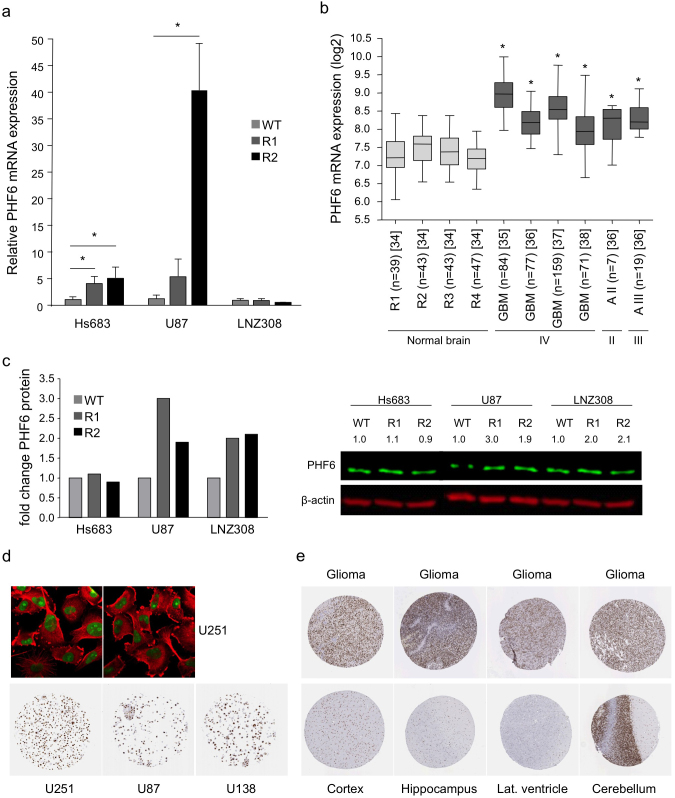
We determined the endogenous PHF6 mRNA expression levels in Hs683, U87, and LNZ308 wild type cells and their TMZ-resistant subclones by qRT-PCR to further evaluate the findings from the mirConnX integration network (Fig. 4a). We observed a >4-fold increase in PHF6 expression in both TMZ-resistant subclones of Hs683 and U87. PHF6 was even >40-fold increased in the second subclone U87-R2 compared to the wild type cells. No change on PHF6 mRNA expression level was observed in the resistant subclones of LNZ308. In addition, using the microarray analysis and visualization platform R2 (http://r2.amc.nl), we found that the PHF6 mRNA expression level is significantly increased in glioblastoma tissue as well as in grade II and III astrocytoma tissue compared to different regions of normal brain tissue (p<0.01) (Fig. 4b)34,35,36,37,38. Since miRNAs function as post-transcriptional regulators, the effect of these miRNAs on PHF6 mRNA translation is likely to be visible on the protein level. Therefore, we performed Western blotting for PHF6 to determine the protein expression levels in the wild type and resistant cell lines (Fig. 4c). We observed a modest increase in PHF6 protein expression in the TMZ-resistant subclones of U87 and LNZ308. However, we did not reproducibly observe an increased PHF6 protein expression in Hs683-R1 and -R2 cells, although we did measure an increase in PHF6 mRNA expression in these subclones. These results indicate that TMZ resistance in the glioblastoma subclones of U87 seems to correlate with increased PHF6 mRNA and protein expression, in subclones of Hs683 with increased PHF6 mRNA expression only, and in subclones of LNZ308 with increased PHF6 protein expression only, although the differences observed on protein level were at most modest. Furthermore, PHF6 protein was high to moderately expressed in three glioblastoma cell lines (Fig. 4d). Finally, we evaluated PHF6 protein expression in high-grade glioma samples and normal human brain tissues, using the Human Protein Atlas (Fig. 4e, Table 1)39. Most glioma samples showed high to medium PHF6 protein expression. Cell types in different normal brain regions showed variable PHF6 expression (Table 1). Neuronal cells in the cortex showed high PHF6 expression. Glial cells in the cortex showed medium PHF6 expression while these cells showed low expression of PHF6 in the hippocampus and lateral ventricle wall. In normal cerebellum, PHF6 was found to be highly expressed.
