Abstract
Disorders of sex development (DSD) are rare disorders in which there is discordance between chromosomal, gonadal, and phenotypic sex. Only a minority of patients clinically diagnosed with DSD obtains a molecular diagnosis, leaving a large gap in our understanding of the prevalence, management, and outcomes in affected patients. We created a novel DSD-genetic diagnostic tool, in which sex development genes are captured using RNA probes and undergo massively parallel sequencing. In the pilot group of 14 patients, we determined sex chromosome dosage, copy number variation, and gene mutations. In the patients with a known genetic diagnosis (obtained either on a clinical or research basis), this test identified the molecular cause in 100% (7/7) of patients. In patients in whom no molecular diagnosis had been made, this tool identified a genetic diagnosis in two of seven patients. Targeted sequencing of genes representing a specific spectrum of disorders can result in a higher rate of genetic diagnoses than current diagnostic approaches. Our DSD diagnostic tool provides for first time, in a single blood test, a comprehensive genetic diagnosis in patients presenting with a wide range of urogenital anomalies.
Keywords: endocrinology, genetic testing, high-throughput, DNA sequencing, sexual development
Disorders of sex development (DSD) are defined as a rare set of conditions in which the chromosomal, gonadal, and phenotypic sex is atypical. DSD has a prevalence of 0.1–0.5% of live births, yet only 13% of patients will ever receive a definitive genetic diagnosis (this percentage is based on a systematic electronic medical chart review targeting patients categorized as DSD at one major mid-western academic medical center) (1, 2). The uncertainty regarding the child’s gender and future psychosocial and psychosexual development is extraordinarily stressful for the child’s family (1, 3, 4). From the time of initial presentation, patients with DSD undergo a wide spectrum of clinical and endocrine tests, from which life-altering decisions are made about gender assignment, medical treatments, and surgery. Yet, to date, evidence is lacking to justify support for specific management strategies of these patients (5).
The promise of next-generation sequencing in the clinical arena is hindered by the difficulty in differentiating between an inconsequential sequence polymorphism and a disease-causing mutation. Although the first predictive test for BRCA1 and BRCA2 mutations had numerous detractors, genetic testing for BRCA1 and BRCA2 has transformed the management of high-risk patients and in the process, researchers have discovered a vast number of gene variants, which are now classified based on their cancer risk (6, 7).
Unlike traditional genetic diagnostic tests that at most sequence a handful of genes or target a panel of known mutations, we have combined multiple genetic tests and put forward a novel and integrated role for comprehensive molecular genetic diagnostics in the clinical realm. Our test combines multiple genetic testing modalities routinely ordered in DSD patients, including sex chromosome complement determination, copy number variant (CNV) analysis, and gene sequencing. Currently, gene sequencing is done on a gene-by-gene basis. Many genes, particular those for rare or complex disorders, are only offered on a research basis, further complicating the genetic diagnostic process. This strategy replaces multiple single-gene sequencing tests with a unified test, thereby drastically improving the odds of identifying a high-risk variant and of assigning the appropriate management based on the individual’s genetic risk.
We propose a novel diagnostic process allowing clinicians to initially identify a genetic mutation, which would be followed by relevant metabolic, endocrine, and imaging tests for functional assessment of the gene mutation. This diagnostic approach can eliminate non-indicated clinical tests, sparing the patient unnecessary stress and saving healthcare system’s resources. Finally, by pinpointing the genetic diagnosis at the beginning of the diagnostic process, we can more accurately analyze and predict both future developmental issues in the child and the risk of recurrence within the family. In our pilot group of patients, we have shown that this novel targeted diagnostic approach can accurately diagnose the genetic basis of DSD in the majority of patients. This new test shifts the paradigm of the diagnostic processes and ultimately has the potential to increase the rate of genetic diagnosis, provide more cost-effective care, and allow for more informed clinical management in patients with DSD.
