Here, Park et al. used 3D imaging and ChIP-3C techniques to investigate the topographies of the immunoglobulin (Ig) genes and transcripts during B-cell development. The authors show that active Ig genes residing on three different chromosomes colocalize in transcription factories, often near the nuclear periphery. Furthermore, active Ig genes display trans-chromosomal enhancer interactions and frequently share interchromatin trafficking channels. These results reveal tight interconnections between nuclear organization and gene expression during maximal levels of antibody production in plasma cells.
Keywords: immunoglobulin genes, transcription factories, nuclear organization, enhancers, long-range interactions, RNA trafficking
Abstract
To understand the relationships between nuclear organization and gene expression in a model system, we employed three-dimensional imaging and chromatin immunoprecipitation (ChIP)-chromosome conformation capture (3C) techniques to investigate the topographies of the immunoglobulin (Ig) genes and transcripts during B-cell development. Remarkably, in plasma cells, when antibody synthesis peaks, active Ig genes residing on three different chromosomes exhibit pronounced colocalizations in transcription factories, often near the nuclear periphery, and display trans-chromosomal enhancer interactions, and their transcripts frequently share interchromatin trafficking channels. Conceptually, these features of nuclear organization maximize coordinated transcriptional and transcript trafficking control for potentiating the optimal cytoplasmic assembly of the resulting translation products into protein multimers.
During the course of B-cell development, the genes encoding immunoglobulin (Ig) heavy (H) and light (L) chains become activated by the regulated gene rearrangement of variable (V), diverse (D), and joining (J) region gene segments (Schatz and Ji 2011; Bossen et al. 2012). The hallmark of successful B-cell terminal differentiation is the plasma cell stage, in which each cell is estimated to secrete its own mass in antibody molecules every day (Hendershot and Sitia 2004). The mechanisms behind such remarkably high levels of gene expression used by these “workhorses” of the adaptive immune system have remained elusive. We investigated this question by examining the nuclear organization of transcribing Igκ, IgH, and IgJ genes and their resulting transcripts in mouse plasma cells and their B-cell progenitors and discovered an unsuspected striking order in these components. These genes reside on three different chromosomes, and in plasma cells together encode secreted IgM pentamers (H2L2)5(J)1 (Shimizu et al. 2009).
We report here the results of studies employing three-dimensional (3D) imaging and chromatin immunoprecipitation (ChIP)-chromosome conformation capture (3C) on the nuclear organization of active Ig genes and their transcripts in plasma cells. Strikingly, actively transcribing Igκ, IgH, and IgJ genes from three different chromosomes exhibit pronounced colocalizations at the same transcription factories, which are often located near the nuclear periphery. Long-range trans-chromosomal enhancer interactions accompanied such Ig gene pairing, nuclear positioning, and transcriptional activity. Furthermore, functionally rearranged IgH gene alleles preferentially colocalize with other functional Ig genes. Moreover, transcripts from peripherally positioned Ig genes exhibit restricted intranuclear accumulations, and transcripts originating from interiorly colocalized Ig genes largely share common interchromatin channels for their trafficking to the cytoplasm through nuclear pores. These results reveal tight interconnections between nuclear organization and gene expression during maximal levels of antibody production in plasma cells.
