Abstract
IQGAP1 has emerged as a key component in the regulation of cytoskeleton dynamics during cell migration, maintenance of adherens junctions, microbial pathogenesis and intracellular trafficking. IQGAP1 is known to localize to the protruding edge of lamellipodia in a variety of cell types and interact with regulators of actin dynamics. Here, we provide evidence suggesting a novel role of IQGAP1 in cell motility through cell edge retraction. In some of the cell lines examined, IQGAP1 was markedly separated from WAVE localization suggesting IQGAP1 may localize to retracting edges. B16F10 mouse melanoma cells exhibited the most restricted separation in which the appearance of GFP-IQGAP1 correlated with cell edge retraction velocity and the disappearance of mCherry-Arp3. These results demonstrate that in some cell types IQGAP1 may function to promote cell retraction not lamellipodium edge protrusion. In addition, we examined co-localization of IQGAP1 with adhesion site markers, myosin IIA, calmodulin and IQGAP2. In areas rich in IQGAP1 there was decreased immunofluorescence staining of vinculin, paxillin and phosphorylated-tyrosine indicating adhesion site disassembly. Interestingly, calmodulin, but not myosin IIA or IQGAP2, co-localized with IQGAP1 in areas of cell retraction. Overall these results suggest a new role of IQGAP1, distinct form IQGAP2, in cell migration through up regulation of contractility and downregulation of adhesion sites potentially through calmodulin interaction.
Keywords: IQGAP1, IQGAP2, Calmodulin, Actin, Cell Adhesion, Cell Retraction
1. Introduction
IQGAP1 is a 190 kDa, multi-functional protein first cloned and characterized as a Ras GTPase-activating protein (RasGAP) related protein with four IQ motifs [1] responsible for binding to calmodulin and calmodulin-like proteins [2]. Through the calponin homology domain (CHD), a WW motif, IQ repeats, and the C-terminal RasGAP related protein domain, IQGAP1 interacts with numerous binding partners to mediate a multitude of cellular and biological functions [3, 4]. Many of the interactions likely underlie the mechanism of IQGAP1 in cancer development and progression [5, 6].
Among the diverse functions, IQGAP1 plays a role in cell-matrix interaction and actin dynamics at the cell leading edge during motility. Actin stress fibers and focal adhesions in fibroblasts induced by hyaluronan are dependent on the presence of IQGAP1 [7]. In the leading edge of vascular smooth muscle cells, PDGF stimulation recruits IQGAP1 which is necessary for focal adhesion formation and cell migration [8]. At β1 integrin activation sites, Rac1 and RhoA activities are suppressed and enhanced, respectively, through a pathway involving IQGAP1 association with RacGAP1 [9, 10]. IQGAP1 has been shown to co-localize with several other proteins in actin ruffles and in the leading edge of lamellipodia. IQGAP1 co-localizes with S100P in membrane ruffles following stimulation with epidermal growth factor [11]. At the leading edge, IQGAP1 co-localizes with phosphorylated VEGF receptor [12], protein 4.1R [13], CLASP2 [14], APC, Rac1, CDC42 [15], and N-WASP and Arp3 in actin-rich structures [16, 17]. In our current studies, we observed varying subcellular localization of IQGAP1 that was cell-type dependent. Surprisingly, the subcellular localization of IQGAP1 was restricted to actively retracting areas in some cell lines. Overall, our study points to a new role for IQGAP1 in cell migration, namely retraction of cell edges potentially through up regulation of contractility and downregulation of cell-matrix interactions.
