Autophagy is crucial for cellular homeostasis and plays important roles in tumorigenesis. FIP200 is an essential autophagy gene that promotes mammary tumorigenesis. Here, Wei et al. find that ablation of FIP200 reduces growth of established tumors. p62 knockdown or deficiency in established FIP200-null tumors dramatically impairs tumor growth, and this is associated with the up-regulated activation of the NF-κB pathway by p62. This study demonstrates that p62 and autophagy synergize to promote tumor growth.
Keywords: tumor growth, p62, autophagy, mouse models
Abstract
Autophagy is crucial for cellular homeostasis and plays important roles in tumorigenesis. FIP200 (FAK family-interacting protein of 200 kDa) is an essential autophagy gene required for autophagy induction, functioning in the ULK1–ATG13–FIP200 complex. Our previous studies showed that conditional knockout of FIP200 significantly suppressed mammary tumorigenesis, which was accompanied by accumulation of p62 in tumor cells. However, it is not clear whether FIP200 is also required for maintaining tumor growth and how the increased p62 level affects the growth in autophagy-deficient FIP200-null tumors in vivo. Here, we describe a new system to delete FIP200 in transformed mouse embryonic fibroblasts as well as mammary tumor cells following their transplantation and show that ablation of FIP200 significantly reduced growth of established tumors in vivo. Using similar strategies, we further showed that either p62 knockdown or p62 deficiency in established FIP200-null tumors dramatically impaired tumor growth. The stimulation of tumor growth by p62 accumulation in FIP200-null tumors is associated with the up-regulated activation of the NF-κB pathway by p62. Last, we showed that overexpression of the autophagy master regulator TFEBS142A increased the growth of established tumors, which correlated with the increased autophagy of the tumor cells. Together, our studies demonstrate that p62 and autophagy synergize to promote tumor growth, suggesting that inhibition of both pathways could be more effective than targeting either alone for cancer therapy.
Autophagy is an essential, conserved lysosomal degradation pathway that controls the homeostasis of the cytoplasm by bulk degradation of unnecessary or dysfunctional cellular organelles and protein aggregates (Yang and Klionsky 2010; Mizushima and Komatsu 2011; Rubinsztein et al. 2012). In addition to the previously described tumor-suppressive roles of autophagy (Liang et al. 1999; Qu et al. 2003; Yue et al. 2003; Mathew et al. 2009), the prosurvival function of autophagy under stress conditions (e.g., nutrient deprivation, hypoxia, and therapeutic stress) has been found to promote tumor growth and progression. Pharmacological or genetic inhibition of autophagy was shown to sensitize tumor cells to cytotoxic effects and chemotherapy and ionizing irradiation to enhance cancer treatments (Levine and Kroemer 2008; Rubinsztein et al. 2012; White 2012). Furthermore, several recent studies demonstrated that disruption of autophagy genes led to decreased proliferation and survival in oncogene transformed tumor cells and caused reduced tumor growth in vivo (Guo et al. 2011; Wei et al. 2011; Yang et al. 2011), further suggesting that the inhibition of autophagy may benefit cancer treatment. Nevertheless, still relatively little is known about the requirement and mechanisms of autophagy for tumor initiation and/or tumor progression under different oncogenic contexts or various stages of tumorigenesis (White 2012; Chen and Guan 2013).
The FIP200 (FAK family-interacting protein of 200 kDa) gene encodes an evolutionarily conserved protein characterized by a large coiled-coil region containing a leucine zipper motif (Ueda et al. 2000; Abbi et al. 2002; Chano et al. 2002). FIP200 is an essential autophagy protein, forming a ULK1–ATG13–FIP200–ATG101 complex to initiate autophagosome formation (Hara et al. 2008; Ganley et al. 2009; Hosokawa et al. 2009; Jung et al. 2009; Behrends et al. 2010). p62/SQSTM1 (also known as sequestosome-1, referred to as p62 here) is an adaptor protein that localizes to sites of autophagosome formation and can associate with autophagosome-localizing protein LC3 and ubiquitinated proteins (Bjorkoy et al. 2005). p62 itself is also an autophagy substrate and accumulates as protein aggregates in autophagy-deficient cells. As it also interacts with proteins in a number of intracellular signaling pathways, p62 plays important roles at the crossroads of autophagy, apoptosis, and cancer (Moscat and Diaz-Meco 2009; Rubinsztein et al. 2012; White 2012).
In contrast to data showing both tumor promotion and suppression roles for autophagy (Kimmelman 2011; White 2012; Chen and Guan 2013), p62 has been shown to play protumorigenesis functions in several previous studies (Duran et al. 2008; Guo et al. 2011). Mathew et al. (2009) found that p62 accumulation upon autophagy inhibition in apoptosis-deficient cells increased tumorigenesis through increased oxidative stress and deregulation of NF-kB signaling. Conversely, p62−/− mice exhibited significant resistance to Ras-induced lung adenocarcinomas due to decreased NF-kB activation (Duran et al. 2008). Similarly, RasV12 transformed, p62-null iBMK (immortal baby mouse kidney epithelial) cells showed reduced survival under starvation conditions and decreased tumorigenesis when compared with RasV12 transformed, p62+/+ iBMK cells (Guo et al. 2011). However, our previous studies showed that decreased mammary tumorigenesis caused by FIP200 deletion and consequent autophagy inhibition was also associated with a significant increase in p62 accumulation in the FIP200-null tumor cells (Wei et al. 2011). These results raise the interesting possibility that p62 may also play a tumor suppression function under some conditions, such as in autophagy-deficient tumor cells (i.e., after FIP200 deletion). Moreover, while previous studies revealed that deletion of FIP200 or other autophagy genes inhibited tumorigenesis (Guo et al. 2011; Wei et al. 2011; Yang et al. 2011), it is not clear whether FIP200-mediated autophagy is also required in maintaining growth of established tumors, which is an important question in the future design of therapies targeting autophagy genes for treatment.
Here we developed a novel system that allows deletion of FIP200 as well as expression of ectopic genes in tumor cells within an established and growing tumor in an inducible manner in vivo. Using this sophisticated system, we showed that autophagy disruption by FIP200 deletion significantly impeded the growth of established tumors, and either p62 knockdown or p62 deficiency in established FIP200-null tumors further impaired tumor growth. We further found that overexpression of the autophagy master regulator TFEBS142A stimulated the growth of established tumors, which correlated with the increased autophagy of the tumor cells. Therefore, knockout/knockdown of p62 and suppression of autophagy by FIP200 deletion can synergize to inhibit tumor growth, providing new insights for the future design of anti-cancer treatment.
Results
Autophagy disruption by deletion of FIP200 impairs growth of established tumors
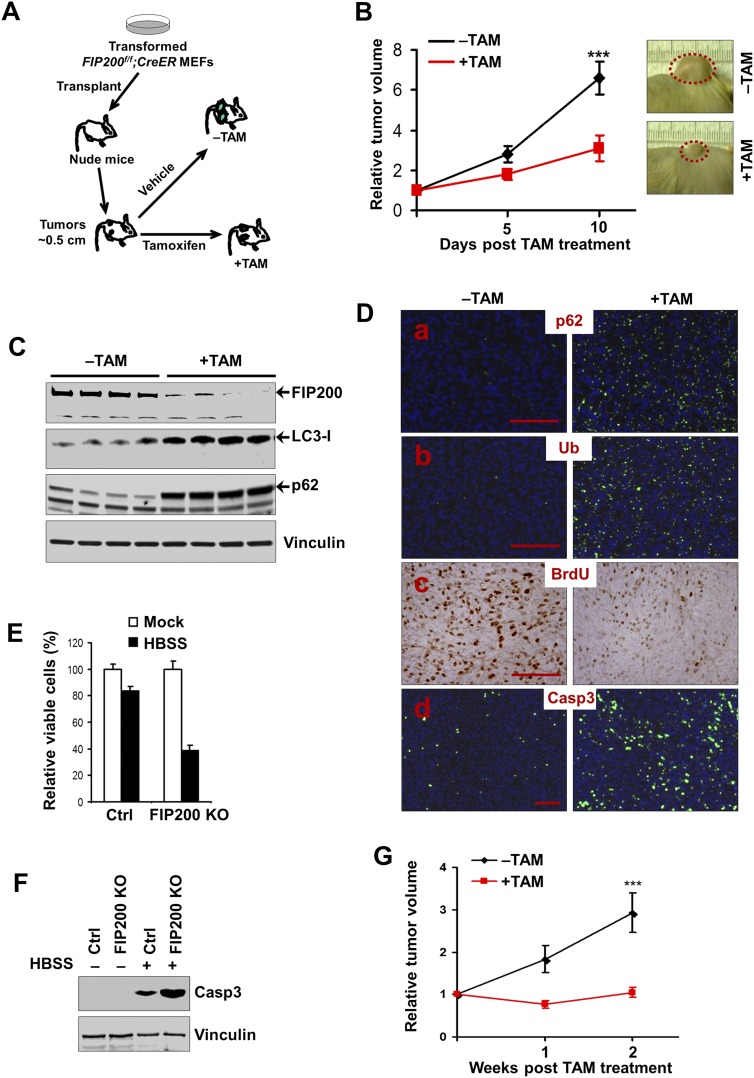
Our previous studies showed that autophagy inhibition by FIP200 deletion decreased cell growth of E1A/H-RasV12 transformed mouse embryonic fibroblasts (MEFs) (Wei et al. 2011; Wei and Guan 2012). To evaluate whether autophagy is required to maintain the growth and/or viability of established tumors in vivo, MEFs were isolated from FIP200-floxed mice (Gan et al. 2006) and then transformed by E1A/H-RasV12 and infected with MSCV·CreERT2, which expresses Cre recombinase in a tamoxifen (TAM)-dependent manner (Kumar et al. 2009; Supplemental Fig. S1A). As expected, treatment of these transformed FIP200f/f;CreER MEFs with 4-hydroxytamoxifen (4-OHT) led to efficient deletion of the floxed FIP200 alleles (Supplemental Fig. S1B). The transformed FIP200f/f;CreER MEFs were then transplanted into the flanks of athymic nude mice. Animals with advanced tumors (0.5 cm in diameter) were treated with TAM to induce the deletion of FIP200 in the tumor cells to examine the effects on the growth of established tumors in vivo (Fig. 1A). As shown in Figure 1B, autophagy deficiency by FIP200 deletion resulted in a significantly decreased tumor growth in TAM-treated mice compared with control recipient mice without TAM treatment. In additional control experiments, we found that TAM treatment did not affect the growth of tumors derived from E1A/H-RasV12 transformed FIP200f/f MEFs in the recipient mice (Supplemental Fig. S2), indicating that reduced tumor growth by transformed FIP200f/f;CreER MEFs was due to FIP200 deletion (induced by CreER after TAM treatment) rather than TAM treatment per se.
