Abstract
We investigated systematically the effects of Tet-induced oxidation products of 5-methylcytosine (5mC) on Dnmt1- and DNMT3a-mediated cytosine methylation in synthetic duplex DNA. We found that the replacement of 5mC at CpG site with a 5-hydroxymethylcytosine, 5-formylcytosine, 5-carboxylcytosine or 5-hydroxymethyluracil resulted in altered methylation of cytosine at both the opposite and the neighboring CpG sites. Our results provided important new knowledge about the implications of the 5-methylcytosine oxidation products in maintenance cytosine methylation.
Methylation of cytosine residues at CpG dinucleotide site in DNA is a key epigenetic mark that plays crucial roles in multiple cellular processes.1 Dynamic DNA methylation is crucial for mammalian development, and aberrant cytosine methylation pattern is associated with many human diseases.2 DNA (cytosine- 5)-methyltransferases (DNMTs) catalyse the methylation process. In mammals, DNMT3a and DNMT3b establish the initial methylation patterns during embryonic development.3 DNMT1, on the other hand, methylates the unmethylated cytosine at a hemimethylated CpG site following DNA replication, thereby maintaining the DNA methylation patterns during cell division.4 This division of labour between maintenance and de novo DNA methylation is not absolute; emerging evidence showed that DNMT3a and DNMT3b may also be involved in maintenance DNA methylation by correcting errors that are left behind by DNMT1 during DNA replication.5
Although enzymes catalysing DNA methylation have been well characterized, the DNA demethylation is much more complicated.6,7 Recent discovery of the oxidation of 5-methyl-2’-deoxycytidine (5mC) by ten-eleven translocation (Tet) proteins provides some novel mechanistic insights about cytosine demethylation in mammals.8,9 TET proteins sequentially oxidize 5mC to 5-hydroxymethyl-2’-deoxycytidine (5hmC), 5-formyl-2’- deoxycytidine (5fC) and 5-carboxyl-2’-deoxycytidine (5caC) (Fig. 1). It was also postulated that 5hmC may be deaminated to yield 5-hydroxymethyl-2’-deoxyuridine (5hmU) by the action of some deaminases.10 5hmU is present in mammalian tissues at a frequency of 1 per 105 nucleosides, though some of 5hmU may also form from oxidatively induced damage of thymidine in DNA.11 TET-mediated demethylation may occur via active and passive mechanisms. The active demethylation proceeds through the combined action of TET proteins with thymine DNA glycosylase (TDG) and the subsequent action by the base excision repair (BER) machinery.12,13 Nevertheless, active demethylation is not the major mechanism for genome-wide demethylation.14 Passive demethylation is based on replication-dependent dilution of 5mC, and it occurs only when DNMT1 fails to methylate the newly synthesized DNA sequence. In vitro, the replacement of a 5mC at CpG site with 5hmC can reduce the efficiency of DNMT1-mediated methylation of the cytosine residue at the opposite CpG site by approximately 60-fold,15 suggesting that 5hmC can induce the loss of cytosine methylation in vivo through a process that is dependent on DNA replication.16 However, it remains unexplored how other TET products (i.e., 5fC, 5caC and 5hmU) affect passive DNA demethylation. Herein, we set out to assess how substitution of 5mC at a hemimethylated CpG site with a 5fC, 5caC or 5hmU alters the efficiencies of mouse Dnmt1 and human DNMT3a in methylating the cytosine residue in the opposing and adjacent 5’ CpG sites.
Fig. 1.
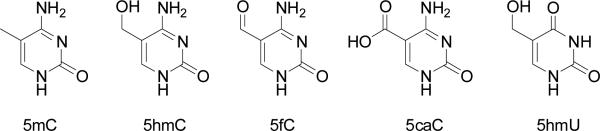
The chemical structures of 5-methylcytosine and its oxidised derivatives.
