Based on transplantation and lineage tracing studies, a hierarchy of stem and progenitor cells has been shown to exist among the mammary epithelium. In this review, Visvader and Stingl integrate recent data on the mammary stem cell differentiation hierarchy and its control at the transcriptional and epigenetic levels. They also discuss the relevance of the evolving hierarchy to the identification of “cells of origin” of breast cancer.
Keywords: mammary stem cell, development, breast cancer, steroid hormone, epigenome, lineage tracing
Abstract
The mammary epithelium is highly responsive to local and systemic signals, which orchestrate morphogenesis of the ductal tree during puberty and pregnancy. Based on transplantation and lineage tracing studies, a hierarchy of stem and progenitor cells has been shown to exist among the mammary epithelium. Lineage tracing has highlighted the existence of bipotent mammary stem cells (MaSCs) in situ as well as long-lived unipotent cells that drive morphogenesis and homeostasis of the ductal tree. Moreover, there is accumulating evidence for a heterogeneous MaSC compartment comprising fetal MaSCs, slow-cycling cells, and both long-term and short-term repopulating cells. In parallel, diverse luminal progenitor subtypes have been identified in mouse and human mammary tissue. Elucidation of the normal cellular hierarchy is an important step toward understanding the “cells of origin” and molecular perturbations that drive breast cancer.
The mammary gland is a remarkably adaptive organ whose development closely reflects the physiological stage (Hennighausen and Robinson 2005). The epithelium of the mammary gland is composed of two main cellular lineages: luminal cells that surround a central lumen and highly elongated myoepithelial cells that are located in a basal position adjacent to the basement membrane. Collectively, these cells are organized into a series of branching ducts that terminate in secretory alveoli during lactation. The mammary epithelium exists in a highly dynamic state, undergoing profound changes during the morphogenetic cycle (Fig. 1). Dissection of the normal epithelial differentiation hierarchy is fundamental to understanding breast cancer heterogeneity. Breast cancer is not a single disease but comprises multiple different pathological and molecular subtypes. Although the stratification of breast cancer has dramatically impacted on treatments and outcomes, patient response to targeted therapy or chemotherapy remains highly unpredictable. Intertumor heterogeneity is widely believed to reflect “cells of origin” of cancer as well their genetic mutational profiles (for review, see Visvader 2011). The longevity and extensive self-renewal properties of stem cells suggest that they are strong candidates for cells of origin of cancer, but there is also considerable evidence implicating progenitors or transit-amplifying cells as targets of transformation. In this review, we have attempted to integrate recent data on the mammary stem cell (MaSC) differentiation hierarchy and its control at the transcriptional and epigenetic levels, including the influence of different hormonal environments. Finally, we discuss the relevance of the evolving hierarchy to the identification of “cells of origin” in breast cancer.
Figure 1.
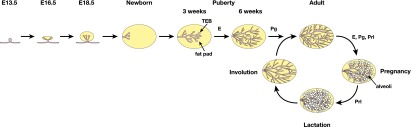
Schematic diagram of the primary stages of mammary gland ontogeny in the embryo and adult. In the mouse embryo, the placodes (visible at embryonic day 11.5 [E11.5]) evolve into mammary buds that penetrate the underlying mesenchyme around E13.5. These buds sprout by E15.5 and develop a lumen. By E18.5, a small arborized gland that has invaded the developing fat pad is evident. In the postnatal animal, development of the mammary gland remains relatively dormant until puberty at 3 wk, when profound morphogenesis occurs, largely under the control of estrogen (E). In the young adult gland, progesterone (Pg) regulates side branching, while in pregnancy, the steroid hormones estrogen, progesterone, and prolactin (Prl) all play roles in alveolar expansion. In the late stages of pregnancy and during lactation, the peptide hormone prolactin plays a key role in establishing the secretory state. After lactation, the gland involutes and returns to a state that resembles the virgin gland.
Prospective isolation of mouse MaSCs
In 1959, DeOme et al. (1959) published a seminal paper that described the use of de-epithelialized (cleared) mammary fat pads of mice as a site for transplantation of normal, preneoplastic, and malignant mammary tissue. When portions of the normal mammary epithelium of donor mice were transplanted into the cleared fat pads of recipient mice, the segment could regenerate the entire ductal epithelial tree. Subsequent studies demonstrated that successful engraftment could be obtained with any segment of the mammary epithelial tree (Hoshino 1962; Daniel et al. 1968; Smith and Medina 1988), thereby indicating that the repopulating cells, inferred to be MaSCs, are widely distributed. A number of morphologically distinct cells were hypothesized to represent MaSCs. Most notable among these are the cap cells that line the terminal end buds (TEBs) located at the tips of ducts during pubertal development (Williams and Daniel 1983) and the small light cells (SLCs) visualized by electron microscopy (Chepko and Smith 1997).
The ability of a single cell to regenerate a ductal lobular outgrowth complete with both luminal and myoepithelial cells upon serial transplantation in mice has been traditionally considered the gold standard assay for the detection of MaSCs. A clonal analysis of retrovirally marked epithelial cells first suggested that a single cell was capable of reconstituting the entire mammary epithelium (Kordon and Smith 1998). Subsequent studies, which used flow cytometry to identify mammary epithelial cell subpopulations, provided functional evidence that a single cell could reconstitute the mammary epithelium when transplanted into a cleared mammary fat pad (Shackleton et al. 2006; Stingl et al. 2006) and that the vast majority (∼99%) of these cells have a CD49fhiCD29hiCD24+/modSca1low phenotype (Shackleton et al. 2006; Stingl et al. 2006; Sleeman et al. 2007; Shehata et al. 2012). Notably, this immunophenotype characterizes the wider basal population, which comprises MaSCs (frequency of about one in 50 basal cells), mature myoepithelial cells, and presumptive basal progenitor intermediates. Microscopic visualization of single cells prior to implantation, a process that ensures selection of viable cells, indicated that approximately one in 18 cells within the basal population have the potential to generate an entire mammary gland (Shackleton et al. 2006). MaSCs can be preferentially enriched over other basal cells based on their higher expression levels of CD24 and EpCAM (Shehata et al. 2012; Smalley et al. 2012). Moreover, size can also provide a degree of enrichment whereby larger cells from the basal fraction (>10 µm) were capable of mammary reconstitution, whereas cells <10 µm lacked this capacity (Machado et al. 2013). Based on these data, it could be deduced that the SLC (∼8 µm) does not function as a stem cell.
Although it has proven difficult to segregate stem cells from myoepithelial and progenitors in the basal population, some progress has been made in identifying “stem cell” genes through the analysis of knock-in and transgenic reporter mice. Expression of the phosphatase gene s-Ship is exclusively restricted to the outer cap cell layer of the TEBs in puberty (Bai and Rohrschneider 2010). While Ship-GFP+ basal cells were enriched for MaSCs relative to GFP− cells, this study also revealed that not all MaSCs in pubertal glands are cap cells. Lgr5 reporter mice have stimulated considerable interest, as Lgr5 is a key Wnt/β-catenin target gene in intestinal stem cells and also marks stem cells in other organs (Barker et al. 2013). However, the analysis of Lgr5-EGFP-IRES-creERT2 reporter activity in the mammary gland has yielded disparate data. While one group (Plaks et al. 2013) showed that a rare subset of Lgr5+ cells in the basal fraction had dramatically enriched repopulating activity upon transplantation compared with Lgr5− cells, two other studies (de Visser et al. 2012; Rios et al. 2014) using the same reporter strain demonstrated that both Lgr5+ and Lgr5− cells harbor repopulating potential. These data imply that the MaSC compartment is heterogeneous, comprising at least Lgr5+ and Lgr5− subsets. Interestingly, self-renewing Lgr5+ stem cells and quiescent Lgr5− stem cells have been described in the intestine and stomach, where they lie in distinct niches (Takeda et al. 2011). It is noteworthy that Lgr5 may not be a target of the Wnt/β-catenin pathway in the mammary gland, as Lgr5+ cells were not enriched for expression of the Wnt pathway gene Axin2 (Plaks et al. 2013). The small population of Axin2+ cells restricted to the basal population exhibited only twofold higher repopulating activity than Axin2− cells, indicating that MaSCs are not restricted to the Wnt-responsive subset despite the clonal expansion of MaSCs elicited by Wnt3A (Zeng and Nusse 2010).
Prospective isolation of human MaSCs
The most widely used approaches to date for detecting putative human mammary stem and progenitor cells have relied on in vitro and in vivo assays to interrogate the growth and differentiation of phenotypically distinct subsets of mammary epithelial cells. However, these approaches have led to conflicting data. Several studies indicate that cells with repopulating capacity in vivo and bipotent differentiation capacity in vitro and characterized by an EpCAMloCD49fhi phenotype are restricted to the basal cell compartment (Stingl et al. 1998, 2001; Eirew et al. 2008; Lim et al. 2009). This contrasts with another report (Keller et al. 2012) suggesting that both the luminal and basal cell populations contain bipotent progenitors and repopulating cells (Keller et al. 2012). Adding to the confusion, undifferentiated ductal luminal/suprabasal cells expressing bilineage markers have been postulated to be the most potent mammary epithelial cell population (Ginestier et al. 2007; Villadsen et al. 2007; Pece et al. 2010). These discrepancies are likely explained by the different strategies used for dissociation of breast tissue by various groups as well the assays adopted to assess “stemness.” For example, aldehyde dehydrogenase 1 (ALDH1) was reported to identify human breast stem cells, since only ALDH1+ cells could generate mammary structures in humanized mouse mammary fat pads (Ginestier et al. 2007). However, another study found that outgrowths under the renal capsule were derived only from the ALDHlo (basal) epithelial subset (Eirew et al. 2012).
Evidence for slow-cycling and quiescent stem cells
The cycling status of MaSCs in the adult mammary gland has been difficult to study owing to the low frequency of these cells in the epithelium and a paucity of suitable markers for their purification. One perceived property of adult stem cells is that they are slowly dividing and thereby have the ability to retain synthetic DNA nucleosides. Compatible with this notion, the MaSC/basal population was found to be enriched for long-lived label-retaining cells (Shackleton et al. 2006). Another perceived feature of adult tissue stem cells is that they retain their template DNA strands during mitosis. In the mouse mammary gland, sequential administration of 3H-thymidine and BrdU identified cells that retain their template DNA strand (Smith 2005). Interestingly, 30%–40% of label-retaining cells also expressed the estrogen receptor (ER) and progesterone receptor (PR) (Booth and Smith 2006), which is somewhat counterintuitive given that ER expression is usually associated with epithelial cell differentiation.