Figure 1.
Autophagy disruption by deletion of FIP200 impairs growth of established tumors. (A) Schematics for the deletion of floxed FIP200 induced by TAM after transplantation of transformed FIP200f/f;CreER MEFs, as described in the Materials and Methods. (B) Transformed FIP200f/f;CreER MEFs were injected subcutaneously into athymic nude mice, animals (n = 4 each) were treated with vehicle control (−TAM, black line) or TAM (+TAM, red line) by intraperitoneal (i.p.) injection, and single tumor growth was measured at the indicated time points. Data points represent means of fold change and SD in tumor volume relative to 1 d after the last TAM injection. Representative tumor burden images at the final time point are shown at the right. (***) P < 0.001. (C) Tumor lysates harvested from the recipient mice at the final time point were analyzed by Western blotting for various proteins using vinculin as a loading control, as indicated. (D) Representative images of tumor sections harvested from the recipient mice at the final time point and analyzed by immunohistochemistry or immunofluorescence with anti-p62, anti-ubiquitin (anti-Ub), anti-BrdU, and anti-caspase-3 (Casp3) antibodies. Bar, 100 μm. (E,F) Transformed FIP200f/f;CreER MEFs were treated with 4-OHT to activate Cre recombinase to produce FIP200 knockout (KO) cells or with mock medium to generate control (Ctrl) cells, respectively (see Supplemental Fig. S1). Cell viability was determined after 6 h with or without HBSS starvation in E. Cell lysates were analyzed by Western blotting for proteins as indicated after 3 h with or without HBSS starvation in F. (G) Immortalized FIP200f/f;PyMT;CreER tumor cells were injected into the mammary fat pads of nude mice. Animals (n = 4 each) were treated with vehicle control (black line) or TAM (red line) by i.p. injection, and single tumor growth was measured at the indicated time points. Data points represent means of fold change and SD in tumor volume relative to 1 d after the last TAM injection. (***) P < 0.001.
We next analyzed FIP200 deletion in the tumors developed in the transformed FIP200f/f;CreER MEF-transplanted recipient mice that had been treated with or without TAM. Figure 1C shows a significantly diminished expression of FIP200 in the tumor samples from the recipient mice treated with TAM compared with those from control mice, suggesting efficient deletion of FIP200 by the strategy outlined in Figure 1A, as expected. Furthermore, increased accumulation of LC3-I (i.e., likely caused by reduced LC3-I-to-LC3-II conversion) was observed in lysates prepared from tumors with FIP200 deletion induced by TAM treatment compared with those in control mice, suggesting defective autophagy in these cells. Likewise, starvation-induced puncta formation was decreased upon the induction of FIP200 deletion by 4-OHT treatment of transformed FIP200f/f;CreER MEFs (Supplemental Fig. S3). Consistent with these observations, an increased amount of p62 was also found in FIP200-null tumor cells, as measured by both Western blotting analysis of the tumor lysates (Fig. 1C) and immunofluorescent staining of sections from the tumors developed in TAM-treated recipient mice (Fig. 1D, panel a). Similar to our previous findings in mammary tumor cells and neurons (Liang et al. 2010; Wei et al. 2011), large ubiquitin-positive aggregates were also detected in sections of tumors in TAM-treated recipient mice but not in those from control mice (Fig. 1D, panel b). Together, these results demonstrate defective autophagy in the established tumors from transformed FIP200f/f;CreER MEFs upon deletion of FIP200 by TAM administration to the recipient mice.
Previous studies have shown that FIP200 deletion decreased proliferation of transformed MEFs as well as skin epithelial cells in vitro (Wei et al. 2009, 2011). Consistent with the reduced tumor growth, we found significantly reduced cell proliferation in tumors from the recipient mice following TAM treatment compared with that treated with vehicle control, as measured by BrdU incorporation assays (Fig. 1D, panel c). We also detected increased apoptosis in FIP200-null tumors in vivo, as measured by immunofluorescent staining using cleaved caspase-3 (Fig. 1D, panel d), although our previous studies showed that FIP200 deletion had no effect in vitro on apoptosis of E1A/H-RasV12 transformed MEFs under normal growth conditions (Wei et al. 2011). We then examined the survival of transformed FIP200f/f;CreER MEFs after treatment of 4-OHT to delete FIP200 in vitro. Similar to previous results (Wei et al. 2011), FIP200 deletion did not affect apoptosis of transformed FIP200f/f;CreER MEFs in nutrient-replete culture medium. However, it significantly decreased the survival of these cells under nutrient starvation conditions (i.e., in Hank’s balanced salt solution [HBSS] medium) (Fig. 1E,F).
To further elucidate the role of FIP200 in tumor growth, we prepared spontaneously immortalized mammary tumor cells from FIP200f/f;MMTV-PyMT mice (Wei et al. 2011) and then infected them with recombinant retroviruses encoding CreER. The resulting FIP200f/f;PyMT;CreER tumor cells were then transplanted into nude mice to allow tumor growth until ∼0.5 cm, followed by TAM treatment to induce deletion of the floxed FIP200 genes (Supplemental Fig. S4A). We found that ablation of FIP200 in the PyMT-driven mammary tumor cells also significantly decreased their growth compared with those tumors in the recipient mice not treated with TAM (Fig. 1G). As a control for TAM effect, we showed that similar treatment with TAM of recipient mice that had been transplanted with FIP200f/f;PyMT tumor cells (that were infected by empty control retroviruses) did not alter mammary tumor growth in these mice (Supplemental Fig. S4B,C). Collectively, these results suggest that FIP200-mediated autophagy plays a critical role for established tumor growth by promoting tumor cell proliferation as well as survival under nutrient depletion stress conditions in vivo.
