Histone post-transcriptional modifications play important roles in the regulation of replication, transcription, and DNA repair. Key questions remain regarding the role of the acetyltransferase complex Hat1p/Hat2p in chromatin remodeling. High-resolution structural studies by Li et al. now reveal how Hat1p/Hat2p recognizes and facilitates the modification of N-terminal lysine residues on the newly assembled histone H3/H4 heterodimer prior to its incorporation into chromatin. This study delineates the mechanism of substrate recognition by the Hat1p/Hat2p complex, which is critical to DNA replication and chromatin remodeling processes.
Keywords: acetyltransferase, Hat1p/Hat2p, histone modification
Abstract
Post-translational modifications of histones are significant regulators of replication, transcription, and DNA repair. Particularly, newly synthesized histone H4 in H3/H4 heterodimers becomes acetylated on N-terminal lysine residues prior to its incorporation into chromatin. Previous studies have established that the histone acetyltransferase (HAT) complex Hat1p/Hat2p medicates this modification. However, the mechanism of how Hat1p/Hat2p recognizes and facilitates the enzymatic activities on the newly assembled H3/H4 heterodimer remains unknown. Furthermore, Hat2p is a WD40 repeat protein, which is found in many histone modifier complexes. However, how the WD40 repeat proteins facilitate enzymatic activities of histone modification enzymes is unclear. In this study, we first solved the high-resolution crystal structure of a Hat1p/Hat2p/CoA/H4 peptide complex and found that the H4 tail interacts with both Hat1p and Hat2p, by which substrate recruitment is facilitated. We further discovered that H3 N-terminal peptides can bind to the Hat2p WD40 domain and solved the structure of the Hat1p/Hat2p/CoA/H4/H3 peptide complex. Moreover, the interaction with Hat2p requires unmodified Arg2/Lys4 and Lys9 on the H3 tail, suggesting a novel model to specify the activity of Hat1p/Hat2p toward newly synthesized H3/H4 heterodimers. Together, our study demonstrated the substrate recognition mechanism by the Hat1p/Hat2p complex, which is critical for DNA replication and other chromatin remodeling processes.
As the key building blocks of chromatin, histones are subjected to a variety of post-translational modifications that have critical roles in replication, transcription, heterochromatin maintenance, and DNA repair (Nakayama et al. 2001; Grewal and Moazed 2003; Kouzarides 2007; Bannister and Kouzarides 2011). Histone acetylation, which represents the transfer of an acetyl moiety from acetyl-coenzyme A (AcCoA) to the ε-amino group of target lysine residues, is catalyzed by histone acetyltransferase (HAT) families (Brownell and Allis 1996; Wu et al. 2012). Hat1p, the first HAT identified in yeast, was originally isolated from cytosolic extract and therefore characterized as a type B HAT (Parthun et al. 1996), although further studies showed that Hat1p is also present in the nucleus (Verreault et al. 1998; Imhof and Wolffe 1999; Poveda et al. 2004). As its unique feature, Hat1p is specific for the acetylation of newly synthesized H4 at Lys5 and Lys12 residues before the incorporation of H3/H4 heterodimer into nucleosomes (Kleff et al. 1995; Parthun et al. 1996). Genetic deletion of Hat1p in yeast or its homolog, Hat1, in mice resulted in the loss of acetylation on H4K5 and H4K12 on the nascent histone (Parthun et al. 1996; Nagarajan et al. 2013), supporting that Hat1p is the sole enzyme responsible for this evolutionarily conserved histone modification.
Several binding partners of Hat1p have been identified, including Hat2p and Hif1 (Hat1-interacting factor 1). Hat2p and its human homolog, retinoblastoma-binding protein RbAp46, significantly enhanced the enzymatic activity of Hat1p by facilitating H4 association with Hat1p (Parthun et al. 1996). Furthermore, Hat2p can modulate the substrate specificity of Hat1p; i.e., while the recombinant Hat1p acetylated both H4K5 and H4K12 in vitro, the Hat1p/Hat2p complex purified from yeast extracts was highly specific for H4K12 (Parthun et al. 1996). Therefore, it has been suggested that Hat1p/Hat2p forms a holoenzyme complex (Ai and Parthun 2004). In addition, Hif1, which is exclusively a nuclear protein, has been shown to bind the Hat1p/Hat2p complex and function as a histone chaperone to facilitate chromatin assembly (Ai and Parthun 2004; Poveda et al. 2004).
In support of its critical roles on chromatin structure, Hat1 deficiency in mice caused neonatal lethality associated with an array of developmental defects, especially of the lungs and skeleton (Nagarajan et al. 2013). In addition, Hat1 knockout mouse embryonic fibroblasts (MEFs) displayed a significant increase in genomic instability as well as defects in DNA repair. Moreover, not only the acetylation on H4, the direct target of Hat1, but the acetylation on histones H3 were also altered during replication-coupled chromatin assembly in cells, possibly due to the potential cross-regulation of distinct histone modifications (Nagarajan et al. 2013). Similarly, although the genetic deletion of Hat1p alone revealed a relatively modest phenotype in yeast, Hat1p deletion combined with mutations of H3 N-terminal tails resulted in the loss of telomeric silencing and also sensitivity to DNA-damaging agents (Kelly et al. 2000; Qin and Parthun 2002).
Although studies have begun to elucidate the roles of Hat1p/Hat2p in chromatin remodeling processes, several important questions remain to be addressed. First, human RbAp46 can bind to the α helix immediately following the N-terminal tail of H4, and it has been suggested that such interaction might facilitate the association between human Hat1 and its substrate (Verreault et al. 1998; Vermaak et al. 1999; Furuyama et al. 2006). However, it is unclear whether this H4/RbAp46 interaction is evolutionarily conserved and, more importantly, whether this interaction could function in the context of the Hat1p/Hat2p catalytic complex. Furthermore, it was proposed that the methylation of the H4 tail (e.g., on H4K8 and H4K16) could interfere with binding to the catalytic cleft on Hat1p and therefore help to control the substrate specificity of Hat1p toward the newly synthesized H4 protein (Dutnall et al. 1998; Makowski et al. 2001; Benson et al. 2007). How specificity is achieved for newly synthesized H3/H4 heterodimers and how Hat2p contributes to this selectivity remain unknown.