To exploit the putative quiescent state of MaSCs, cells were labeled with the lipophilic fluorescent dye PKH26, and label-retaining stem-like cells were selected through mammosphere culture (Cicalese et al. 2009; Pece et al. 2010). This resulted in the enrichment of human mammary repopulating cells by several log orders of magnitude. Subsequent gene expression profiling of purified PKH26+ cells revealed a CD49f+DLL1hiDNERhi phenotype, and cells purified on the basis of these markers exhibited a >500-fold higher frequency of mammosphere-initiating cells. A similar strategy was used for the mouse mammary gland, with enrichment resulting in one MaSC in every three PKH26hi cells (Cicalese et al. 2009). Moreover, analysis of partitioning of the cell fate determinant Numb showed that PKH26hi cells in the MaSC/basal population predominantly divide through asymmetric division (Cicalese et al. 2009). In addition, β1 integrin and a Notch-dependent Aurora A pathway have been implicated in regulating the cell division axis in the mammary gland (Taddei et al. 2008; Regan et al. 2013). The recent evaluation of mice harboring an inducible histone 2b (H2B) promoter linked to a GFP reporter (K5tTA-H2b-GFP) (Dos Santos et al. 2013) revealed a small subset (0.2%) of slowly cycling H2b-GFPhi cells within the MaSC/basal population. CD1d, a MHC-like molecule expressed by antigen-presenting cells, was identified as a marker of these cells and provided an approximately fivefold enrichment of repopulating units, although the proportion of all repopulating units that are CD1d+ remains unclear. The lower concentration of reactive oxygen species (ROS) in the MaSC/basal subset further suggests the presence of quiescent MaSCs (Diehn et al. 2009).
In contrast to these subsets of slow-cycling MaSCs, it is presumed that activated stem cells drive morphogenesis during puberty and alveologenesis. Indeed, the cell cycle status of s-SHIP GFP+ cells suggested that s-SHIP marks actively dividing MaSCs in puberty and early pregnancy (Bai and Rohrschneider 2010). Moreover, recent three-dimensional (3D) imaging of intact mammary ducts in pubertal glands revealed that EdU+ cells colocalize with K5-expressing cells in the cap cell layer of the TEBs (Rios et al. 2014). These proliferative basal cells appeared to undergo asymmetric division and generate luminal cells that moved toward the inner body layer of the TEB. Within the adult gland, basal cells undergoing division also could be readily visualized throughout the ducts by 3D imaging, and these cells had the same elongated morphology as mature myoepithelial cells (Rios et al. 2014). It is presumed that these cells correspond to either MaSCs or basal-restricted progenitors.
Heterogeneity within the MaSC population
Stem cells in multiple systems exhibit a high degree of heterogeneity, in part due to the developmental stage. The same may hold true for the MaSC compartment (Fig. 2), as highlighted by the recent isolation of fetal MaSCs (Spike et al. 2012; Makarem et al. 2013). These cells display a phenotype intermediate between luminal and basal cells, since they express high levels of CD24, EpCAM, and CD49f, and comprise a relatively high frequency of cells that coexpress luminal and basal keratins (Spike et al. 2012). Moreover, these repopulating cells are inferred to be ER−, since this receptor is not expressed in the embryonic mammary bud (Wansbury et al. 2011). Comparison with the adult MaSC/basal population revealed that fetal cells were much more potent than their adult counterparts, with 50-fold to 100-fold higher clonogenic activity in vitro and a fourfold higher repopulating capacity in vivo (Makarem et al. 2013). Likewise, fetal neural and hematopoietic stem cells demonstrate superior self-renewal activity, determined in part by intrinsic molecular pathways (Molofsky et al. 2003; Bowie et al. 2007). By analogy with these systems, the adult MaSC is likely to derive from the primordial MaSC resident in the embryo. Not surprisingly, fetal MaSCs have a gene expression signature distinct from their adult counterparts. They express high levels of Elf5 and Aldh1a3, genes whose expression is restricted to the luminal lineage in the adult mammary gland (Makarem et al. 2013). Concordantly, the gene expression signatures of fetal MRUs most closely resemble that of basal-like breast cancers (Spike et al. 2012), a tumor type believed to originate from ER− luminal progenitor cells. It will be of interest to determine whether there is any relationship between fetal MaSCs and the recently identified subset of pluripotent stem cells defined by CD73+CD90− in adult human breasts (Roy et al. 2013). The presence of pluripotent cells in the adult gland (Hassiotou et al. 2012; Roy et al. 2013) or the possible persistence of fetal MaSCs into adulthood has profound implications for breast oncogenesis. It is relevant that an embryonic stem cell-like expression profile may be associated with poorly differentiated, aggressive breast tumors (Ben-Porath et al. 2008).
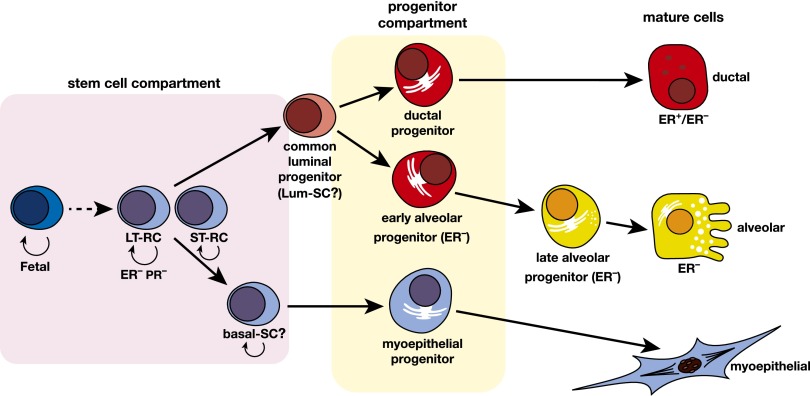
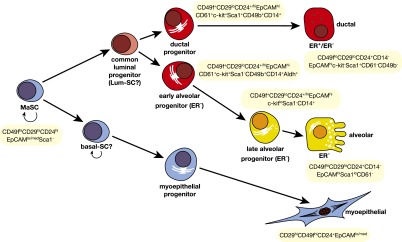
Figure 2.
Hypothetical model of the mammary epithelial hierarchy. A multipotent fetal MaSC has been identified. In the adult mammary gland, the stem cell compartment is heterogeneous and appears to comprise long-term and short-term repopulating cells (LT-RCs and ST-RCs, respectively), both of which are multipotent. These in turn give rise to committed progenitor cells for the myoepithelial and luminal (ductal and alveolar) epithelial lineages, but the precise number of progenitor cells is yet to be determined. Luminal progenitors are restricted to either a ductal or alveolar cell fate: The ductal progenitor possibly comprises both hormone receptor (HR)-positive and HR-negative cells, while the early and late alveolar-restricted progenitors are likely to be HR-negative. There may be a common luminal progenitor for these sublineages. The prospective isolation of cellular subsets from mouse and human mammary tissue provide support for the depicted hierarchical organization. In addition, two types of unipotent cells (lum-SC and myo-SC) may exist; current lineage tracing data are also consistent with long-lived progenitors performing these functions in vivo.
Distinct types of stem cells also exist in the adult mammary gland. During pregnancy, a variant population of MaSCs undergoes a dramatic but transient expansion to coordinate alveologenesis (Asselin-Labat et al. 2010). These cells have significantly lower self-renewal capacity on serial transplantation compared with those from virgin glands, reminiscent of the short-term repopulating cell in the hematopoietic system. Furthermore, the molecular expression profiles of the two MaSC/basal pools differed profoundly, with the induction of alveolar-restricted genes during pregnancy. The presence of a “lineage priming” signature suggests that these cells are poised for alveolar differentiation (Pal et al. 2013). Another subset with short-term repopulating potential was identified within the luminal H2Bhi subset of young mice and shown to significantly expand during pregnancy (Kaanta et al. 2013). It is not clear whether these cells are similar to those in the basal population, but the close proximity of the two populations (Kaanta et al. 2013) raises the possibility that these cells lie at the interface between the basal and luminal subsets. It is tempting to speculate that these short-term repopulating cells contribute to the transient increase in breast cancer associated with pregnancy, perhaps by facilitating expansion of a mutant cell that arose in puberty.
Lineage tracing studies unravel long-lived bipotent and unipotent cells
Although transplantation assays are useful for quantifying the repopulating capacity of defined subpopulations, lineage tracing is essential to track stem cells and their progeny in vivo and ascertain their relative contributions to development, homeostasis, and cancer. Ideally, both spatial and temporal levels of control are used to track cell fate, in combination with well-characterized gene promoters to drive expression in the cell type of interest. Two primary inducible systems have been used by stem cell biologists: a tamoxifen-regulatable version of cre that is engineered behind an endogenous or transgenic promoter or a tetracycline-responsive reverse transactivator (rtTA) linked to a cell-specific promoter, incorporating a separate TetO-cre allele.
In the first inducible cell fate mapping study in the mammary gland (Van Keymeulen et al. 2011), unipotent cells were tracked over substantial periods of time throughout the different stages of development, leading to the conclusion that unipotent stem cells rather than bipotent stem cells drive development and homeostasis in the postnatal mammary gland. Although embryonic K14-expressing cells gave rise to all epithelial cell types in the adult gland, the control of the basal (K14+/K5+) and luminal (K18+/K8+) lineages was uncoupled in the postnatal mammary gland; that is, unipotent luminal and basal cells were found to independently contribute to expansion of the ductal tree in puberty and pregnancy and maintain ductal homeostasis in adulthood. Notably, the basal-restricted cells were long-lived, as they could be traced over two cycles of pregnancy, lactation, and involution. These cells may represent basal-restricted stem cells but could conceivably correspond to long-lived progenitors (Fig. 2). The life span of a mature myoepithelial cell may also be relevant here, as cell turnover is much lower among basal compared with luminal cells (Zeps et al. 1999; R Giraddi and J Stingl, unpubl.).
The recent tracking of bipotent stem cells in vivo has indicated that the differentiation hierarchy may not be so complex (Rios et al. 2014). A model for the emerging hierarchy is presented in Figure 2. Using a novel 3D imaging technique for cell fate mapping studies combined with a multicolor Confetti reporter and a doxycycline-inducible system, bipotent MaSCs could be traced at a clonal level in situ. Importantly, the 3D imaging strategy enables visualization of extensive portions of the intact ductal tree at single-cell resolution. The entire basal cell lineage was labeled using the K5 promoter and validated through installation of an IRES-GFP cassette. Imaging of hundreds of small isolated clones in 3D showed that K5-labeled cells generated both myoepithelial and luminal cells. Myoepithelial-only clones were also prominent, compatible with labeling of the basal lineage. Two additional models (the K14creERT2 strain and a different K5-creERT2 strain) established the existence of bipotent MaSCs (Rios et al. 2014). These cells could be equally termed multipotent stem cells, as they also contributed to the alveolar branch of the luminal lineage during pregnancy. Interestingly, the presence of different-colored myoepithelial cells surrounding the alveoli suggests that these cells migrate from adjacent ductal regions during alveolar cell expansion. In addition, labeled cells in both lineages were efficiently traced over one or two rounds of pregnancy–lactation–involution (8- or 16-wk chases, respectively) following induction in adulthood. The extensive ductal labeling by these stem cells during involution further indicated that they orchestrate remodeling of the epithelial tree. Notably, Lgr5 also marked bipotent stem cells in the adult gland (Rios et al. 2014). In contrast to the prepubertal mammary gland, where Lgr5 is restricted to the proximal portion (Van Keymeulen et al. 2011; de Visser et al. 2012), Lgr5-labeled cells were distributed throughout the epithelial tree of the adult gland (Rios et al. 2014). The low proportion of cells labeled in this model suggests that it is difficult to achieve efficient tamoxifen-induced recombination in these cells.