p62 synergizes with autophagy for tumor growth
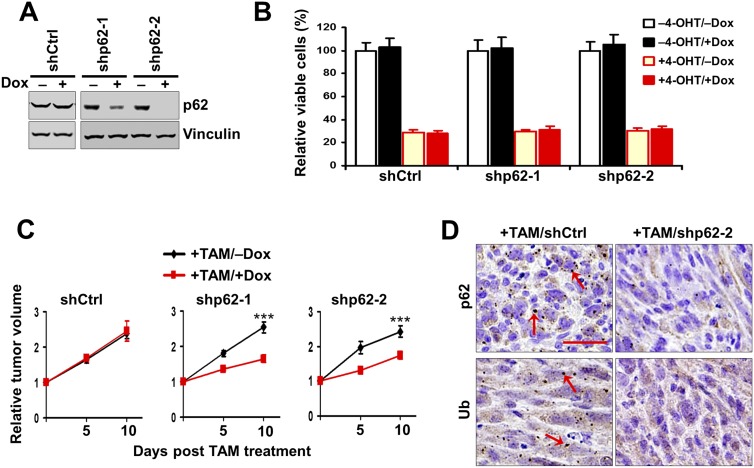
Accumulation of p62 aggregates is a common feature of autophagy disruption (Moscat and Diaz-Meco 2009; Rubinsztein et al. 2012; White 2012). Previous studies showed that increased p62 accumulation promoted tumorigenesis in apoptosis-defective tumor cells upon autophagy inhibition by Atg5 deletion (Mathew et al. 2009). However, our above results suggested that p62 accumulation correlated with reduced tumor growth in FIP200-null transformed MEFs, raising the potentially alternative role of p62 in the inhibition of growth of established tumors in vivo. To evaluate such a possibility directly, we introduced inducible p62 shRNAs under the control of doxycycline (Dox) into the transformed FIP200f/f;CreER MEFs. Cells derived from two out of three different shRNAs (designated as shp62-1 and shp62-2 cells) showed efficient suppression of p62 levels upon Dox treatment to induce shRNA expression (Fig. 2A). However, although FIP200 deletion induced by 4-OHT treatment caused a significant decrease in cell survival as observed earlier (+4-OHT vs. −4-OHT), p62 knockdown did not rescue the deficient survival of FIP200 knockout cells (+Dox vs. −Dox in +4-OHT condition) (Fig. 2B), suggesting that increased p62 upon FIP200 deletion did not contribute to the decreased survival of these cells. p62 knockdown did not affect the survival of control cells (+Dox vs. −Dox in −4-OHT condition) either.
Figure 2.
p62 Knockdown decreases the growth of autophagy-deficient tumors. (A) Various transformed MEFs were treated with 250 nM 4-OHT to delete FIP200 and then cultured for 2 d with or without 1 μg/mL Dox, as indicated. Cell lysates were analyzed by Western blotting for the indicated proteins. (B) Various transformed MEFs (5 × 104) were seeded into six-well plates, and the percentage (normalized to cells under −4-OHT/−Dox conditions) of the number of cells was determined after 5 d in DMEM + 10% FBS culture medium under various conditions, as indicated. (C) Various transformed MEFs were injected subcutaneously into athymic nude mice, all animals (n = 4 each) were treated with TAM by i.p. injection for FIP200 deletion and fed without (black line) or with (red line) Dox-containing food, and single tumor growth was measured at the indicated time points. Data points represent means of fold change and SD in tumor volume relative to 1 d after the last TAM injection. (***) P < 0.001. (D) Representative images of tumor sections harvested from the recipient mice at the final time point and analyzed by immunohistochemistry with anti-p62 and anti-ubiquitin (anti-Ub) antibodies. Arrows mark p62-positive and ubiquitin-positive aggregates. Bar, 100 μm.
We next examined the effect of p62 knockdown on transformed MEFs following FIP200 deletion in established tumors in vivo. shp62-1 and shp62-2 cells were transplanted into recipient nude mice, which were then treated with TAM and with or without Dox after tumors reached the size of ∼0.5 cm in diameter to induce FIP200 deletion with or without p62 knockdown. Surprisingly, p62 knockdown did not promote tumor growth (i.e., restore the reduced growth of transformed MEFs following TAM-induced FIP200 deletion) but rather further reduced it in recipient mice transplanted with shp62-1 or shp62-2 cells (Fig. 2C). As expected, expression of a control shRNA (shCtrl) did not affect growth of tumors derived from transformed MEFs following FIP200 deletion (i.e., +TAM). Immunohistochemical staining of tumor sections verified decreased p62-positive aggregations in tumor cells with p62 knockdown compared with those expressing shCtrl (Fig. 2D, top panels). A partial decrease in the ubiquitin-positive aggregations was also observed in tumors with p62 knockdown (Fig. 2D, bottom panels). Together, these results suggest that p62 accumulation is not responsible for the decreased tumor growth in transformed MEFs following FIP200 deletion in established tumors and that down-regulation of p62 further impairs the growth of autophagy-deficient tumors.
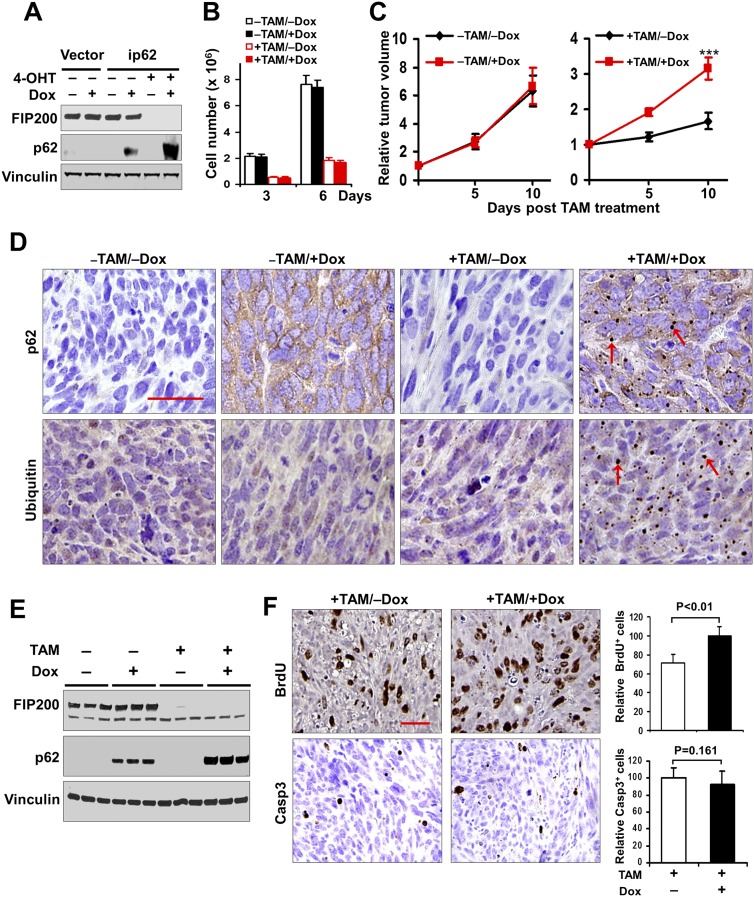
To further evaluate the role of p62 in regulation of tumor growth and exclude the possibility that the observed effects in shp62-1 or shp62-2 cells were due to off-target effects of p62 shRNAs, we generated FIP200f/f,p62−/− mice by crossing floxed FIP200 (Gan et al. 2006) and p62−/− (Komatsu et al. 2007) mice and then prepared transformed FIP200f/f;p62−/−CreER MEFs using a strategy similar to that for the transformed FIP200f/f;CreER MEFs (see Supplemental Fig. S1A). We next introduced an inducible p62 expression vector under the control of Dox into the transformed FIP200f/f;p62−/−CreER MEFs to obtain transformed FIP200f/f;p62−/−CreER;ip62 MEFs (designated as ip62 cells). Western blotting analysis verified the expression of p62 upon Dox treatment both with and (to a higher level) without FIP200 expression (i.e., −4-OHT and +4-OHT, respectively) in these cells (Fig. 3A). Similar to results of p62 knockdown in transformed FIP200f/f;CreER MEFs, re-expression of p62 in transformed FIP200f/f;p62−/−CreER MEFs did not affect their survival in vitro (Fig. 3B).
Figure 3.
Ectopic p62 expression increases the growth of autophagy-deficient tumor. (A) Various transformed MEFs were treated with or without 250 nM 4-OHT and then cultured for 2 d with or without 1 μg/mL Dox, as indicated. Cell lysates were analyzed by Western blotting for the indicated proteins. (B) ip62 cells (5 × 104) were seeded into six-well plates, and the number of cells was determined after 3 or 6 d in DMEM + 10% FBS culture medium under various conditions, as indicated. (C) ip62 cells were injected subcutaneously into athymic nude mice, animals (n = 4 each) were treated with or without TAM by i.p. injection for FIP200 deletion and fed without (black line) or with (red line) Dox-containing food, and single tumor growth was measured at the indicated time points. Data points represent means of fold change and SD in tumor volume relative to 1 d after the last TAM injection. (***) P < 0.001. (D) Representative images of tumor sections harvested from the recipient mice at the final time point and analyzed by immunohistochemistry with anti-p62 and anti-ubiquitin (anti-Ub) antibodies. Arrows mark p62-positive and ubiquitin-positive aggregates. Bar, 100 μm. (E) Lysates were prepared from mammary tumors of recipient mice and then analyzed by Western blotting for the indicated proteins. (F) The proliferation and apoptosis of tumor cells in recipient mice were analyzed by BrdU incorporation assays as described in Materials and Methods (top panels) or immunohistochemistry using anti-cleaved caspase-3 (bottom panels), respectively. Bar, 100 μm. Data at the right represent mean ± SD (n = 4 for each sample) of relative numbers of BrdU-positive or cleaved caspase-3-positive cells per field.