2. Materials and Methods
2.1. Cell culture and reagents
A375, CHO, NIH 3T3, B16F10 and B16F1 cell lines were purchased from American Type Culture Collection (Manassas, VA, USA) and maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA, USA) and antibiotics. Trypsin/EDTA solution (Mediatech, Manassas, VA, USA) was used for cell detachment. Fugene 6 transfection reagent was purchased from Roche Diagnostics. Mouse laminin and fibronectin from bovine plasma were purchased from Invitrogen and Sigma Aldrich (Saint Louis, MO, USA), respectively. Primary antibodies for immunofluorescence staining were purchased from the following vendors: rabbit polyclonal anti-WAVE2 and mouse monoclonal anti-IQGAP2 from Santa Cruz Biotechnology Inc (Santa Cruz, CA, USA); mouse monoclonal anti-IQGAP1 and mouse monoclonal anti-paxillin from BD Transduction Laboratories; mouse monoclonal anti-vinculin and rabbit monoclonal anti-calmodulin from Abcam (Cambridge, England); mouse monoclonal anti-phosphorylated tyrosine from EMD Millipore (Billerica, MA, USA); rabbit polyclonal anti-myosin IIA from Sigma Aldrich. Secondary anti-rabbit-Alexa 488, anti-mouse-Alexa 546 and anti-mouse Alexa-647 were from Invitrogen. pEGFP-IQGAP1 (plasmid 30112 deposited by David Sacks [18]) and Arp3-pmCherryC1 (plasmid 27682 deposited by Christien Merrifield [19]) were obtained from Addgene (Cambridge, MA, USA).
2.2. Immunofluorescence microscopy
Glass coverslips coated with 30 μg/ml mouse laminin or 30 μg/ml bovine serum fibronectin for 24 hours at 4 °C were placed in 35 mm-diameter dishes containing DMEM with freshly thawed 10% FBS. CHO and NIH 3T3 cells were added to dishes with fibronectin-coated coverslips; A375, B16F10 and B16F1 cells were added to dishes with laminin-coated coverslips. Cells were incubated for 30-60 minutes at 37°C with 5% CO2. For all antibodies, except anti-IQGAP2, coverslips were fixed for 60 minutes at 22 °C in cytoskeleton-stabilizing buffer (80 mM PIPES, 2 mM EGTA, 3 mM MgCl2, pH=6.9) with 4% paraformaldehyde and 0.1% Triton-X 100. For anti-IQGAP2 staining, coverslips were fixed for 20 minutes at 22 °C in phosphate-buffered saline containing 1% glutaraldehyde and 0.1% Triton-X 100, and then incubated in 1% sodium borohydride solution for 10 minutes. After fixation, coverslips were washed in water, blocked with 2% bovine serum albumin for 15 minutes and incubated with primary antibodies for 20 minutes at 37°C. Coverslips were incubated with secondary antibodies and mounted onto glass slides using Aqua Poly/Mount (Polysciences, Warrington, PA, USA). Images were acquired using a Leica DMIRE2 HC inverted epifluorescence microscope fitted with a 12-bit grayscale CCD camera.
2.3. Live-cell imaging
Delta T Dish microscope culture dishes from Bioptechs (Butler, PA, USA) were coated with 25 μg/ml laminin. Approximately 24 hours post-transfection of the B16F10 cells with the GFP-IQGAP1 and mCherry-Arp3 plasmids, cells were re-seeded into the laminin-coated dishes. Cells were incubated for 30 minutes at 37 °C prior to initiating the imaging. During the acquisition of images, the chamber was maintained at 37 °C using the Delta T heated-lid controller system (Bioptechs) and infused with humidified, 5% CO2-air mixture.
2.4. Video Analysis
GFP-IQGAP1, mCherry-Arp3 and phase contrast image sets were acquired at 13-second intervals. For the live cell studies, a video of cell movement over 150 seconds was constructed from individual image sets. Positions of the cell edge, GFP-IQGAP1 intensities and mCherry-Arp3 intensities were tracked frame-by-frame using Metamorph image analysis software. GFP and mCherry intensities were corrected for photobleaching by determining the signal decay rate in whole cells. The photobleaching rate constant was 0.003/sec for both GFP and mCherry in our experimental conditions.