Figure 4. PHF6 mRNA and/or protein expression is increased in TMZ-resistant cells and primary glioblastoma samples.
(a), PHF6 mRNA levels in wild type and TMZ-resistant subclones as determined by qRT-PCR. (b), in silico analysis of PHF6 mRNA expression in normal brain tissue (grey) of different brain regions (R1 = entorhinal cortex, R2 = hippocampus, R3 = post central gyrus, R4 = superior frontal gyrus)34, in glioblastoma (grade IV) tissue, and in grade II (II) and III astrocytoma (III) tissue (dark grey)35,36,37,38 using R2 analysis software. (c), PHF6 protein expression in Hs683, U87, and LNZ308 wild type and TMZ-resistant subclones as determined by Western blotting. (d), immunofluorescent staining of PHF6 (green) in the nucleolus of U251 glioblastoma cells. Microtubules are stained in red. PHF6 is highly expression in U251 and U138 glioblastoma cell lines and moderately expressed in U87 glioblastoma cell line. (e), PHF6 protein expression in normal brain tissue (cortex, hippocampus, lateral ventricle wall, and cerebellum) and in high-grade glioma tissue (the Human Protein Atlas). These images were derived from the Human Protein Atlas. Shown are averages, error bars indicate SD. *p<0.05 student's t test.
Table 1. PHF6 protein expression in different cell types in different brain regions and in high-grade glioma cells as shown in Fig. 4d (the Human Protein Atlas). Light grey is n.a. for these brain regions.
| Cerebral cortex | Hippocampus | Lateral ventricle wall | Cerebellum | High-grade glioma | |
|---|---|---|---|---|---|
| Neuronal cells | High | Medium | Low | ||
| Glial cells | Medium | Low | Low | ||
| Endothelial cells | Medium | ||||
| Neuropil | Not detected | ||||
| Purkinje cells | Medium | ||||
| Cells in granular layer | High | ||||
| Cells in molecular layer | High | ||||
| Tumor cells | High (5/12) | ||||
| Medium (6/12) | |||||
| Low (1/12) |
PHF6 knockdown in combination with TMZ treatment decreases cell viability
As proof of concept of potential drug target confirmation, we performed functional analysis of PHF6 in TMZ resistance by PHF6 knockdown in Hs683-R1, U87-R1, and LNZ308-R1 cells using siRNAs in the presence or absence of TMZ. Cells were transfected with siRNAs against PHF6 (siPHF6) or with a non-targeting siRNA (siNT) as control. After transfection, we first performed qRT-PCR on PHF6 to determine if the transfection was successful (Fig. 5a). PHF6 mRNA levels were significantly reduced in U87-R1 and Hs683-R1 cells transfected with siPHF6 compared to cells transfected with the control siRNA (p<0.05), however, knockdown of PHF6 mRNA in Hs683-R1 was less pronounced (<2-fold). Next, the effect of combined PHF6 knockdown and TMZ treatment on cell viability was determined (Fig. 5b and c). DAPI-stained nuclei were quantified, in order to determine cell viability after treatment (Fig. 5b and c). Treatment with TMZ had a minor effect on cell viability in the non-transfected cells. Transfection with the control siRNA had no effect in U87-R1 cells and a minor effect in LNZ308-R1 cells while transfection toxicity was observed in Hs683-R1 cells. Transfection with siPHF6 resulted in a significant reduction in the number of cells compared to cells transfected with the control siRNA in U87-R1 and LNZ308-R1 cells (p<0.05). Addition of TMZ 24 hrs after transfection resulted in enhanced cell kill as compared to TMZ or siPHF6 alone in U87-R1 and LNZ308-R1 cells (p<0.05) (Fig. 5c). In conclusion, these data show that PHF6 knockdown was successful and in combination with TMZ it resulted in a modest but significant effect on viability in two of three TMZ-resistant glioblastoma cells tested.