Results
Overall structure of the Hat1p/Hat2p/CoA/H4 complex
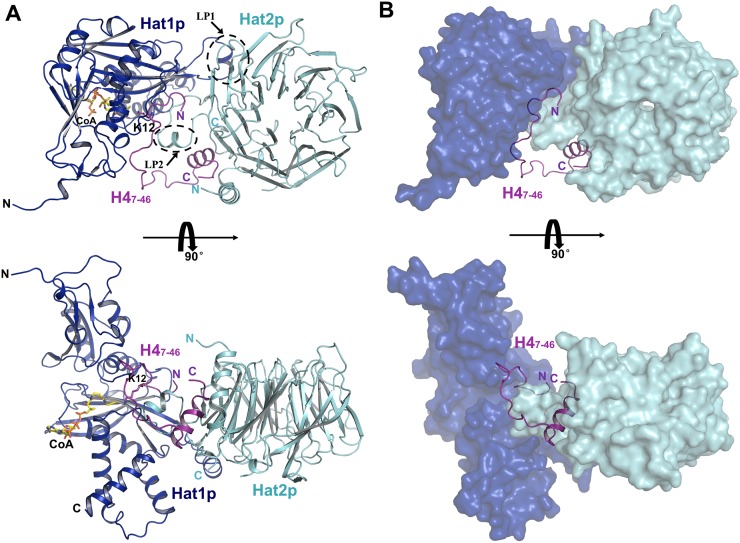
To unravel those questions, we solved the crystal structure of a Hat1p/Hat2p/CoA/H4 peptide complex at 2.0 Å resolution (Fig. 1; Supplemental Table S1). In the structure, each Hat1p/Hat2p/CoA/H4 heterotetrameric complex contains four components, with the stoichiometry of 1:1:1:1 (Fig. 1A). Although the segment of yeast histone H4 used for crystallization contained residues 1–48, only residues 7–46 could be well defined in the structure (Supplemental Fig. S1A). Residues 1–7 and 319–320 of Hat1p and residues 1–8, 86–105, and 387–401 of Hat2p cannot be located, likely due to the flexibility of these regions.
Figure 1.
Crystal structure of yeast Hat1p/Hat2p/H4 peptide/CoA. (A) Overall structure of the Hat1p/Hat2p/H4 peptide/CoA complex. The protein structure is shown in the cartoon, and CoA is shown in stick representation, with Hat1p, Hat2p, and H4 peptide colored in blue, cyan, and purple, respectively. (B) H4 peptide in the structure of the Hat1p/Hat2p/H4 peptide/CoA complex. Hat1p and Hat2p are shown in surface mode, with Hat1p colored blue, and Hat2p colored cyan.
Consistent with previous reports, Hat1p was comprised of two domains, an N-terminal and a C-terminal domain (Wu et al. 2012), with a CoA molecule bound to the C-terminal domain (Fig. 1A). Sequence alignments showed that the regions clustered in the Hat1p C-terminal domain were highly conserved among different species (Dutnall et al. 1998). Within these conserved regions, a segment (LP1, residues 200–208) is more diverse in both amino acid composition and segment length (Dutnall et al. 1998). In our structure, this LP1 segment formed a well-ordered helix (Fig. 1A), although its corresponding region was disordered in the human Hat1 structure (Protein Data Bank [PDB] code 1BOB) (Dutnall et al. 1998). Except for the LP1 region, Hat1p in the Hat1p/Hat2p/CoA/H4 complex structure is superposed well with the solo structure of Hat1. The RMSD (root-mean-square deviation) is ∼0.376 Å over the aligned 316 Cα atoms of the Hat1 structure. In particular, the residues forming the active site were well aligned. These analyses suggest that the Hat1p has almost no conformational change during the formation of the Hat1p/Hat2p/CoA/H4 complex.
The Hat2p protein presents a typical compact seven-bladed propeller-like structure similar to other WD40 proteins (Han et al. 2006; Murzina et al. 2008; Song et al. 2008). The N-terminal portion of Hat2p forms a helix adjacent to the seventh blade, and the C-terminal part folds back to the end of this N-terminal helix (Fig. 1A). In addition, a long loop (LP2, residues 324–344) extending from the sixth blade contributes to the interaction of Hat2p with both Hat1p and H4 (Fig. 1A).
Hat1p and Hat2p interaction
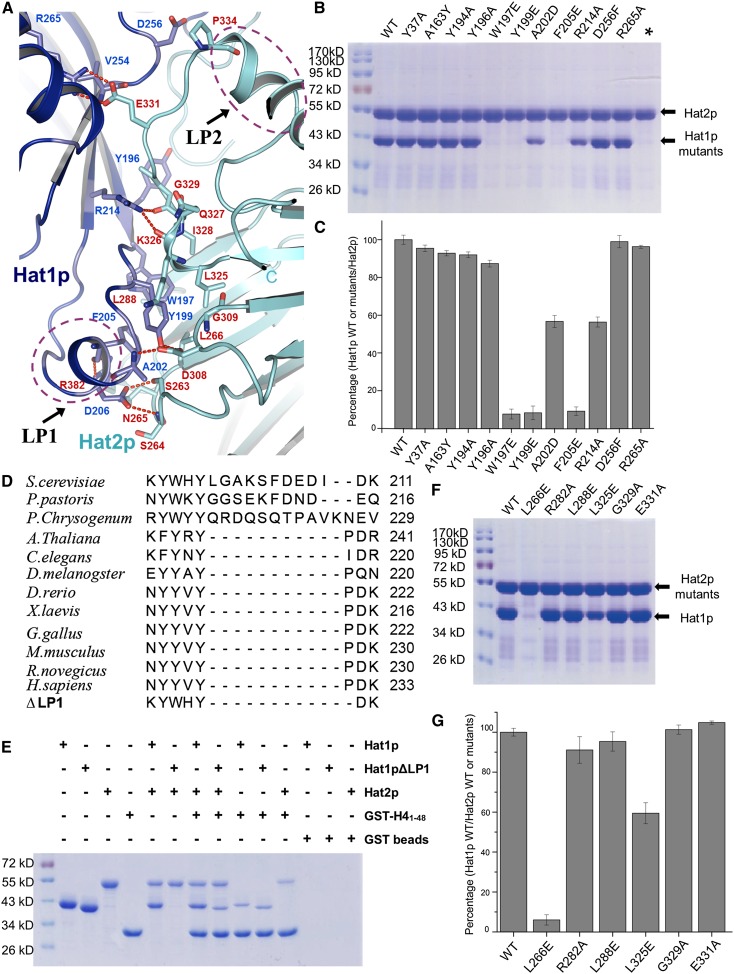
The Hat1p and Hat2p interaction is stabilized by a series of hydrogen bonds, salt bridges, and hydrophobic interactions, and the interface was buried ∼820 Å2 (Fig. 2A; Supplemental Table S2). The LP1 segment of Hat1p contributes most to these interactions. The side chains of Tyr199 of Hat1p and Asp308 of Hat2p form hydrogen bonds with the main chain of Hat1p-Ala202, while Lys211 and Phe205 of Hat1p contact with Leu288 and Arg282 of Hat2p via main chain or side chain hydrogen bonds, respectively. Asp206 of Hat1p also interacts with Ser263 and Ser264 of Hat2p through their side chain hydrogen bonds (Fig. 2A). Furthermore, Trp197, Tyr199, and Phe205 of Hat1p together with Leu266, Leu288, and Leu325 of Hat2p form a hydrophobic core at the interface, which appears to be critical for the complex formation (Figs. 2A–C). In addition, several residues in the LP2 segment of Hat2p also contribute to the Hat1p/Hat2p interaction (Fig. 2A).