3. Results
3.1. Cell type-dependent co-localization of IQGAP1 with WAVE
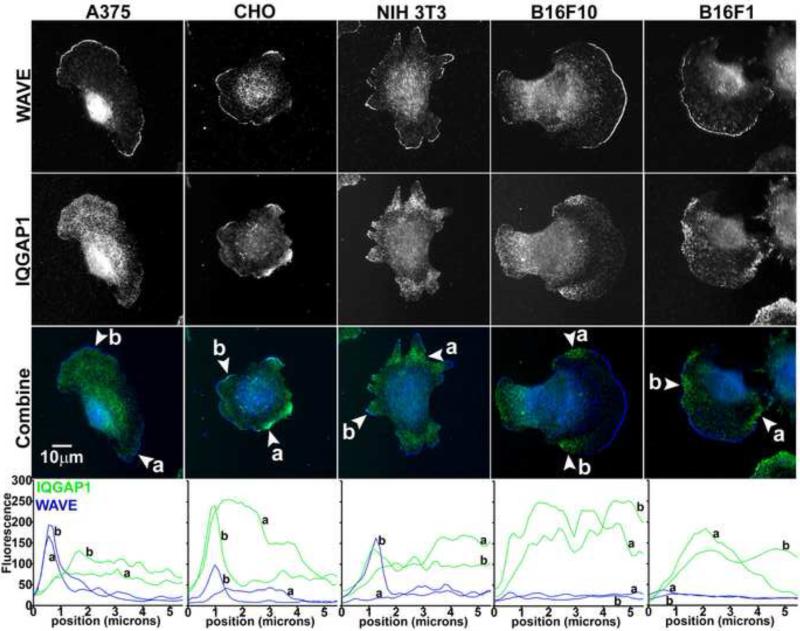
IQGAP1 has been reported to localize to protruding cell edges through association with other protein effectors in the actin protrusion machinery [12-17]. Here we examined co-immunolocalization of IQGAP1 and WAVE in a variety of cell types (Figure 1). In all cell types WAVE localized to the extreme cell edges, consistent with the role of WAVE in polymerizing actin barbed ends [20]. WAVE therefore served as a reliable reference for examination of IQGAP1 among different cell types. Interestingly, we observed varying patterns of IQGAP1 localization within the same cell type and across cell types. In B16F1 and B16F10 mouse melanoma cells, we observed a striking difference in IQGAP1 localization compared to many previous reports [12-17]. In the mouse melanoma cell lines, there was a distinct separation of IQGAP1 from WAVE localization (Figure 1, linescans), indicating that IQGAP1 localizes selectively behind WAVE-negative cell edges in retracting areas. In the A375 human melanoma cell line, IQGAP1 localized throughout the cell lamella and lamellipodium behind WAVE, but not in areas of cell retraction where WAVE was absent. In CHO and NIH 3T3 cells, IQGAP1 localized to areas where WAVE was both present and absent, indicating that IQGAP1 may localize to areas of edge protrusion and retraction within the same cell type. The results show that subcellular localization of IQGAP1 is cell type-dependent, and IQGAP1 localizes and may function in areas of cell retraction.
Fig 1.
Cell type-dependent localization of IQGAP1. A375, CHO, NIH 3T3, B16F10 or B16F1 cells were added to coverslips coated with extracellular matrix protein. After cell attachment and spreading, coverslips were fixed and stained with anti-IQGAP1 and anti-WAVE2 antibodies. Representative images for each cell type are shown. Arrowheads in the combined images (WAVE blue, IQGAP1 green) indicate locations of linescans perpendicular to the cell edge. Bottom of figure are linescans of fluorescence intensity that extended 5.5 microns into the cell from the edge at locations a and b.