In addition to bipotent MaSCs contributing to all of the major stages of morphogenesis in the postnatal gland, they also actively participate in the maintenance of the ductal tree throughout adult life. Extensive ductal labeling was observed even after a 52-wk chase (Rios et al. 2014). The presence of unicolored ducts further suggested that only one or two stem cells underpin the replenishment of cells along large ductal regions, presumably via the generation of restricted luminal and myoepithelial progenitors. These findings indicate that homeostasis is a very active process in the adult and aging mammary gland. The longevity of these cells further suggests that they are prime targets for breast carcinogenesis. It is noteworthy that the increased risk of breast cancer imposed by exposure to ionizing radiation in teenage women is evident many years following exposure, thus indicating the persistence of long-lived cells (Land and McGregor 1979).
Matters arising in lineage tracing and transplantation studies
The approach of examining the cell fate of freshly dissociated mammary epithelial cells using transplantation and ex vivo assays has received considerable criticism in recent reports (Van Keymeulen et al. 2011; de Visser et al. 2012; van Amerongen et al. 2012). Lineage tracing studies have suggested that the transplantation assay may not reflect the true behavior of stem cells, since the process of tissue dissociation and transplantation into a cleared fat pad may impart properties upon cells that they would not normally exhibit in intact tissues. However, there is reasonable concordance between lineage tracing assays and the transplantability of prospectively isolated cells. Labeled luminal cells implanted alone could not engraft in the mammary fat pad (Van Keymeulen et al. 2011; van Amerongen et al. 2012), while bipotent cells induced and labeled for 24 h could reconstitute a bilayered mammary tree upon transplantation, whereas luminal-restricted progenitor cells could not (NY Fu and JE Visvader, unpubl.). Nonetheless, basal lineage-restricted cells tracked in prepubescent glands of Axin2 mice exhibited repopulating activity upon transplantation (van Amerongen et al. 2012). It is possible that the full potential of some basal cellular subsets is only manifest at a specific developmental time point in vivo but that the transplantation assay unleashes the complete lineage potential of a given cell. It is also important to note that the basal population is heterogeneous and that the activity of bipotent basal stem cells and basal-restricted progenitor cells may be uncoupled during development depending on the tissue requirements and microenvironmental cues. In the case of long-lived progenitor cells, one question to consider is how long-lived a progenitor is before it is coined a stem cell. A more detailed in vivo kinetic analysis of labeled cells representing the different epithelial subtypes may shed light on these issues.
How does one reconcile the discrepancies between different cell lineage tracing studies using keratin gene promoters to drive expression in the basal compartment? First, identical cells may not be labeled in vivo due to promoter infidelity or low transcriptional activity being insufficient to reach the threshold of cre necessary for recombination. In the neural system, it has been suggested that the level of cre obtained from certain promoters in early stem or progenitor subtypes may not be sufficient to trigger recombination and that higher levels are likely to be achieved through the use of BAC transgenic rather than targeted knock-in mice (Anthony and Heintz 2008). Second, multipotent stem cells could be more difficult to label than progenitors, perhaps reflecting their cell cycle status or quiescence. In the case of the K14-rtTA transgenic strain used by Van Keymeulen et al. (2011) the K14 promoter may be more strongly expressed in basal progenitors than bipotent stem cells, thereby predominantly tracing long-lived basal progenitor cells in vivo. Third, both transgenic and knock-in models are prone to insertion site effects that could alter the level or timing of expression from the selected promoter. Although this pertains more to transgenic mouse strains, it can apply to knock-in strains. Last, the dose of tamoxifen for induction of expression in the creERT2 mouse model is critical. Not only is ER indispensable for mammary development, where it plays a key role in the luminal compartment, but antagonism of ER also profoundly affects MaSC function (Asselin-Labat et al. 2010). Therefore, it is essential to use transient and low doses of the anti-estrogen tamoxifen, since higher doses grossly impair mammary gland development in puberty, adulthood, and pregnancy (Rios et al. 2014).
Another layer of complexity lies in the apparent cell fate switches observed at different points of the morphogenetic cycle (van Amerongen et al. 2012). Integral to resolving this will be our understanding of the markers and properties of primitive fetal and adult MaSCs. In the Axin2, Lgr5, and K14 promoter-driven models, labeling of cells at the embryonic stage (or day 1 post-partum) versus the adult leads to very different outcomes. For example, induction of Axin2-expressing cells in the mammary anlage predominantly yielded cells of the luminal lineage in the adult mammary gland, whereas cells labeled in prepubescent mice gave rise to only basal cells. Cell fate switching from the luminal to basal lineage was also reported during the first 12 d of postnatal development in the case of Lgr5-expressing cells (de Visser et al. 2012). In adulthood, induction of Axin2-labeled cells resulted in tracing of only the basal lineage, yet in pregnancy, they could generate both luminal and myoepithelial cells of the alveolar structures. These data suggest that basal-restricted stem cells in the virgin gland might be switched to bipotent stem cells in a different hormonal context. It is likely that the fate of the majority of cells at a given developmental time point is read out in lineage tracing experiments, but in cases where the degree of labeling of cells is low, the fate of most cells in that lineage remains questionable. Of relevance, in a heterogeneous population comprising only a small percentage of stem cells, sufficient recombination must be achieved in order to target the stem cell; otherwise, only the most prevalent cell type in that lineage will be labeled.
In summary, although lineage tracing is a very powerful technique to interrogate cell fate in vivo, we should be mindful of its potential pitfalls. A negative result in lineage tracing does not necessarily mean that the cell type in question does not exist in vivo, particularly in relation to a heterogeneous cell lineage. In the future, nonbiased approaches for tracking cell fate in vivo that are not reliant on targeting with a cell-specific promoter and avoid the use of tamoxifen could be considered. Along these lines, Winton and colleagues (Kozar et al. 2013) have studied intracrypt dynamics in the intestinal epithelium through a continuous clonal labeling approach to track a genetic clonal mark acquired during DNA replication. The recent development of cellular barcoding technology offers an alternative approach to investigate the diversity of differentiation potential, although this requires an intervening culture period. The transplantation of barcoded basal cells into cleared fat pads resulted in many bilineage cells within the primary outgrowths, compatible with the existence of a common stem cell (Nguyen et al. 2014).
Elucidation of distinct subtypes of mouse luminal cells
Multiple distinct subsets within the luminal compartment of the mammary epithelium have been identified at the morphological and functional levels. In mice, luminal progenitors can be discriminated from differentiated luminal cells using a variety of cell surface markers (Fig. 3); these include CD61 (Asselin-Labat et al. 2007), CD49b (Shehata et al. 2012), CD14 (Asselin-Labat et al. 2011; Shehata et al. 2012), and c-Kit (Asselin-Labat et al. 2011; Regan et al. 2012). These markers, however, are not functionally equivalent, since some markers (e.g., CD14 and CD49b) are expressed by all luminal progenitors, whereas others (e.g., c-kit and CD61) have more restricted expression patterns. For instance, the CD61+ population encompasses a small ER+ (<10%) and larger ER− subset (Asselin-Labat et al. 2007) but does not contain alveolar-restricted progenitors, and, moreover, this marker is down-regulated during pregnancy. It should be also noted that the utility of c-Kit and CD61 is dependent on the mouse strain, since they effectively mark luminal progenitors from FVB/N mice (Asselin-Labat et al. 2011; Regan et al. 2012) but not C57Bl6/J mice (Shehata et al. 2012; ML Asselin-Labat, unpubl.). Sca-1 has proven to be a useful marker, since its expression identifies a subpopulation of luminal cells that express high levels of the luminal differentiation proteins ER, K8, and K18 (Sleeman et al. 2007; Shehata et al. 2012).
Figure 3.
Markers of prospectively identified epithelial subsets in the mouse mammary gland. Summary of cell surface markers used for the isolation of epithelial cell subsets from the mouse mammary gland. ER denotes ERα.
By combining different markers, a number of luminal cell subpopulations have been resolved. The subpopulation defined by CD49b−CD14−Sca-1+ expression is nonclonogenic and corresponds to mature luminal cells (Sleeman et al. 2007; Shehata et al. 2012). Among the luminal progenitors, the largest subset displays a CD49b+CD14+Sca-1−Aldh1+ phenotype and is enriched for undifferentiated colony-forming cells (CFCs) (Shehata et al. 2012). This population can be further subdivided on the basis of c-kit expression, with c-Kitlo cells perceived to represent relatively mature alveolar progenitors, whereas c-Kithi cells likely correspond to more primitive alveolar progenitors. Although most of the CFCs detected in the mouse mammary epithelium have an ER− phenotype, a rare subset of ER+ progenitors can be detected and purified on the basis of their CD49b+c-Kit+Sca-1+ phenotype (Regan et al. 2012; Shehata et al. 2012). Moreover, rare proliferating ER+ cells have been detected in adult mice (Booth et al. 2008a), although the role of these cells in the homeostatic mammary gland remains obscure.
Diverse subtypes of luminal cells in human breasts
A similar diversity of luminal cell types has emerged in human breast tissue. Luminal and basal cells isolated from human breasts can be discriminated on the basis of the expression of EpCAM and CD49f, with luminal cells displaying an EpCAMhiCD49flo-med phenotype and basal cells displaying an EpCAMloCD49fhi phenotype (Stingl et al. 2001; Eirew et al. 2008; Lim et al. 2009). Among the luminal cells, progenitors can be resolved from differentiated luminal cells on the basis of expression of CD49f and c-KIT (Stingl et al. 2001; Lim et al. 2009). This progenitor population (CD49f+c-KIT+) can be subfractionated according to ALDH1 activity (Eirew et al. 2012; Shehata et al. 2012). The ALDH1+ fraction contains most of the CFCs, which express Elf-5 but lower levels of luminal cell differentiation markers, including ERα. A variant of ERα, termed ERα36, which mediates nongenomic rapid estrogen signaling, also may be expressed in this subset, given the recent observation that ALDH1+ERα− cells in hormone receptor (HR)-positive breast tumors express this gene (Deng et al. 2014). The EpCAMhiCD49f− subset expresses genes typical of mature ductal cells (Lim et al. 2009; Shehata et al. 2012). Curiously, luminal progenitor cells have much shorter telomeres than mature cells, perhaps rendering them more susceptible to DNA damage (Kannan et al. 2013).
Discordant data have arisen during the characterization of CFCs within the mammary gland. For instance, it has been found that CFCs within the luminal cell populations are restricted in their fate (Eirew et al. 2008; Lim et al. 2009), yet others have reported that these cells exhibit multilineage potential in vitro (Deng et al. 2014; Liu et al. 2014). Moreover, the differentiation potential of ALDH1+ luminal progenitor cells is controversial, since some studies demonstrate that these cells are luminal-restricted when cultured in vitro and do not provide long-term engraftment in vivo (Eirew et al. 2012; Shehata et al. 2012), whereas other reports suggest that these cells have multilineage potential both in vitro and in vivo (Ginestier et al. 2007; Liu et al. 2014). These conflicting results are likely due to the use of different in vitro culture conditions, which highlight the difficulty in inferring the role of these cells in normal tissue homeostasis.