Results and Discussion
Active Ig genes of plasma cells exhibit pronounced colocalizations in transcription factories, often near the nuclear periphery
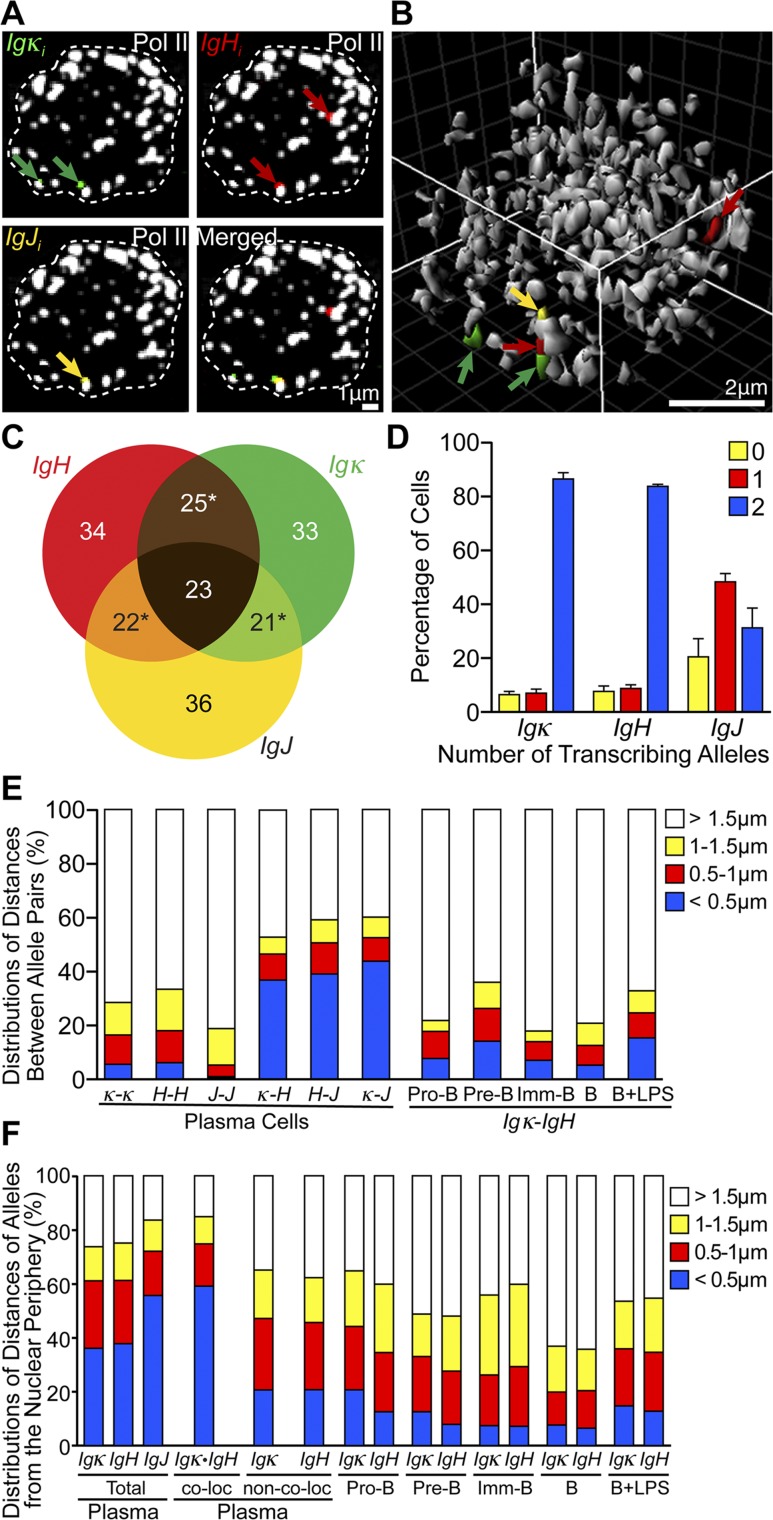
Transcription of genes by RNA polymerase II (Pol II) occurs at transcription factories, which are clusters of approximately eight Pol II molecules that colocalize as nuclear foci in fixed or living cells (Cook 1999; Edelman and Fraser 2012; Cisse et al. 2013; Ghamari et al. 2013). We wondered whether the different actively transcribing Ig genes of plasma cells might co-occupy the same transcription factories at frequencies markedly enhanced compared with the levels of gene colocalization seen in earlier B-cell developmental stages or those previously observed in less active expression systems (Osborne et al. 2007; Schoenfelder et al. 2010). To investigate these questions, we used multicolor, 3D RNA immunofluorescent in situ hybridization (immuno-FISH) using antisense riboprobes complementary to intronic sequences of Ig gene primary transcripts and antibodies against Pol II. Analyses of confocal optical sections and 3D image reconstructions of single plasma cell nuclei revealed a remarkable degree of colocalization of different Ig genes’ primary transcripts at or near the same transcription factories (Fig. 1A,B), which we quantified among hundreds of plasma cell nuclei (Supplemental Table 1). Strikingly, for the subset of these nuclei that transcribed at least one allele of each of the Igκ, IgH, and IgJ genes (73%), at least 44% exhibited colocalization of transcript signals between two different Ig genes, while 23% manifested colocalization of such signals between three different Ig genes (Fig. 1C). Importantly, by performing sequential 3D RNA and DNA FISH for each gene as well as multicolor 3D DNA FISH or immuno-FISH, we demonstrated that these primary transcript signals actually reflect gene locus positions within transcription factories (Fig. 2; Supplemental Fig. 2). This frequency of triplet gene colocalization is >20-fold higher than ever noticed before for genes transcribed by Pol II, and transcribing α-globin and β-globin genes in mouse erythroblasts only exhibit ∼7% colocalization in transcription factories (Schoenfelder et al. 2010). Furthermore, although the Igκ and IgH genes were mainly biallelically transcribed (Fig. 1D; Supplemental Table 2), their homologous alleles exhibited statistically significant much lower colocalizations (Fig. 1E, left, <0.5 μm apart class, blue coding; Supplemental Table 3A,C). Hence, there must be a sorting mechanism in plasma cells that gathers together transcribing heterologous Ig genes into the same transcription factories or precludes significant colocalization of homologous alleles (or both). Moreover, Igκ and IgH genes showed a statistically significant lower colocalization in earlier B-cell stages, when their expression was much lower (Fig. 1E, right; Supplemental Table 3B,C).
Figure 1.