3.2. IQGAP1 and Arp3 correlation with cell edge retraction velocity
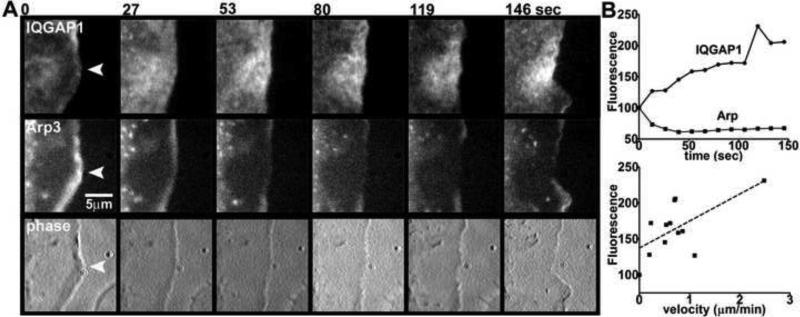
Among the variety of cell lines studied, we observed a clear intracellular separation of IQGAP1 from WAVE in B16F10 mouse melanoma cells (see Figure 1). In order to correlate cell edge movement with IQGAP1 on a time scale of seconds, we performed live experiments with B16F10 cells co-transfected with mCherry-Arp3 and GFP-IQGAP1. Arp2/3 is part of the complex that initiates actin filament branching in actively protruding lamellipodia, and thus localizes in a pattern similar to WAVE [20]. At time=0 sec, Arp3 fluorescence intensity was at the highest point, while IQGAP1 intensity was at the lowest point indicating that the cell edge was in a state of protrusion at the beginning of the time course (Figure 2). While the cell edge began to retract, we observed accumulation of GFP-IQGAP1 and a concomitant decrease in mCherry-Arp3 fluorescence (Figure 2A, B). Moreover, there was a positive correlation between fluorescence intensity of GFP-IQGAP1 and velocity of cell edge retraction (Figure 2B).
Fig 2.
Correlation of cell edge retraction velocity with IQGAP1. (A) B16F10 cells were cotransfected with GFP-IQGAP1 and mCherry-Arp3, and imaged on laminin for a total time of 146 sec at 13-sec intervals. Select time points show IQGAP1, Arp3 and phase images of an area of cell edge retraction. (B) Frames were analyzed for GFP-IQGAP1 and mCherry-Arp3 intensities, and edge velocity changes in the areas marked by the arrowheads. Top graph is a plot of IQGAP1 or Arp3 fluorescence intensities over time. Bottom graph is correlation of IQGAP1 fluorescence intensity with cell edge retraction velocity (Pearson's r = 0.64, n = 12, p = 0.025).
3.3. Downregulation of adhesion sites is associated with IQGAP1
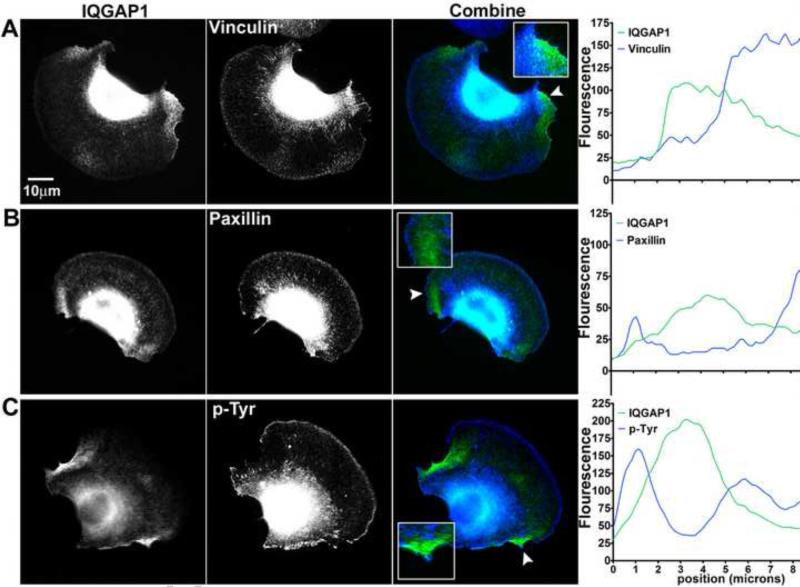
IQGAP1 is known to regulate cell-cell interactions, cell-matrix adhesions [4], and Rho family GTPases activity at β1-integrin activation sites [9, 10]. We determined if IQGAP1 localization was associated with a decrease or an increase in adhesion site activity. In these experiments, we performed co-immunolocalization studies of IQGAP1 with three adhesion markers in B16F10 mouse melanoma cells on laminin substrate. After 30 minutes on laminin, the cells typically formed broad lamellipodia with retracting edges rich in IQGAP1. We performed linescan intensity analysis of vinculin, paxillin and phosphorylated tyrosine (p-Tyr) in regions of cell retraction (Figure 3). Linescan analysis revealed 2-3 micron separation in the peak fluorescence intensities between IQGAP1 and the adhesion site markers (Figure 3A-C, linescans). These results suggest that the micron-range separation of IQGAP1 from cell-matrix adhesion sites is the result of downregulation through IQGAP1 signaling coordinated with cell edge retraction.