Figure 5. PHF6 knockdown in combination with TMZ treatment decreases cell viability.
(a), PHF6 mRNA expression in U87-R1 and Hs683-R1 cells 24 hrs after transfection with siPHF6 or siNT as determined by qRT-PCR. (b), representative images of the different treatment conditions (CTRL, TMZ, siNT/CTRL, siNT/TMZ, siPHF6/CTRL, siPHF6/TMZ) in Hs683-R1, U87-R1, and LNZ308-R1 cells four days after treatment with TMZ. The panels show DAPI-stained nuclei. Size bar, 20 µm. (c), quantification of cell viability of Hs683-R1, U87-R1, and LNZ308-R1 cells after treatment with siPHF6 or siNT in the presence or absence of 50 µM (Hs683-R1), 300 µM (U87-R1) or 400 µM (LNZ308-R1) TMZ. Shown are averages, error bars indicate SD. *p<0.05 student's t test. * in c indicates statistical significance of combination of PHF6 knockdown and TMZ compared to PHF6 knockdown or TMZ treatment alone.
Discussion
The aim of this study was to determine whether integrative miRNA/mRNA network analysis is a feasible technique for the identification of drug resistance factors in glioblastoma. We used TMZ-sensitive glioblastoma cell lines and their TMZ-resistant subclones as a proof of concept to identify potential TMZ-sensitizing factors. mirConnX was used to generate integrative interaction networks of miRNA/mRNA expression data of these TMZ-resistant and -sensitive glioblastoma cells, and allowed for the identification of PHF6 as a potential TMZ-sensitizing drug target in glioblastoma. This tool was used before in a study performed by Kusko et al.29 in which it demonstrated its applicability for the identification of miRNAs that co-vary with differentially expressed p53/hypoxia genes in idiopathic pulmonary fibrosis and emphysema.
A limitation of mirConnX is that separate interaction networks have to be built in order to compare two different conditions, as shown here for the wild type and TMZ-resistant conditions. This factor has been taken into account in the algorithm Classification based Analysis of Paired Expression data of RNA (CAPE RNA), developed by Hecker et al.40. This tool puts the emphasis on differentially occurring interactions between different samples in a collective dataset but does not seem to have an integrated transcription factor function as for mirConnX. Furthermore, Bar-Joseph et al.41 reported that miRNA and mRNA interaction networks can shift over time suggesting that it is a dynamic process. Schulz et al.42 developed the algorithm Development of MiRNA Dynamic Regulatory Events Miner (mirDREM), a tool that allows reconstruction of dynamic regulatory networks that model the effects of miRNAs and TFs on their targets over time. It could be of value to determine the dynamics of these interactions in the TMZ-sensitive and TMZ-resistant cells used in this study, e.g. in the presence of TMZ.
Here, mirConnX identified PHF6 to be targeted by the largest set of miRNAs, namely miR-143, miR-93, miR-183, miR-96, and miR-214, in our TMZ-resistant subclones. In each individual TMZ-resistant cell line a different combination of these miRNAs seems to control PHF6 regulation. This could be an explanation for the variation in PHF6 expression observed in the different resistant cell lines. Using independent databases, we confirmed the strong binding potential between miR-143, miR-93, miR-183, and miR-214 and the 3′UTR of PHF6. Of these miRNAs, only miR-96 was not identified by these prediction programs to strongly interact with PHF6, which could be due to its close proximity to miR-183. The miRNA-183/96/182 cluster has been previously implicated to play a role in TMZ resistance. However, in that study21 overexpression of the cluster resulted in TMZ resistance while our study suggests that the interaction of miR-183 and miR-96 and PHF6 mRNA is involved in induced TMZ resistance.