Different transcribing Ig genes of plasma cells exhibit a pronounced colocalization in transcription factories, often near the nuclear periphery. (A) Representative four-color 3D RNA immuno-FISH results of the same 0.3-μm optical section of a plasma cell nucleus, outlined by white dashed lines. (Green) Igκi; (red) IgHi; (yellow) IgJi; (white) α-Pol II. As expected, ≥89% of Ig gene transcription was associated with transcription factories (Supplemental Fig. 1A), which averaged 155 ± 17 in number per plasma cell nucleus (Supplemental Fig. 1B). (B) Reconstructed 3D image of the same plasma cell nucleus shown in A, with the same color coding. (C) The percentages of cells that possess the indicated primary transcript localized <0.5 μm from a different Ig gene’s primary transcript are depicted in the overlapping images as quantified from 3D RNA FISH results for the 342 plasma cells examined that transcribed at least one allele of each of the Igκ, IgH, and IgJ genes. Data are from 470 cells analyzed in three independent experiments (see Supplemental Table 1 for primary data). The sums of the percentages in any one-gene circle total >100% because the same cells sometimes transcribed two different pairs of colocalized Ig genes (asterisks). (D–F) Bar graphs of 3D RNA FISH results from 607 pro-B cells, 618 pre-B cells, 608 imm-B cells, 625 B cells, 614 B + LPS cells, and 614 plasma cells analyzed from three independent experiments (see also Supplemental Tables 2–4 for primary data and P-values from χ2 statistical tests). (D) The total number of transcribing alleles of Igκ, IgH, and IgJ genes per plasma cell and the percentage of cells for each case (data are represented as means ± SD). (E) Distributions of distances between transcribing homologous and/or heterologous allele gene pairs in plasma and other B cells. For homologous allele pairing, 374, 362, and 169 plasma cells that exhibited biallelic transcription for Igκ, IgH, and IgJ, respectively, were analyzed from three independent experiments. (F) Transcribed allele separation distances from the nuclear periphery in plasma and other B cells with and/or without consideration of allele pairing. For the distribution of distances with consideration of allele pairing, 307 plasma cells that had one colocalized Igκ•IgH allele pair were analyzed from three independent experiments.
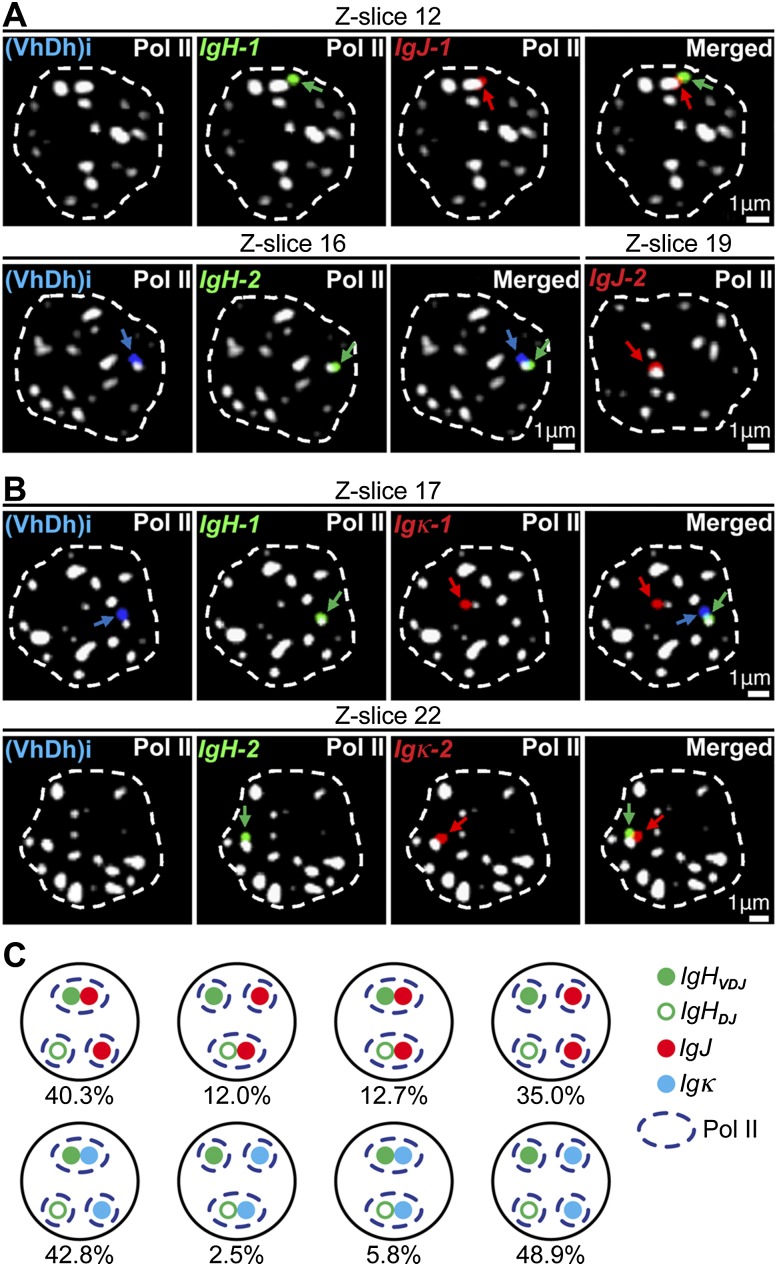
Figure 2.
Functional Ig genes preferentially colocalize in transcription factories. (A,B) Four-color 3D DNA immuno-FISH images of selected 0.3-μm-thick optical sections (Z-slices) of the same plasma cell nuclei (outlined by white dashed lines) from either wild-type (WT) (A) or homozygous Vκ8-Jκ5 knock-in mice (B). (Blue) IgH (VhDh)i; (green) IgH; (red) IgJ or Igκ; (white) α-Pol II. (C) Cartoon summarizing the quantification of DJ and VDJ rearranged IgH alleles that were localized within <1.0 μm from both Pol II and either IgJ or Igκ alleles. Data are from 300 and 306 plasma cells analyzed from wild-type and homozygous Vκ8-Jκ5 knock-in mice, respectively, in three independent experiments (see also Supplemental Table 5 for primary data and P-values from χ2 statistical tests).