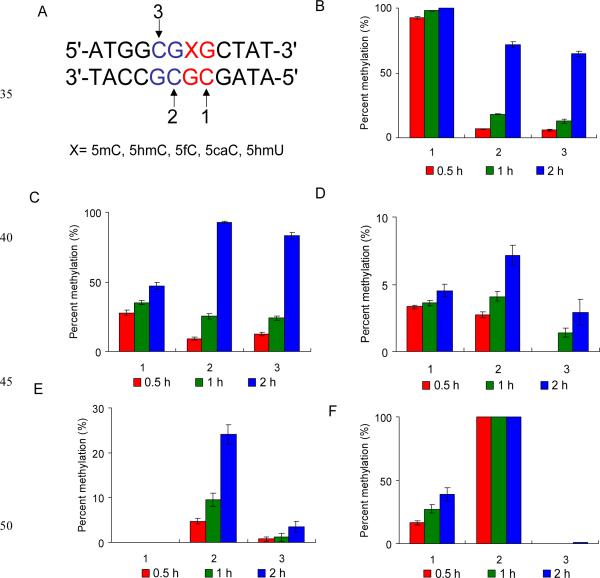
The X-ray crystal structure of Dnmt1-DNA complex revealed that at least 3 base pairs of DNA are directly involved in Dnmt1 binding.17 In addition, the CGCG tetranucleotide sequences are clustered in CpG islands at a frequency of 5 or more per CpG island in the human genome;18 thus, it is necessary to examine whether the modified cytosine derivatives affect methylation of cytosine residues at the adjacent CpG site. To assess how the presence of modified cytosine derivatives perturbs methylation, we designed a 12-mer duplex oligodeoxyribonucleotide (ODN) comprised of two adjacent CpG sites in the middle of the DNA sequence (Fig. 2A). At the 3’ side CpG site, the cytosine in the top strand is substituted with a 5mC, 5hmC, 5fC, 5caC or 5hmU (these ODNs were characterized by ESI-MS and MS/MS, see Fig. S1–S5). In the viewpoint that the native substrate for DNMT1 is a hemimethylated CpG site, the 5mC-containing duplex serves as the control. In this duplex, there are two methylation sites in the bottom strand and one in the top strand (Fig. 2A). In this context, we employed a relatively short duplex DNA substrate so that the methylation products can be analysed directly by LC-MS. Our results showed that the 12-mer duplex substrates are long enough for the binding and methylation by Dnmt1 or DNMT3a (vide infra). We used online LC-MS/MS experiments to identify the sites and quantify the levels of cytosine methylation, where MS/MS also facilitated us to differentiate the methylation at sites 1 or 2 (Fig. S6–S12). For instance, the MS/MS for the [M – 3H]3- ion of the bottom strand with methylation at site 1 or 2 revealed a series of [an – Base] and wn ions (Fig. S12), which arise from the neutral loss of a nucleobase followed by chain cleavage at the 3’ C-O bond of the same nucleoside.19 As displayed in Fig. S12B, the mass difference between the w5 (m/z 1541.2) and w6 (m/z 1844.2) ions corresponds to the residue mass of a 5mC-5’-phosphate (303 Da), whereas the mass difference between w 2-7 (m/z 1086.3) and w 2-8 (m/z 1230.8) ions is in keeping with the residue mass of an unmethylated 2’-deoxycytidine-5’-phosphate (289 Da). Therefore, this tandem mass spectrum supports the methylation of site 2, but not site 1. On the other hand, the MS/MS shown in Fig. S12A supports the methylation of site 1, but not site 2. We observed previously that the efficiency for the formation of abundant fragment ions from the methylated substrate is almost the same as that from the corresponding unmethylated sequence.20 In addition, the area ratios of peaks found in the selected-ion chromatograms for monitoring the formation of specific fragment ions of ODNs follow an excellent linear relationship with the molar ratios of the ODNs in the mixture, and a molar ratio of ODNs that is as low as 0.2% could be quantified.21 Therefore, we quantified the methylation level by assuming that the ionisation and fragmentation efficiencies for the methylated ODNs are the same as their unmethylated counterparts.
Fig. 2.