Further analysis of PHF6 expression in our TMZ-resistant and -sensitive cells showed mRNA upregulation of PHF6 in the resistant subclones of Hs683 and U87 but not in the resistant subclones of LNZ308. On the protein level, both subclones of U87 and LNZ308 showed a modest increase of PHF6. As mentioned above, PHF6 expression is potentially regulated by a different combination of miRNAs in each of the resistant cell lines and could explain the variation in PHF6 expression. Functional analysis of PHF6 expression in the context of TMZ resistance was performed using siRNAs against PHF6 in combination with TMZ treatment. We observed a modest effect on cell viability when we combined the siRNA against PHF6 with TMZ in comparison to TMZ treatment or transfection with PHF6 siRNA alone in U87-R1 and LNZ308-R1 cells. This effect was not detected in the TMZ-resistant subclone Hs683-R1 in which a relatively high transfection toxicity was observed. Furthermore, Hs683-R1 showed no increase in PHF6 protein expression compared to its wild type counterpart, which could be an additional explanation for the lack of effect observed in these cells. In addition, knockdown of PHF6 in Hs683-R1 cells is less pronounced compared to PHF6 knockdown in U87-R1 cells and could also add to the lack of effect on cell viability observed in this cell line.
The PHF6 gene is reported to be involved in cell cycle control and genomic maintenance43. It is a nucleolus, ribosomal RNA promoter-associated protein that contains two PHD-like zinc finger domains. PHF6 interacts with its PHD1 domain with upstream binding factor (UBF)43. UBF specifically interacts with the ribosomal RNA (rRNA) promoter region, thereby activating rRNA transcription. PHF6 is a negative regulator of UBF and, therefore, suppresses rRNA transcription43,44. It has also been reported that PHF6 interacts with the nucleosome remodelling and deacetylation (NuRD) complex45. The NuRD complex includes two histone deacetylases (HDAC1/2) and possesses nucleosome remodelling activity. Thereby, the PHF6-NuRD complex reveals a role for PHF6 in chromatin structure modification45,46. Moreover, it has been shown that down regulation or loss of function, by miRNAs or somatic mutations, of PHF6 contributes to the development of T-cell acute lymphoblastic leukemia (T-ALL) and acute myeloid leukemia (AML) suggesting that PHF6 functions as a tumour suppressor gene in these hematologic malignancies43,47,48,49.
The mechanism through which PHF6 could result in TMZ resistance in glioblastoma cells is not clear. It is possible that treatment with TMZ results in a genotoxic stress response causing the induction of PHF6. As mentioned previously, PHF6 has been reported to contribute to genomic maintenance and cell cycle control43. PHF6 inhibits rRNA synthesis, resulting in a decrease in ribosome biogenesis, which may ultimately lead to ribosomal stress. It has been reported that upon ribosomal stress, the ribosomal proteins (RPs) L5 and L11 are translocated from the nucleolus to the nucleoplasm50,51. In the nucleoplasm, L5 and/or L11 could inhibit MDM2, a partner of p53, resulting in the induction of a p53 response50. This response will force cells into senescence, apoptosis, or cell cycle arrest52. However, this mechanism would only be applicable to cells that have a wild type p53, as is the case in U87 cells but not in LNZ308 and Hs683 cells, which harbour mutant p53. Therefore, the mechanism via which PHF6 could lead to TMZ resistance warrants further investigation.
Previously, we identified TMZ resistance factors solely using the mRNA expression profiles of these cells33. In that study, PHF6 was not identified when only considering differences in mRNA expression levels between TMZ-sensitive wild type cells and their TMZ-resistant subclones. Since differential regulation of miRNA and mRNA pairs plays an important role in multiple cellular processes, including drug resistance in cancer, taking into account gene expression regulation by miRNAs in drug resistance can add to the understanding of TMZ resistance in glioblastoma. Therefore, this study shows the added value of a systems biology approach using software tools such as mirConnX to examine drug resistance in glioblastoma. This approach may complement the studies exploring for TMZ resistance factors in glioblastoma based on gene expression only and can provide novel insights in mechanisms underlying TMZ resistance. In addition, integration of miRNA and mRNA profiles to examine drug resistance has been applied before in anti-estrogen-resistance in breast cancer cells53. There, they showed the usefulness of this approach in the identification of miRNA-related network clusters that contribute to anti-estrogen resistance.