Interestingly, at the beginning of B-cell development, when the Ig genes are silent, they are located at the nuclear periphery, while in pro-B cells poised for V(D)J joining and transcription, these genes are located at the nuclear interior (Kosak et al. 2002). However, in plasma cells, when the Ig genes are fully transcriptionally activated, we found that the transcribing Ig genes were localized preferentially near the nuclear periphery, which was more pronounced for colocalized gene pairs and reduced in earlier B-cell stages in a statistically significant fashion (Fig. 1F; Supplemental Table 4). Although peripheral nuclear locations are commonly associated with heterochromatin and gene silencing, with characteristic sequence-dependent interactions at the nuclear lamina (Zullo et al. 2012), certain active genes are known to exhibit close proximities to nuclear pores as opposed to the nuclear lamina in this locale (Takizawa et al. 2008; Blobel 2010). Notably, both gene colocalization and peripheral location could be induced several-fold by lipopolysaccharide (LPS) treatment of cultured splenic B cells (Fig. 1E,F), which also increases Ig gene transcription several-fold but at levels <30-fold those exhibited by plasma cells because such culturing only partially differentiates B cells into antibody-secreting plasmablasts. We conclude that the pronounced colocalization of Ig genes in transcription factories (often near the nuclear periphery) that is seen in plasma cells correlates with their ultrahigh levels of transcription for antibody production. Possibly, the observed peripheral targeting of the Ig genes may be linked to the specialized transcription and splicing apparati associated with the generation of mRNA species encoding secreted molecules (Martincic et al. 2009; Benson et al. 2012). Furthermore, the high frequency of peripheral gene colocalizations exhibited by plasma cells is specific to the Ig genes because control 3D RNA FISH experiments that examine primary transcripts from the β-actin (ACTB) gene or plasma cell-specific BLIMP-1 (PRDM1) and syndecan-1 (SDC1) genes revealed no peripheral enrichment and only very low colocalization with the actively transcribed Ig genes, with the exception of SDC1, which resides on chromosome 12, ∼100 Mb away from IgH (Supplemental Fig. 3).
Functionally rearranged IgH alleles preferentially colocalize in transcription factories with other functional Ig genes
Interestingly, >80% of the IgH and Igκ genes are biallelically transcribed in plasma cells (Fig. 1D). However, because of allelic exclusion, only one of the two alleles of each IgH and Igκ gene will be expressed as functional antibody chains in single cells (Vettermann and Schlissel 2010), while the transcripts arising from incompletely or nonfunctionally rearranged alleles will be selectively degraded in the cytoplasm (Tinguely et al. 2012). If pairing of different Ig genes in the same transcription factories is functionally linked to optimization of antibody production, then one would predict that functionally rearranged Ig genes would be preferred partners in these structures. To address this prediction, we used four-color 3D DNA immuno-FISH to examine the frequencies of colocalization of IgH genes with either functional IgJ or Igκ alleles in transcription factories (Fig. 2A,B). We focused on the subset of plasma cells that possessed one DJ and one VDJ rearranged IgH allele using one hybridization probe that detects all IgH alleles and another that was specific for the Vh–Dh intervening region [(VhDh)i], which is deleted upon V-to-DJ rearrangement (Bossen et al. 2012). In addition, for these experiments, we employed homozygous Vκ8-Jκ5 knock-in mice to insure that both Igκ alleles were functional, whereas IgJ alleles do not undergo gene rearrangement and are functional “as is.” Strikingly, 40.3% and 42.8% of these nuclei, respectively, exhibited colocalization of the VDJ rearranged IgH allele with a functional IgJ and Igκ allele in transcription factories, whereas only 12% and 2.5% exhibited corresponding pairing of the DJ but not the VDJ rearranged allele with these other Ig genes (Fig. 2C). Statistical analysis of these results supports the conclusion that functional Ig genes colocalize preferentially (Supplemental Table 5).