Curiously, approximately one-quarter of normal human breast tissue samples contain an additional luminal cell population that is characterized by low expression of ERBB3 (Shehata et al. 2012), and, in some samples, this population can be the dominant luminal subpopulation. These cells are unusual in that they have an undifferentiated luminal progenitor gene signature and yet are almost entirely devoid of colony-forming and engrafting potential. It is not clear why these ERBB3− cells are present in some patient samples and not others, but these cells may be remnants of alveoli from previous pregnancies, since ERBB3 is required for ductal formation but not alveologenesis (Lahlou et al. 2012).
It is important to note that when mammary tissue is dissociated and analyzed by flow cytometry, information regarding the geographical location of cells within the mammary epithelial tree is lost. Elegant studies by Petersen and colleagues (Villadsen et al. 2007) have demonstrated that most epithelial cells with proliferative potential in the human breast are restricted to the ducts and not lobules of the epithelial tree. This includes luminal progenitor cells, which were largely restricted to the ducts, while the mature EpCAMhiCD49f− luminal subset was mainly found in the lobules. Similarly, within the basal cell population, EpCAMhi cells resided in the ducts, and those with low EpCAM levels were found in the lobules.
Interestingly, both human and mouse mammary luminal progenitors can display a remarkable degree of “plasticity” under nonphysiological conditions, suggesting that cell fate decisions are not irreversible. Exposure of luminal progenitor cells to different microenvironments or culturing of these cells can reprogram them to a stem-like state. For example, both ER+ and ER− luminal progenitors implanted into cleared mammary fat pads could generate structures containing all mammary epithelial cell types, including stem cells capable of repopulation, albeit at a low frequency (Sleeman et al. 2007; Vaillant et al. 2011; Shehata et al. 2012), while the injection of cells in the presence of Matrigel enhanced this phenomenon (Vaillant et al. 2011). In addition, there is evidence for bidirectional interconversion among stem and more differentiated human breast epithelial cells cultured in vitro (Chaffer et al. 2011). Although altered microenvironments can elicit dedifferentiation of committed luminal cells, it is important to note that in vivo evidence for interconversion between the luminal and basal lineages is lacking.
Identification of luminal progenitor cells via lineage tracing
Early studies by Wagner et al. (2002) demonstrated the feasibility of using Cre-lox-based technology to genetically mark subsets of mammary epithelial cells. Through mapping the progeny of whey acidic protein (WAP)-expressing cells, largely activated in late pregnancy and lactation, a subset of cells was discovered to survive the involution process and function as self-renewing alveolar precursors in subsequent pregnancies. These were termed parity-induced mammary epithelial cells (PI-MECs) (Wagner et al. 2002). Interestingly, PI-MECs were recently demonstrated to reside exclusively within the luminal layer of ducts and to directly contribute to the secretory alveoli that arise during pregnancy (Chang et al. 2014). Moreover, these cells could generate all ER− cells within the alveolus but did not contribute to the hormone-sensing ER+ cells or the myoepithelial lineage.
Luminal progenitor cells have been recently tracked in the mouse mammary gland using a number of inducible lineage tracing models. In a doxycycline-regulatable mouse strain controlled by the promoter of the luminal progenitor gene Elf5 (Lim et al. 2010), virtually all progenitor activity coincided with Elf5 expression in pubertal and adult glands (Rios et al. 2014). Elf5-expressing cells contributed solely to the luminal lineage throughout the different stages of development. Indeed, the luminal progenitor population appears to be the key driver of morphogenesis during puberty and alveologenesis, with numerous progenitor cells recruited throughout the TEBs, subtending ducts, and alveolar units. Analogous to the WAPcre-driven reporter system (Chang et al. 2014), multiple Elf5 progenitors also contributed to the formation of each alveolus but not the myoepithelium. Even though Elf5-expressing luminal cells could be traced for 8–10 wk and thus are relatively long-lived, only a very small proportion of labeled cells remained after a 20-wk chase, thereby distinguishing them from bipotent stem cells (Rios et al. 2014). Furthermore, the Elf5-labeled pool was diminished at the end of each involution cycle, indicating that a new pool of progenitors is recruited for each round of alveologenesis. These data are compatible with the findings of Chang et al. (2014) (although inducible cell fate mapping could not be used) who showed that some lobules generated during the second pregnancy were from previously labeled cells, whereas others derived from unlabeled cells that likely represent new alveolar progenitors.
K8-creER targeted cells also contributed specifically to the luminal lineage in vivo (Van Keymeulen et al. 2011). In this case, however, the proportion of labeled cells appeared relatively stable over two rounds of pregnancy (<6%), thus differing from Elf5-marked cells. Although different progenitors may have been targeted by the K8 and Elf5 promoters, the low proportion of labeling in the case of the K8 model precludes any definitive conclusions at this point. It is also perplexing that the K8 and K18 promoters targeted different luminal cells (progenitors and mature cells, respectively) when these keratins are coexpressed in the luminal lineage (Van Keymeulen et al. 2011).
The Notch signaling pathway plays a critical role in luminal lineage determination (see below). Lineage tracing of rare Notch3-expressing cells (0.1%–4%) revealed that these cells are transiently quiescent, luminal progenitor cells that can generate both ER+ and ER− ductal progeny (Lafkas et al. 2013). Such cells also contributed to alveolar structures in a polyclonal fashion and could survive multiple rounds of pregnancy, reminiscent of PI-MECs. Lineage tracing studies of another member of this gene family, Notch2, identified two previously unrecognized cell types in the luminal lineage (Sale et al. 2013). The large (L) and small (S) cells described were both exceedingly rare, since they constituted <1% of the total mammary epithelium. It is notable that S cells do not correspond to the previously reported SLCs (∼9 µm) identified by electron microscopy, as they are substantially smaller. Imaging in 3D will be necessary to resolve their identity in the future.
Hormonal signaling between epithelial subtypes by paracrine mechanisms
The mammary epithelium is highly sensitive to the effects of the ovarian steroid hormones estrogen and progesterone, which are integral to puberty, estrus cycling, and pregnancy (Brisken and O’Malley 2010). The receptors for these nuclear steroid hormones (ERα and PR) remain critical prognostic markers and therapeutic targets in breast cancer. MaSCs were found to be exquisitely responsive to steroid hormones despite the size of the MaSC/basal subset being unaltered by hormone deprivation (Asselin-Labat et al. 2010; Joshi et al. 2010). These findings are of particular interest, as the MaSC/basal subset lacks expression of ERα and PR in both mouse and human mammary glands, thus invoking an indirect mechanism of action (Fig. 4; Asselin-Labat et al. 2006; Lim et al. 2009). Nevertheless, the cycling properties of the hormone-deprived MaSC subset suggested that they had entered a more quiescent state (Asselin-Labat et al. 2010). Conversely, progesterone promotes a transient but profound expansion of the MaSC population in the diestrus phase of each cycle, presumably through amplifying estrogen signaling (Joshi et al. 2010).
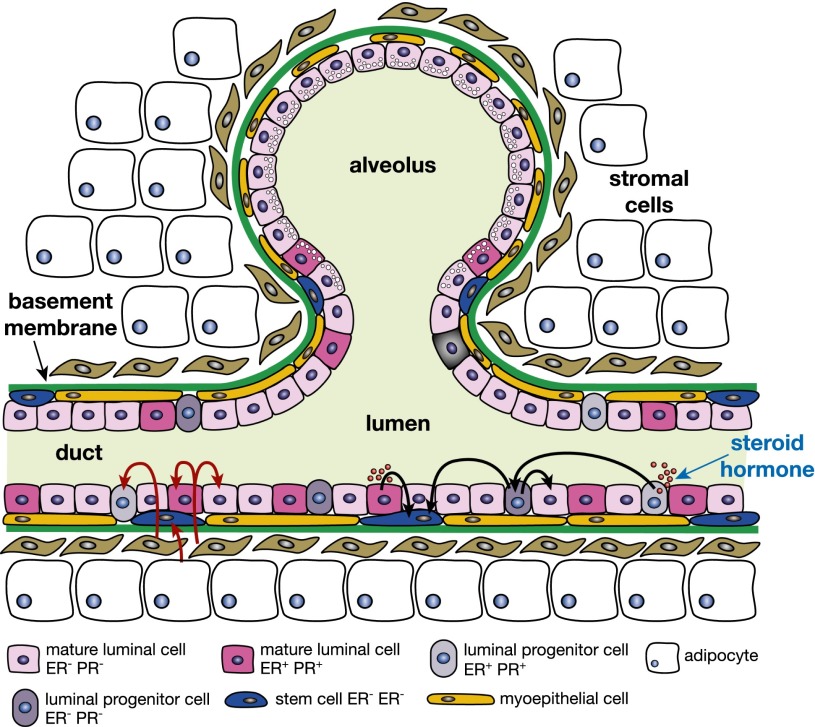
Figure 4.
Schematic depiction of potential regulatory cross-talk between different ductal mammary cells in response to steroid hormone stimulation. Estrogen or progesterone (red circles in lumen) activate ER+ epithelial cells (either mature cells or progenitors), which secrete paracrine factors that activate HR-negative stem cells or luminal progenitor cells and/or mature luminal cells. The black arrows indicate signals from steroid hormones, including those between HR-negative progenitors that have been indirectly activated by hormones, and their signaling to stem and other cells. The red arrows depict a further layer of interaction between stromal cells (fibroblasts and adipocytes) and mammary epithelial cells lining the ducts.
It is well recognized that estrogen and progesterone exert their mitogenic effects primarily through paracrine signaling (Fig. 4). In terms of paracrine effectors of signaling to MaSCs, two key candidates have emerged, RANKL and Wnt4, both of which are direct targets of PR (Fernandez-Valdivia and Lydon 2012). Expression of these genes was dramatically elevated in luminal cells from mammary glands of mice treated with estrogen plus progesterone (Joshi et al. 2010). Moreover, the levels of RANK and the Wnt coreceptor Lrp5 were augmented in the MaSC/basal population, suggesting that they are important paracrine effectors in the steady-state gland. In the milieu of pregnancy, where progesterone and the RANKL–RANK signaling axis are important regulators of alveologenesis (Fata et al. 2000), only RANKL was directly implicated in controlling MaSC activity in vivo (Asselin-Labat et al. 2010). In mouse models of tumorigenesis, treatment with the synthetic progestin medroxyprogesterone acetate (MPA) increased the size of the MaSC/basal population prior to carcinogen exposure (Gonzalez-Suarez et al. 2010; Schramek et al. 2010). Importantly, blockade of RANKL signaling suppressed these MPA-mediated effects and revealed that they were intrinsic to epithelial cells, thus highlighting the RANK pathway as a potential target for breast cancer prevention.