The ip62 cells were then transplanted into nude mice to determine the effect of re-expression of p62 on the growth of established tumors in vivo. After tumors reached ∼0.5 cm, mice were treated with TAM for FIP200 deletion and/or Dox for p62 re-expression, and tumor growth was then monitored. We found that re-expression of p62 in tumors with FIP200 expression (i.e., −TAM) did not significantly affect the growth of tumors under these conditions (compare +Dox vs. −Dox) (Fig. 3C, left panel). In contrast, for tumors with FIP200 deletion (i.e., +TAM), re-expression of p62 significantly increased tumor growth compared with those without p62 re-expression (compare +Dox vs. −Dox) (Fig. 3C, right panel). Immunohistochemical staining of the tumor sections showed expression of p62 after Dox induction and the presence of p62-containing aggregates in FIP200 knockout tumors (Fig. 3D, top panels). Similarly, ubiquitin-positive aggregations were also observed only in autophagy-deficient FIP200 knockout tumor cells with p62 re-expression (Fig. 3D, bottom panels). Analyses of lysates from the tumors with four different genotypes verified the efficiency of FIP200 deletion and p62 re-expression by treatment with TAM and Dox, respectively, and that FIP200 deletion further enhanced the level of p62 expression (Fig. 3E). BrdU incorporation assays showed a significantly increased proliferation of tumor cells upon p62 re-expression in FIP200 knockout tumors (Fig. 3F, top panels and graph). In contrast, no significant differences for apoptosis were observed between tumor cells with or without p62 re-expression, as measured by staining with cleaved caspase-3 (Fig. 3F, bottom panels and graph). Taken together, these results provide further support that p62 is still required for the growth of established tumors derived from autophagy-deficient transformed MEFs and that p62 promotes tumor growth primarily through its regulation of cell proliferation.
Regulation of tumor growth by p62 through the NF-kB pathway in FIP200 knockout tumor cells
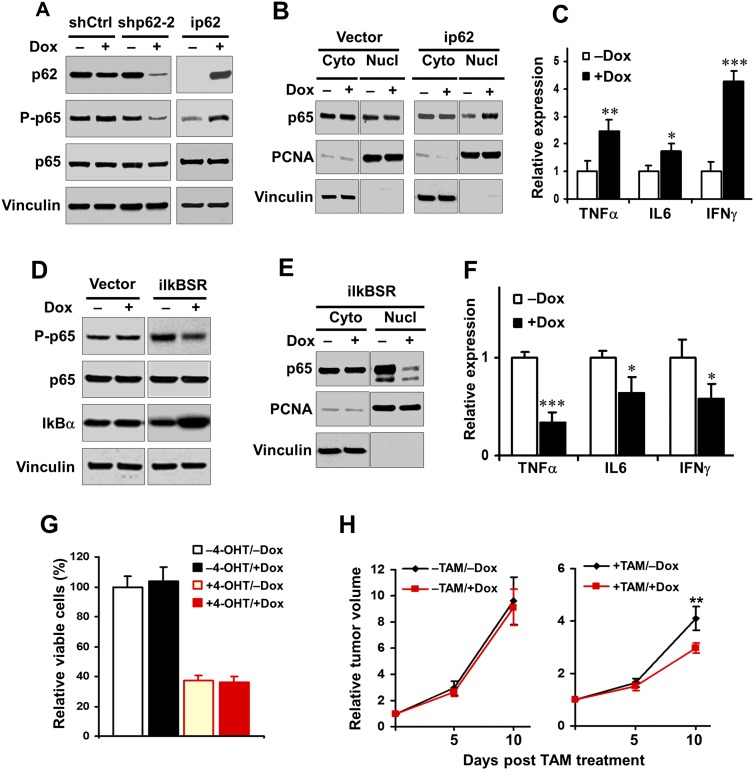
Previous studies demonstrated biochemically and genetically that p62 is required for activation of the NF-kB pathway (Duran et al. 2004; Martin et al. 2006; Duran et al. 2008; Ling et al. 2012). We therefore tested whether p62 also regulates the NF-kB pathway in FIP200-null autophagy-deficient cells and whether this may contribute to p62 promotion of tumor growth derived from these cells. We found that knockdown of p62 in FIP200 knockout cells (i.e., +Dox vs. −Dox in shp62-2 cells following FIP200 deletion by 4-OHT) decreased NF-kB signaling, as assessed by the phosphorylation of p65 (Fig. 4A, left four lanes). Conversely, re-expression of p62 in FIP200 knockout/p62 knockout cells (i.e., +Dox vs. −Dox in ip62 cells following FIP200 deletion by 4-OHT) increased NF-kB signaling (Fig. 4A, right two lanes). Furthermore, induction of p62 re-expression promoted p65 nuclear translocation (Fig. 4B, right panels), whereas just treatment of the tumor cells with vector alone by Dox did not affect p65 nuclear translocation as expected (Fig. 4B, left panels). Similarly, we found that several endogenous targets of NF-kB signaling, including TNFα, IL6, and IFNγ, were up-regulated upon re-expression of p62 in FIP200 knockout/p62 knockout tumor cells (Fig. 4C) but not by Dox treatment of tumor cells with vector alone (Supplemental Fig. S5). These studies provide strong support that p62 regulates NF-kB signaling in FIP200 knockout autophagy-deficient tumor cells.
Figure 4.
p62 regulates NF-kB activation, and inhibition of NF-kB impairs the tumor growth under autophagy-deficient condition. (A) Various transformed MEFs were treated with 250 nM 4-OHT to delete FIP200 and then cultured for 2 d with or without 1 μg/mL Dox to induce p62 knockdown (left four lanes) or ectopic p62 expression (right two lanes), as indicated. Cell lysates were analyzed by Western blotting for the indicated proteins. (B) Various transformed MEFs were treated with 250 nM 4-OHT to delete FIP200 and then cultured for 2 d with or without 1 μg/mL Dox, as indicated. Cytoplasmic (Cyto) and nuclear (Nucl) fractions were then prepared and analyzed by Western blotting for the indicated proteins. (C) ip62 cells were treated with 250 nM 4-OHT for deletion of FIP200 and then cultured for 2 d with or without 1 μg/mL Dox. RNA was isolated from cells and subjected to the analysis by quantitative RT–PCR (qRT–PCR) to detect the expression of NF-kB target genes, as indicated. The mean ± SD of relative levels (normalized to no Dox treatment) is shown. (D) Various transformed MEFs were treated with 4-OHT to delete FIP200 and then cultured overnight with or without 1 μg/mL Dox, as indicated. Cell lysates were analyzed by Western blotting for the indicated proteins. (E) iIkBSR cells were treated with 250 nM 4-OHT to delete FIP200 and then cultured for 2 d with or without 1 μg/mL Dox, as indicated. Cytoplasmic (Cyto) and nuclear (Nucl) fractions were prepared and analyzed by Western blotting for the indicated proteins. (F) iIKBSR cells were treated with 250 nM 4-OHT for deletion of FIP200 and then cultured for 2 d with or without 1 μg/mL Dox. RNA was isolated from cells and subjected to the analysis by qRT–PCR to detect the expression of NF-kB target genes, as indicated. The mean ± SD of relative levels (normalized to no Dox treatment) is shown. (G) iIkBSR cells (5 × 104) were seeded into six-well plates, and the percentage (normalized to cells under −4-OHT/−Dox conditions) of the number of cells was determined after 5 d in DMEM + 10% FBS culture medium under various conditions, as indicated. (H) iIkBSR cells were injected subcutaneously into athymic nude mice, animals (n = 4 each) were treated with or without TAM by i.p. injection for FIP200 deletion and fed without (black line) or with (red line) Dox-containing food, and single tumor growth was measured at the indicated time points. Data points represent means of fold change and SD in tumor volume relative to 1 d after the last TAM injection. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
To directly assess the potential role of NF-kB signaling regulated by p62 (as shown in the above experiments) in FIP200 knockout cells, we introduced into transformed FIP200f/f;CreER MEFs a Dox-controlled inducible expression vector encoding IkBα superrepressor (IkBSR), a nonphosphorylatable, dominant-negative inhibitor protein for the NF-kB pathway (Meylan et al. 2009). The resulting cells were designated as iIkBSR cells (i.e., transformed FIP200f/f;CreER;iIkBSR MEFs). Western blotting analysis of lysates from these cells verified that Dox-induced overexpression of IkBSR reduced NF-kB signaling, as assessed by the phosphorylation of p65 (Fig. 4D, right two lanes). Furthermore, expression of IkBSR also decreased nuclear translocation of p65 (Fig. 4E) and expression of NF-kB targets, including TNFα, IL6, and IFNγ (Fig. 4F), providing further validation that IkBSR inhibits NF-kB signaling in FIP200 knockout autophagy-deficient tumor cells. However, we found that IkBSR expression did not affect the survival of iIkBSR cells either with or without FIP200 deletion (i.e., see +Dox vs. −Dox in +4-OHT cells or +Dox vs. −Dox in −4-OHT cells) in vitro (Fig. 4G). We next examined the effect of IkBSR on the growth of tumors derived from the transplantation of iIkBSR cells in vivo. We found that induced expression of IkBSR did not affect the growth of the tumors with FIP200 (i.e., +Dox vs. −Dox in −TAM tumors) (Fig. 4H, left panel), although previous studies showed inhibition of lung tumor growth by IkBSR (Meylan et al. 2009). Such a discrepancy could be due to different cells used or possibly more rapid growth of tumors from transformed MEFs masking the effect of NF-kB pathway inhibition in these cells. Interestingly, however, induction of IkBSR expression reduced the growth of the established tumors from the iIkBSR cells following FIP200 deletion (i.e., +Dox vs. −Dox in +TAM tumors) (Fig. 4D, right panel). Together, these results suggest that p62 promotes the growth of autophagy-deficient tumors in vivo at least partially through its stimulation of the NF-kB signaling pathway.