Ig genes colocalizing in the same transcription factories exhibit trans-chromosomal enhancer interactions
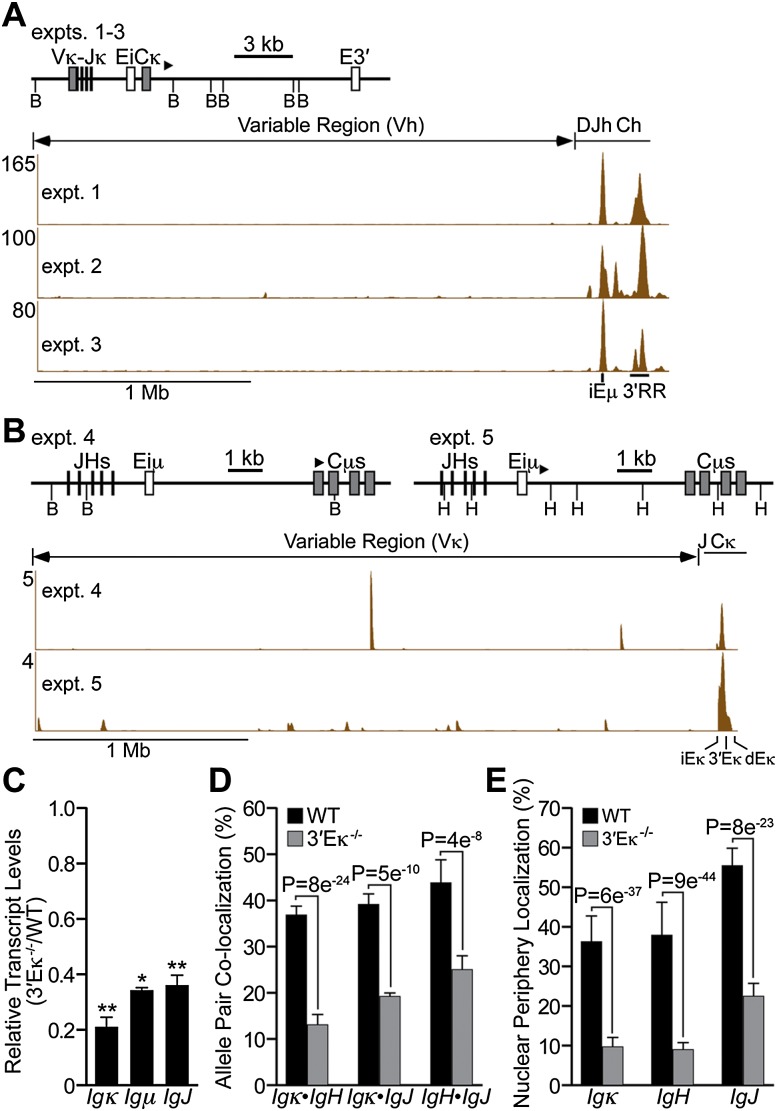
To identify genomic sequences that may be responsible for localizing the Igκ and IgH genes in the same transcription factories, we performed ChIP experiments with antibodies against Pol II followed by 3C and high-throughput DNA sequencing (Pol II ChIP-3C-seq) (Dekker et al. 2002; Schoenfelder et al. 2010). This approach revealed proximity between the Igκ gene and the IgH gene’s enhancer regions (iEμ and 3′RR) (Fig. 3A) and between the IgH gene and sequences overlapping with all three of the Igκ gene’s enhancers as well as different Vκ regions among the experimental samples (Fig. 3B). Collectively, these results demonstrate that neighboring trans-chromosomal sequences between the Ig genes in the same transcription factories include their enhancer elements. Furthermore, as a control, we performed Pol II ChIP-3C-seq experiments with pre-B cells and found that trans-chromosomal enhancer interactions made between the Igκ and IgH genes correlated with the differentiation-dependent functions of these elements (Supplemental Fig. 4).
Figure 3.
The Ig gene enhancers mediate trans-chromosomal interactions at transcription factories, and deletion of 3′Eκ affects the transcript levels and nuclear organization of multiple Ig genes. (A,B) Plasmablasts (LPS-stimulated splenic B cells) were subjected to α-Pol II ChIP-3C-seq analyses (samples used in experiments [expt.] 1 and 2 were from homozygous Vκ8-Jκ5 knock-in mice; samples used in experiments 3–5 were from wild-type (WT) mice; enzyme cleavages used BamHI in experiments 1–4 and HindIII in experiment 5). The biotinylated bait primers used for extensions are shown as bold arrowheads above the restriction maps. (A) Igκ sequences used as bait with interrogation of IgH locus-interacting sequences. Shown on the X-axis are the positions of Vh, Dh, Jh, and Ch regions (top) and iEμ and 3′RR enhancers (bottom). Depicted on the Y-axis are normalized sequence counts per million reads mapped to the IgH locus from the indicated experimental samples. (B) IgH sequences used as bait with interrogation of Igκ locus-interacting sequences. Shown on the X-axis are the positions of the Vκ and JCκ regions (top) and κ enhancers (bottom). Depicted on the Y-axis are normalized sequence counts per million reads mapped to the Igκ locus from the indicated experimental samples. (C–E) Analyses of plasma cells from wild-type and 3′Eκ−/− mice (data are represented as means ± SD from three independent experiments). (C) Real-time PCR assays were performed to measure the levels of Ig gene transcripts. SD; (*) P < 0.05; (**) P < 0.01. (D,E) Bar graphs of 3D RNA FISH results from 614 and 629 cells analyzed from wild-type and 3′Eκ−/− mice, respectively (see also Supplemental Table 6 for primary data and P-values from χ2 statistical tests). (D) Colocalization frequencies between transcribing heterologous allele gene pairs (<0.5 μm). (E) Positioning for alleles separated <0.5 μm from the nuclear periphery.
To further investigate the functional significance of trans-chromosomal enhancer interactions in plasma cells, we examined the importance of the 3′Eκ enhancer on the transcription and nuclear organization of the Ig genes. Previous studies have shown that 3′Eκ is not essential for Igκ gene rearrangement, normal activated B-cell transcription (Gorman et al. 1996), or normal expansion of plasma cells (Zhou et al. 2012). Nevertheless, relative to wild-type controls, each Ig gene of plasma cells from 3′Eκ−/− mice exhibited reduced transcriptional activity, colocalization in transcription factories, and positioning near the nuclear periphery (Fig. 3C–E; Supplemental Table 6). Furthermore, the results of 3C experiments confirmed the importance of 3′Eκ in fostering proximity between the Igκ locus and iEμ and 3′RR in the IgH locus (Supplemental Fig. 5). These results suggest that trans-chromosomal enhancer interactions contribute to the steady-state mRNA levels of the Ig genes by regulating their nuclear organization. However, other mechanisms that depend on post-transcriptional feedback may exist to regulate the proportionalities of Ig gene mRNA levels either exclusively or in combination with such nuclear organization events.