Fig 3.
Downregulation of adhesion site markers associated with IQGAP1 localization. B16F10 cells transfected with GFP-IQGAP1 were stained with antibodies against (A) vinculin, (B) paxillin or (C) phosphorylated tyrosine (p-Tyr). Fluorescence intensity in the combined images of IQGAP1 (green) and adhesion sites (blue) were quantified using linescan analysis. Arrowheads in the combined images indicate location of linescan that extended 8.3 microns into the cell from the edge.
3.4 Cellular localization of IQGAP1, myosin IIA, calmodulin and IQGAP2 in retracting versus protruding edges
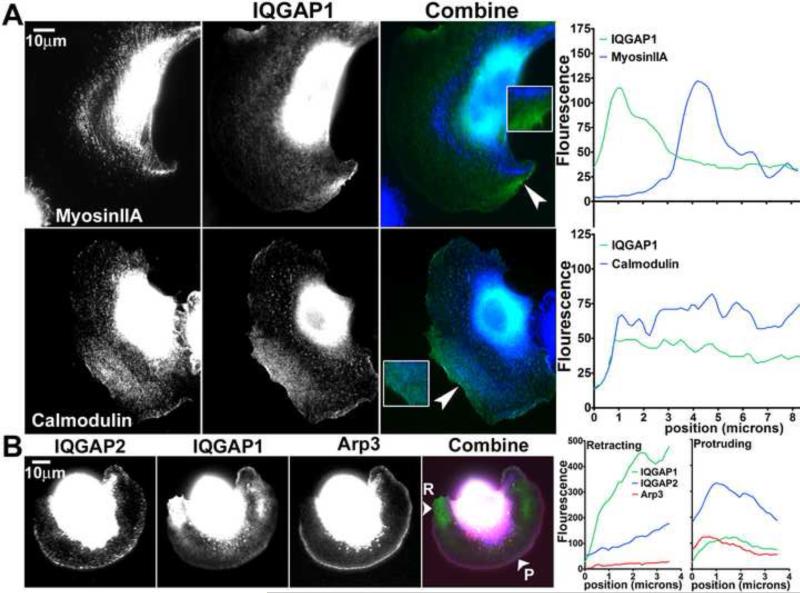
Since IQGAP1 is known to bind calmodulin in a calcium-dependent manner and interact with myosin essential light chain [2], we examined whether IQGAP1 is associated with calmodulin in retracting areas in B16F10 cells. Interestingly, calmodulin, but not myosin IIA, co-localized with IQGAP1 in retracting cell areas (Figure 4A). The peak fluorescence intensity of myosin IIA was separated from the peak intensity of IQGAP1 by approximately 3 microns (Figure 4A, linescan). Since IQGAP2 and IQGAP1 are known to have overlapping functions [3], we examined if IQGAP2 localizes with IQGAP1 in B16F10 cells. In these experiments, IQGAP1 was detected by localization of transfected GFP-IQGAP1, and IQGAP2 was detected by immunofluorescence. Where localization of IQGAP1 was highest at retracting edges, there was little localization of IQGAP2 (Figure 4B, linescans). While relatively little GFP-IQGAP1 localized at the protruding cell edge, IQGAP2 intensity was highest at the protruding edge along with mCherry-Arp3 (Figure 4B, linescans). These results indicate that calmodulin, but not myosin IIA, localizes with IQGAP1 in retracting cell edges while IQGAP2 localizes to protruding edges.
Fig 4.
Subcellular localization of IQGAP1, myosin IIA, calmodulin and IQGAP2 in B16F10 cells. (A) Cells transfected with GFP-IQGAP1 were stained with anti-myosin IIA or anti-calmodulin antibodies. Fluorescence intensity of the IQGAP1 image (green) combined with the myosin IIA or calmodulin image (blue) was quantified using linescan analysis. Arrowheads in the combined images indicate location of linescan that extended 8.5 microns into the cell from the edge. (B) Cells co-transfected with GFP-IQGAP1 and mCherry-Arp3 were stained with anti-IQGAP2 antibodies. The IQGAP2 (blue), IQGAP1 (green) and Arp3 (red) images were combined and fluorescence intensity in retracting (R) and protruding (P) edges was quantified by linescan analysis. Arrowheads in the combined image indicate location of linescan that extended 3.5 microns into the cell from the edge.