Increased autophagy by enforced overexpression of TFEBS142A correlates with enhanced tumor growth
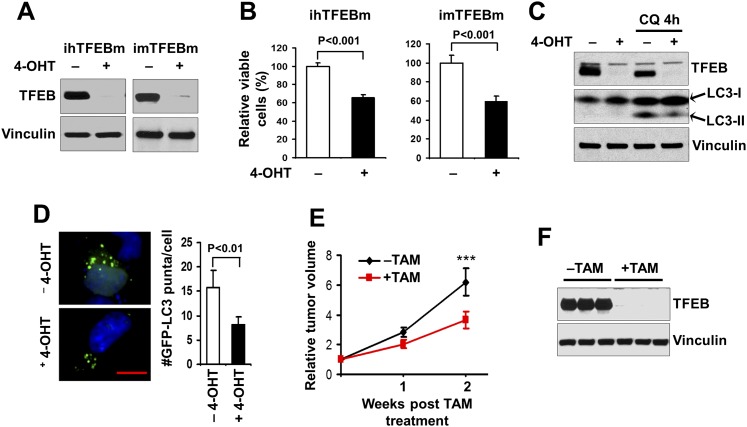
To complement our studies of autophagy inhibition by FIP200 deletion and further explore the role of autophagy in tumor growth in vivo, we examined the effect of enforced overexpression of TFEBS142A, the S142A mutant of transcription factor TFEB that has been shown recently to stimulate autophagy (Settembre et al. 2011), in breast cancer cells using an inducible strategy. MDA-MB-231 human breast cancer cells or the spontaneously immortalized mouse mammary tumor cells derived from MMTV-PyMT mice were infected by recombinant retroviruses encoding floxed TFEBS142A as well as that encoding MSCV-CreER (Supplemental Fig. S6A). The resulting cells were designated as ihTFEBm (for inducible human breast cancer cells with TFEB mutant) and imTFEBm (for inducible mouse breast cancer cells with TFEB mutant), respectively. As expected, treatment with 4-OHT induced Cre expression, leading to the removal of the floxed TFEBS142A, which generates corresponding control cells for the tumor cells overexpressing TFEBS142A in both ihTFEBm (Fig. 5A, left two lanes) and imTFEBm (Fig. 5A, right two lanes) cells (Supplemental S6A). The effect of TFEB overexpression in breast cancer cells was then evaluated by examining survival of these cells in vitro. We found that both ihTFEBm (Fig. 5B, left graph) and imTFEBm (Fig. 5B, right graph) cells showed significantly increased cell survival compared with the respective control cells (i.e., −4-OHT vs. +4-OHT) (Fig. 5B).
Figure 5.
Enhanced autophagy by ectopic expression of the TFEBS142A mutant correlates with increased tumor growth. (A) ihTFEBm (left) or imTFEBm (right) cells were treated with 4-OHT for 2 d to delete the ectopic floxed TFEBS142A mutant TFEB and then cultured overnight in growth medium, as indicated. Cell lysates were analyzed by Western blotting for the indicated proteins. (B) ihTFEBm (left) or imTFEBm (right) cells (5 × 104) were seeded into six-well plates, and the percentage (normalized to cells under −4-OHT conditions) of the number of cells was determined after 6 d in culture medium under various conditions, as indicated. (C) imTFEBm cells were treated with 4-OHT as described in A and then replated in growth medium with (right two lanes) or without (left two lanes) 50 μM chloroquine (CQ) for 4 h. Cell lysates were analyzed by Western blotting for the indicated proteins. (D) ihTFEBm cells were infected by recombinant retroviruses encoding GFP-LC3 and then treated with 250 nM 4-OHT to delete floxed TFEBS142A. The cells were then replated and cultured in serum-starved medium overnight. Representative images (left panels) and the mean ± SD (right graph) of GFP-LC3 puncta (green) are shown, respectively. Slides were counterstained with DAPI (blue). Bar, 5 μm. (E) imTFEBm tumor cells were injected into the mammary fat pads of nude mice. Animals (n = 5 each) were treated with vehicle control (black line) or TAM (red line) by i.p. injection, and single tumor growth was measured at the indicated time points. Data points represent means of fold change and SD in tumor volume relative to 1 d after the last TAM injection. (***) P < 0.001. (F) Lysates were prepared from mammary tumors of recipient mice and then analyzed by Western blotting for the indicated proteins.
We further evaluated the effect of TFEB overexpression on basal and starvation-induced autophagy in these cells. Compared with control cells, tumor cells with TFEBS142A overexpression showed an increased ratio of LC3-II/LC3-I under basal conditions in the presence of chloroquine (CQ) (Fig. 5C), which inhibits autolysosome acidification and subsequent LC3-II degradation (Mizushima 2004). Moreover, starvation-induced puncta formation was also significantly higher in ihTFEBm cells with TFEBS142A overexpression compared with the control cells (i.e., −4-OHT vs. +4-OHT) (Fig. 5D). These results are consistent with previous findings of promotion of autophagy by TFEB (Settembre et al. 2011). Last, imTFEBm cells were transplanted into recipient nude mice and used for examination of the role of TFEB-promoted autophagy in tumor growth in vivo (Supplemental Fig. S6B). After the tumors reached 0.5 cm in diameter, the recipient mice were treated with TAM to induce the removal of TFEBS142A overexpression in the tumor cells. Comparison of tumor growth in mice with or without TAM treatment showed a significantly increased growth rate of tumors with TFEBS142A overexpression (i.e., in recipient mice without TAM treatment) compared with those control tumors (i.e., the floxed TFEBS142A is deleted in tumors by TAM treatment) (Fig. 5E). Western blotting analyses of lysates of the tumors in these two groups of mice verified the deletion of TFEB in tumors from recipient mice that had been treated with TAM (Fig. 5F). Together, complementary with the finding that autophagy disruption by FIP200 deletion impairs the growth of established tumors, these results indicate that enhanced autophagy by TFEBS142A overexpression correlates with increased tumor growth rate for established tumors.