Materials and methods
Clinical diagnosis of all DSD patients is outlined in Table 1 and clinical features are described in detail in Appendix S1. Patients with known genetic diagnoses were diagnosed in either clinical laboratories or on a research basis. Patients 45, X and 47, XXY, and DSDPt7 were diagnosed in a clinical laboratory using karyotype and/or Sanger sequencing. DSDPts 2, 3, 8 and 9 were genetically diagnosed on a research basis only after extensive endocrine work-up. All other patients did not have a genetic diagnosis. The clinical and genetic diagnoses were blinded to the investigators. This study was approved by the Institutional Review Board at the University of California, Los Angeles. Capture was performed using custom Sure Select Target Enrichment System Kit (Agilent) (8). We designed oligonucleotide baits tiled against exonic and intronic regions of 35 known genes of sex development, up to 10 kb regions upstream and downstream of all known genes in sex development, and 3–10 kb spaced every 10 Mb along the X- and Y-chromosomes (Table S1A,B). All clinically associated genes reported in the literature (as of December 2009) in both sex determination and sex differentiation were included. We also included a subset of genes for ovarian insufficiency. Up to six custom bar-coded samples were pooled, captured with the baits designed for one reaction and sequenced on a single lane of Illumina GAIIx for 76 cycles or HiSeq2000 for 50 cycles. The reads were aligned to the whole genome using Novoalign (http://www.novocraft.com/index.html) and the aligned reads were processed using sam tools (http://samtools.sourceforge.net/) and Picard (http://picard.sourceforge.net/) (9) to remove potential polymerase chain reaction duplicates. Both single-nucleotide variants (SNVs) and small insertions and deletions (INDELs) within the captured coding exonic and splice-site intervals of the DSD genes were called using sam tools pileup tool and annotated using the SeqWare project (http://seqware.sourceforge.net) (10). The SNVs and INDELs were further filtered to include only those resulting in non-synonymous nucleotide substitution, frameshift, in-frame INDELs, splice-site, or early-termination mutations. Finally, in order to minimize the risk of false-positive SNV findings, only the variants called with SNV Phred score ≥30, total coverage 10× and percent of non-reference call 15 were further≥analyzed (Fig. S1). All variants with≥coding consequences were analyzed against the public Human Gene Mutation Database (HGMD), dbSNP132 (common variants present at ≥1% frequency), and SNVs were run through three independent protein pathogenicity predictors: Polyphen-2, sift, and Mutation Assessor in order to determine whether they were likely to be disease-causing (11-13). When used together, the three independent in silico pathogenicity predictors have a higher positive predictive value and any of the predictors alone (14). If two of the three algorithms predicted a tolerable/benign effect of the mutation this was considered ‘likely benign’. All bioinformatic analysis was performed blinded to the patient’s chromosomal sex, phenotype, and diagnosis.
Table 1.
Clinical diagnosis of patients with DSDa
| Identification | Clinical diagnosis | Genetic diagnosis known (Y/N) |
Genetic diagnosis identified by targeted sequencing (Y/N) |
|---|---|---|---|
| 46, XY male | Control XY male | – | – |
| 46, XX female | Control XX female | – | – |
| 47, XXY KS | Klinefelter syndrome | Y | Y |
| 45, XO TS | Turner syndrome | Y | Y |
| DSDPt1 | 5-Alpha reductase deficiency | N | Y (SRD5A2, E200K) |
| DSDPt2 | 46, XY gonadal dysgenesis | Y [NROB1 (DAX1) duplication] | Y |
| DSDPt3 | 46, XY gonadal dysgenesis | Y (SRY, Y127C) | Y |
| DSDPt4 | 46, XY gonadal dysgenesis + campomelic dysplasia |
N | N |
| DSDPt5 | 46, XY gonadal dysgenesis + galactosemia | N | N |
| DSDPt6 | 46, XX testicular DSD | N | N |
| DSDPt7 | 46, XY DSD | Y (AR, M788T) | Y |
| DSDPt8 | 46, XY female + AHC | Y (DAX1, Y121*) | Y |
| DSDPt9 | 46, XY DSD severe combined adrenal and gonadal deficiency |
Y (CYP11A1) | Y |
| DSDPt10 | 46, XX testicular DSD | N | N |
| DSDPt11 | 46, XY gonadal dysgenesis | N | N |
| DSDPt12 | 46, XY gonadal dysgenesis | N | Y (ATRX, K1045E) |
AHC, adrenal hypoplasia congenita; DSD, disorders of sex development; N, no; Y, yes.
All patients in this study were clinically diagnosed with a DSD.
SNV validation
To determine the accuracy of our sequencing pipeline, we compared DSDPt12 to previously acquired Illumina IM SNP Genotyping data. This array was run and analyzed as per manufacturer’s protocols at the Southern California Genotyping Consortium and had a call rate of 0.9963. Six hundred ninety one SNPs within the targeted region were also genotyped on this array and compared to our SNP and INDEL variant callers. Four of 691 SNPs were discordant between the genotyping data and the Illumina Sequencing data, giving a false-negative rate of 0.57%. These false negatives were four SNVs that the genotyping data called heterozygous while the sequencing data did not. Conversely, the sequencing did not identify any SNVs at base positions where the genotyping data did not call an SNV.
Sex chromosome dosage and CNV analysis
Our sex chromosome complement analysis comprised of two normalization steps. First, we normalize for differences in coverage levels because starkly different coverage levels between samples is a common issue, known to hinder CNV analysis of targeted sequencing data (15). Second, we normalize the X- and Y-chromosome coverage to the sample’s autosomes in order to perform inter-sample comparisons.
More precisely, any sample whose depth of coverage (DOC) was 0.5 standard deviation higher than the mean DOC of all the samples was subjected to normalization of the DOC by reanalyzing a randomly selected subset of reads. Once the DOC was similar among all samples, we further normalized the samples, by dividing the mean DOC on the X and the Y chromosomes, Ci(chrX) and Ci(chrY) respectively, with the mean DOC of the patient’s autosomes, Ci(chrAut). Since there are two copies of every autosome, and 0, 1, or 2 copies of the X and Y chromosomes, the ratio derived (0, 0.5, or 1) allows us to estimate the number of X and a Y chromosome per sample.