Figure 2.
Binding interface between Hat1p and Hat2p. (A) Close-up view of the interface between Hat1p and Hat2p. The interaction residues of Hat1p and Hat2p are labeled and shown in stick representation and colored in blue and yellow, respectively. The salt bridges are shown as red dashed lines. LP1 of Hat1p and LP2 of Hat2p are circled by purple dashed lines. (B) Pull-down of different Hat1p mutants by wild-type (WT) Hat2p. Each mutation is labeled above the line. (C) The interaction between Hat1p wild type or mutants and Hat2p wild type was evaluated by quantifying the intensity of the bands in SDS-PAGE using the AlphaEaseFC software and normalizing it to the wild type. The presented data are the average of three experiments. The error bars correspond to one standard deviation. The mutants are labeled as in B. (D) The sequence alignment of the LP1 region among species. The deletion of the Hat1p LP1 region is also shown at the bottom. (E) Pull-downs of Hat1p, LP1-deleted Hat1p, Hat2p, and GST-H41–48. (F) Pull-downs of wild-type Hat1p and different Hat2p mutants. Each mutation is labeled above the line. (G) The interaction between Hat2p wild type or mutants and Hat1p wild type was evaluated by quantifying the intensity of the bands in SDS-PAGE using the AlphaEaseFC software and normalizing it to the wild type. The presented data are the average of three experiments. The error bars correspond to one standard deviation. The mutants are labeled as in F.
To confirm the structural observation, we used the in vitro pull-down assay to determine the residues of Hat1p that are critical for Hat1p/Hat2p heterodimer formation. The mutations of W197E, Y199E, and F205E in Hat1p, each of which eliminates a hydrophobic interaction, abolished the interaction with Hat2p (Figs. 2B,C). Although the LP1 segment is not very conserved in higher organisms (Fig. 2D; Supplemental Fig. S2), the structure reveals that the LP1 segment might be important for the Hat1p/Hat2p heterodimer formation. Deletion of the LP1 segment (ΔLP1) of Hat1p abolished the interaction between Hat1p and Hat2p in vitro (Fig. 2E; Supplemental Fig. S3), which suggested that some other protein might involve in the interaction of Hat1p and Hat2p. Indeed, consistent with the structural observation, the N terminus of histone H4 might function as a “linker” protein that interacts with both Hat1p and Hat2p (Fig. 1), which could spatially interact with each other even without the contribution of LP1 (Fig. 2E). For higher organisms, the Hat1p/Hat2p complex formation may be more complicated than in yeast. Consistent with this hypothesis, although the W197E and W199E mutants of Hat1p would abolish the interaction between the Hat1p and Hat2p heterodimeric formation in vitro, the W197E and W199E alleles of HAT1 were able to complement the DNA damage sensitivity in the hat1Δ yeast, suggesting that the Hat1p/Hat2p interaction is likely to be stabilized by multiple interactions or other proteins in vivo (Supplemental Fig. S4).
On the other hand, the mutations of Y37A, A163Y, Y194A, Y196A, and A202D of Hat1p showed little effect on the interaction in vitro (Fig. 2B,C). Similarly, we also tested a set of mutants of Hat2p and found that the L266E of Hat2p lost its ability to bind Hat1p, while other mutations showed little effect (Fig. 2F,G).
H4 interacts with both Hat1p and Hat2p
The binding of the N-terminal segment of Histone H4 with Hat1p/Hat2p could be divided into three regions (residues 8–18, 19–30, and 31–45) (Fig. 3A–C). The N-terminal region (residues 7–18) of H4 was embedded in the cave between Hat1p and Hat2p and mainly interacted with Hat1p, which was similar to the reported Hat1/H4 structure (Wu et al. 2012). Lys20 in the second region (residues 19–30) formed a salt bridge with the Asp357 from LP2 of Hat2p (Fig. 3A). The C-terminal region (residues 29-45), which formed the helix 1 of H4, inserted into the groove formed by the Hat2p LP2 extension, the N-terminal helix 1, and the C terminus (Fig. 3B,C), which is consistent with the structural observation between H4 and RbAp46/8 (the human homologs of Hat2p) (Fig. 3D; Murzina et al. 2008). Overall, the interface areas of the N-terminal and C-terminal regions of H4 covered ∼608 Å2 and 802 Å2, respectively. The interaction between H4 and the Hat1p/Hat2p complex was strongly stabilized by salt bridge bonds formed by the positively charged residues of H4, such as Lys16, Lys20, Arg35, Arg39, Arg40, and Lys44, with the conserved acidic residues (Asp335, Asp336, Glu338, and Asp339) of Hat2p LP2 (Fig. 3B; Supplemental Fig. S5).
Figure 3.
H4 could recognize both Hat1p and Hat2p. (A) Close-up interview of the binding sites of the H4 peptide between Hat1p and Hat2p. The side chains from Hat1p and Hat2p are labeled and shown as blue sticks and yellow sticks, respectively. The salt bridges are shown as dashed lines. (B,C) Close-up interview of the binding sites of the H4 peptide on Hat2p. The binding interfaces on Hat2p are shown in a charged surface and stick representation in an electrostatic surface (B) and cartoon (C), respectively. For B, white, blue, and red indicate neutral, positive, and negative surfaces, respectively. The H4 peptide is shown as purple sticks. The residues are labeled. (D) Structure alignment of the Hat2p/H41–48 in the Hat1p/Hat2p/H4 peptide/CoA complex with the RbAp46/H425–42 structure (PDB code 3CFS). (E) GST pull-down assay of different GST-tagged mutant H4 peptides and the Hat1p/Hat2p complex. Each mutation is labeled above the gel. (F) The interaction between the Hat1p/Hat2p complex and GST-tagged mutant H4 peptides was evaluated by quantifying the intensity of the bands in SDS-PAGE using the AlphaEaseFC software and normalizing it to the wild type (WT). The presented data are the average of three experiments. The error bars correspond to one standard deviation. The mutants are labeled as in E.