Discussion
Autophagy has been shown to play complex roles in the regulation of cancer development and progression in different stages of tumorigenesis and oncogenic events (Mathew et al. 2009; Kon et al. 2011; Lock et al. 2011; Takamura et al. 2011; Wei et al. 2011; Yang et al. 2011; Chen and Guan 2013; Choi et al. 2013). Our previous studies showed that deletion of an essential autophagy gene, FIP200, significantly decreased mammary tumorigenesis in a mouse model of human breast cancer in vivo (Wei et al. 2011; Wei and Guan 2012). By creating and analyzing unique models that allow us to manipulate FIP200 and p62 in the established tumors, we show here that FIP200-mediated autophagy is also required for maintaining tumor growth in vivo and that the accumulated p62 upon autophagy blockage did not contribute to the reduced tumor growth but stimulated the growth of autophagy-deficient tumors. Therefore, our results reveal a synergistic mechanism of p62 signaling and autophagy in the promotion of cancer growth, implicating a potentially more effective therapeutic strategy by targeting both pathways.
p62 is a multifunctional adaptor protein implicated in selective autophagy, serving as an autophagy receptor, which shows abnormal accumulation in aggregates upon autophagy inhibition by deletion of FIP200 or other autophagy genes in a variety of cells (Bjorkoy et al. 2005; Komatsu et al. 2007; Liang et al. 2010; Mizushima and Komatsu 2011). However, the pathophysiological significance of this increased p62 aggregation remains to be clarified, as previous studies suggested that p62 ablation rescued liver injury but not the neuronal-degenerative phenotype induced by Atg7 conditional knockout (Komatsu et al. 2007). Although a number of previous studies suggested that p62 plays a positive role in the promotion of cancer development and progression (Duran et al. 2008; Guo et al. 2011), such a potential role for the accumulated p62 in FIP200-null tumor cells is not immediately apparent, as these cells showed a reduced tumorigenesis compared with those with wild-type FIP200, suggesting a potential contribution of p62 aggregation to the reduced tumorigenesis of autophagy-deficient tumor cells (Guo et al. 2011; Wei et al. 2011; Yang et al. 2011). Surprisingly, our analysis of FIP200, p62 double-knockout transformed cells here showed not only that p62 ablation did not rescue the defective tumor growth phenotype but that it actually further decreased tumor growth compared with FIP200 single-knockout cells. These results are unlike the case for Atg7 conditional knockout in either the liver (rescued) or neurons (no effect) observed earlier (Komatsu et al. 2007) but suggested an additional relationship of p62 with autophagy in that they can synergize to promote tumor growth in vivo.
It was found recently that deficiency in p62 decreased tumorigenesis of oncogenic K-ras transformed cells in xenograft assays (Guo et al. 2011). This inhibitory effect was suggested to be caused by interference of autophagosome cargo delivery or autophagosome formation after p62 deletion in these cells. However, such a mechanism is unlikely to explain our observation in FIP200-null transformed cells, as autophagy is already inhibited in these cells by FIP200 deletion. Although it plays a role in autophagy as FIP200 (and other Atg genes such as Atg5 and Atg7), p62 may not affect autophagy to the same extent as autophagy genes like FIP200 and has also been shown to regulate a number of other intracellular signaling pathways. Therefore, p62 could potentially affect tumor growth through a different mechanism than affecting autophagy per se. In support of such a possibility, whereas mice with knockout of autophagy genes (e.g., Beclin-1, Atg5, Atg7, and FIP200) displayed embryonic or perinatal lethality (Yue et al. 2003; Kuma et al. 2004; Komatsu et al. 2005; Gan et al. 2006), p62-null mice can survive for more than 1 year but with defects in adult mice such as adult-onset obesity and diabetes (Rodriguez et al. 2006). In addition to its role as an autophagy receptor, p62 has been shown to regulate other signaling pathways such as Keap1–Nrf2 and NF-kB pathways, which may contribute to the regulation of tumor growth by p62 (Duran et al. 2008; Komatsu et al. 2010; Inami et al. 2011; Takamura et al. 2011), particularly in FIP200 knockout autophagy-deficient tumor cells observed in the present study.
The Keap1–Nrf2 system is one of the major cellular defense mechanisms against oxidative and electrophilic stresses (Motohashi and Yamamoto 2004; Hayes and McMahon 2009; Villeneuve et al. 2010). Recently, somatic mutations in Keap1 and Nrf2 leading to constitutive activation of Nrf2 were identified in multiple types of cancers, such as lung, gall bladder, and head and neck cancers (Padmanabhan et al. 2006; Singh et al. 2006; Shibata et al. 2008a,b; Hayes and McMahon 2009; Sporn and Liby 2012). Recent studies showed that p62 accumulation upon autophagy deficiency can displace the inhibitory binding of Keap1 to Nrf2 by its direct interaction at the same site, leading to Nrf2 activation and increased expression of target anti-oxidative response genes such as NAD(P)H dehydrogenase quinone 1 (Nqo1) and glutathione S-transferase μ1 (Gstm1) (Copple et al. 2010; Jain et al. 2010; Komatsu et al. 2010; Lau et al. 2010; Riley et al. 2010; Inami et al. 2011; Takamura et al. 2011). Interestingly, previous studies showed that persistent activation of Nrf2 by excess accumulation of p62 in liver-specific Atg7 knockout mice accompanied by hepatacellular adenoma development (Inami et al. 2011) and p62-dependent induction of Nrf2 targets such as Gstm1 and Nqo1 played important roles in the liver defects of autophagy-deficient mice (Komatsu et al. 2007). However, unlike the previous studies (Komatsu et al. 2007; Inami et al. 2011; Takamura et al. 2011), we did not find consistent and significant up-regulation of Nqo1 in ip62 cells after Dox treatment to induce p62 expression in FIP200 knockout cells (Supplemental Fig. S7), which possibly promoted tumor growth (i.e., +Dox vs. −Dox, in +TAM conditions) (see Fig. 3C). Therefore, future studies will be necessary to further determine whether the Keap1–Nrf2 pathway may play a role in mediating p62 regulation of tumor growth in FIP200-null autophagy-deficient tumors.
Although the mechanisms by which p62 regulates tumor growth in FIP200-null autophagy-deficient cells are not completely understood at present, our results suggest that p62 promoted tumor growth at least partially through its stimulation to NF-kB activation. These results are consistent with a previous finding that p62 is an important NF-kB mediator in lung tumorigenesis (Duran et al. 2008). They are also supported by previous studies showing that p62 is essential for maintaining NF-kB activation by preventing the turnover of K63-polyubiquitinated proteins, including TRAF6 in the NF-kB signaling pathway (Sanz et al. 2000; Wooten et al. 2005). Furthermore, NF-kB signaling has recently been shown to regulate p62 expression, which could act in an autoregulatory loop to extend NF-kB activation in the development of pancreatic cancer (Ling et al. 2012). Given the critical role of NF-kB in a variety of cancers (Meylan et al. 2009), it would be very interesting to determine whether such an autoregulatory loop between p62 and NF-kB signaling is also important for the growth of FIP200-null autophagy-deficient tumors in vivo.
It should be noted that while both Duran et al. (2008) and our current studies support a role of p62 in promoting tumor growth through NF-kB signaling, we found that restoration of p62 in p62 knockout tumor cells only affected tumor growth of autophagy-deficient cells but not autophagy-competent cells. Moreover, our p62 knockdown as well as p62 re-expression studies were performed in autophagy-deficient cells and established tumors (i.e., changes in p62 were induced after tumor formation), as we were interested in determining the role of increased p62 accumulation in these cells. Therefore, the differential setting and experimental conditions could potentially explain the apparently different results for the effect of p62 on tumor growth in the autophagy-competent cells in these two studies. Future studies will be required to examine these interesting possibilities.
It will also be interesting to examine in future studies whether other autophagy genes are also required for maintaining tumor growth in a manner similar to FIP200 and, more interestingly, whether p62 also synergizes with other autophagy genes using similar mechanisms. Likewise, as TFEB also promotes lysosomal biogenesis besides inducing autophagy, future studies will be necessary to determine a specific role for autophagy versus other lysosomal functions that are affected in the tumor cells with or without TFEBS142A expression.
In summary, our studies identify a role for FIP200 in promoting the growth of established tumors and also show a requirement for p62 at least in part through the downstream NF-kB signaling in the growth of autophagy-deficient tumors in vivo. The synergistic functions of the p62 pathway and autophagy in maintaining tumor growth suggest potentially more effective cancer therapies through targeting both autophagy and consequently elevated p62 accumulations.
Materials and methods
Mice, genotyping, and cell culture
FIP200f/f, FIP200f/f;MMTV-PyMT, and MMTV-PyMT transgenic mice were as described previously (Wei et al. 2011). p62−/− mice were kindly provided by Dr. Tetsuro Ishii of the University of Tsukuba (Komatsu et al. 2007) and were used to cross with FIP200f/f mice to produce FIP200f/+;p62+/− and then FIP200f/f;p62+/− mice. Genotyping for the FIP200, PyMT, and p62 alleles were as described previously (Komatsu et al. 2007; Wei et al. 2011). Mice were housed and handled by following the local, state, and federal regulations. All procedures relating to the care and treatment of the animals were performed in accordance with the National Institutes of Health guidelines.
HEK293 cells, MEFs, and MDA-MB-231 cells were maintained in DMEM supplemented with 10% FBS. Cell viability was measured by “measured by trypan blue” exclusion using cell viability analyzer (Vi-CELL, Beckman-Coulter, Inc.).
Preparation of transformed MEFs and mammary tumor cells capable of inducible deletion of floxed FIP200
Embryos at day 13.5 (E13.5) were isolated from the timed intercrosses of FIP200f/f;p62+/− mice and used for the preparation of MEFs with FIP200f/f and FIP200f/f;p62−/− genotypes essentially as described previously (Wei et al. 2011). They were then transformed by E1A/H-RasV12 as previously described (Wei et al. 2006) and subsequently infected by MSCV·CreERT2.puro or MSCV vector alone (Kumar et al. 2009) to produce transformed FIP200f/f;CreER MEFs, transformed FIP200f/f MEFs, and transformed FIP200f/f;p62−/−CreER MEFs, respectively.