In conclusion, in this study we show, using a relatively small sample size, that network analysis programs such as mirConnX can be useful tools to examine miRNA/mRNA interactions in order to identify potential drug targets in glioblastoma. In addition, integrative miRNA/mRNA network analysis using such software tools may be considered for the identification of drug resistance factors in other diseases as well.
Methods
Cells
Generation of TMZ-resistant glioblastoma subclones was described previously33. Briefly, parental Hs683, U87, and LNZ308 glioblastoma cell lines were treated twice a week in duplicate with 33 µM TMZ for multiple weeks until two independent resistant subclones were generated. Hs683 (WT, R1 and R2), U87 (WT, R1 and R2), and LNZ308 (WT, R1 and R2) cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM; PAA Laboratories, GmbH, Pasching, Austria)) supplemented with 10% fetal bovine serum (FBS) (PAA Laboratories, GmbH, Pasching, Austria), and 1 mg/ml penicillin-streptomycin (both PAA Laboratories, GmbH, Pasching, Austria) at 37°C and 5% CO2 in a humidified incubator.
mRNA and miRNA gene expression analysis
Total RNA was isolated using mirVana miRNA isolation kit (Life Technologies). mRNA microarray expression analysis was performed previously, as described elsewhere33, using Agilent 4×44K Whole Human GE arrays (Agilent Technologies), according to the manufacturer's instructions. The same RNA samples were used for miRNA profiling. Samples were hybridized to Agilent SurePrint G3 Human v16 miRNA 8×60k arrays (Agilent Technologies). Scanning of the slides was performed with Agilent Microarray scanner (G2565BA). Image analysis was performed using feature extraction software version 10.5 (Agilent Technologies, Inc., CA, USA). The Agilent miRNA_105_Dec08_ protocol was applied using default settings. For each wild type and resistant subclone, a normalization factor was calculated by dividing the sum of all intensities of the WT by the sum of all intensities of the WT and the resistant subclones. Subsequently, each intensity value was multiplied by its corresponding normalization factor. Intensity values <2-fold of background intensity level were regarded as not detectable. Relative miRNA expression in R1 or R2 cells compared to WT cells was determined by calculating the resistant/wild type expression ratio. A miRNA with an expression ratio of ≥1.5 was considered upregulated while a miRNA with an expression ratio of ≤0.67 was considered downregulated.
mirConnX
The web interface mirConnX (http://www.benoslab.pitt.edu/mirconnx) was used to generate mRNA/miRNA interaction networks of the wild type cells and of the resistant subclones24. The normalized miRNA and mRNA microarray expression data each containing six matched columns representing different samples were used as input files for the mirConnX software. Next, we specified the parameters of choice. We selected human_h19_(GRCh37)_20111109 for organism type. Gene symbol and ID were selected for gene ID and miRNA ID, respectively. In the analysis options section, we selected Pearson's correlation as the association measure of choice. The regulation threshold for the minimum integrated regulation score was set at 0.95. The regulation score is calculated using the weighted sum of both the user-specified weight of the prior static network and the weight of the association measure, in this case Pearson's correlation, which determined correlations between miRNA and mRNA interactions in user-provided expression data24. We selected the option to calculate the presence of feed-forward loops. For the prior weight factor we selected 0.3. To further analyze the specific miRNA/mRNA interactions, we downloaded the data from the network as spreadsheets. We compared the miRNA/mRNA interaction lists of the TMZ-sensitive and the TMZ-resistant network manually to determine which mRNA depicted in the TMZ-resistant network is potentially targeted by the largest set of either upregulated or downregulated miRNAs and is absent in the TMZ-sensitive network.
In silico analysis of PHF6 mRNA and protein expression
For assessing PHF6 mRNA expression in glioblastoma and different regions of normal brain tissue, we used R2, the microarray analysis and visualization tool provided by the Department of Human Genetics of the Academic Medical Centre, Amsterdam, The Netherlands (http://r2.amc.nl). MAS5.0 normalized microarray datasets of glioblastoma35,36,37,38 were compared to different regions of normal brain34.