Previous structural and biochemical studies suggested that the Lys12 of H4 (H4K12) inserts into the active site of Hat1p to gain access to AcCoA (Dutnall et al. 1998; Wu et al. 2012). In our structure, H4K12 is aligned in the active site, with the CoA molecule coming into a concave groove from the opposite side of H4K12, which enables the ε-amino group of H4K12 and the SH group of CoA to contact each other (Fig. 3A; Supplemental Fig. S6). Our structure was consistent with the catalytic mechanism that had been proposed with the structure of human Hat1 (Wu et al. 2012). Hat1p/Hat2p complex purified from yeast extracts specifically acetylated H4K12 residue, while the recombinant yeast Hat1p was not only able to acetylate H4K12 but also exhibited low activity to H4K5 (Parthun et al. 1996). In vitro acetylation assays showed that the recombinant Hat1p/Hat2p complex specifically acetylated the H4K12 residue, in agreement with the idea that Hat2p regulates the substrate specificity of Hat1p (Supplemental Fig. S7). It seems that Hat2p could exert such an effect potentially through binding to helix 1 of H4, which helps to preferentially stabilize H4K12 in the active site.
Previous studies of Hat1p using a variety of histone substrates identified a putative glycine-rich consensus sequence that Hat1p targets for acetylation (G−3X−2G−1KX+1G+2) (Parthun et al. 1996). We constructed mutations in this consensus sequence in the context of GST-H4 tail fusions to determine whether these residues influence binding of the Hat1p/Hat2p complex to its substrate. As seen in Figure 3, E and F, the glycine residues in the −1 and −3 position could influence Hat1p/Hat2p complex binding, while the +2 glycine has little effect. Acetylation of Lys16 is a highly abundant chromatin mark (Dang et al. 2009). In the structure, a salt bridge could be observed between H4K16 and E338 of Hat2p (Fig. 3A). The isothermal titration calorimetry (ITC) results show that the acetylation of Lys16 could decrease the binding affinity between the Hat1p/Hat2p complex and the H4 peptide (Supplemental Fig. S8), which indicates that the Hat1p/Hat2p complex prefers to bind newly synthesized H4.
Hat2p could bind the different methylated states of H3
Our structure of the Hat1p/Hat2p/CoA/H4 complex demonstrated that the Hat2p has a critical role in facilitating the binding of H4 and the enzymatic specificity of Hat1p (Supplemental Fig. S7). However, previous studies suggested that the newly synthesized H3 and H4 proteins form a heterodimer before the acetylation of H4, and it was believed that the free form of the H4 protein was only transiently present after translation (Ruiz-Carrillo et al. 1975; Jackson et al. 1976). Therefore, it is very important to understand how Hat1p/Hat2p recognizes newly assembled H3/H4 heterodimers and, in particular, whether H3 has any function in this process.
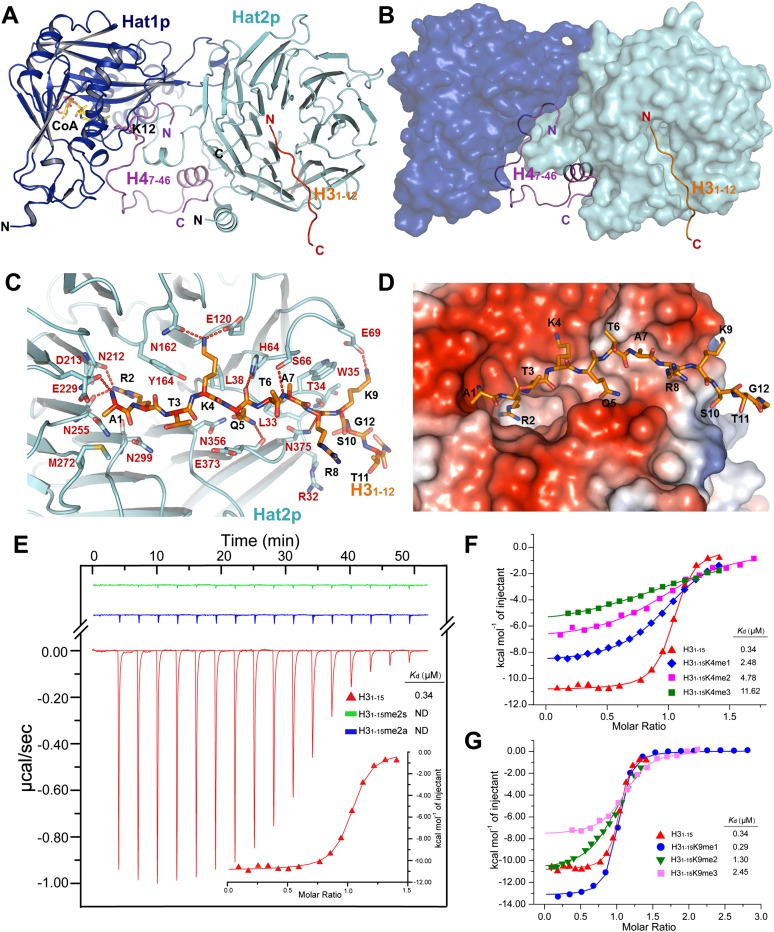
Previous studies have reported that the proteins that contain the WD40 domain could bind to histone tails, such as WDR5 and NURF55 (Couture et al. 2006; Han et al. 2006; Schmitges et al. 2011), but in the Hat1p/Hat2p/CoA/H4 complex, no direct interface was observed between the hole of the WD40 domain of Hat2p and the H4 tail. We propose that the Hat2p WD40 domain might be able to bind to the H3 tail in H3/H4 heterodimer. Indeed, our in vitro ITC analyses indicated that the H3 peptide containing the first 15 amino acids could interact with the Hat1p/Hat2p/H4 peptide complex with a ratio of 1:1 and with an apparent affinity with a Kd of ∼0.34 μM (Supplemental Fig. S9).