Pregnancy is one of the most significant factors to influence breast cancer risk. A single full-term pregnancy before the age of 20 is highly protective against ER-positive disease. Although a number of factors have been implicated in this phenomenon (Schedin 2006), one current hypothesis is that an early pregnancy reduces the number or activity of MaSCs. Functional studies in mice have indicated that MaSC activity is diminished following an early pregnancy when compared with their counterparts from nulliparous glands (Siwko et al. 2008; Meier-Abt et al. 2013), although a separate study found no difference (Britt et al. 2009). Nevertheless, epithelial cell fate may be altered by a single early pregnancy, since molecular profiling studies of parous versus nulliparous glands revealed an altered Wnt:Notch signaling ratio in the MaSC/basal subset. Decreased expression of Wnt target genes was accompanied by decreased proliferative potential (Meier-Abt et al. 2013). Moreover, early pregnancy decreased the proportion of HR-positive luminal cells. Reduced responsiveness to ovarian hormones presumably contributes to the observed shift in expression toward differentiation.
The mammary epigenome and its hormone responsiveness
Epigenetic programs play a fundamental role in adult stem cells and their progressive restriction to committed progenitors and differentiated cells (Margueron and Reinberg 2011). In the mammary gland, a growing number of chromatin regulators have been recently implicated in controlling the balance between self-renewal and differentiation. The histone methylation reader Pygo-2, a coactivator of the Wnt pathway, is necessary for suppressing luminal and alveolar differentiation of the MaSC-enriched population by coordinating the activity of the Wnt and Notch pathways (Gu et al. 2013). Interestingly, Pygo2 facilitates binding of β-catenin to the Notch3 gene in MaSC/basal cells in order to maintain this gene in a “poised” bivalent chromatin state. The polycomb-repressive complex-1 (PRC1) gene Bmi1 (Liu et al. 2006; Pietersen et al. 2008) and PRC2 gene Ezh2 (Pal et al. 2013) influence mammary repopulating potential but play distinct roles in the developing gland. Bmi-1 is important for preventing precocious alveolar differentiation of committed cells in the adult gland, whereas Ezh2 exerts a fundamental role in regulating the proliferation of progenitor cells along the hierarchy. Conditional targeting of Ezh2 in the mouse mammary gland impaired ductal morphogenesis, concomitant with a drastic decrease in the clonogenic activity of all basal and luminal progenitor populations as well as MaSC activity in vivo (Pal et al. 2013). Similarly, mice expressing a doxycycline-regulatable shRNA against Ezh2 exhibited a delay in ductal elongation, which was linked to a decrease in the luminal progenitor pool in early puberty (Michalak et al. 2013). Interestingly, Ezh2 is highly expressed in the fetal MaSC-enriched subset in contrast to Bmi-1 levels, which declined during the late stages of embryogenesis (Spike et al. 2012).
To gain insight into the global epigenetic signatures of human breast epithelial cells, CD44+ (enriched for putative progenitors) and CD24+ (luminal cell-enriched) populations were sorted, and their histone methylation and DNA methylation patterns were determined. H3K27me3 (frequently localized to K27 blocks in gene-poor domains) and DNA methylation patterns were distinct in the two subsets, suggesting that gene expression programs in the different cell types are controlled by epigenetic mechanisms (Maruyama et al. 2011). More recently, the histone methylation profiles of three functionally distinct mouse epithelial populations (MaSC/basal, luminal progenitor, and mature luminal cells) were established in different hormonal contexts (Pal et al. 2013). In the steady-state gland of young adult mice, comparative chromatin and gene expression analyses revealed that H3K4me3 and H3K27me3 modifications were tightly correlated with transcriptional activity, whereas H3K9me2 was not. Intriguingly, H3K27me3 modifications increased dramatically during restriction of the MaSC/basal population to the luminal lineage, suggesting that cell fate decisions in the mammary gland are orchestrated by polycomb complex-mediated repression.
The mammary epigenome undergoes highly specific changes in different hormonal contexts (Pal et al. 2013). The most profound change was observed in the global H3K27me3 map of luminal cells during pregnancy but not that of the MaSC/basal population (Fig. 5). Neither H3K4me3 nor H3K9Ac exhibited substantial changes in their distribution or intensity as a function of gene expression. Notably, the expression and phosphorylation of the key H3K27me3 methyltransferase Ezh2 strongly correlated with H3K27me3 levels, and progesterone was identified as an important in vivo regulator of Ezh2, thus linking hormonal cues to changes in chromatin structure. Ezh2 phosphorylation by the cell cycle kinase CDK1 emerged as one potential mechanism by which hormones influence luminal progenitor expansion in pregnancy. Intriguingly, the Rank signaling axis was again implicated in progesterone-mediated induction of Ezh2 that selectively occurred in PR− progenitor cells in vivo (Pal et al. 2013). One corollary of these findings is that sustained hormonal signaling may initiate breast carcinogenesis through globally altering the epigenetic landscape of progenitor cells. Indeed, epigenetic alterations could account for the increase in breast cancer risk that has been reported following in utero exposure to diethylstilbestrol (Hoover et al. 2011).
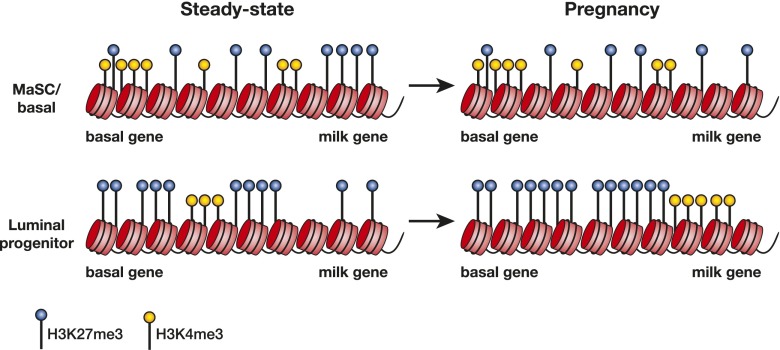
Figure 5.
Regulation of the epigenetic state by steroid hormones and a dominant role for H3K27me3 modification. In the MaSC/basal population of the steady-state (virgin) gland, genes are epigenetically marked by H3K4me3 and H3K27me3, but as cells restrict to the luminal lineage, H3K27me3 modifications increase, suppressing gene expression. During pregnancy, where progesterone is a key hormone, only small changes occur in the H3K4me3-modified landscape of the MaSC/basal and luminal subsets, whereas a profound increase in H3K27me3 modifications occurs within the luminal subset, implying that H3K27me3 marks are important for regulating alveologenesis. Hypothetical histone modifications on putative basal and milk genes in the different subsets and hormonal states are shown.
Molecular regulators of MaSCs
There has been an exponential growth in our understanding of transcription factors and modulators that govern the mammary epithelial hierarchy over recent years, largely through the characterization of mammary epithelial subsets in targeted mice. A few genes that have been more recently explored are highlighted here. Both Slug and Sox9 have emerged as key determinants of the MaSC state. Perturbation of either of these genes impaired MaSC activity (Guo et al. 2012), and Slug-deficient mice exhibited a delay in morphogenesis in puberty. Consistent with a functional role for Slug in the maintenance of the “stem-like” state, the expression of a number of luminal lineage genes was aberrantly induced in Slug−/− mammary epithelial cells (Nassour et al. 2012). The MaSC/basal population also expresses several other EMT (epithelial–mesenchymal transition) genes besides Snai2/Slug, such as Twist, which together have been postulated to confer “stemness” on epithelial cells (Mani et al. 2008). The basal-restricted transcription factor p63 has been long suspected as a master regulator of stem cells, since p63-null mice lack rudimentary mammary glands (Mills et al. 1999). Reminiscent of that observed for Slug, overexpression of p63 in luminal cells was sufficient to elicit a basal phenotype. Indeed, p63 and Notch were found to have opposing roles in mammary epithelial cells: ΔN-p63 maintains a basal cell fate, while Notch signaling down-regulates p63 expression prior to luminal lineage commitment (Yalcin-Ozuysal et al. 2010), a restriction point where Notch1 and Notch3 play a crucial role (Bouras et al. 2008; Raouf et al. 2008). Other potential transcriptional regulators of MaSCs include the transcription factors Stat3, CCAAT/enhancer-binding protein-β, and c-myc, all of which affect mammary repopulating ability in vivo (LaMarca et al. 2010; Staniszewska et al. 2012; Moumen et al. 2013). Conversely, the tumor suppressor p53 serves a critical role in restricting the renewal of MaSCs and regulating their asymmetric division (Cicalese et al. 2009; Chiche et al. 2013).
Regulators of the luminal lineage
A network of crucial transcriptional regulators has been unraveled for the luminal lineage. Although the ETS transcription factor Elf5 is an important determinant of the secretory cell lineage (Oakes et al. 2008), it also plays a broader role in dictating luminal cell fate. In accordance, Elf5 is a top-ranking signature gene of luminal progenitor cells (Lim et al. 2010). Targeted deletion of Elf5 in mice leads to cells acquiring an EMT-like phenotype in pregnancy and lactation, and Elf5 was found to directly repress the transcription of Slug, suggesting that it suppresses a basal lineage-determining program while promoting luminal lineage specification (Oakes et al. 2008; Chakrabarti et al. 2012a,b). Interestingly, RANKL was identified as a paracrine effector of progesterone-induced expression of Elf5 in CD61+ luminal progenitors, and inhibition of RANKL prevented the expansion of Elf5+ luminal cells in the secretory lineage (Lee et al. 2013).
Several other transcription factors orchestrate functional development of the alveolar secretory epithelium, which is required for successful lactation. Gata-3 is a crucial regulator of the luminal lineage and is sufficient to induce milk protein gene expression in the MaSC/basal subset in the absence of a lactogenic stimulus. Despite Gata-3 being an important component of the ERα signaling axis (Eeckhoute et al. 2007), it is essential for the maturation of both ER+ and ER− luminal progenitor cells into ductal and secretory alveolar cells (Kouros-Mehr et al. 2006; Asselin-Labat et al. 2007, 2011). More recently, FoxM1 was established as a transcriptional repressor of Gata-3, whereby FoxM1 coordinates methylation of the Gata-3 promoter via its association with DNMT3b and blocks luminal progenitor cell differentiation (Carr et al. 2012). The reduced pool of luminal progenitor cells accompanying loss of Stat5a suggests a role for this gene in the generation or expansion of alveolar progenitor cells from MaSCs (Yamaji et al. 2009). Interestingly, the Stat6-regulated zinc finger protein Zfp157 is required to establish the balance of Gata-3- versus Stat5a-expressing luminal cell subtypes during alveologenesis (Oliver et al. 2012). Physiologically, C/EBPβ also specifies luminal cell fate in the mammary gland, and its loss induces misexpression of basal markers in the luminal cell compartment (LaMarca et al. 2010).