After the relative ratio of sex chromosome to autosome for each sample was calculated, samples were grouped by their estimated karyotype: XX, XY, XXY or XO. Two-sample t-test was used to assess the significance of the separation between different copy number groups: 1 vs 2 X chromosomes and 0 vs 1 Y chromosome. The X and the Y chromosomes were tested separately as their copy number states can be considered to be independent of each other’s.
The same approach was taken for the copy number assessment of the DSD genes, except that instead of taking the mean DOC of the chromosomes, the mean DOC of each gene (G) was calculated [Ci(Gj)] and normalized by dividing with the mean DOC of the autosomes [Ci(chrAut)].
To detect CNVs at genic or exonic level for sample i, gene Gj or exon Ej, the normalized DOC Ci(Gj)/Ci(chrAut) or Ci(Ej)/Ci(chrAut), was compared to those of the rest of the samples to determine if it was significantly greater or less. For the CNV analysis of the genes or the exons, outlier for each gene was determined by assessing how the normalized coverage of a sample is significantly different from the rest of the samples. The Z-score of a known duplication was used to determine the lower bound (LB) and upper bound (UB) for each gene or exon and a gene or exon falling outside the LB or UB was considered an outlier.
Results
Targeted sequencing achieved a capture efficiency of ~51.5% and a mean coverage of ×48.3 per sample with 92.6% of the targeted base positions being sequenced at ≥×10. A total of 16 individuals were sequenced in our targeted approach, 2 unaffected individuals and 14 patients clinically diagnosed with DSD (Table 1). Some of the targeted regions were not covered due to the presence of repetitive regions, which comprised 2.3% of the total regions covered. Repetitive regions are historically difficult to sequence and map back to the reference genome and these findings are consistent with previously published results of targeted sequencing (8, 16). However, none of the genes sequenced are known to have mutations dependent on the size of these repetitive regions. To estimate the rates of false-positive and false-negative SNV calls, we compared SNP genotyping data to our SNV and INDEL calls in DSDPt12 and calculated a false-negative call rate of <1% and found 0 false positives.
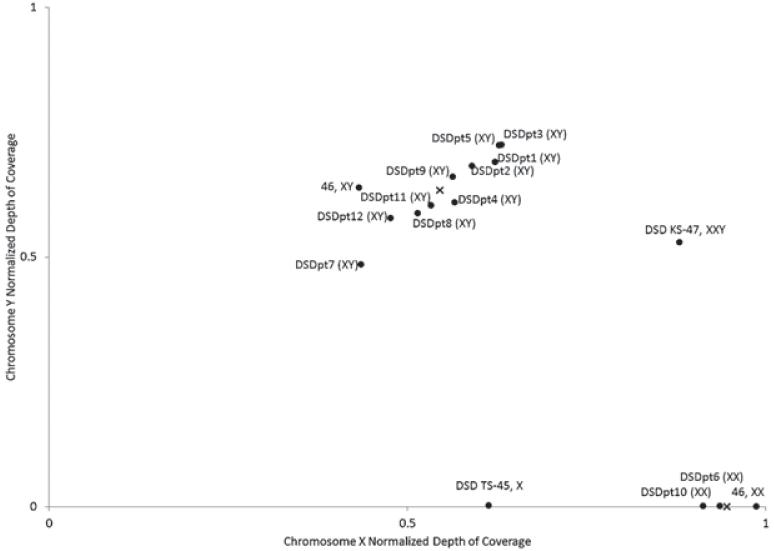
In order to analyze sex chromosome dosage, the normalized DOCs for both chromosomes, Ci(chrX)/Ci(chr-Aut) and Ci(chrY)/Ci(chrAut), were calculated and independently examined (Fig. 1). First, we reliably distinguish between samples with one or two X chromosomes (p < 0.001). Calling the number of Y chromosomes was also clear as the four samples with no Y chromosome had nearly null coverage while the 10 samples with Y chromosomes had close to half the coverage compared to the (diploid) autosomes (p < 0.001). Sample 47, XXY was properly clustered with the XX samples on the x-axis and the XY samples on the y-axis. Sample 45, X was properly clustered with the XY samples on the x-axis and had null coverage on the Y-chromosome. All of our called sex chromosome dosage matched clinically performed cytogenetic karyotype tests.
Fig. 1.
Sex chromosome complement. Depth of coverage (DOC) along the X- and Y-chromosomes was normalized to the autosomal DOC and plotted to determine sex chromosome complement. All XY samples (upper left) clustered together and had normalized coverage ranging from 0.43 to 0.63 for the X-chromosome and 0.58 to 0.72 for the Y-chromosome. All XX samples (lower right) clustered together, had null Y-chromosome coverage, and had close to 1 for normalized X-chromosome coverage, indicating an absence of the Y-chromosome and two copies of the X-chromosome. An X marks the mean coverage for the 46, XY cluster or the 46, XX cluster. The p-values for separating the two clusters were <0.001 for both directions, and DSDpt3 that had the highest DOC (0.63) along the X-chromosome was separated from the 46, XX cluster with a p-value of <0.001.