We next set out to cocrystallize the H3 peptide (residues 1–15) with the Hat1p/Hat2p/H4 complex and CoA molecule and solved the structure of the Hat1p/Hat2p/H3/H4/CoA complex at a resolution of 2.5 Å (Fig. 4A,B). The omit Fo–Fc electron density clearly defined the first 12 amino acids of the H3 peptide in the structure (Supplemental Fig. S1B), which is located in a narrow channel on the surface of the WD40 domain (Fig. 4C,D). Arg2 of H3 inserts into the small hole of the WD40 domain of Hat2p. The interaction between H3 and Hat2p is stabilized through highly conserved residues in Hat2p by forming a series of hydrogen bonds and salt bridges (Fig. 4C; Supplemental Fig. S5). The overall binding surface is negatively charged on Hat2p, corresponding with the positively charged N-terminal tail of H3 (Fig. 4D). Of note, the structure of the Hat1p/Hat2p/H3/H4/CoA complex closely resembles the structure of Hat1p/Hat2p/H4/CoA, with an RMSD of 0.663 Å for Hat1p and 0.227 Å for Hat2p, suggesting that the binding of the H3 N-terminal tail to Hat2p does not induce significant conformational change on the Hat1p/Hat2p complex (Supplemental Fig. S10).
Figure 4.
Hat2p could bind different methylation statuses of histone H3. (A) Overall structure of the Hat1p/Hat2p/H3/H4/CoA complex. The protein structure is shown in cartoon, and CoA is shown in stick representation, with Hat1p, Hat2p, the H3 peptide, and the H4 peptide colored in blue, cyan, orange, and purple, respectively. (B) The H3 peptide and H4 peptide in the structure of the Hat1p/Hat2p/H3 peptide/H4 peptide/CoA complex. Hat1p and Hat2p are shown in surface mode, with Hat1p colored blue, and Hat2p colored cyan. (C) Close-up interview of the binding sites of the H3 peptide on the Hat2p surface. The interaction residues of Hat2p and the H3 peptide are labeled in chartreuse and orange stick representation, respectively, and the salt bridges are shown as dashed lines. (D) Electrostatic potential surface of the H3-binding sites on Hat2p. Negatively and positively charged surfaces are colored in red and blue, respectively. (E) ITC curves of free H31–15 and methylation of H3R2 (H31–15R2me2s and H31–15R2me2a) peptides titrated into the Hat1p/Hat2p/H4 peptide complex. (ND) Not detectable. The first peak in the thermogram has not been used for analysis. (F,G) ITC fitting curves of the Hat1p/Hat2p/H4 peptide complex with histone H3 peptides of different K4 methylation states (F) and different K9 methylation states (G). Peptide sequences and complete thermodynamic parameters are listed in Supplemental Table S3. The first peak in the thermogram has not been used for analysis.
It is quite interesting that the side chain of H3R2 inserts into the small hole of the WD40 domain of Hat2p. The Arg2 forms hydrogen bonds with Asn212 and Asp213 of Hat2p. In addition, the side chains of Arg123 and Arg125 of Hat2p show a large conformational change upon binding with H3 (Supplemental Fig. S11), which might help to make more space for the side chain of H3R2. H3R2 can be subjected to symmetric or asymmetric dimethylation in cells, which regulates transcriptional activity (Kirmizis et al. 2007). The structure reveals that the methylation of Arg2 might disrupt the corresponding interactions and also result in the spatial constraint for insertion into the pocket. The size of the small hole appears to be conserved among different species (Supplemental Fig. S12). As predicted, in contrast to the unmethylated H3 peptide, symmetric or asymmetric dimethylation of Arg2 abolished the interaction between H3 and the Hat1p/Hat2p/H4 complex in the ITC assay (Fig. 4E).
The different methylation states on H3K4 and H3K9 are common events that occur in cells and have different biological functions for regulating the gene transcription and chromatin assembly (Stewart et al. 2005; Sims and Reinberg 2006). Our structure reveals that the side chain of H3K4 forms two hydrogen bonds with the side chains of Asn162 and Glu120 of Hat2p, and the side chain of H3K9 forms a salt bridge with the side chain of Glu69 of Hat2p (Fig. 4C), which indicates that the modified H3K4 or H3K9 might influence the reorganization of H3 with Hat2p. Indeed, the ITC results showed that H3K4me1, H3K4me2, and H3K4me3 have binding affinity to the Hat1p/Hat2p/H41–48 complex with a Kd of 2.48, 4.78, and 11.62 μM, and H3K9me1, H3K9me2, and H3K9me3 bind to the Hat1p/Hat2p/H41–48 complex with a Kd of 0.29, 1.30, and 2.45 μM, respectively (Fig. 4F,G; Supplemental Fig. S13). Compared with the free H3 tail binding to the Hat1p/Hat2p/H41–48 complex with a Kd ∼0.34 μM, the trimethylation on H3K4 destabilizes the interaction between H3 and Hat2p, and methylation of H3K9 has a weaker effect on the binding affinity. Previous studies have reported that the newly synthesized histone H3 has the monomethylation on H3K9 (Loyola et al. 2006). Our results suggest that Hat2p could recognize the histone H3 tails with/without this modification (Fig. 4G), which indicates that the H3K9 might be monomethylated either before or after the H4K12 acetylation on the newly synthesized histone H3/H4 heterodimer. Definitely, this needs further investigation to know the sequential orders. Together, our data indicated that the interaction of H3 with Hat2p depends on the modification status of its tail.
Discussion
Histone acetylation is a highly dynamic post-translational modification that critically regulates replication, transcription, and heterochromatin silencing (Shahbazian and Grunstein 2007). Hat1p, which is the founding member of type B HATs identified in Saccharomyces cerevisiae, functions with its cofactor, Hat2p, to acetylate N-terminal lysine residues of histone H4 before its incorporation into chromatin (Ai and Parthun 2004). In the present study, we revealed that a hydrophobic core at the interface of Hat1p and Hat2p, formed by an array of conserved residues, contributes most to the interaction between the two proteins. Furthermore, the mutations of W197E, Y199E, and F205E on Hat1p, and L266E on Hat2p, which disrupt this hydrophobic interaction, abolished the formation of the Hat1p/Hat2p complex (Figs. 1, 2).
Previous studies suggested that residues 5–19 of H4 could bind directly to the active site of Hat1p (Wu et al. 2012). Our data further showed that the N-terminal portion of H4 not only binds to Hat1p but also directly interacts with Hat2p, especially through residues 29–41 (Fig. 3). This region, which is normally unfolded (Dutnall et al. 1998; Wu et al. 2012), folds into a helix upon interaction with Hat2p. Therefore, Hat2p contributes to the substrate specificity for Hat1p/Hat2p by stabilizing the access of H4K12 to the active site of Hat1p. Consistent with this notion, while recombinant human and yeast Hat1 can acetylate both H4K5 and H4K12, we showed that the complex of recombinant Hat1p/Hat2p specifically acetylates H4K12 (Supplemental Fig. S7).