4. Discussion
The wide range of cellular activities mediated through IQGAP1 protein is attributed to multiple interacting partners [4]. In particular, numerous reports show that IQGAP1 localizes to actin-rich sites at the leading edge of cells [12-17] and in membrane ruffles [11]. In these current studies we examined intracellular distribution of IQGAP1 and found cell-type dependent localization throughout lamellipodia and lamella, at the leading edge, and surprisingly, actively retracting cell areas. Using immunolocalization and GFP-IQGAP1 expression approaches, we observed the most restricted distribution of IQGAP1 to retracting areas in mouse melanoma cells. In CHO cells, IQGAP1 appeared to localize to both protruding and retracting cell edges suggesting IQGAP1 may play a dual role in actin dynamics within the same cells type. In A375 human melanoma cells, IQGAP1 localized throughout lamellipodia and lamella behind protruding edges, but not in retracting edges. These results demonstrate for the first time that IQGAP1 plays a role in cell retraction signaling potentially through downregulation of adhesion sites and up regulation of myosin-induced retraction.
The results of our study raise important questions about IQGAP1 signaling in actin dynamics during cell motility. In particular, these questions concern the identity of the domain in IQGAP1 that is responsible for localization and the details of signaling in areas of cell retraction. Another important point to address is the experimental conditions that may influence IQGAP1 localization such as expression levels, cell density and presence of growth factors. Overexpression of intracellular IQGAP1 may force localization to lower affinity sites on the actin cytoskeleton. Indeed, in our studies we did observe localization to the leading edge in B16F10 cells expressing GFP-IQGAP1; however, the intensity was still much lower compared to retracting areas of the cell. IQGAP1 is known to stabilize [21, 22] as well as disassemble [23-25] adherens junctions at sites of cell-cell contact; thus, cell density and whether cells form contacts will likely determine subcellular localization of IQGAP1. For example, leading edge localization may be observed in a scratch wound system, where cells polarize and the leading edge moves into the scratch wound while the opposite side maintains cell-cell contact. Other studies have shown leading edge association induced by growth factor receptor signaling [11, 12, 16], further underscoring the effect of experimental conditions on the intracellular localization of IQGAP1. In our studies we observed strong IQGAP1 co-localization with calmodulin, but not myosin IIA or IQGAP2. These results not only provide evidence for differential function between IQGAP1 and IQGAP2 in cell retraction and protrusion, respectively, but suggest IQGAP1 cellular distribution and perhaps function involves calmodulin interaction. There have been several studies detailing the binding interactions between calmodulin and myosin essential light chain with IQGAP1. If IQGAP1 binds to calmodulin in retracting cell edges, then localization may require the IQ [2] or CHD [26] domains and be calcium dependent [2]. Binding of IQGAP1 to calmodulin in areas of retraction may displace other binding partners from IQGAP1 such as B-Raf [27] or Cdc42 [28] which in turn could mediate downstream effects on myosin activity and adhesion site turnover. Alternatively, IQGAP1 could mediate retraction through recruitment of calmodulin and calcium [29], or local interaction with Rho family GTPases [30]. Overall, our studies show that intracellular localization of IQGAP1 is variable, ranging from restriction to leading or retracting cell edges depending on cell type, to both leading and retracting edges within the same cell type. In those cell types where IQGAP1 localization is restricted to retracting cell areas, IQGAP1 likely plays novel roles in cell motility through regulation of adhesion site dynamics and myosin retraction.
Article highlights.