Levels of cytosine methylation in different substrates methylated by Dnmt1 at three different CpG sites. (A) The sequence of the duplex DNA employed for the in vitro methylation assay. 1, 2 and 3 indicate the potential methylation sites. The substrates are duplex ODNs containing a 5mC (B), 5hmC (C), 5fC (D), 5caC (E) or 5hmU (F). The data represent the
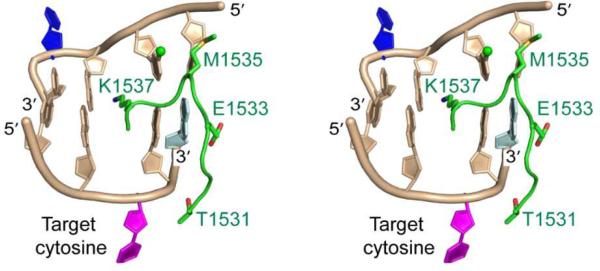
We first examined how the presence of 5mC oxidation products alters the Dnmt1-mediated methylation reaction. Our quantification data, as summarized in Fig. 2, revealed that the incorporation of 5hmC, 5fC, 5caC and 5hmU into the CpG site affected the methylation of not only the opposite CpG site but also the adjacent 5’ CpG site. We found that the substitution of 5mC at CpG site with a 5hmC led to a significant reduction in the methylation level of the cytosine residue in the opposite CpG site (site 1 in Fig. 2A), which is consistent with previous findings (Fig. 2B&C).15 The replacement of 5mC with 5fC led to an even greater decline in methylation efficiency at site 1, where less than 5% methylation was detected at this site after 2 h. Likewise, we observed marked decreases in the methylation rates at sites 2 and 3, where the methylation levels after 2 h were approximately 7% and 2%, respectively (Fig. 2D). For the 5caC-carrying substrate, we observed that, at 2 h after reaction, sites 2 and 3 were methylated at 25% and 5%, respectively, whereas no methylation was detectable at site 1 (Fig. 2E). Usually the methylation rates at sites 2 and 3 should be similar because the CpG site becomes hemi-methylated after one of these cytosines is methylated, which subsequently facilitates the efficient methylation of the cytosine at the opposite CpG site. This is indeed what we found for the 5mC- and 5hmC-containing substrates (Fig. 2B&C). The preferential methylation of site 2 over site 3 for the 5fC- and 5caC-harboring substrates suggests that site 3 is highly blocked even after site 2 is methylated. Notably, the structure of the covalent Dnmt1-DNA complex reveals that the guanine two bases downstream of the target cytosine is in close proximity with a target recognition domain (TRD) loop of Dnmt1 that interacts extensively with the DNA major groove (Fig. 3).17,22 Replacement of this downstream nucleoside with a 5caC or 5fC may alter the orientation of the TRD loop, thereby affecting its interaction with DNA. Taken together, these results demonstrated that 5fC and 5caC, aside from serving as intermediates of active cytosine demethylation,8,9 block maintenance methylation to a much greater extent than 5hmC. This would lead to the passive demethylation of 5mC. Moreover, our results suggest that 5fC and 5caC may spread the loss of methylation signal through blocking methylation of the neighbouring 5’ CpG sites.
Fig. 3.
Stereo view of intermolecular contacts between one TRD loop of Dnmt1 and the DNA major groove in the productive covalent Dnmt1-DNA complex. The 5-methyl group from the 5mC in the parental strand is coloured in green. The flipped-out target cytosine is in purple, the flipped-out cytosine in the parental strand is in dark blue, and the guanine two bases downstream of the target cytosine is in light blue. Amino acid residues that are in contact with DNA are labelled.