Unexpectedly, we discovered that Hat2p in the Hat1p/Hat2p complex can directly interact with the N-terminal tail of H3 (Fig. 4) in addition to stabilizing the association with substrate H4. The analysis of surface electrostatic potential on Hat2p showed that its H3-binding surface in the WD40 domain is highly negatively charged, which helps the interaction with the basic residues of the H3 N-terminal tail (Fig. 4C,D). More interestingly, we observed that the binding of H3 to Hat2p depends on the methylation status of H3; i.e., dimethylation of H3R2 abolished the interaction and different methylation status of H3K4 and H3K9 and also affected the peptide-binding affinity (Fig. 4E–G). Many histone modification enzyme complexes, such as the Nurd/Sin3, MLL/Set1, and PRC2 complexes, contain WD40 repeat proteins as core subunits. Several studies have reported that certain WD40 domains recognize methylated histone tails (Couture et al. 2006; Han et al. 2006; Margueron et al. 2009). It is possible that complementary amino acid changes in the catalytic and WD40 subunits can accommodate their robust association. Alternatively, other subunits in the enzyme complex may facilitate such association (Ahringer 2000; Schmitges et al. 2011; Dharmarajan et al. 2012). Our studies demonstrate that a subset of proteins with WD40 domains could function as a reader for histone methylation to regulate distinct chromatin-associated events. In addition to the methylation of H3R2 and H3K4, the N terminus of H3 can be acetylated in cells on multiple lysine residues (K9, K14, K18, K23, and K27). Based on the charge complementarity, such acetylation would be predicted to disrupt the interaction between H3 and Hat2p. Therefore, it is tempting to speculate that by discriminating against unmethylated/unacetylated H3 N-terminal tails, Hat2p helps to establish the specificity of Hat1p/Hat2p toward newly synthesized H3/H4 heterodimers and away from heavily modified pre-existing histones. Such a mechanism could be essential for the proper maintenance of histone modification markers during chromatin assembly in DNA replication or transcription.
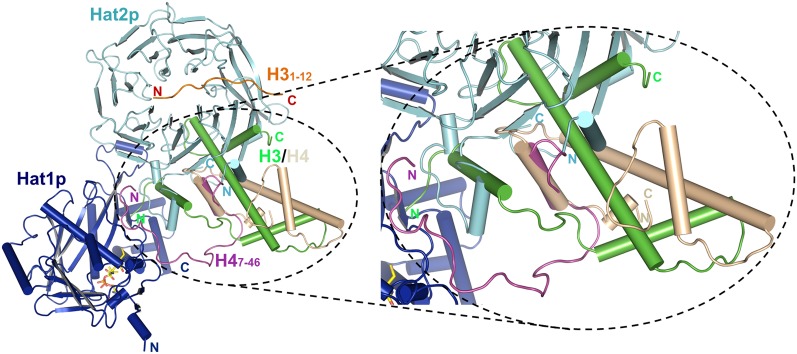
It is intriguing to note that in a model based on our structures, due to spatial constraint, H3/H4 in the Hat1p/Hat2p/H3/H4 complex cannot adopt the heterodimer conformation like that observed in nucleosomes (Fig. 5). This is in agreement with the fact that Hat1p cannot acetylate histone H4 in nucleosomes (Kleff et al. 1995; Parthun et al. 1996). It is possible that upon their synthesis in the cytosol, H3/H4 might adopt a transition state conformation, and the association with certain histone chaperone proteins, such as Hif1, helps to stabilize the native heterodimer. Alternatively, Hat1p/Hat2p themselves might function as histone chaperones, and this hypothesis was supported by the fact that H3/H4 remains associated with Hat1p/Hat2p even after acetylation (Imhof and Wolffe 1999; Lusser et al. 1999; Vermaak et al. 1999; Ai and Parthun 2004; Poveda et al. 2004; Wu et al. 2012), and the Hat1p/Hat2p has been reported to interact with the origin replication complex (Suter et al. 2007; Ge et al. 2011). After acetylation by the Hat1p/Hat2p complex, the H3/H4 heterodimer is then transferred to other chromatin-assembling factors that rearrange the H3/H4 conformation so that it becomes ready to be incorporated into chromatin. For instance, Asf1 has been shown to bind H3/H4 following acetylation and could cause a conformation change of the heterodimer (Zhou et al. 2011). Further structural and biochemical analyses are required to address these possibilities.
Figure 5.
The predicted model of the Hat1p/Hat2p complex binding with the free form of the H3 and H4 proteins. Hat1p, Hat2p, H3, and H4 in the predicted model are colored in blue, cyan, green, and wheat, respectively. The structure is shown in cartoon representation. The H3 and H4 N-terminal peptides, which are crystallized in the complex, are highlighted with orange and purple. The left figure shows the close-up view of the predicted H3 and H4 conformation.
In summary, with the high-resolution structure of the HAT Hat1p with its cofactor, Hat2p, and substrate, H4, we revealed that Hat2p is important for the substrate specificity of Hat1p. We further discovered that although H3 is not a direct substrate for Hat1p/Hat2p, the N-terminal tail of H3 binds with the WD40 domain of Hat2p, and this interaction depends on the different methylated status of H3. Based on the structure of the Hat1p/Hat2p/H3/H4 complex (Fig. 4A), we propose a model in which the simultaneous binding of H3 and H4 by Hat1p/Hat2p specifies acetylation toward newly synthesized H3/H4 heterodimers (Fig. 5), which could be critical for various chromatin-associated processes.
Materials and methods
Protein expression and purification
Yeast Hat2p (full length) with an engineered N-terminal 6xHis tag was cloned into vector pFastBac 1 (Invitrogen) and expressed in Hi-5 insect cells for 36 h at 28°C. After harvesting, Hat2p was initially purified using Ni-NTA (Novagen) and washed three times with buffer containing 25 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 15 mM imidazole. Yeast Hat1p (1–320) with a stop codon at the end was ligated into vector pET30a and expressed in Escherichia coli strain BL21 (DE3). The precleared cell extract with overexpressed Hat1p was loaded to the Ni column prepared before, which had bound Hat2p. After an extensive wash, the Hat1p/Hat2p complex was eluted with 25 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 250 mM imidazole and further fractionated by anion ion exchange column chromatography (Source-Q, Pharmacia). In order to obtain the Hat1p/Hat2p/H4 peptide complex, the purified Hat1p/Hat2p complex was applied to a column containing GST-H41–48 bound to glutathione sepharose (Pharmacia), which was overexpressed in Rosetta (DE3).
After washing with 25 mM Tris-HCl (pH 8.0) and 150 mM NaCl, the Hat1p/Hat2p/H41–48 complex was released by cleavage of histone H4 from GST using PreScission protease. The complex was further purified by anion ion exchange and gel filtration chromatography (Superdex-200, Pharmacia). For crystallization, the complex was finally prepared in a buffer containing 10 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 3 mM DTT.