Primary mammary tumor cells were isolated from the tumors with diameters of 2–3 cm developed from FIP200f/f;MMTV-PyMT or MMTV-PyMT mice and then cultured continuously for >5 mo in DMEM/F12 medium to obtain spontaneously immortalized FIP200f/f;PyMT or PyMT tumor cells, as described previously (Wei et al. 2011). FIP200f/f;PyMT tumor cells were then infected by MSCV·CreERT2.puro (Addgene plasmid 22776) or MSCV vector alone (Kumar et al. 2009) to produce FIP200f/f;PyMT;CreER or control FIP200f/f;PyMT tumor cells.
In some experiments, the transformed MEFs were treated with 250 nM 4-OHT (Sigma) to delete floxed FIP200 alleles via Cre induction, as described (Kumar et al. 2009). In other experiments, these cells or mammary tumor cells were transplanted into recipient nude mice followed by treatment with TAM to delete floxed FIP200 alleles via Cre induction in vivo (see details below).
Tumor cell transplantation, treatment with TAM or Dox, and tumor size monitoring
Transformed MEFs or mammary tumor cells were harvested, washed twice in DMEM, and then injected (5 × 105 cells in 1:1 PBS:Matrigel [BD Bioscience]) into the flank region (for transformed MEFs) or the number 4 inguinal mammary fat pads (for mammary tumor cells) of 7-wk-old female athymic nude mice (Frederick Cancer Research Facility of U.S. National Cancer Institute) under isofluorane anesthesia. After tumors appeared, tumor size was monitored, as described previously (Wei et al. 2011). Once the tumors reached ∼0.5 cm in diameter, mice were randomly assigned and continuously treated for 4 d with TAM (100 µL at 10 mg/mL in corn oil) or an equal volume of vehicle. For Dox-inducible expression in vivo, mice were fed with Dox-containing food (625 mg/kg; Harlan Laboratories). One day after the last TAM treatment or the first day of feeding with Dox-containing food was set as day 0; tumor size was then measured in the indicated time points. Tumor volume was calculated using the following formula: volume = (width2 × length)/2.
cDNA expression and knockdown
The plasmids TRE·IkBSR-Flag Pgk·Cre encoding IkBSR (Addgene plasmid 22504; Dr. Tyler Jacks), TRMPVIR (Addgene plasmid 27994; Dr. Scott Lowe), HA-p62 (Addgene plasmid 28027; Dr. Qing Zhong), and pEGFP-N1-TFEB (Addgene plasmid 38119; Dr. Andrea Ballabio) were all obtained from Addgene. cDNA fragments for p62 and IkBSR were prepared by PCR using the respective plasmids as templates and then inserted into the BamHI/EcoRI sites of TRMPVIR, which allows inducible expression of these genes upon Dox treatments. Recombinant retroviruses encoding p62 and IkBSR were prepared as described previously (Wei et al. 2006) and used to infect transformed FIP200f/f;p62−/−CreER MEFs and FIP200f/f;CreER MEFs to generate transformed FIP200f/f;p62−/−CreER;ip62 MEFs (i.e., ip62 cells) and FIP200f/f;CreER;iIkBSR MEFs (i.e., iIkBSR cells), respectively.
Two different shRNAs targeting mouse p62 (CGAGGTTGACATTGATGTGGAA and CGCTGACAATGGCTATGTCCTA) were also inserted into TRMPVIR at the XhoI/EcoRI sites to generate vectors to knock down p62 in a Dox-inducible manner, as described previously (Zuber et al. 2011). They were used to prepare corresponding recombinant retroviruses for infection into transformed FIP200f/f;CreER MEFs to generate shp62-1 and shp62-2 cells, respectively.
The cDNA fragment containing the TFEBS142A mutant and flanked by two loxP sites was prepared using pEGFP-N1-TFEB as a template by PCR. TFEB-S142A-5 (GCTGGCAACAGTGCTCCCAATgcaCCCATGGCCATGCTGCACATTG) and TFEB-S142A-3 (CAATGTGCAGCATGGCCATGGGtgcATTGGGAGCACTGTTGCCAGC) were used as mutagenesis primers, and TFEB-Lox-RI5 (CCGCGAATTCATAACTTCGTATAGCATACATTATACGAAGTTATCCCACCATGGCGTCACGCATAGGG) and TFEB-Lox-Bgl3 (GCGCAGATCTATAACTTCGTATAATGTATGCTATACGAAGTTATTCACAGCACATCGCCCTCCTC) were used for the two loxP sites. This fragment was then inserted into the MSCVneo vector (Clontech) to allow for Cre-mediated deletion of the TFEBS142A mutant overexpression. Recombinant retroviruses encoding the floxed TFEBS142A mutant were used to infect MDA-MB-231 human breast cancer cells or PyMT tumor cells to generate ihTFEBm and imTFEBm cells, respectively, as described previously (Wei et al. 2006).
GFP-fused LC3 vector was produced by cloning a fragment containing LC3 cDNA into pLEGFP-C1 (Clontech) vector to generate pLEGFP-LC3, as described previously (Wei et al. 2006). This vector was then used to infect transformed MEFs or mammary tumor cells to monitor puncta formation.
Western blotting
Cell lysates were prepared, and Western blotting was carried out as described previously (Wei et al. 2006, 2009). We used the following primary antibodies: LC3B (#2775), cleaved caspase-3 (#9661), Phospho-p65 (Ser536) (#3033), p65 (#8242), Nqo1 (#3187), IkBα (#4814), and TFEB (#4240), all from Cell Signaling Technology; FIP200 (#10043-2-AP) from Proteintech Group; p62 (#BML-PW9860) from Enzo Life Sciences; PCNA (sc-56) from Santa Cruz Biotechnology; and vinculin (#V4505) from Sigma. In some experiments, the cytoplasmic and nuclear fractions were prepared before subjecting the lysates to Western blotting analysis.
RNA isolation and semiquantitative RT–PCR
Total RNA was isolated from various transformed MEFs using TRIzol according to the manufacturer’s instructions (Invitrogen). Equal RNA amounts were added to reverse transcriptase reaction mix (Themo Scientific), with oligo-dT as primer. The resulting templates were subjected to PCR using the following specific primers: mouse TNFα F (5′-CATCTTCTCAAAATTCGAGTGACAA-3′), mouse TNFα R (5′-TGGGAGTAGACAAGGTACAACCC-3′), mouse IL-6 F (5′-GAGGATACCACTCCCAACAGACC-3′), mouse IL-6 R (5′-AAGTGCATCATCGTTGTTCATACA-3′), mouse IFNγ F (5′-TCAAGTGGCATAGATGTGGAAGAA-3′), and mouse IFNγ R (5′-TGGCTCTGCAGGATTTTCATG-3′).
Immunohistochemistry, immunofluorescence, and BrdU incorporation assays
Tumor samples were fixed overnight in 10% phosphate-buffered formalin (Fisher Scientific) and embedded in paraffin. They were then sectioned at 5-μm depth and subjected to immunohistochemistry or immunofluorescence as previously described (Wei et al. 2009, 2011). The primary antibodies used include p62 (Enzo Life Sciences), ubiquitin (#Z0458, DAKO), and cleaved caspase-3 (Cell Signaling). Sections were counterstained with haematoxylin for immunohistochemistry or DAPI for immunofluorescence.
For BrdU incorporation assays, mice were injected intraperitoneally with BrdU (100 μg per gram body weight; Sigma). After 3 h, mice were sacrificed, and tumor samples were prepared, fixed overnight in formalin, and embedded in paraffin. BrdU uptake was detected using the BrdU staining kit (Invitrogen).
Statistical analysis
Statistical significance was evaluated by paired t-test, using P < 0.05 as indicative of statistical significance.
Acknowledgments
We are grateful to Dr. Jianbin Wang and Dr. Rick Cerione of Cornell University and Dr. Scott Lowe of Memorial Sloan-Kettering Cancer Center for generously providing Ha-RasV12 and E1A plasmids, respectively; Dr. Tetsuro Ishii of the University of Tsukuba in Japan for p62 knockout mice; and Dr. Yongqiang Chen, Dr. Ming Luo, Dr. Huaping Fan, and Dr. Shaogang Sun at the University of Michigan and Dr. Ri Cui, Dr. Zhenghua Luo, Dr. Yong Peng, Dr. Hui-Lung Sun, Dr. Taewan Kim, Yuntao Dai, Dr. Nicola Zanesi, and Dr. Sulhinder Sandhu at Ohio State University for discussion of and assistance with different methods used in this study. We thank both C.M.C.’s and J.-L.G.’s laboratory colleagues for helpful comments on the manuscript. This research was supported by National Institutes of Health grants to C.M.C. and J.-L.G. (CA150926).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.237354.113.