The Human Protein Atlas was used to check for protein expression levels of PHF6 in high-grade glioma tissue and different regions of normal brain tissue39.
qRT-PCR
Total RNA was isolated using TRIzol RNA isolation protocol (Life Technologies, CA, USA), according to the manufacturer's protocol. RNA concentrations were determined using the NanoDrop 1000 (Thermo Scientific, MA, USA) and, subsequently, equal amounts of RNA were converted into cDNA using Omniscript kit (Qiagen, Valencia, CA, USA). QuantiTect Primer Assay (Qiagen, Valencia, CA, USA) including forward and reverse primers for PHF6 were used to measure PHF6 mRNA levels. Forward and reverse primers (Biolegio, Nijmegen, The Netherlands) for GAPDH were used for GAPDH mRNA detection. Next, qRT-PCR was performed using the 7500 real-time PCR system (Applied Biosystems, Life Technologies, CA, USA). The data were normalized to GAPDH expression levels. The Ct values were used to calculate the relative fold difference in mRNA levels (ΔΔCt method).
Western blot
Expression levels of PHF6 were assessed by Western blot analysis. After cell lysis, 40 μg of protein was loaded on gel and was transferred to a polyvinylidene difluoride (PVDF) membrane after electrophoresis. Next, the membranes were incubated with the primary antibodies rabbit anti-PHF6 (1:1,000; Sigma-Aldrich, Missouri, USA) and mouse anti-β-actin (1:10,000; Santa Cruz Biotechnology, Texas, USA) over night (O/N) at 4°C. Subsequently, the membranes were incubated with green goat-anti-rabbit and red goat-anti-mouse IR dye-labeled secondary antibodies (1:10,000; IRDye LI-COR Biosciences, Nebraska, USA) for 2 hours at room temperature. Protein detection and visualization of PHF6 and β-actin proteins were performed with Odyssey Infrared Imaging System (LI-COR® Biosciences, Nebraska, USA). Analysis and quantification of protein expression were performed with Image Studio (LI-COR® Biosciences, Nebraska, USA) and ImageJ. PHF6 protein expression was normalized to β-actin protein expression levels.
Cell viability assay
Cells were plated in 96-well plates and transfected after 24 hrs with 66 nM of siPHF6 (Qiagen) or siNT (Thermo Scientific Dharmacon) using lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The next day, cells were treated with TMZ and incubated for another 72 hrs. Cells were then washed with PBS and fixed in 3.6% formaldehyde. Permeabilization of the cell membrane was performed with PBS containing 0.1% Trition X-100. Next, we used 4′,6-diamidino-2-phenylindole (DAPI; 0.3 μg/ml in PBS) to stain the nuclei of the cells. Cell number was assessed by counting the number of DAPI-stained cells using the Acumen Ex3 laser scanning cytometer (TTP LabTech, Royston, UK).
Statistical analysis
Differences in biological properties between treated and untreated cells were analysed using two-sided Student's t-test. Differences between groups were determined using one-way ANOVA. The p values <0.05 were considered statistically significant. Data were analyzed using GraphPad Prism version 5 software (GraphPad Software, La Jolla, CA, USA).
Author Contributions
T.W. and J.J. conceived this study. T.W., L.H. and R.S.R. contributed to the experimental design. L.H. and R.S.R. performed the experiments. T.W., L.H. and R.S.R. contributed to data analysis. The manuscript was written by L.H., R.S.R. and T.W., which was edited and reviewed by all the authors (L.H., R.S.R., T.W., J.J., E.H., D.P.N., G.J.L.K., W.P.V., P.W.).
Acknowledgments
This work was funded by Stichting STOPhersentumoren.nl (TW, JJ), KiKa (TW), and NWO (TW).