The Hat1p/Hat2p/H41–48/H31–15 complex was obtained by mixing Hat1p/Hat2p/H41–48 with H31–15 in 1:10 molar ratio.
Crystallization, data collection, and structure determination
Before crystallization, CoA was added to the Hat1p/Hat2p/H41–48 complex and the Hat1/Hat2/H41–48/H31–15 complex solutions with 2:1 molar ratio. Crystals of the Hat1p/Hat2p/H41–48/CoA complex or the Hat1p/Hat2p/H41–48/H31–15/CoA complex were generated by mixing 1 μL of protein solution with 1 μL of well buffer using the hanging drop vapor diffusion method at 4°C. Diffraction quality crystals were obtained at condition of 0.2 M potassium sodium tartrate and 20% (w/v) polyethylene glycol 3350. Thin plate-shaped crystals appeared overnight and reached the maximal size in ∼1 wk. The crystals were cryoprotected in reservoir solution containing 15%–20% glycerol and flash-frozen in liquid nitrogen prior to data collection.
The Hat1p/Hat2p/H47–46/CoA complex structure was solved using molecular replacement with the Hat1 (PDB ID code 1BOB) structure and the RbAp46/RbAp48 (PDB ID code 3CFV or 3CFS) structure as search models. All of the data were collected at the Shanghai Synchrotron Radiation Facility BL17U and integrated and scaled using the HKL2000 package (Otwinowski and Minor 1997). Further processing was carried out using programs from the CCP4 suite (Collaborative Computational Project, Number 4 1994). Data collection statistics are summarized in Supplemental Table S1. The final model rebuilding was performed using COOT (Emsley and Cowtan 2004), and the protein structure was refined with PHENIX (Adams et al. 2002) using noncrystallographic symmetry (NCS) and stereochemistry information as restraints. The structure factors of the Hat1p/Hat2p/H47–46/CoA complex have been deposited in the PDB (ID code 4PSW). The structure of the Hat1p/Hat2p/H47–46/H31–12/CoA complex was solved with molecular replacement using the structure of the Hat1p/Hat2p/H47–46/CoA complex as a model, and the structure factors were deposited in the PDB (ID code 4PSX). Structural figures were generated in PyMOL (http://www.pymol.org).
In vitro acetylation assays and mass spectrometry (MS)
In vitro HAT reactions were performed for 1 h at 30°C in a 50-μL reaction mixture containing ∼8 μg of the Hat1p/Hat2p/H41–48 complex, 1 mM AcCoA (Sigma), 5 mM nicotinamide (Sigma), 1 mM PMSF, and 1mM DTT in HAT buffer (final 50 mM Tris-HCl at pH 7.5, 5% glycerol, 0.1 mM EDTA, 50 mM KCl) (Grant et al. 1997). For identification of acetylated lysine, samples were separated on one-dimensional SDS-PAGE under the reducing condition. The gel bands corresponding to the targeted protein were excised followed by incubation with 50 mM DTT for 30 min at 55°C, treated by 25 mM indole-3-acetic acid (IAA) for 20 min at room temperature in the dark, and digested by the sequence grade modified trypsin (Promega) in 50 mM ammonium bicarbonate overnight at 37°C. The peptides were extracted twice with 0.1% trifluoroacetic acid in 50% acetonitrile aqueous solution for 30 min. The extractions were then centrifuged in a speedvac to reduce the volume. For liquid chromatography-tandem MS (LC-MS/MS) analysis, the digestion product was separated by a 65-min gradient elution at a flow rate of 0.250 µL/min by using the EASY-nLCII integrated nano-high-performance LC (nano-HPLC) system (Proxeon), which was directly interfaced with the Thermo LTQ-Orbitrap mass spectrometer. The analytical column was a homemade fused silica capillary column (75 µm inner diameter, 150 mm length; Upchurch) packed with C-18 resin (300 Å, 5 µm; Varian). Mobile phase A consisted of 0.1% formic acid, and mobile phase B consisted of 100% acetonitrile and 0.1% formic acid. The LTQ-Orbitrap mass spectrometer was operated in the data-dependent acquisition mode using Xcalibur 2.1 software, and there was a single full-scan mass spectrum in the Orbitrap (400–1800 m/z, 30,000 resolution) followed by 15 data-dependent MS/MS scans in the ion trap at 35% normalized collision energy. The MS/MS spectra from each LC-MS/MS run were searched against the selected database using a Proteome Discovery searching algorithm.
ITC
Calorimetric experiments were conducted at 25°C with a MicroCal iTC200 instrument (GE Healthcare). The protein samples (Hat1p, Hat1p-ΔLP1, Hat2p, and the Hat1p/Hat2p/H41–48complex) were dialyzed against the buffer containing 150 mM NaCl and 20 mM HEPES (pH 7.5). The data were fitted using the Origin 7 software package (Microcal).
Pull-down assay
Hat1p, Hat2p, and H4 mutations were generated by two-step PCR. Hat1p and H4 mutants were cloned into the pGEX6p-1 (Novagen) vector and purified as wild type. Hat2p mutants were cloned into pFASTBAC-1 and purified as wild type. For GST pull-down experiments, GST-H4-bound glutathione sepharose beads (2.0 mg of protein each; GE Healthcare) were incubated with equal amounts of the recombinant Hat1p/Hat2p complex, Hat1p, Hat1pΔLP1, or Hat2p in the binding buffer (20 mM Tris at pH 7.5, 200 mM NaCl) for 30 min at room temperature. Protein-bound glutathione resins were washed six times with the binding buffer and then boiled in sample buffer and subjected to SDS-PAGE analysis. For His tag pull-down experiments, his-tagged wild-type Hat2p or Hat2p mutants were used to pull down nontagged Hat1p mutants or wild-type Hat1p, respectively.
DNA damage sensitivity assay
A fragment containing the HAT1 ORF plus 1 kb of the upstream and downstream sequence was cloned into pRS426 (pRs426-HAT1). HAT1 alleles were constructed by site-directed mutagenesis using pRS426 as the template. The HAT1 expression plasmids were transformed into UCC6584 (wild type) and MPY201 (hat1Δ) (Kelly et al. 2000). These strains also express an H3K9,14R allele as the sole copy of histone H3. Cells were grown in liquid synthetic medium lacking uracil to retain the HAT1 expression vector, and an equal quantity of cells were harvested from each strain. Tenfold serial dilutions were made, and cells were spotted on synthetic medium in the absence or presence of methyl methane sulfonate (MMS).