To identify both rare and common variants that might result in DSD, we employed a number of filters. First, to identify variants previously identified in the literature as causative of DSD we compared all coding SNVs and INDELs against HGMD public, which includes both rare and more ‘common’ causes of DSD. We then filtered out common variants using dbSNP132 (≥1% frequency) and then ran all novel variants through in silico protein pathogenicity predictors to determine if they were likely benign and causative (see Fig. S1).
An average of 30 SNVs and INDELs was called along all coding exons ±3 bp and in the testis-specific SOX9 enhancer (17) in each patient. Few variants led to protein-level changes (frameshift, in-frame INDELs, early-termination, missense, and splice-site) in each sample. Of 19 high-quality protein-changing variants in all patients, five were reported to be causal variants for a similar phenotype in the HGMD public version (Table 2) (18). In the DSD patients without sex chromosome abnormalities, four patients (DSDPts 3, 7, 8, and 9) had a previously identified genetic diagnosis, all of which were identified through screening of HGMD. This approach also identified a genetic diagnosis of 5-alpha reductase deficiency in DSDPt1. None of the mutations identified in HGMD were present in dbSNP132 (≥1% frequency), indicating that these are rare variants. Additionally, we also identified a genetic mutation not present in HGMD public in DSDPt12, which is described below.
Table 2.
Filtered SNVs and INDELs identified in DSD patientsa
| Sample | Chr | Start | End | Gene | SNV INDEL |
SNP quality |
Protein change | Causative variant? |
In HGMD public? |
In dbSNP132 (1 %)? |
Polyphen-2 | sift | Mutation Assessor |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 47, XXY KS | chr10 | 104586872 | 104586873 | CYP17A1 | T→G | 34 | H79P | – | N | N | Possibly damaging |
Tolerated | Low |
| 47, XXY KS | chr8 | 106870217 | 106870218 | ZFPM2 | G→C | 114 | S210T | – | N | N | Benign | Tolerated | Neutral |
| DSDPtl | chr2 | 31607980 | 31607981 | SRD5A2 | C→T | 225 | E200K | Y | Y | N | – | – | Medium |
| DSDPt3 | chr8 | 11651992 | 11651993 | GATA4 | G→A | 228 | V380M | – | N | rs114868912 | Benign | Tolerated | Neutral |
| DSDPt3 | chr9 | 1046731 | 1046732 | DMRT2 | G→A | 194 | R382Q | – | N | rs72703237 | Benign | Tolerated | Neutral |
| DSDPt3 | chr17 | 75373191 | 75373192 | CBX2 | C→T | 228 | A452V | – | N | rs76915888 | Benign | Tolerated | Neutral |
| DSDPt3 | chr9 | 1046397 | 1046398 | DMRT2 | C→G | 100 | P271A | – | N | rs72703236 | Probably damaging |
Damaging | Medium |
| DSDPt3 | chrY | 2715264 | 2715265 | SRY | T→C | 225 | Y127C | Y | Y | N | Probably damaging |
– | – |
| DSDPt5 | chr12 | 52104848 | 52104849 | AMHR2 | A→C | 136 | T108P | – | N | N | Possibly damaging |
Tolerated | Neutral |
| DSDPt5 | chr8 | 106870217 | 106870218 | ZFPM2 | G→C | 228 | S210T | – | N | N | Benign | Tolerated | Neutral |
| DSDPt6 | chr17 | 75373191 | 75373192 | CBX2 | C→T | 228 | A452V | – | N | rs76915888 | Benign | Tolerated | Neutral |
| DSDPt6 | chrX | 139414764 | 139414765 | S0X3 | C→T | 228 | A43T | – | N | N | Benign | – | Neutral |
| DSDPt7 | chrX | 66858443 | 66858444 | AR | T→C | 225 | M788T | Y | Y | N | Probably damaging |
Damaging | Medium |
| DSDPt8 | chrX | 30237128 | 30237129 | NR0B1 | G→T | 212 | Y121 * | Y | Y | N | Termination | Termination | Termination |
| DSDPt9 | chr9 | 884196 | 884197 | DMRT1 | T→G | 46 | V→Gb | – | N | rs116766038 | Non-coding by CCDS |
Non-coding by CCDS |
Non-coding by CCDS |
| DSDPt9 | chr15 | 72424436 | 72424437 | CYP11A1 | INS:→A | 208 | Intronic splice-site mutation |
Y | N | N | INDELc | INDEL | INDEL |
| DSDPt9 | chr15 | 72422525 | 72422526 | CYP11A1 | DEL:T→- | 726 | Frameshift, early-termination |
Y | Y | N | INDEL | INDEL | INDEL |
| DSDPtl 1 | chr15 | 72446744 | 72446745 | CYP11A1 | C→T | 204 | V79I | – | N | N | Benign | Tolerated | Low |
| DSDPtl 2 | chrX | 76824270 | 76824271 | ATRX | T→C | 139 | K1045E | Y | N | N | Probably damaging |
Tolerated | Low |
CCDS, consensus coding sequence; DSD, disorders of sex development; HGMD, Human Gene Mutation Database; INDELs, insertions and deletions; N, no; SNV, single-nucleotide variant; Y, yes.