For 5hmU-containing duplex DNA, to our surprise, site 2 becomes fully methylated within 30 min, at a rate that is even greater than methylation at site 1 for the 5mC-bearing substrate (Fig. 2F). The structure of the covalent Dnmt1-DNA complex revealed that the Dnmt1-catalyzed reaction requires not only the target cytosine for methylation, but also the adjacent nucleobase on the 3’ side of the cytosine in the opposite strand, to be flipped out of duplex DNA (Fig. 3).17 Thus, Dnmt1-mediated methylation of site 2 for the 5hmU-bearing substrate also requires the flipping of 5hmU out of duplex DNA. The 5hmU:G mispair is much less stable than the 5mC:G base pair, rendering it much easier for 5hmU to be flipped out of the duplex, which may contribute to the elevated methylation efficiency at site 2. To further substantiate this finding, we used two other DNA substrates, where 5hmU is substituted with a T or A, and conducted the same methylation reactions. The results showed that site 2 in these DNA substrates could also be efficiently methylated (Fig. S13). Therefore, nucleotide mismatch in DNA strand perturbs Dnmt1-mediated DNA methylation. For the 5hmU-bearing substrate, even though site 2 was completely methylated, methylation at site 3 was barely detectable, showing that methylation of site 3 was highly blocked. This can again be attributed to the compromised interaction between the modified nucleoside (5hmU) and the TRD loop of DNMT1, as discussed above.
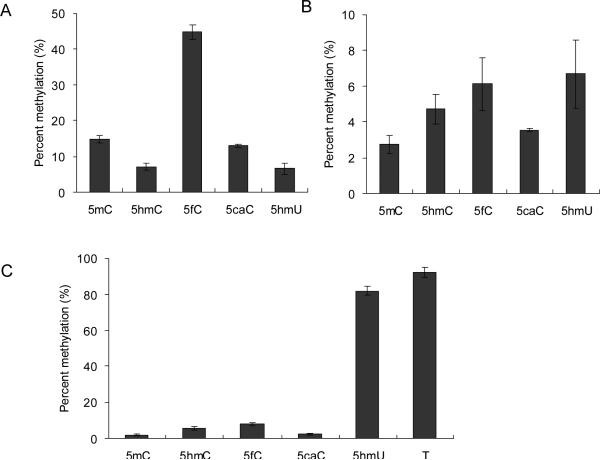
5hmC was proposed to be involved in passive demethylation in vivo.16 However, transfected hydroxymethylated plasmids in human cells retain maintenance methylation through cycles of plasmid replication as efficiently as the methylated plasmid, suggesting that 5hmC does not completely block maintenance methylation in cells.23 This may be attributed to the fact that DNMT3a possesses similar activity on hemi-5hmC and hemi-5mC CpG sites involved in maintenance methylation.15 We employed human DNMT3a to assess its activity on the above-described substrates. Our results showed that the 5caC-bearing substrate is indistinguishable from the corresponding 5mC-containing substrate in the methylation efficiencies at all three sites (Fig. 4). The substitution of 5mC with 5fC, however, results in elevated methylation efficiencies at all three sites. 5fC and 5caC are considered as new epigenetic modifications and, in conjunction with potential protein partner(s), they may assume distinct regulatory function at some specific sites.21 Cells may keep the sites methylated by DNMT3a same as the maintenance of fully hydroxymethylated CpG sites during cell division.15 The replacement of 5mC with 5hmC or 5hmU, on the other hand, led to elevated methylation at sites 2 and 3, but diminished methylation at site 1 (Fig. 3). It is worth noting that DNMT3a displays a markedly higher activity in methylating site 3 when the 5mC at the hemimethylated CpG site is substituted with a 5hmU or thymine (Fig. 3C). Thus, the formation of 5hmU:G mispair on the 3’ side of CpG site may introduce inadvertent CpG methylation, which may justify for the needs of DNA glycosylases (i.e. TDG and MBD4) of the efficient removal of 5hmU mispaired with guanine.22 These observations indicated that DNMT3a may interact with DNA differently from Dnmt1, and future structural studies of DNMT3a-DNA complex may provide a better mechanistic understanding of this difference.
Fig. 4.
Levels of cytosine methylation by DNMT3a at sites 1 (A), 2 (B), and 3 (C) in duplex ODNs after a 6 h reaction (The methylation sites are indicated in Fig. 1A). The data represent the mean and standard deviation of results from three independent methylation reactions.