References
- Abbi S, Ueda H, Zheng C, Cooper LA, Zhao J, Christopher R, Guan JL 2002. Regulation of focal adhesion kinase by a novel protein inhibitor FIP200. Mol Biol Cell 13: 3178–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW 2010. Network organization of the human autophagy system. Nature 466: 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171: 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chano T, Kontani K, Teramoto K, Okabe H, Ikegawa S 2002. Truncating mutations of RB1CC1 in human breast cancer. Nat Genet 31: 285–288 [DOI] [PubMed] [Google Scholar]
- Chen S, Guan JL 2013. Tumor-promoting and -suppressive roles of autophagy in the same mouse model of BrafV600E-driven lung cancer. Cancer Discov 3: 1225–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi AM, Ryter SW, Levine B 2013. Autophagy in human health and disease. N Engl J Med 368: 651–662 [DOI] [PubMed] [Google Scholar]
- Copple IM, Lister A, Obeng AD, Kitteringham NR, Jenkins RE, Layfield R, Foster BJ, Goldring CE, Park BK 2010. Physical and functional interaction of sequestosome 1 with Keap1 regulates the Keap1–Nrf2 cell defense pathway. J Biol Chem 285: 16782–16788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran A, Serrano M, Leitges M, Flores JM, Picard S, Brown JP, Moscat J, Diaz-Meco MT 2004. The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Dev Cell 6: 303–309 [DOI] [PubMed] [Google Scholar]
- Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J 2008. The signaling adaptor p62 is an important NF-κB mediator in tumorigenesis. Cancer Cell 13: 343–354 [DOI] [PubMed] [Google Scholar]
- Gan B, Peng X, Nagy T, Alcaraz A, Gu H, Guan JL 2006. Role of FIP200 in cardiac and liver development and its regulation of TNFα and TSC–mTOR signaling pathways. J Cell Biol 175: 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Lam DH, Wang J, Ding X, Chen S, Jiang X 2009. ULK1·ATG13·FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem 284: 12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, Kamphorst JJ, Chen G, Lemons JM, Karantza V, et al. 2011. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 25: 460–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 181: 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, McMahon M 2009. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci 34: 176–188 [DOI] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. 2009. Nutrient-dependent mTORC1 association with the ULK1–Atg13–FIP200 complex required for autophagy. Mol Biol Cell 20: 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inami Y, Waguri S, Sakamoto A, Kouno T, Nakada K, Hino O, Watanabe S, Ando J, Iwadate M, Yamamoto M, et al. 2011. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J Cell Biol 193: 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T 2010. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem 285: 22576–22591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH 2009. ULK–Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 20: 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmelman AC 2011. The dynamic nature of autophagy in cancer. Genes Dev 25: 1999–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, et al. 2007. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 131: 1149–1163 [DOI] [PubMed] [Google Scholar]
- Komatsu M, Kurokawa H, Waguri S, Taguchi K, Kobayashi A, Ichimura Y, Sou YS, Ueno I, Sakamoto A, Tong KI, et al. 2010. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol 12: 213–223 [DOI] [PubMed] [Google Scholar]
- Kon M, Kiffin R, Koga H, Chapochnick J, Macian F, Varticovski L, Cuervo AM 2011. Chaperone-mediated autophagy is required for tumor growth. Sci Transl Med 3: 109ra117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N 2004. The role of autophagy during the early neonatal starvation period. Nature 432: 1032–1036 [DOI] [PubMed] [Google Scholar]
- Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T 2009. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev 23: 2700–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A, Wang XJ, Zhao F, Villeneuve NF, Wu T, Jiang T, Sun Z, White E, Zhang DD 2010. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol Cell Biol 30: 3275–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G 2008. Autophagy in the pathogenesis of disease. Cell 132: 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B 1999. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402: 672–676 [DOI] [PubMed] [Google Scholar]
- Liang CC, Wang C, Peng X, Gan B, Guan JL 2010. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem 285: 3499–3509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J, Kang Y, Zhao R, Xia Q, Lee DF, Chang Z, Li J, Peng B, Fleming JB, Wang H, et al. 2012. KrasG12D-induced IKK2/β/NF-κB activation by IL-1α and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell 21: 105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, Debnath J 2011. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell 22: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P, Diaz-Meco MT, Moscat J 2006. The signaling adapter p62 is an important mediator of T helper 2 cell function and allergic airway inflammation. EMBO J 25: 3524–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell 137: 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, Jacks T 2009. Requirement for NF-κB signalling in a mouse model of lung adenocarcinoma. Nature 462: 104–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N 2004. Methods for monitoring autophagy. Int J Biochem Cell Biol 36: 2491–2502 [DOI] [PubMed] [Google Scholar]
- Mizushima N, Komatsu M 2011. Autophagy: renovation of cells and tissues. Cell 147: 728–741 [DOI] [PubMed] [Google Scholar]
- Moscat J, Diaz-Meco MT 2009. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 137: 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi H, Yamamoto M 2004. Nrf2–Keap1 defines a physiologically important stress response mechanism. Trends Mol Med 10: 549–557 [DOI] [PubMed] [Google Scholar]
- Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M 2006. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell 21: 689–700 [DOI] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. 2003. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 112: 1809–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BE, Kaiser SE, Shaler TA, Ng AC, Hara T, Hipp MS, Lage K, Xavier RJ, Ryu KY, Taguchi K, et al. 2010. Ubiquitin accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. J Cell Biol 191: 537–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Duran A, Selloum M, Champy MF, Diez-Guerra FJ, Flores JM, Serrano M, Auwerx J, Diaz-Meco MT, Moscat J 2006. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab 3: 211–222 [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Codogno P, Levine B 2012. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov 11: 709–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz L, Diaz-Meco MT, Nakano H, Moscat J 2000. The atypical PKC-interacting protein p62 channels NF-κB activation by the IL-1–TRAF6 pathway. EMBO J 19: 1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, Hirohashi S 2008a. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology 135: 1358–1368.e4 [DOI] [PubMed] [Google Scholar]
- Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S 2008b. Cancer related mutations in NRF2 impair its recognition by Keap1–Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci 105: 13568–13573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, et al. 2006. Dysfunctional KEAP1–NRF2 interaction in non-small-cell lung cancer. PLoS Med 3: e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporn MB, Liby KT 2012. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev Cancer 12: 564–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N 2011. Autophagy-deficient mice develop multiple liver tumors. Genes Dev 25: 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Abbi S, Zheng C, Guan JL 2000. Suppression of Pyk2 kinase and cellular activities by FIP200. J Cell Biol 149: 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve NF, Lau A, Zhang DD 2010. Regulation of the Nrf2–Keap1 antioxidant response by the ubiquitin proteasome system: an insight into cullin–ring ubiquitin ligases. Antioxid Redox Signal 13: 1699–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Guan JL 2012. Pro-tumorigenic function of autophagy in mammary oncogenesis. Autophagy 8: 129–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Wang X, Gan B, Urvalek AM, Melkoumian ZK, Guan JL, Zhao J 2006. Sumoylation delimits KLF8 transcriptional activity associated with the cell cycle regulation. J Biol Chem 281: 16664–16671 [DOI] [PubMed] [Google Scholar]
- Wei H, Gan B, Wu X, Guan JL 2009. Inactivation of FIP200 leads to inflammatory skin disorder, but not tumorigenesis, in conditional knock-out mouse models. J Biol Chem 284: 6004–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Wei S, Gan B, Peng X, Zou W, Guan JL 2011. Suppression of autophagy by FIP200 deletion inhibits mammary tumorigenesis. Genes Dev 25: 1510–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E 2012. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 12: 401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten MW, Geetha T, Seibenhener ML, Babu JR, Diaz-Meco MT, Moscat J 2005. The p62 scaffold regulates nerve growth factor-induced NF-κB activation by influencing TRAF6 polyubiquitination. J Biol Chem 280: 35625–35629 [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ 2010. Eaten alive: a history of macroautophagy. Nat Cell Biol 12: 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, Bause A, Li Y, Stommel JM, Dell’antonio G, et al. 2011. Pancreatic cancers require autophagy for tumor growth. Genes Dev 25: 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N 2003. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci 100: 15077–15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber J, McJunkin K, Fellmann C, Dow LE, Taylor MJ, Hannon GJ, Lowe SW 2011. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat Biotechnol 29: 79–83 [DOI] [PMC free article] [PubMed] [Google Scholar]