The mechanisms that drive the colocalization of multiple genes into single transcription factories are not known, but it has been proposed that such gene colocalizations should favor the coordinate regulation of the cohabiting genes by common shared transcription factors (Schoenfelder et al. 2010). Because Ig gene enhancers use several common transcription factors that include E2A, IRF4, and PU.1 (Lim et al. 2009; Lin et al. 2012; Ochiai et al. 2013), we propose that continual dynamic Ig gene sorting to different transcription factories during transcription cycles may result in eventual high Ig gene colocalization owing to the beneficial cooperative sharing of those factors. This mechanism for driving the colocalization of multiple genes into single factories has been supported by mathematical modeling when essential shared transcription factors are limiting (Kang et al. 2011). This hypothesis predicts that overexpression of a previously limiting transcription factor may result in reduced Ig gene colocalization in the same transcription factories, while underexpression of a previously abundant transcription factor may result in enhanced Ig gene colocalization.
The nuclear organization of different Ig gene transcripts exhibits features favorable for their efficient export to the cytoplasm
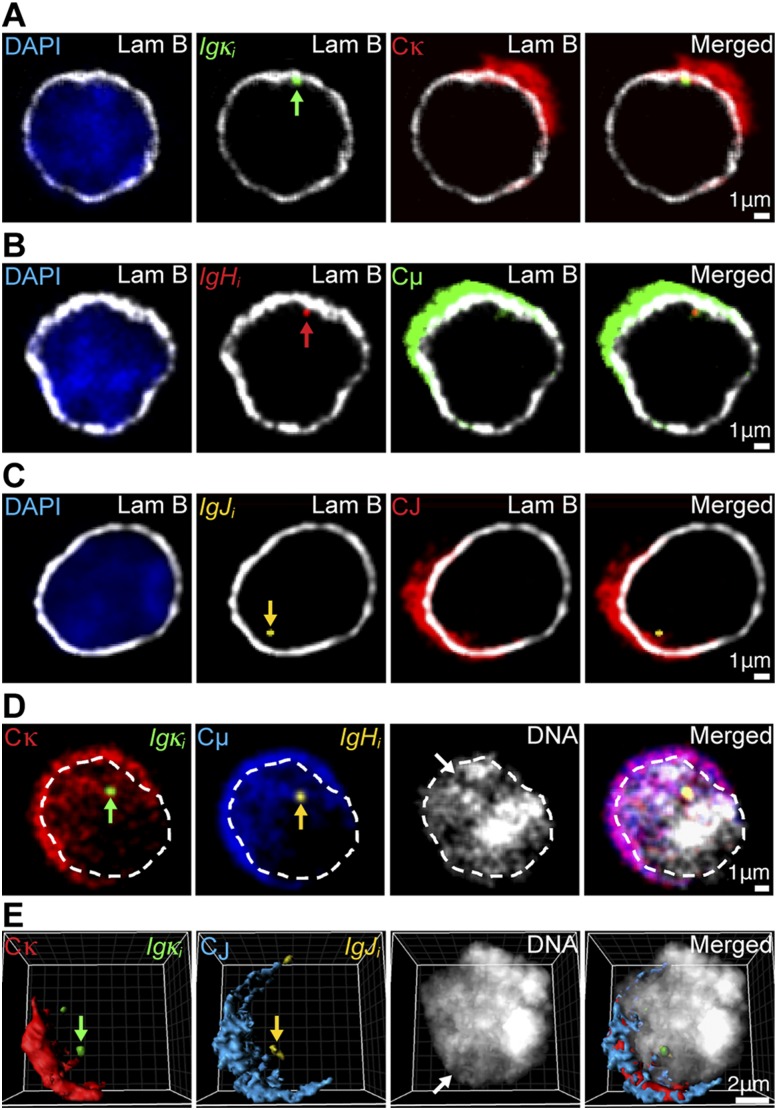
The synthesis of antibody molecules depends on the Ig genes’ transcripts being successfully capped, spliced, polyadenylated, and trafficked from their sites of synthesis to the nuclear pores for transport to the cytoplasm (Grünwald and Singer 2010; Hübner and Spector 2010; Mor et al. 2010; Noble and Wente 2010; Stewart 2010; Pederson 2011). To investigate the possibility that Ig gene nuclear organization may contribute favorably to the trafficking of their transcripts for nuclear export, we performed multicolor 3D RNA immuno-FISH to decorate simultaneously intron- and exon-containing RNA transcripts for specific Ig genes in plasma cells along with the nuclear envelope lamina network or total DNA. This approach revealed crescent-shaped images for the Ig mRNA hybridization patterns in the cells’ cytoplasm (Fig. 4), which corresponded to the endoplasmic reticulum (ER)-rich sides of these cells, as demonstrated by significant colocalization of the ER marker protein disulfide isomerase (PDI) (Fricker et al. 1997), with Ig gene mRNA, poly(A)+ RNA, and rRNA hybridization patterns (Supplemental Fig. 6). Transcripts arising from peripherally localized Ig genes appeared to exhibit only modest intranuclear accumulation around their sites of synthesis (Fig. 4A–C; Supplemental Fig. 7A,B). Interestingly, these genes and even those colocalized throughout the nucleus exhibited statistically significant preferred locations facing the ER-rich sides of cells (Supplemental Fig. 8A–C; Supplemental Tables 7, 8) as if they are gated to neighboring nuclear pores according to the original hypothesis of Blobel (1985). Significantly, transcribing control genes encoding β-actin, BLIMP-1, and SDC1 did not reveal a significant preference to be located in nuclei toward these crescents (Supplemental Fig. 8A,D; Supplemental Table 9). Moreover, nuclear RNAs that arose from nuclear interior-colocalized Igκ and IgH or Igκ and IgJ genes appeared to often share common interchromatin channels for trafficking (Fig. 4D,E; Supplemental Fig. 7C, white arrows). Both inspection and quantification of the intranuclear distributions of Ig gene transcripts relative to DNA, lamin-B, and PDI in selected or serial confocal sections as well as 3D image reconstructions of plasma cells provided direct support for the sharing of common interchromatin RNA trafficking channels between different Ig gene transcripts arising from colocalized genes and for these RNA track signals indeed being within the nucleus and not due to cytoplasmic ER invaginations (Fig. 4; Supplemental Figs. 7, 9, 10; Fricker et al. 1997). Furthermore, as expected, total poly(A)+ RNA also used interchromatin channels for nuclear RNA trafficking (Supplemental Fig. 9B). We conclude that the pronounced colocalization of Ig genes in transcription factories of plasma cells, which can occur near either the periphery or nuclear interior, is a feature favorable for the trafficking of their mRNAs for export on the ER-rich sides of cells, thereby potentiating the optimal assembly of their translation products into antibody molecules in the cytoplasm.