High-quality SNVs with coding consequences identified in the 16 patients were run through a series of filters such as HGMD to identify SNVs and INDELs previously reported in the literature. All INDELs were reported as causative. Remaining SNVs were filtered using dbSNP132 (≥1% frequency) to identify rare variants. The identified rare SNV variants were then run through multiple in silico protein pathogenicity predictors to determining whether the coding variation was likely to be causative of the disease. Two of three pathogenicity predictors were required to indicate the mutation was ‘damaging’ in order for it to be classified as causative.
This mutation lies in the coding region of an isoform of DMRT1.
In silico protein predictors do not provide pathogenicity for INDELs.
Of the remaining 14 variants, six were found to be common polymorphisms recorded in dbSNP132 (≥1% frequency), and therefore classified as likely benign mutations. The remaining eight variants were not present in dbSNP132 (≥1% frequency), indicating they were rare variants, thus potentially pathogenic. Discerning between benign and potentially pathogenic rare variants required a multistep approach.
INDELs that result in out-of-frame coding consequences or lie in canonical splice junctions are automatically classified as ‘likely pathogenic’ (19). A single insertion disrupting a canonical splice-site found in CYP11A1 for DSDPt9 was not identified in HGMD or dbSNP132 (≥1% frequency). This was classified as likely pathogenic and is concordant with the known compound heterozygous genetic diagnosis in CYP11A1.
The remaining seven rare SNV variants were analyzed using two independent methods: (i) SNVs were run through three in silico protein pathogenicity prediction algorithms and (ii) SNVs were manually evaluated based on inheritance of the disease (i.e. sex-limited, recessive, dominant) and patient phenotype. These two analyses ensured that we were not excessively filtering out rare variants solely based on in silico pathogenicity predictors. We opted to be more conservative in our calling of benign variants, requiring that two of three pathogenicity predictors predict a tolerable effect and that the manual analysis did not find the SNV as likely to be causative.
In DSDPt12, diagnosed with 46, XY gonadal dysgenesis, we identified one hemizygous K1045E mutation in ATRX (20). One of the three pathogenicity predictors called the missense mutation ‘probably damaging’ and manual inspection identified SNV in ATRX as causative in DSDPt12 with 46, XY gonadal dysgenesis. Therefore, we called this a likely pathogenic mutation in the patient. All other SNVs were called as benign or tolerable by two of the three predictors. When manually evaluated, these same SNVs were either not pathogenic because of inheritance (e.g. the gene typically requires pathogenic mutations on both alleles to show a phenotype) and/or the phenotype was sex-limited and only displayed in either XY or XX individuals. In combining the data from in silico protein pathogenicity predictors and manual evaluation, the remaining six SNVs were classified as likely benign.
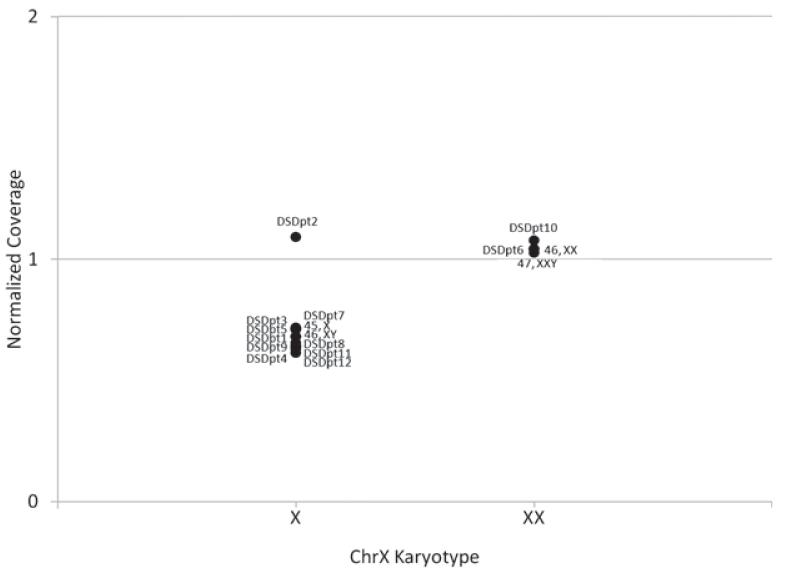
As duplication and deletions can contribute to DSD, we screened all of our patients for causative DSD duplications and deletions (21, 22). In DSDPt2, who has an XY karyotype, the mean DOC of NROB1 (DAX1), CDSDpt2 (NROB1), was elevated to the level of the mean coverage achieved by the samples with XX karyotype (Fig. 2), indicating copy number increase at the locus (21). The normalized coverage of NROB1 gene in DSDPt2 was 2.75 standard deviations away (Z-score = 3.04) from the mean of the normalized coverage of NROB1 of all XY samples (p 0.002). To call CNVs, we have chosen a significance=threshold of Z-score 3.04 away from the mean to call a deletion or duplication involving an entire gene based on the results from DSDPt2, which generates a false-positive rate for CNVs of 0.1%. All other DSD genes were tested in the same manner, with no additional duplications or deletions identified (Appendix S1, Fig. S2).
Fig. 2.