References
- Stupp R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 352, 987–96 (2005). [DOI] [PubMed] [Google Scholar]
- Stupp R. et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 10, 459–66 (2009). [DOI] [PubMed] [Google Scholar]
- Roos W. P. et al. Apoptosis in malignant glioma cells triggered by the temozolomide-induced DNA lesion O6-methylguanine. Oncogene. 26, 186–97 (2007). [DOI] [PubMed] [Google Scholar]
- Caporali S. et al. DNA Damage Induced by Temozolomide Signals to both ATM and ATR: Role of the Mismatch Repair System. Mol Pharmacol. 66, 478–491 (2004). [DOI] [PubMed] [Google Scholar]
- Hegi M. E. et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 352, 997–1003 (2005). [DOI] [PubMed] [Google Scholar]
- Esteller M., Hamilton S. R., Burger P. C., Baylin S. B. & Herman J. G. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 59, 793–797 (1999). [PubMed] [Google Scholar]
- Qian X. C. & Brent T. P. Methylation hot spots in the 5′ flanking region denote silencing of the O6-methylguanine-DNA methyltransferase gene. Cancer Res. 57, 3672–3677 (1997). [PubMed] [Google Scholar]
- Cahill D. P. et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 13, 2038–45 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C. et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 66, 3987–91 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip S. et al. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res. 15, 4622–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujifuku K. et al. miR-195, miR-455-3p and miR-10a(*) are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer Lett. 296, 241–8 (2010). [DOI] [PubMed] [Google Scholar]
- Li P. et al. MiR-181b suppresses proliferation of and reduces chemoresistance to temozolomide in U87 glioma stem cells. J Biomed Res. 24, 436–43 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L. et al. MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Res. 1352, 255–64 (2010). [DOI] [PubMed] [Google Scholar]
- Shi L. et al. MicroRNA-125b-2 confers human glioblastoma stem cells resistance to temozolomide through the mitochondrial pathway of apoptosis. Int J Oncol. 40, 119–29 (2012). [DOI] [PubMed] [Google Scholar]
- Yang Y.-P. et al. Inhibition of cancer stem cell-like properties and reduced chemoradioresistance of glioblastoma using microRNA145 with cationic polyurethane-short branch PEI. Biomaterials. 33, 1462–76 (2012). [DOI] [PubMed] [Google Scholar]
- Wong S. T. S. et al. MicroRNA-21 inhibition enhances in vitro chemosensitivity of temozolomide-resistant glioblastoma cells. Anticancer Res. 32, 2835–41 (2012). [PubMed] [Google Scholar]
- Asuthkar S., Velpula K. K., Chetty C., Gorantla B. & Rao J. S. Epigenetic regulation of miRNA-211 by MMP-9 governs glioma cell apoptosis, chemosensitivity and radiosensitivity. Oncotarget. 3, 1439–54 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Sai K., Chen F. & Chen Z. miR-181b modulates glioma cell sensitivity to temozolomide by targeting MEK1. Cancer Chemother Pharmacol. 72, 147–58 (2013). [DOI] [PubMed] [Google Scholar]
- Comincini S. et al. microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol Ther. 14, 574–86 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz J. L. et al. Delivery of Functional Anti-miR-9 by Mesenchymal Stem Cell-derived Exosomes to Glioblastoma Multiforme Cells Conferred Chemosensitivity. Mol Ther Nucleic Acids. 2, e126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. et al. The miR-183/96/182 cluster regulates oxidative apoptosis and sensitizes cells to chemotherapy in gliomas. Curr Cancer Drug Targets. 13, 221–31 (2013). [DOI] [PubMed] [Google Scholar]
- Chen L. et al. Downregulation of miR-221/222 sensitizes glioma cells to temozolomide by regulating apoptosis independently of p53 status. Oncol Rep. 27, 854–60 (2012). [DOI] [PubMed] [Google Scholar]
- He L. & Hannon G. J. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 5, 522–31 (2004). [DOI] [PubMed] [Google Scholar]
- Huang G. T., Athanassiou C. & Benos P. V. mirConnX: condition-specific mRNA-microRNA network integrator. Nucleic Acids Res. 39, W416–23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales G. et al. MAGIA, a web-based tool for miRNA and Genes Integrated Analysis. Nucleic Acids Res. 38, W352–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S. et al. MicroRNA and mRNA integrated analysis (MMIA): a web tool for examining biological functions of microRNA expression. Nucleic Acids Res. 37, W356–62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. C., Morris Q. D. & Frey B. J. Bayesian inference of MicroRNA targets from sequence and expression data. J Comput Biol. 14, 550–63 (2007). [DOI] [PubMed] [Google Scholar]