Figure 4.
Transcripts originating from different Ig genes exhibit features of nuclear organization favorable for export to the cytoplasm. (A–C) Representative four-color 3D RNA immuno-FISH images of the same 0.3-μm optical sections of individual plasma cells with peripherally localized transcribing Ig genes. Primary transcripts are indicted by green, red, or yellow arrows. (A) (Blue) DAPI; (white) α-Lam B; (green) Igκi; (red) Cκ. (B) (Blue) DAPI; (white) α-Lam B; (red) IgHi; (green) Cμ. (C) (Blue) DAPI; (white) α-Lam B; (yellow) IgJi; (red) CJ. (D,E) (Red) Cκ; (green) Igκi; (blue) Cμ or CJ; (yellow) IgHi or IgJi; (white) DAPI. White arrows depict interchromatin channels occupied by processed Ig gene transcripts. Primary transcripts of colocalized alleles are indicated by green or yellow arrows. (D) Representative five-color 3D RNA FISH images of the same 0.3-μm optical section of a plasma cell, with interiorly colocalized Ig genes and the nucleus outlined by white dashed lines. (E) Representative reconstructed five-color 3D RNA FISH image of a plasma cell with interiorly colocalized Ig genes.
In conclusion, our results reveal tight interconnections between nuclear organization and gene expression, which correlate with maximal antibody production specifically in plasma cells during B-cell development (schematically presented in Supplemental Fig. 11). The functional significance of colocalizing multiple genes encoding different subunits of a multimeric protein in transcription factories is likely important for coordinating not only their transcriptional control (Schoenfelder et al. 2010) but also the trafficking of their transcripts for export in close juxtaposition to the cytoplasm. Such coordination of transcript trafficking might occur by some form of a nuclear pore gene gating mechanism (Blobel 1985) or may simply proceed by multiple Ig gene transcripts emanating from shared transcription factories flowing through common interchromatin channels on their way to the cytoplasm. The preferential location of Ig genes near the nuclear periphery on the ER-rich sides of plasma cells likely further contributes to maximizing antibody synthesis. Conceptually, these features should provide a favorable biological microenvironment in the nucleus for the efficient synthesis of any heteromeric protein.
Materials and methods
Mouse lines
All strains were used in accordance with protocols approved by the University of Texas Southwestern Medical Center Institutional Animal Care and Use Committee (IACUC).
3D RNA immuno-FISH
Multicolor 3D RNA FISH or immuno-FISH used published techniques (Osborne et al. 2007; Schoenfelder et al. 2010), which were modified as detailed in the Supplemental Material.
ChIP-3C-seq
Pol II ChIP-3C-seq was performed by modifying a protocol kindly provided by Peter Fraser (Schoenfelder et al. 2010).
Additional materials and methods are provided in the Supplemental Material.
Acknowledgments
We are indebted to Peter Fraser, Katherine Luby-Phelps, and Joachim Seemann for contributing advice, antibodies, and protocols; Martin Weigert and Mark Schlissel for providing animal models; and Jose Cabrera for expert graphic illustrations. This investigation was supported by grants GM29935 and AI067906 from the National Institutes of Health and grant I-0823 from the Robert A. Welch Foundation to W.T.G., and by grant 31270928 from the National Natural Science Foundation of China to Y.X.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.237479.114.