Copy number variant analysis. To determine CNV status for NROB1 (DAX1 ), coverage for each gene was normalized to autosomal coverage of each sample and all samples plotted based on normalized coverage. Since NROB1 is an X-chromosome gene, normalized coverage was plotted separately based on the X-chromosome karyotype. DSDpt2, with 46, XY GD, had significantly higher coverage than other XY individuals, indicating a duplication of the gene (p = 0.002). An X indicates the mean coverage for the gene.
Discussion
The proposed method of broad-scale sequencing of all known DSD genes offers significant advantages over current diagnostic procedures for the assessment of DSD. The vast majority of disease-causing mutations can be attributed to sequence and copy number variations affecting the coding regions of genes. We limited the targeted genes to those with known roles in sex development, as pathogenic mutations in these genes can be confidently reported back to the clinician and patient. While we cannot identify novel genes in sex development using this approach, limiting the number of genes sequenced streamlines the bioinformatic analysis and restricts the pathogenic variants to those genes relevant to phenotype. Another major advantage of this DSD-specific approach, rather than whole-exome or whole-genome sequencing, is that we decrease the chance of incidental findings unrelated to DSD. For instance, we eliminate the possibility of diagnosing minors with adult-onset diseases unrelated to the reason for genetic testing. Genetic testing for adult-onset diseases is ethically questionable in children, and under current guidelines is only performed in exceptional circumstances (23). However, because the targeted method is readily scalable, inclusion of novel sex development genes or expansion of the targeted region to also include all genes resulting in ovarian insufficiency or male infertility can be easily updated in future capture designs.
Intensive study of important disease genes such as CFTR and BRCA1/BRCA2 has taught us the complexity of single-gene disorders. The vast majority of disease genes show remarkable mutational heterogeneity in the general population (as opposed to the more restricted mutation sets found in certain ethnic groups), with mutations scattered across all exons of the genes with no particular ‘hot-spots’. In such situations, there is really no alternative to whole-gene sequencing (at least of the exons and intron-exon junctions) if one is to entertain any hope of identifying the majority of causative mutations in affected individuals. The current price for full-gene sequencing of BRCA1 and BRCA2, for example, is, at time of this writing, about $3600. The price for other individual genes offered in the clinical setting ranges from about $1500 to $3000, depending on the exonic size and other factors such as overall demand and test exclusivity. For those genetic disorders caused by many different genes, such as DSD, the cost of sequencing them is prohibitive and is rarely done.
The majority of the patients with congenital adrenal hyperplasia, for example, have mutations in CYP21A2, while a smaller proportion of patients with the same condition have mutations in one of the four other genes that give rise to similar phenotypes (24) (POR, STAR, HSD3B2, CYP11B1, and CYP17A1). Sequencing all six genes by current methods would cost over $10,000. Assuming that we pool a minimum of seven bar-coded samples for one reaction worth of targeted baits and sequence on one lane of an Illumina HiSeq2000 flow cell with 50-bp paired-end reads, our comprehensive sequencing approach would cost less than $1000 per sample. Included in this cost per sample is the labor and reagents for library preparation ($350), cost of targeted baits ($150), bioinformatic analysis ($200), and the full-sequencing service ($300). Even with the additional costs that need to be factored in if performed within a clinical setting, such as hospital overhead, maintenance of CLIA, CAP and state certifications, and the higher labor costs of licensed medical technologists, the proposed method would come out to be significantly cheaper while providing more information than current one-gene-at-a-time Sanger sequencing approaches. While endocrine testing and radiological imaging can help to prioritize the order in which genes are sequenced, these tests may lengthen the diagnostic process, increase costs, and are sometimes invasive. Sequencing all the genes upfront requires only a blood draw, and the entire process can be completed in less than 3 weeks, which is shorter than the turnaround time for clinical molecular genetic sequencing tests for single genes.
Several groups have explored similar targeted sequencing approaches to encompass all known genes conferring an inherited risk of breast/ovarian cancer, congenital ocular disorders, or hereditary hearing loss (16, 25, 26). Both commercial and academic reference laboratories have realized the utility of such panels, and now offer sequencing services for all genes causing various cardiomyopathies, and other phenotypic traits associated with 20 or more genes (27). All of these approaches simply replace the current one-gene-at-a-time sequencing tests traditionally offered, reducing the cost while increasing the diagnostic yield. However, we have yet to see a systematic reevaluation of the powerful role that such genetic diagnosis can play in early diagnosis and management. By integrating these diagnostic tools into the clinical framework, we might eliminate unnecessary tests and the risks, costs, and diagnostic delays associated with them. Furthermore, with the cost of whole-genome sequencing soon falling below the aggregate cost of performing standard Sanger sequencing on two or three single genes, it is likely that health care systems will be reluctant to cover the latter, when much more comprehensive diagnostic information can be gained for the same price by massively parallel sequencing.