Implications of the differentiation hierarchy for breast cancer
Breast cancer has been stratified into at least five definitive molecular subtypes, although subgroups within subtypes have been recently identified through gene expression profiling studies (Curtis et al. 2012). The major subtypes include luminal A, luminal B, HER2-positive, claudin-low, and basal-like (Herschkowitz et al. 2007; Lehmann et al. 2011; Prat and Perou 2011). Comparative molecular studies of the epithelial gene expression signatures established for the different mammary epithelial populations with those representing the cancer subtypes have suggested that distinct cells of origin may give rise to the different subtypes (Lim et al. 2009; Prat et al. 2010). The MaSC/basal signature is most closely aligned with the expression profile of the claudin-low subtype, while the luminal progenitor signature is closest to that of the basal-like subtype (Fig. 6). Moreover, ALDH+ER− luminal progenitor cells in human breast tissue exhibit a signature that most strongly correlates with the basal-like subtype (Shehata et al. 2012). The cells of origin for the remaining subtypes (luminal A, luminal B, and HER2-positive), however, remain elusive. Although the luminal A profile was most concordant with the signature of mature luminal cells, it is presumed that a small progenitor subset within this population is the likely target. In cases where dedifferentiation or cell plasticity occurs during neoplastic progression, the cell of origin cannot be defined.
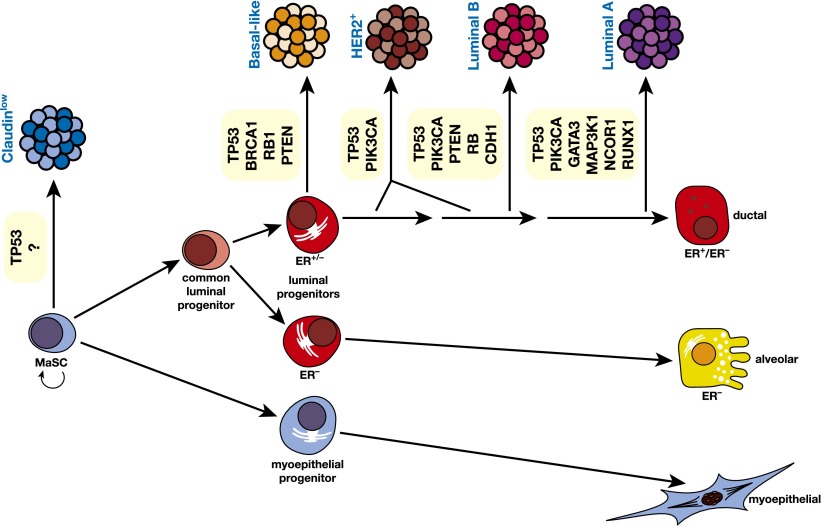
Figure 6.
Schematic model of the human breast epithelial hierarchy and potential relationships with breast tumor subtypes. The five major tumor types are shown linked to their closest normal epithelial counterpart based on gene expression profiling. The HER2+ subtype could originate through amplification of the HER2 locus in a luminal target cell that is either ER+ or ER−. Examples of commonly mutated genes in the different subtypes of breast cancer are indicated.
Biological evidence for these gene expression correlations across normal and cancerous cells has been obtained in the case of BRCA1-associated cancers. Different strategies have revealed that the luminal progenitor is the “cell of origin” for basal-like breast cancers arising in BRCA1 mutation carriers. These studies included the cellular analysis of preneoplastic human breast tissue from BRCA1 mutation carriers (Lim et al. 2009), determination of the proclivity of luminal versus basal breast epithelial cells from BRCA1 heterozygous women toward transformation (Proia et al. 2011), and the generation of specific mouse models of mammary tumorigenesis harboring inactivation of Brca1 (and p53) (Molyneux et al. 2010; Bai et al. 2012). In all of these cases, the luminal progenitor rather than the MaSC emerged as the cell of origin for basal-like cancers developing in these carriers. Thus, the genetic background of these individuals appears to have preprogrammed cells specifically within the luminal lineage toward a basal-like phenotype.
Genetic lesions also contribute to tumor heterogeneity, although their precise role in the generation of intertumoral breast heterogeneity is yet to be established. Nevertheless, a number of genome-wide sequencing studies of breast cancers have identified a plethora of somatic mutations that are common to specific subtypes of breast cancer (for review, see Ellis and Perou 2013). Figure 6 indicates some of the recurrent somatic alterations associated with breast cancer subtypes. Of note, mutations in the tumor suppressor p53 and activating mutations in PIK3CA are commonly found across the different subtypes. The tumor suppressor retinoblastoma Rb is frequently associated with luminal B and triple-negative tumors, and targeted mouse models have shown that inactivation of Rb1 in mammary epithelial progenitor cells can recapitulate these subtypes, thus supporting a role for genetic lesions in directly influencing tumor histopathology (Jiang et al. 2010). The recent observation that different cancer subtypes could arise from BLG-cre targeted cells in different genetic backgrounds indicated that genetic drivers are important determinants of phenotype, but the precise cell types in which the genes were inactivated are yet to be defined (Melchor et al. 2014). Although both the initiating genetic events and the “cell of origin” inevitably act in concert to determine tumor pathology and behavior, the microenvironment also plays a pivotal role in influencing the course of tumorigenesis (Polyak and Kalluri 2010).
Concluding remarks
Many questions and challenges lie ahead for the mammary gland field. What is the complexity within the MaSC compartment? Can quiescent stem cells be prospectively isolated, given that only cycling cells can be tracked in vivo by lineage tracing? What are the constituents of the niches for putative quiescent and activated stem cells? There is increasing evidence that signals from the mammary microenvironment can contribute to mammary epithelial cell specification (Booth et al. 2008b), but the nature of these instructive signals and the cell types from which they emanate remain elusive. What is the functional interrelationship between ER+ and ER− progenitor cells? Are ER+ cells stochastically generated from ER− cells, and do they exist in a state of flux? Do ductal and alveolar luminal cells arise from their own lineage-restricted precursors? Are unipotent lineage-restricted stem cells a separate entity, or do committed progenitor cells account for these functions? Further insight into these questions, including the number of distinct stem and progenitor subtypes and the degree of plasticity inherent within the hierarchy, will demand the continued isolation of highly purified populations, transplantation assays, and in vivo cell fate mapping studies. These studies hold the promise of eventually identifying the “cells of origin” of cancer and novel biomarkers expressed by them, which may enable earlier detection of breast cancer and the development of effective preventive therapies.
Acknowledgments
We are grateful to G. Lindeman for critical reading of the manuscript, and P. Maltezos for figure preparation. J.V. is supported by a National Health and Medical Research Council (NHMRC) Australia Fellowship and thanks the NHMRC Independent Research Institutes Infrastructure Support Scheme (IRIISS), the Victorian State Government through Operational Infrastructure Support, the Australian Cancer Research Foundation, and the National Breast Cancer Foundation. J.S. acknowledges the support of The University of Cambridge, The Breast Cancer Campaign, Cancer Research UK, Hutchison Whampoa Limited, and National Institutes for Health Research (NIHR) Cambridge Biomedical Research Centre.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.242511.114.
References
- Anthony TE, Heintz N 2008. Genetic lineage tracing defines distinct neurogenic and gliogenic stages of ventral telencephalic radial glial development. Neural Dev 3: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Labat ML, Shackleton M, Stingl J, Vaillant F, Forrest NC, Eaves CJ, Visvader JE, Lindeman GJ 2006. Steroid hormone receptor status of mouse mammary stem cells. J Natl Cancer Inst 98: 1011–1014 [DOI] [PubMed] [Google Scholar]
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, et al. 2007. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol 9: 201–209 [DOI] [PubMed] [Google Scholar]
- Asselin-Labat ML, Vaillant F, Sheridan JM, Pal B, Wu D, Simpson ER, Yasuda H, Smyth GK, Martin TJ, Lindeman GJ, et al. 2010. Control of mammary stem cell function by steroid hormone signalling. Nature 465: 798–802 [DOI] [PubMed] [Google Scholar]
- Asselin-Labat ML, Sutherland KD, Vaillant F, Gyorki DE, Wu D, Holroyd S, Breslin K, Ward T, Shi W, Bath ML, et al. 2011. Gata-3 negatively regulates the tumor-initiating capacity of mammary luminal progenitor cells and targets the putative tumor suppressor caspase-14. Mol Cell Biol 31: 4609–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Rohrschneider LR 2010. s-SHIP promoter expression marks activated stem cells in developing mouse mammary tissue. Genes Dev 24: 1882–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Smith MD, Chan HL, Pei XH 2012. Germline mutation of Brca1 alters the fate of mammary luminal cells and causes luminal-to-basal mammary tumor transformation. Oncogene 32: 2715–2725 [DOI] [PubMed] [Google Scholar]
- Barker N, Tan S, Clevers H 2013. Lgr proteins in epithelial stem cell biology. Development 140: 2484–2494 [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA 2008. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40: 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth BW, Smith GH 2006. Estrogen receptor-α and progesterone receptor are expressed in label-retaining mammary epithelial cells that divide asymmetrically and retain their template DNA strands. Breast Cancer Res 8: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth BW, Boulanger CA, Smith GH 2008a. Selective segregation of DNA strands persists in long-label-retaining mammary cells during pregnancy. Breast Cancer Res 10: R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth BW, Boulanger CA, Smith GH 2008b. Stem cells and the mammary microenvironment. Breast Dis 29: 57–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouras T, Pal B, Vaillant F, Harburg G, Asselin-Labat ML, Oakes SR, Lindeman GJ, Visvader JE 2008. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell 3: 429–441 [DOI] [PubMed] [Google Scholar]
- Bowie MB, Kent DG, Dykstra B, McKnight KD, McCaffrey L, Hoodless PA, Eaves CJ 2007. Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proc Natl Acad Sci 104: 5878–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisken C, O’Malley B 2010. Hormone action in the mammary gland. Cold Spring Harb Perspect Biol 2: a003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt KL, Kendrick H, Regan JL, Molyneux G, Magnay FA, Ashworth A, Smalley MJ 2009. Pregnancy in the mature adult mouse does not alter the proportion of mammary epithelial stem/progenitor cells. Breast Cancer Res 11: R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr JR, Kiefer MM, Park HJ, Li J, Wang Z, Fontanarosa J, DeWaal D, Kopanja D, Benevolenskaya EV, Guzman G, et al. 2012. FoxM1 regulates mammary luminal cell fate. Cell Rep 1: 715–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K, et al. 2011. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci 108: 7950–7955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R, Hwang J, Andres Blanco M, Wei Y, Lukacisin M, Romano RA, Smalley K, Liu S, Yang Q, Ibrahim T, et al. 2012a. Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat Cell Biol 14: 1212–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R, Wei Y, Romano RA, DeCoste C, Kang Y, Sinha S 2012b. Elf5 regulates mammary gland stem/progenitor cell fate by influencing notch signaling. Stem Cells 30: 1496–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TH, Kunasegaran K, Tarulli GA, De Silva D, Voorhoeve PM, Pietersen AM 2014. New insights into lineage restriction of mammary gland epithelium using parity-identified mammary epithelial cells. Breast Cancer Res 16: R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepko G, Smith GH 1997. Three division-competent, structurally-distinct cell populations contribute to murine mammary epithelial renewal. Tissue Cell 29: 239–253 [DOI] [PubMed] [Google Scholar]