References
- Benson MJ, Aijo T, Chang X, Gagnon J, Pape UJ, Anantharaman V, Aravind L, Pursiheimo P, Oberdoerffer S, Liu XS, et al. 2012. Heterogenous nuclear ribonucleoprotein L-like (hnRNPLL) and elongation factor, RNA polymerase II, 2 (ELL2) are regulators of mRNA processing in plasma cells. Proc Natl Acad Sci 109: 16252–16257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G 1985. Gene gating: a hypothesis. Proc Natl Acad Sci 82: 8527–8529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G 2010. Three-dimensional organization of chromatids by nuclear envelope-associated structures. Cold Spring Harb Symp Quant Biol 75: 545–554 [DOI] [PubMed] [Google Scholar]
- Bossen C, Mansson R, Murre C 2012. Chromatin topology and the regulation of antigen receptor assembly. Annu Rev Immunol 30: 337–356 [DOI] [PubMed] [Google Scholar]
- Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq L, Hajj B, Dahan M, Darzacq X 2013. Real-time dynamics of RNA polymerase II clustering in live human cells. Science 341: 664–667 [DOI] [PubMed] [Google Scholar]
- Cook PR 1999. The organization of replication and transcription. Science 284: 1790–1795 [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N 2002. Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Edelman LB, Fraser P 2012. Transcription factories: genetic programming in three dimensions. Curr Opin Genet Dev 22: 110–114 [DOI] [PubMed] [Google Scholar]
- Fricker M, Hollinshead M, White N, Vaux D 1997. Interphase nuclei of many cell types contain deep, dynamic, tubular membrane-bound invaginations of the nuclear envelope. J Cell Biol 136: 531–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghamari A, van de Corput MPC, Thongjuea S, van Cappellen WA, van Ijcken W, van Haren J, Soler E, Eick D, Lenhard B, Grosveld FG 2013. In vivo live imaging of RNA polymerase II transcription factories in primary cells. Genes Dev 27: 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman JR, van der Stoep N, Monroe R, Cogne M, Davidson L, Alt FW 1996. The Igκ 3′ enhancer influences the ratio of Igκ versus Igλ B lymphocytes. Immunity 5: 241–252 [DOI] [PubMed] [Google Scholar]
- Grünwald D, Singer RH 2010. In vivo imaging of labeled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature 467: 604–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot LM, Sitia R 2004. Immunoglobulin assembly and secretion. In Molecular biology of B cells (ed. T Honjo T, et al.), pp. 261–273. Elsevier, London [Google Scholar]
- Hübner MR, Spector DL 2010. Chromatin dynamics. Ann Rev Biophys 39: 471–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Xu B, Yao Y, Lin W, Hennessy C, Fraser P, Feng J 2011. A dynamical model reveals gene co-localizations in nucleus. Plos Comp Biol 7: e1002094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296: 158–162 [DOI] [PubMed] [Google Scholar]
- Lim J-H, Kim H-G, Park S-K, Kang C-J 2009. The promoter of the immunoglobulin J chain gene receives its authentic enhancer activity through the abutting Mef2 and PU.1 sites in a DNA-looping interaction. J Mol Biol 390: 339–352 [DOI] [PubMed] [Google Scholar]
- Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, Murre C 2012. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol 13: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C 2009. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol 10: 1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, Shav-Tal Y 2010. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat Cell Biol 12: 543–552 [DOI] [PubMed] [Google Scholar]
- Noble KN, Wente SR 2010. Nuclear RNA on the move. Nat Cell Biol 12: 525–527 [DOI] [PubMed] [Google Scholar]
- Ochiai K, Maienschein-Cline M, Simonetti G, Chen J, Rosenthal R, Brink R, Chong AS, Klein U, Dinner AR, Singh H, et al. 2013. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity 38: 918–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P 2007. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol 5: 1763–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. 2011. The nucleus introduced. In The nucleus (ed. Misteli T and Spector DL), pp. 1–16. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schatz DG, Ji Y 2011. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol 11: 251–263 [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42: 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y, Meunier L, Hendershot LM 2009. pERp1 is significantly up-regulated during plasma cell differentiation and contributes to the oxidative folding of immunoglobulin. Proc Natl Acad Sci 106: 17013–17018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M 2010. Nuclear export of mRNA. Trends Biochem Sci 36: 609–617 [DOI] [PubMed] [Google Scholar]
- Takizawa T, Meaburn KJ, Misteli T 2008. The meaning of gene positioning. Cell 135: 9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinguely A, Chemin G, Péron S, Sirac C, Reynaud S, Cogné M, Delpy L 2012. Cross talk between immunoglobulin heavy-chain transcription and RNA surveillance during B cell development. Mol Cell Biol 32: 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettermann C, Schlissel MS 2010. Allelic exclusion of immunoglobulin genes: models and mechanisms. Immunol Rev 237: 22–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Xiang Y, Ding X, Garrard WT 2012. A new hypersensitive site, HS10, and the enhancers, E3′ and Ed, differentially regulate Igκ gene expression. J Immunol 188: 2722–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, et al. 2012. DNA sequence-dependent compartmentalization and silencing at the nuclear lamina. Cell 149: 1474–1487 [DOI] [PubMed] [Google Scholar]