The ability to provide a single test that produces such a variety of genetic information (copy number variation, sex chromosome complement, and sequence variants) has the potential to significantly alter clinical practice. In our cohort of 14 patients, a diagnosis was identified in 9 of 14 (64%). Parents with children afflicted with a genetic disease place a high value on obtaining a genetic diagnosis, even with the knowledge that a diagnosis is not reached in all cases and that identification of the genetic lesion will not necessarily affect medical management (28). By establishing a primary genetic diagnosis, the patient is spared a long and difficult diagnostic process including numerous costly and sometimes invasive tests. For parents dealing with the appreciably greater stress of a child presenting with a DSD, a genetic test may provide a better understanding of the condition’s etiology and outcomes. The next step is to evaluate the impact of genomic sequencing on quality of life in patients with rare genetic disorders such as DSD.
Our novel results show the potential of using next-generation sequencing to reframe the typical diagnosis pipeline within clinical medicine. This is especially true for those patients with rare disorders who have variable phenotypes and multiple genes associated with the phenotype, such as DSD. The development of clinical diagnostic tools targeted toward broader phenotypes will catapult molecular diagnostics from a confirmatory test to a primary diagnostic tool that can diagnose and triage the patient earlier into appropriate management.
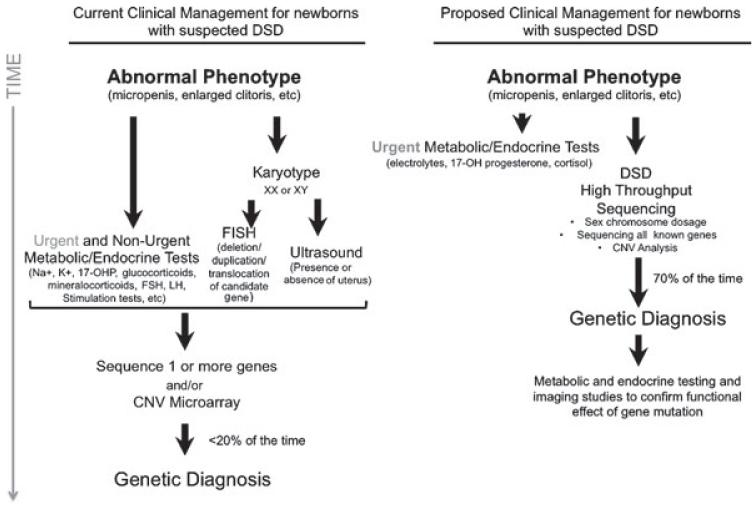
Traditional clinical diagnosis for newborns presenting with atypical phenotypic sex requires karyotype tests, electrolyte measurements, hormone challenges and stimulation tests, and imaging studies to visualize the gonads and internal reproductive structures. For newborns presenting with ambiguous genitalia, the major life-threatening concern is salt-wasting adrenal crisis. Therefore, we propose that all newborns presenting with ambiguous genitalia should be monitored until it can be safely determined that there is no risk of adrenal crisis. At the same time, in lieu of performing all the other subsequent clinical tests enumerated above, we propose that a blood specimen be sent for targeted DSD sequencing to identify the causative gene mutation. Once the involved gene is identified, follow-up functional tests can be performed to direct clinical management (Fig. 3). Establishing a precise genetic etiology early on allows one to predict the likelihood of developmental delay as well as conditions that might not be apparent in the newborn period.
Fig. 3.
Proposed integration of targeted sequencing approach to clinical management of suspected DSD. Current clinical management begins with the identification of an abnormal phenotype and is followed by multiple metabolic and endocrine tests, genetic tests, and imaging studies in order to identify the mostly likely candidate for sequencing. Targeted sequencing approach would prioritize a genetic diagnosis, which would be functionally assayed and confirmed with endocrine and imaging studies of the patient.
The targeted approach is ideal for disorders that have similar phenotypes, typically affecting a single organ system, which can be the result of mutations in many different genes. Rare cases that are not genetically diagnosed by the targeted approach will require a more comprehensive work-up to identify novel gene variants, non-coding variants, or copy number changes. However, these cases are the exception and not the rule and we believe the targeted approach will provide a diagnosis in the majority of DSD patients. Patients in whom a targeted approach is unable to identify a genetic cause of the DSD or patients with rare cases of DSD that have not been associated with a gene, such as agonadism, would be excellent candidates for whole-exome and whole-genome approaches. Because non-targeted approaches identify more novel variants, both within coding and the non-coding regions, these technologies can be more difficult to interpret clinically.
Next-generation sequencing has only begun to illuminate the genetic variants responsible for rare Mendelian diseases. As targeted sequencing approaches become cheaper and generate more data, it is up to the medical community to create sophisticated tests to utilize the technology such that physicians and patients can benefit from this revolutionary technology.
Supplementary Material
Acknowledgement
This work was funded by the Doris Duke Charitable Foundation and NICHD 1R01HD068138 “DSD-TRN” (Disorders of Sex Development-Translational Research Network).
Footnotes
The following Supporting information is available for this article:
Appendix S1. Supplementary methods.
Additional Supporting information may be found in the online version of this article.
Conflict of interest
The authors have declared no conflicting interests.