- Chiche A, Moumen M, Petit V, Jonkers J, Medina D, Deugnier MA, Faraldo MM, Glukhova MA 2013. Somatic loss of p53 leads to stem/progenitor cell amplification in both mammary epithelial compartments, basal and luminal. Stem Cells 31: 1857–1867 [DOI] [PubMed] [Google Scholar]
- Cicalese A, Bonizzi G, Pasi CE, Faretta M, Ronzoni S, Giulini B, Brisken C, Minucci S, Di Fiore PP, Pelicci PG 2009. The tumor suppressor p53 regulates polarity of self-renewing divisions in mammary stem cells. Cell 138: 1083–1095 [DOI] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. 2012. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486: 346–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel CW, De Ome KB, Young JT, Blair PB, Faulkin LJ Jr 1968. The in vivo life span of normal and preneoplastic mouse mammary glands: a serial transplantation study. Proc Natl Acad Sci 61: 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Zhang XT, Wang ML, Zheng HY, Liu LJ, Wang ZY 2014. ER-α36-mediated rapid estrogen signaling positively regulates ER-positive breast cancer stem/progenitor cells. PLoS ONE 9: e88034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeOme KB, Faulkin LJ Jr, Bern HA, Blair PB 1959. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res 19: 515–520 [PubMed] [Google Scholar]
- de Visser KE, Ciampricotti M, Michalak EM, Tan DW, Speksnijder EN, Hau CS, Clevers H, Barker N, Jonkers J 2012. Developmental stage-specific contribution of LGR5(+) cells to basal and luminal epithelial lineages in the postnatal mammary gland. J Pathol 228: 300–309 [DOI] [PubMed] [Google Scholar]
- Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, et al. 2009. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 458: 780–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos CO, Rebbeck C, Rozhkova E, Valentine A, Samuels A, Kadiri LR, Osten P, Harris EY, Uren PJ, Smith AD, et al. 2013. Molecular hierarchy of mammary differentiation yields refined markers of mammary stem cells. Proc Natl Acad Sci 110: 7123–7130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M 2007. Positive cross-regulatory loop ties GATA-3 to estrogen receptor α expression in breast cancer. Cancer Res 67: 6477–6483 [DOI] [PubMed] [Google Scholar]
- Eirew P, Stingl J, Raouf A, Turashvili G, Aparicio S, Emerman JT, Eaves CJ 2008. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med 14: 1384–1389 [DOI] [PubMed] [Google Scholar]
- Eirew P, Kannan N, Knapp DJ, Vaillant F, Emerman JT, Lindeman GJ, Visvader JE, Eaves CJ 2012. Aldehyde dehydrogenase activity is a biomarker of primitive normal human mammary luminal cells. Stem Cells 30: 344–348 [DOI] [PubMed] [Google Scholar]
- Ellis MJ, Perou CM 2013. The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discov 3: 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fata JE, Kong YY, Li J, Sasaki T, Irie-Sasaki J, Moorehead RA, Elliott R, Scully S, Voura EB, Lacey DL, et al. 2000. The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell 103: 41–50 [DOI] [PubMed] [Google Scholar]
- Fernandez-Valdivia R, Lydon JP 2012. From the ranks of mammary progesterone mediators, RANKL takes the spotlight. Mol Cell Endocrinol 357: 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, et al. 2007. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 1: 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez E, Jacob AP, Jones J, Miller R, Roudier-Meyer MP, Erwert R, Pinkas J, Branstetter D, Dougall WC 2010. RANK ligand mediates progestin-induced mammary epithelial proliferation and carcinogenesis. Nature 468: 103–107 [DOI] [PubMed] [Google Scholar]
- Gu B, Watanabe K, Sun P, Fallahi M, Dai X 2013. Chromatin effector Pygo2 Mediates Wnt–notch crosstalk to suppress luminal/alveolar potential of mammary stem and basal cells. Cell Stem Cell 13: 48–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, et al. 2012. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148: 1015–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassiotou F, Beltran A, Chetwynd E, Stuebe AM, Twigger AJ, Metzger P, Trengove N, Lai CT, Filgueira L, Blancafort P, et al. 2012. Breastmilk is a novel source of stem cells with multilineage differentiation potential. Stem Cells 30: 2164–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW 2005. Information networks in the mammary gland. Nat Rev Mol Cell Biol 6: 715–725 [DOI] [PubMed] [Google Scholar]
- Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, et al. 2007. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 8: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover RN, Hyer M, Pfeiffer RM, Adam E, Bond B, Cheville AL, Colton T, Hartge P, Hatch EE, Herbst AL, et al. 2011. Adverse health outcomes in women exposed in utero to diethylstilbestrol. N Engl J Med 365: 1304–1314 [DOI] [PubMed] [Google Scholar]
- Hoshino K 1962. Morphogenesis and growth potentiality of mammary glands in mice. I. Transplantability and growth potentiality of mammary tissue of virgin mice. J Natl Cancer Inst 29: 835–851 [PubMed] [Google Scholar]
- Jiang Z, Deng T, Jones R, Li H, Herschkowitz JI, Liu JC, Weigman VJ, Tsao MS, Lane TF, Perou CM, et al. 2010. Rb deletion in mouse mammary progenitors induces luminal-B or basal-like/EMT tumor subtypes depending on p53 status. J Clin Invest 120: 3296–3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi PA, Jackson HW, Beristain AG, Di Grappa MA, Mote PA, Clarke CL, Stingl J, Waterhouse PD, Khokha R 2010. Progesterone induces adult mammary stem cell expansion. Nature 465: 803–807 [DOI] [PubMed] [Google Scholar]
- Kaanta AS, Virtanen C, Selfors LM, Brugge JS, Neel BG 2013. Evidence for a multipotent mammary progenitor with pregnancy-specific activity. Breast Cancer Res 15: R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan N, Huda N, Tu L, Droumeva R, Aubert G, Chavez E, Brinkman RR, Lansdorp P, Emerman J, Abe S, et al. 2013. The luminal progenitor compartment of the normal human mammary gland constitutes a unique site of telomere dysfunction. Stem Cell Reports 1: 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller PJ, Arendt LM, Skibinski A, Logvinenko T, Klebba I, Dong S, Smith AE, Prat A, Perou CM, Gilmore H, et al. 2012. Defining the cellular precursors to human breast cancer. Proc Natl Acad Sci 109: 2772–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordon EC, Smith GH 1998. An entire functional mammary gland may comprise the progeny from a single cell. Development 125: 1921–1930 [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z 2006. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 127: 1041–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozar S, Morrissey E, Nicholson AM, van der Heijden M, Zecchini HI, Kemp R, Tavare S, Vermeulen L, Winton DJ 2013. Continuous clonal labeling reveals small numbers of functional stem cells in intestinal crypts and adenomas. Cell Stem Cell 13: 626–633 [DOI] [PubMed] [Google Scholar]
- Lafkas D, Rodilla V, Huyghe M, Mourao L, Kiaris H, Fre S 2013. Notch3 marks clonogenic mammary luminal progenitor cells in vivo. J Cell Biol 203: 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahlou H, Muller T, Sanguin-Gendreau V, Birchmeier C, Muller WJ 2012. Uncoupling of PI3K from ErbB3 impairs mammary gland development but does not impact on ErbB2-induced mammary tumorigenesis. Cancer Res 72: 3080–3090 [DOI] [PubMed] [Google Scholar]
- LaMarca HL, Visbal AP, Creighton CJ, Liu H, Zhang Y, Behbod F, Rosen JM 2010. CCAAT/enhancer binding protein β regulates stem cell activity and specifies luminal cell fate in the mammary gland. Stem Cells 28: 535–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land CE, McGregor DH 1979. Breast cancer incidence among atomic bomb survivors: implications for radiobiologic risk at low doses. J Natl Cancer Inst 62: 17–21 [PubMed] [Google Scholar]
- Lee HJ, Gallego-Ortega D, Ledger A, Schramek D, Joshi P, Szwarc MM, Cho C, Lydon JP, Khokha R, Penninger JM, et al. 2013. Progesterone drives mammary secretory differentiation via RankL-mediated induction of Elf5 in luminal progenitor cells. Development 140: 1397–1401 [DOI] [PubMed] [Google Scholar]
- Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA 2011. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121: 2750–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E, Vaillant F, Wu D, Forrest NC, Pal B, Hart AH, Asselin-Labat ML, Gyorki DE, Ward T, Partanen A, et al. 2009. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 15: 907–913 [DOI] [PubMed] [Google Scholar]
- Lim E, Wu D, Pal B, Bouras T, Asselin-Labat ML, Vaillant F, Yagita H, Lindeman GJ, Smyth GK, Visvader JE 2010. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res 12: R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Dontu G, Mantle ID, Patel S, Ahn NS, Jackson KW, Suri P, Wicha MS 2006. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res 66: 6063–6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, et al. 2014. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Reports 2: 78–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado HL, Kittrell FS, Edwards D, White AN, Atkinson RL, Rosen JM, Medina D, Lewis MT 2013. Separation by cell size enriches for mammary stem cell repopulation activity. Stem Cells Transl Med 2: 199–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarem M, Kannan N, Nguyen LV, Knapp DJ, Balani S, Prater MD, Stingl J, Raouf A, Nemirovsky O, Eirew P, et al. 2013. Developmental changes in the in vitro activated regenerative activity of primitive mammary epithelial cells. PLoS Biol 11: e1001630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133: 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Reinberg D 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama R, Choudhury S, Kowalczyk A, Bessarabova M, Beresford-Smith B, Conway T, Kaspi A, Wu Z, Nikolskaya T, Merino VF, et al. 2011. Epigenetic regulation of cell type-specific expression patterns in the human mammary epithelium. PLoS Genet 7: e1001369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Abt F, Milani E, Roloff T, Brinkhaus H, Duss S, Meyer DS, Klebba I, Balwierz PJ, van Nimwegen E, Bentires-Alj M 2013. Parity induces differentiation and reduces Wnt/Notch signaling ratio and proliferation potential of basal stem/progenitor cells isolated from mouse mammary epithelium. Breast Cancer Res 15: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchor L, Molyneux G, Mackay A, Magnay FA, Atienza M, Kendrick H, Nava-Rodrigues D, Lopez-Garcia MA, Milanezi F, Greenow K, et al. 2014. Identification of cellular and genetic drivers of breast cancer heterogeneity in genetically engineered mouse tumour models. J Pathol 10.1002/path.4345 [DOI] [PubMed] [Google Scholar]