Conclusions
Passive DNA demethylation may constitute the major mechanism of demethylation, but it occurs only when DNMTs fail to methylate the newly synthesized DNA sequence. Our results showed that, aside from their possible roles as intermediates for active cytosine demethylation, 5fC and 5caC can also block the activity of Dnmt1, suggesting the potential involvement of these two modified nucleosides in passive DNA demethylation. We also found that 5fC and 5caC could block the methylation at the adjacent 5’ CpG site. On the other hand, DNMT3a activity varies substantially with the nature of the 5mC oxidation products. Moreover, our results showed that a 5hmU:G mispair can increase the activity of Dnmt1 toward methylating neighboring 5’ CpG site in the same strand, and that of DNMT3a toward methylating the adjacent 5’ CpG site in the opposite DNA strand. This result suggests different substrate recognition mechanisms of Dnmt1 and DNMT3a.
Supplementary Material
Acknowledgments
The authors would like to thank the National Institute of Health for supporting this research (R01 CA101864 to Y.W.).
Footnotes
† Electronic Supplementary Information (ESI) available: General methods and MS data. See DOI: 10.1039/b000000x/
References
- 1.Jaenisch R, Bird A. Nat. Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 2.Robertson KD. Nat. Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 3.Okano M, Bell DW, Haber DA, Li E. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 4.Bestor TH. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 5.Jones PA, Liang G. Nat. Rev. Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu SC, Zhang Y. Nat. Rev. Mol. Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng G, Fu Y, He C. Chem. Rev. 2014 doi: 10.1021/cr400432d. DOI: 10.1021/cr400432d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He YF, Li B-Z, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, Sun Y, Li X, Dai Q, Song CX, Zhang K, He C, Xu GL. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo JU, Su Y, Zhong C, Ming G. l., Song H. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu S, Wang J, Su Y, Guerrero C, Zeng Y, Mitra D, Brooks PJ, Fisher DE, Song H, Wang Y. Nucleic Acids Res. 2013;41:6421–6429. doi: 10.1093/nar/gkt360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, Abramowitz LK, Bartolomei MS, Rambow F, Bassi MR, Bruno T, Fanciulli M, Renner C, Klein-Szanto AJ, Matsumoto Y, Kobi D, Davidson I, Alberti C, Larue L, Bellacosa A. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, Lu X, Lu J, Liang H, Dai Q, Xu GL, Luo C, Jiang H, He C. Nat. Chem. Biol. 2012;8:328–330. doi: 10.1038/nchembio.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue A, Shen L, Dai Q, He C, Zhang Y. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Nucleic Acids Res. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen L, Zhang Y. Curr. Opin. Cell Biol. 2013;25:289–296. doi: 10.1016/j.ceb.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song J, Teplova M, Ishibe-Murakami S, Patel DJ. Science. 2012;335:709–712. doi: 10.1126/science.1214453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avril N, Deschavanne P, Bellis M, Filipski J. Biochem. Biophys. Res. Commun. 1995;213:147–153. doi: 10.1006/bbrc.1995.2109. [DOI] [PubMed] [Google Scholar]
- 19.McLuckey SA, Vanberkel GJ, Glish GL. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Wang Y. Biochemistry. 2009;48:2290–2299. doi: 10.1021/bi801467z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong H, Cao H, Wang Y. Nucleic Acids Res. 2007;35:7118–7127. doi: 10.1093/nar/gkm851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song J, Rechkoblit O, Bestor TH, Patel DJ. Science. 2011;331:1036–1040. doi: 10.1126/science.1195380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubosaki A, Tomaru Y, Furuhata E, Suzuki T, Shin JW, Simon C, Ando Y, Hasegawa R, Hayashizaki Y, Suzuki H. Biochem. Biophys. Res. Commun. 2012;426:141–147. doi: 10.1016/j.bbrc.2012.08.053. [DOI] [PubMed] [Google Scholar]
- 24.Song CX, He C. Trends Biochem. Sci. 2013;38:480–484. doi: 10.1016/j.tibs.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morera S, Grin I, Vigouroux A, Couve S, Henriot V, Saparbaev M, Ishchenko AA. Nucleic Acids Res. 2012;40:9917–9926. doi: 10.1093/nar/gks714. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.