References
- 1.Lux A, Kropf S, Kleinemeier E, Jürgensen M, Thyen U, DSD Network Working Group Clinical evaluation study of the German network of disorders of sex development (DSD)/intersexuality: study design, description of the study population, and data quality. BMC Public Health. 2009;9:110. doi: 10.1186/1471-2458-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes IA, Houk C, Ahmed SF, Lee PA, Lawson Wilkins Pediatric Endocrine Society. European Society for Paediatric Endocrinology Consensus Group Consensus statement on management of intersex disorders. J Pediatr Urol. 2006;2:148–162. doi: 10.1016/j.jpurol.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Stein MT, Sandberg DE, Mazur T, Eugster E, Daaboul J. A newborn infant with a disorder of sexual differentiation. J Dev Behav Pediatr. 2003:115–119. doi: 10.1097/00004703-200304000-00008. [DOI] [PubMed] [Google Scholar]
- 4.Meyer-Bahlburg HFL. Gender assignment and psychosocial management. In: Martin L, editor. Encyclopedia of endocrine diseases. Vol. 1. Elsevier; Amsterdam: 2004. pp. 125–134. [Google Scholar]
- 5.Warne GL. Long-term outcome of disorders of sex development. Sex Dev. 2008;2:268–277. doi: 10.1159/000152043. [DOI] [PubMed] [Google Scholar]
- 6.Easton DF, Steele L, Fields P, et al. Cancer risks in two large breast cancer families linked to BRCA2 on chromosome 13q12-13. Am J Hum Genet. 1997;61:120–128. doi: 10.1086/513891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldgar DE, Easton DF, Deffenbaugh AM, et al. Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am J Hum Genet. 2004;75:535–544. doi: 10.1086/424388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAM tools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Connor BD, Merriman B, Nelson SF. SeqWare Query Engine: storing and searching sequence data in the cloud. BMC Bioinformatics. 2010;11(Suppl. 12):S2. doi: 10.1186/1471-2105-11-S12-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 13.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan PA, Duraisamy S, Miller PJ, et al. Interpreting missense variants: comparing computational methods in human disease genes CDKN2A, MLH1, MSH2, MECP2, and tyrosinase (TYR) Hum Mutat. 2007;28:683–693. doi: 10.1002/humu.20492. [DOI] [PubMed] [Google Scholar]
- 15.Sathirapongsasuti JF, Lee H, Horst BA, et al. Exome sequencing-based copy-number variation and loss of heterozygosity detection: exome CNV. Bioinformatics. 2011;27:2648–2654. doi: 10.1093/bioinformatics/btr462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shearer AE, DeLuca AP, Hildebrand MS, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107:21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- 18.Finkielstain GP, Chen W, Mehta SP, et al. Comprehensive genetic analysis of 182 unrelated families with congenital adrenal hyperplasia due to 21-hydroxylase deficiency. J Clin Endocrinol Metab. 2011;96:E161–E172. doi: 10.1210/jc.2010-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tavtigian S. Unclassified variants in the breast cancer susceptibility genes BRCA1 and BRCA2. In: Welcsh P, editor. The role of genetics in breast and reproductive cancers. Springer; New York, NY: 2009. pp. 49–73. [Google Scholar]
- 20.Ion A, Telvi L, Chaussain JL, et al. A novel mutation in the putative DNA helicase XH2 is responsible for male-to-female sex reversal associated with an atypical form of the ATR-X syndrome. Am J Hum Genet. 1996;58:1185–1191. [PMC free article] [PubMed] [Google Scholar]
- 21.White S, Ohnesorg T, Notini A, et al. Copy number variation in patients with disorders of sex development due to 46,XY gonadal dysgenesis. PLoS One. 2011;6:e17793. doi: 10.1371/journal.pone.0017793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tannour-Louet M, Han S, Corbett ST, et al. Identification of de novo copy number variants associated with human disorders of sexual development. PLoS One. 2010;5:e15392. doi: 10.1371/journal.pone.0015392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holtzman NA, Murphy PD, Watson MS, Barr PA. Predictive genetic testing: from basic research to clinical practice. Science. 1997;278:602–605. [PubMed] [Google Scholar]
- 24.Krone N, Arlt W. Genetics of congenital adrenal hyperplasia. Best Pract Res Clin Endocrinol Metab. 2009;23:181–192. doi: 10.1016/j.beem.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walsh T, Lee MK, Casadei S, et al. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107:12629–12633. doi: 10.1073/pnas.1007983107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raca G, Jackson C, Warman B, Bair T, Schimmenti LA. Next generation sequencing in research and diagnostics of ocular birth defects. Mol Genet Metab. 2010;100:184–192. doi: 10.1016/j.ymgme.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wheeler M, Pavlovic A, DeGoma E, Salisbury H, Brown C, Ashley EA. A new era in clinical genetic testing for hypertrophic cardiomyopathy. J Cardiovasc Transl Res. 2009;2:381–391. doi: 10.1007/s12265-009-9139-0. [DOI] [PubMed] [Google Scholar]
- 28.Geelhoed EA, Harrison K, Davey A, Walpole IR. Parental perspective of the benefits of genetic testing in children with congenital deafness. Public Health Genomics. 2009;12:245–250. doi: 10.1159/000203780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.