- Michalak EM, Nacerddine K, Pietersen A, Beuger V, Pawlitzky I, Cornelissen-Steijger P, Wientjens E, Tanger E, Seibler J, van Lohuizen M, et al. 2013. Polycomb group gene Ezh2 regulates mammary gland morphogenesis and maintains the luminal progenitor pool. Stem Cells 31: 1910–1920 [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A 1999. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398: 708–713 [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ 2003. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425: 962–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux G, Geyer FC, Magnay FA, McCarthy A, Kendrick H, Natrajan R, Mackay A, Grigoriadis A, Tutt A, Ashworth A, et al. 2010. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 7: 403–417 [DOI] [PubMed] [Google Scholar]
- Moumen M, Chiche A, Decraene C, Petit V, Gandarillas A, Deugnier MA, Glukhova MA, Faraldo MM 2013. Myc is required for β-catenin-mediated mammary stem cell amplification and tumorigenesis. Mol Cancer 12: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassour M, Idoux-Gillet Y, Selmi A, Come C, Faraldo ML, Deugnier MA, Savagner P 2012. Slug controls stem/progenitor cell growth dynamics during mammary gland morphogenesis. PLoS ONE 7: e53498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LV, Makarem M, Carles A, Moksa M, Kannan N, Pandoh P, Eirew P, Osako T, Kardel M, Cheung AM, et al. 2014. Clonal analysis via barcoding reveals diverse growth and differentiation of transplanted mouse and human mammary stem cells. Cell Stem Cell 14: 253–263 [DOI] [PubMed] [Google Scholar]
- Oakes SR, Naylor MJ, Asselin-Labat ML, Blazek KD, Gardiner-Garden M, Hilton HN, Kazlauskas M, Pritchard MA, Chodosh LA, Pfeffer PL, et al. 2008. The Ets transcription factor Elf5 specifies mammary alveolar cell fate. Genes Dev 22: 581–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver CH, Khaled WT, Frend H, Nichols J, Watson CJ 2012. The Stat6-regulated KRAB domain zinc finger protein Zfp157 regulates the balance of lineages in mammary glands and compensates for loss of Gata-3. Genes Dev 26: 1086–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal B, Bouras T, Shi W, Vaillant F, Sheridan JM, Fu N, Breslin K, Jiang K, Ritchie ME, Young M, et al. 2013. Global changes in the mammary epigenome are induced by hormonal cues and coordinated by Ezh2. Cell Rep 3: 411–426 [DOI] [PubMed] [Google Scholar]
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP 2010. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell 140: 62–73 [DOI] [PubMed] [Google Scholar]
- Pietersen AM, Evers B, Prasad AA, Tanger E, Cornelissen-Steijger P, Jonkers J, van Lohuizen M 2008. Bmi1 regulates stem cells and proliferation and differentiation of committed cells in mammary epithelium. Curr Biol 18: 1094–1099 [DOI] [PubMed] [Google Scholar]
- Plaks V, Brenot A, Lawson DA, Linnemann JR, Van Kappel EC, Wong KC, de Sauvage F, Klein OD, Werb Z 2013. Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep 3: 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polyak K, Kalluri R 2010. The role of the microenvironment in mammary gland development and cancer. Cold Spring Harb Perspect Biol 2: a003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Perou CM 2011. Deconstructing the molecular portraits of breast cancer. Mol Oncol 5: 5–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM 2010. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 12: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proia TA, Keller PJ, Gupta PB, Klebba I, Jones AD, Sedic M, Gilmore H, Tung N, Naber SP, Schnitt S, et al. 2011. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell 8: 149–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raouf A, Zhao Y, To K, Stingl J, Delaney A, Barbara M, Iscove N, Jones S, McKinney S, Emerman J, et al. 2008. Transcriptome analysis of the normal human mammary cell commitment and differentiation process. Cell Stem Cell 3: 109–118 [DOI] [PubMed] [Google Scholar]
- Regan JL, Kendrick H, Magnay FA, Vafaizadeh V, Groner B, Smalley MJ 2012. c-Kit is required for growth and survival of the cells of origin of Brca1-mutation-associated breast cancer. Oncogene 31: 869–883 [DOI] [PubMed] [Google Scholar]
- Regan JL, Sourisseau T, Soady K, Kendrick H, McCarthy A, Tang C, Brennan K, Linardopoulos S, White DE, Smalley MJ 2013. Aurora A kinase regulates mammary epithelial cell fate by determining mitotic spindle orientation in a Notch-dependent manner. Cell Rep 4: 110–123 [DOI] [PubMed] [Google Scholar]
- Rios AC, Fu NY, Lindeman GJ, Visvader JE 2014. In situ identification of bipotent stem cells in the mammary gland. Nature 506: 322–327 [DOI] [PubMed] [Google Scholar]
- Roy S, Gascard P, Dumont N, Zhao J, Pan D, Petrie S, Margeta M, Tlsty TD 2013. Rare somatic cells from human breast tissue exhibit extensive lineage plasticity. Proc Natl Acad Sci 110: 4598–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale S, Lafkas D, Artavanis-Tsakonas S 2013. Notch2 genetic fate mapping reveals two previously unrecognized mammary epithelial lineages. Nat Cell Biol 15: 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedin P 2006. Pregnancy-associated breast cancer and metastasis. Nat Rev Cancer 6: 281–291 [DOI] [PubMed] [Google Scholar]
- Schramek D, Leibbrandt A, Sigl V, Kenner L, Pospisilik JA, Lee HJ, Hanada R, Joshi PA, Aliprantis A, Glimcher L, et al. 2010. Osteoclast differentiation factor RANKL controls development of progestin-driven mammary cancer. Nature 468: 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ, Visvader JE 2006. Generation of a functional mammary gland from a single stem cell. Nature 439: 84–88 [DOI] [PubMed] [Google Scholar]
- Shehata M, Teschendorff A, Sharp G, Novcic N, Russell A, Avril S, Prater M, Eirew P, Caldas C, Watson CJ, et al. 2012. Phenotypic and functional characterization of the luminal cell hierarchy of the mammary gland. Breast Cancer Res 14: R134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwko SK, Dong J, Lewis MT, Liu H, Hilsenbeck SG, Li Y 2008. Evidence that an early pregnancy causes a persistent decrease in the number of functional mammary epithelial stem cells–implications for pregnancy-induced protection against breast cancer. Stem Cells 26: 3205–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman KE, Kendrick H, Robertson D, Isacke CM, Ashworth A, Smalley MJ 2007. Dissociation of estrogen receptor expression and in vivo stem cell activity in the mammary gland. J Cell Biol 176: 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley MJ, Kendrick H, Sheridan JM, Regan JL, Prater MD, Lindeman GJ, Watson CJ, Visvader JE, Stingl J 2012. Isolation of mouse mammary epithelial subpopulations: a comparison of leading methods. J Mammary Gland Biol Neoplasia 17: 91–97 [DOI] [PubMed] [Google Scholar]
- Smith GH 2005. Label-retaining epithelial cells in mouse mammary gland divide asymmetrically and retain their template DNA strands. Development 132: 681–687 [DOI] [PubMed] [Google Scholar]
- Smith GH, Medina D 1988. A morphologically distinct candidate for an epithelial stem cell in mouse mammary gland. J Cell Sci 90: 173–183 [DOI] [PubMed] [Google Scholar]
- Spike BT, Engle DD, Lin JC, Cheung SK, La J, Wahl GM 2012. A mammary stem cell population identified and characterized in late embryogenesis reveals similarities to human breast cancer. Cell Stem Cell 10: 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staniszewska AD, Pensa S, Caffarel MM, Anderson LH, Poli V, Watson CJ 2012. Stat3 is required to maintain the full differentiation potential of mammary stem cells and the proliferative potential of mammary luminal progenitors. PLoS ONE 7: e52608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J, Eaves CJ, Kuusk U, Emerman JT 1998. Phenotypic and functional characterization in vitro of a multipotent epithelial cell present in the normal adult human breast. Differentiation 63: 201–213 [DOI] [PubMed] [Google Scholar]
- Stingl J, Eaves CJ, Zandieh I, Emerman JT 2001. Characterization of bipotent mammary epithelial progenitor cells in normal adult human breast tissue. Breast Cancer Res Treat 67: 93–109 [DOI] [PubMed] [Google Scholar]
- Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, Li HI, Eaves CJ 2006. Purification and unique properties of mammary epithelial stem cells. Nature 439: 993–997 [DOI] [PubMed] [Google Scholar]
- Taddei I, Deugnier MA, Faraldo MM, Petit V, Bouvard D, Medina D, Fassler R, Thiery JP, Glukhova MA 2008. β1 integrin deletion from the basal compartment of the mammary epithelium affects stem cells. Nat Cell Biol 10: 716–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA 2011. Interconversion between intestinal stem cell populations in distinct niches. Science 334: 1420–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant F, Lindeman GJ, Visvader JE 2011. Jekyll or Hyde: does Matrigel provide a more or less physiological environment in mammary repopulating assays? Breast Cancer Res 13: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Bowman AN, Nusse R 2012. Developmental stage and time dictate the fate of Wnt/β-catenin-responsive stem cells in the mammary gland. Cell Stem Cell 11: 387–400 [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C 2011. Distinct stem cells contribute to mammary gland development and maintenance. Nature 479: 189–193 [DOI] [PubMed] [Google Scholar]
- Villadsen R, Fridriksdottir AJ, Ronnov-Jessen L, Gudjonsson T, Rank F, LaBarge MA, Bissell MJ, Petersen OW 2007. Evidence for a stem cell hierarchy in the adult human breast. J Cell Biol 177: 87–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvader JE 2011. Cells of origin in cancer. Nature 469: 314–322 [DOI] [PubMed] [Google Scholar]
- Wagner KU, Boulanger CA, Henry MD, Sgagias M, Hennighausen L, Smith GH 2002. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development 129: 1377–1386 [DOI] [PubMed] [Google Scholar]
- Wansbury O, Mackay A, Kogata N, Mitsopoulos C, Kendrick H, Davidson K, Ruhrberg C, Reis-Filho JS, Smalley MJ, Zvelebil M, et al. 2011. Transcriptome analysis of embryonic mammary cells reveals insights into mammary lineage establishment. Breast Cancer Res 13: R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JM, Daniel CW 1983. Mammary ductal elongation: differentiation of myoepithelium and basal lamina during branching morphogenesis. Dev Biol 97: 274–290 [DOI] [PubMed] [Google Scholar]
- Yalcin-Ozuysal O, Fiche M, Guitierrez M, Wagner KU, Raffoul W, Brisken C 2010. Antagonistic roles of Notch and p63 in controlling mammary epithelial cell fates. Cell Death Differ 17: 1600–1612 [DOI] [PubMed] [Google Scholar]
- Yamaji D, Na R, Feuermann Y, Pechhold S, Chen W, Robinson GW, Hennighausen L 2009. Development of mammary luminal progenitor cells is controlled by the transcription factor STAT5A. Genes Dev 23: 2382–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng YA, Nusse R 2010. Wnt proteins are self-renewal factors for mammary stem cells and promote their long-term expansion in culture. Cell Stem Cell 6: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeps N, Bentel JM, Papadimitriou JM, Dawkins HJ 1999. Murine progesterone receptor expression in proliferating mammary epithelial cells during normal pubertal development and adult estrous cycle. Association with erα and erβ status. J Histochem Cytochem 47: 1323–1330 [DOI] [PubMed] [Google Scholar]