- Cascione L. et al. Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer. PLoS One. 8, e55910 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusko R. L. et al. Comprehensive genomic profiling of the lung transcriptome in emphysema and idiopathic pulmonary fibrosis using RNA-Seq. BMC Proc. 6, P21 (2012). [Google Scholar]
- Winter J., Jung S., Keller S., Gregory R. I. & Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 11, 228–34 (2009). [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A. & Slack F. J. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 6, 259–69 (2006). [DOI] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S. N. & Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 9, 102–14 (2008). [DOI] [PubMed] [Google Scholar]
- Hiddingh L. et al. EFEMP1 induces γ-secretase/Notch-mediated temozolomide resistance in glioblastoma. Oncotarget. 5, 363–74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold N. C. et al. Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci U S A. 105, 15605–10 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murat A. et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 26, 3015–24 (2008). [DOI] [PubMed] [Google Scholar]
- Sun L. et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 9, 287–300 (2006). [DOI] [PubMed] [Google Scholar]
- Gravendeel L. A. M. et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 69, 9065–72 (2009). [DOI] [PubMed] [Google Scholar]
- Freije W. A. et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 64, 6503–10 (2004). [DOI] [PubMed] [Google Scholar]
- Uhlen M. et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 28, 1248–50 (2010). [DOI] [PubMed] [Google Scholar]
- Hecker N. et al. A new algorithm for integrated analysis of miRNA-mRNA interactions based on individual classification reveals insights into bladder cancer. PLoS One. 8, e64543 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Joseph Z., Gitter A. & Simon I. Studying and modelling dynamic biological processes using time-series gene expression data. Nat Rev Genet. 13, 552–64 (2012). [DOI] [PubMed] [Google Scholar]
- Schulz M. H. et al. Reconstructing dynamic microRNA-regulated interaction networks. Proc Natl Acad Sci U S A. 110, 15686–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. et al. PHF6 regulates cell cycle progression by suppressing ribosomal RNA synthesis. J Biol Chem. 288, 3174–83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantzen H. M., Admon A., Bell S. P. & Tjian R. Nucleolar transcription factor hUBF contains a DNA-binding motif with homology to HMG proteins. Nature. 344, 830–6 (1990). [DOI] [PubMed] [Google Scholar]
- Todd M. A. M. & Picketts D. J. PHF6 interacts with the nucleosome remodeling and deacetylation (NuRD) complex. J Proteome Res. 11, 4326–37 (2012). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13, 1924–35 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis K. J. et al. A cooperative microRNA-tumor suppressor gene network in acute T-cell lymphoblastic leukemia (T-ALL). Nat Genet. 43, 673–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P. et al. PHF6 mutations in T-cell acute lymphoblastic leukemia. Nat Genet. 42, 338–42 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P. et al. PHF6 mutations in adult acute myeloid leukemia. Leukemia. 25, 130–4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. & Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 16, 369–77 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursać S. et al. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc Natl Acad Sci U S A. 109, 20467–72 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A. J. p53, the cellular gatekeeper for growth and division. Cell. 88, 323–31 (1997). [DOI] [PubMed] [Google Scholar]
- Nam S., Long X., Kwon C., Kim S. & Nephew K. P. An integrative analysis of cellular contexts, miRNAs and mRNAs reveals network clusters associated with antiestrogen-resistant breast cancer cells. BMC Genomics. 13, 732 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]