Acknowledgments
We thank Dr. Shilong Fan and the staff at the SSRF BL17U beamline for assistance in data collection, and Dr. Haitao Li for providing the modified histone peptides. This work was supported by the Ministry of Science and Technology of China (2011CB910501 and 2012CB911101), the National Natural Science Foundation of China (31030020 and 31170679), and the National Institutes of Health (R01GM062970 to M.R.P.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.240531.114.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Ahringer J 2000. NuRD and SIN3 histone deacetylase complexes in development. Trends Genet 16: 351–356 [DOI] [PubMed] [Google Scholar]
- Ai X, Parthun MR 2004. The nuclear Hat1p/Hat2p complex: a molecular link between type B histone acetyltransferases and chromatin assembly. Mol Cell 14: 195–205 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T 2011. Regulation of chromatin by histone modifications. Cell Res 21: 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson LJ, Phillips JA, Gu Y, Parthun MR, Hoffman CS, Annunziato AT 2007. Properties of the type B histone acetyltransferase Hat1: H4 tail interaction, site preference, and involvement in DNA repair. J Biol Chem 282: 836–842 [DOI] [PubMed] [Google Scholar]
- Brownell JE, Allis CD 1996. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev 6: 176–184 [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Couture JF, Collazo E, Trievel RC 2006. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat Struct Mol Biol 13: 698–703 [DOI] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL 2009. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459: 802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmarajan V, Lee JH, Patel A, Skalnik DG, Cosgrove MS 2012. Structural basis for WDR5 interaction (Win) motif recognition in human SET1 family histone methyltransferases. J Biol Chem 287: 27275–27289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutnall RN, Tafrov ST, Sternglanz R, Ramakrishnan V 1998. Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily. Cell 94: 427–438 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Furuyama T, Dalal Y, Henikoff S 2006. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc Natl Acad Sci 103: 6172–6177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Wang H, Parthun MR 2011. Nuclear Hat1p complex (NuB4) components participate in DNA repair-linked chromatin reassembly. J Biol Chem 286: 16790–16799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11: 1640–1650 [DOI] [PubMed] [Google Scholar]
- Grewal SI, Moazed D 2003. Heterochromatin and epigenetic control of gene expression. Science 301: 798–802 [DOI] [PubMed] [Google Scholar]
- Han Z, Guo L, Wang H, Shen Y, Deng XW, Chai J 2006. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol Cell 22: 137–144 [DOI] [PubMed] [Google Scholar]
- Imhof A, Wolffe AP 1999. Purification and properties of the Xenopus Hat1 acetyltransferase: association with the 14-3-3 proteins in the oocyte nucleus. Biochemistry 38: 13085–13093 [DOI] [PubMed] [Google Scholar]
- Jackson V, Shires A, Tanphaichitr N, Chalkley R 1976. Modifications to histones immediately after synthesis. J Mol Biol 104: 471–483 [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Qin S, Gottschling DE, Parthun MR 2000. Type B histone acetyltransferase Hat1p participates in telomeric silencing. Mol Cell Biol 20: 7051–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A, Santos-Rosa H, Penkett CJ, Singer MA, Vermeulen M, Mann M, Bähler J, Green RD, Kouzarides T 2007. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature 449: 928–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleff S, Andrulis ED, Anderson CW, Sternglanz R 1995. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem 270: 24674–24677 [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Loyola A, Bonaldi T, Roche D, Imhof A, Almouzni G 2006. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol Cell 24: 309–316 [DOI] [PubMed] [Google Scholar]
- Lusser A, Eberharter A, Loidl A, Goralik-Schramel M, Horngacher M, Haas H, Loidl P 1999. Analysis of the histone acetyltransferase B complex of maize embryos. Nucleic Acids Res 27: 4427–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski AM, Dutnall RN, Annunziato AT 2001. Effects of acetylation of histone H4 at lysines 8 and 16 on activity of the Hat1 histone acetyltransferase. J Biol Chem 276: 43499–43502 [DOI] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, et al. 2008. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure 16: 1077–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan P, Ge Z, Sirbu B, Doughty C, Agudelo Garcia PA, Schlederer M, Annunziato AT, Cortez D, Kenner L, Parthun MR 2013. Histone acetyl transferase 1 is essential for mammalian development, genome stability, and the processing of newly synthesized histones H3 and H4. PLoS Genet 9: e1003518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Parthun MR, Widom J, Gottschling DE 1996. The major cytoplasmic histone acetyltransferase in yeast: links to chromatin replication and histone metabolism. Cell 87: 85–94 [DOI] [PubMed] [Google Scholar]
- Poveda A, Pamblanco M, Tafrov S, Tordera V, Sternglanz R, Sendra R 2004. Hif1 is a component of yeast histone acetyltransferase B, a complex mainly localized in the nucleus. J Biol Chem 279: 16033–16043 [DOI] [PubMed] [Google Scholar]
- Qin S, Parthun MR 2002. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol Cell Biol 22: 8353–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Carrillo A, Wangh LJ, Allfrey VG 1975. Processing of newly synthesized histone molecules. Science 190: 117–128 [DOI] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341 [DOI] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M 2007. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem 76: 75–100 [DOI] [PubMed] [Google Scholar]
- Sims RJ 3rd, Reinberg D 2006. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev 20: 2779–2786 [DOI] [PubMed] [Google Scholar]
- Song JJ, Garlick JD, Kingston RE 2008. Structural basis of histone H4 recognition by p55. Genes Dev 22: 1313–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart MD, Li J, Wong J 2005. Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol Cell Biol 25: 2525–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter B, Pogoutse O, Guo X, Krogan N, Lewis P, Greenblatt JF, Rine J, Emili A 2007. Association with the origin recognition complex suggests a novel role for histone acetyltransferase Hat1p/Hat2p. BMC Biol 5: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Wade PA, Jones PL, Shi YB, Wolffe AP 1999. Functional analysis of the SIN3–histone deacetylase RPD3–RbAp48–histone H4 connection in the Xenopus oocyte. Mol Cell Biol 19: 5847–5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B 1998. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol 8: 96–108 [DOI] [PubMed] [Google Scholar]
- Wu H, Moshkina N, Min J, Zeng H, Joshua J, Zhou MM, Plotnikov AN 2012. Structural basis for substrate specificity and catalysis of human histone acetyltransferase 1. Proc Natl Acad Sci 109: 8925–8930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Feng H, Zhou BR, Ghirlando R, Hu K, Zwolak A, Miller Jenkins LM, Xiao H, Tjandra N, Wu C, et al. 2011. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature 472: 234–237 [DOI] [PMC free article] [PubMed] [Google Scholar]