Intracellular localization of IQGAP1 protein is variable and cell type dependent
IQGAP1, but IQGAP2, localizes to retracting cell areas along with calmodulin
Localization of IQGAP1 is associated with down regulation of cell-matrix interactions
Acknowledgements
This work was supported by a grant from the National Institute of General Medical Sciences (J.M.S. R15GM093288) and funds from the Department of Pharmaceutical Sciences at Southern Illinois University Edwardsville.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weissbach L, Settleman J, Kalady MF, et al. Identification of a human rasGAP-related protein containing calmodulin-binding motifs. J Biol Chem. 1994;269:20517–20521. [PubMed] [Google Scholar]
- 2.Pathmanathan S, Hamilton E, Atcheson E, et al. The interaction of IQGAPs with calmodulin-like proteins. Biochem Soc Trans. 2011;39:694–699. doi: 10.1042/BST0390694. [DOI] [PubMed] [Google Scholar]
- 3.Briggs MW, Sacks DB. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep. 2003;4:571–574. doi: 10.1038/sj.embor.embor867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.White CD, Erdemir HH, Sacks DB. IQGAP1 and its binding proteins control diverse biological functions. Cell Signal. 2012;24:826–834. doi: 10.1016/j.cellsig.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.White CD, Brown MD, Sacks DB. IQGAPs in cancer: a family of scaffold proteins underlying tumorigenesis. FEBS Lett. 2009;583:1817–1824. doi: 10.1016/j.febslet.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson M, Sharma M, Henderson BR. IQGAP1 regulation and roles in cancer. Cell Signal. 2009;21:1471–1478. doi: 10.1016/j.cellsig.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 7.Kozlova I, Ruusala A, Voytyuk O, et al. IQGAP1 regulates hyaluronan-mediated fibroblast motility and proliferation. Cell Signal. 2012;24:1856–1862. doi: 10.1016/j.cellsig.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 8.Kohno T, Urao N, Ashino T, et al. IQGAP1 links PDGF receptor-beta signal to focal adhesions involved in vascular smooth muscle cell migration: role in neointimal formation after vascular injury. Am J Physiol Cell Physiol. 2013;305:C591–600. doi: 10.1152/ajpcell.00011.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacquemet G, Morgan MR, Byron A, et al. Rac1 is deactivated at integrin activation sites through an IQGAP1-filamin-A-RacGAP1 pathway. J Cell Sci. 2013;126:4121–4135. doi: 10.1242/jcs.121988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacquemet G, Green DM, Bridgewater RE, et al. RCP-driven alpha5beta1 recycling suppresses Rac and promotes RhoA activity via the RacGAP1-IQGAP1 complex. J Cell Biol. 2013;202:917–935. doi: 10.1083/jcb.201302041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heil A, Nazmi AR, Koltzscher M, et al. S100P is a novel interaction partner and regulator of IQGAP1. J Biol Chem. 2011;286:7227–7238. doi: 10.1074/jbc.M110.135095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaoka-Tojo M, Ushio-Fukai M, Hilenski L, et al. IQGAP1, a novel vascular endothelial growth factor receptor binding protein, is involved in reactive oxygen species--dependent endothelial migration and proliferation. Circ Res. 2004;95:276–283. doi: 10.1161/01.RES.0000136522.58649.60. [DOI] [PubMed] [Google Scholar]
- 13.Ruiz-Saenz A, Kremer L, Alonso MA, et al. Protein 4.1R regulates cell migration and IQGAP1 recruitment to the leading edge. J Cell Sci. 2011;124:2529–2538. doi: 10.1242/jcs.083634. [DOI] [PubMed] [Google Scholar]
- 14.Watanabe T, Noritake J, Kakeno M, et al. Phosphorylation of CLASP2 by GSK-3beta regulates its interaction with IQGAP1, EB1 and microtubules. J Cell Sci. 2009;122:2969–2979. doi: 10.1242/jcs.046649. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe T, Wang S, Noritake J, et al. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev Cell. 2004;7:871–883. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 16.Bensenor LB, Kan HM, Wang N, et al. IQGAP1 regulates cell motility by linking growth factor signaling to actin assembly. J Cell Sci. 2007;120:658–669. doi: 10.1242/jcs.03376. [DOI] [PubMed] [Google Scholar]
- 17.Le Clainche C, Schlaepfer D, Ferrari A, et al. IQGAP1 stimulates actin assembly through the N-WASP-Arp2/3 pathway. J Biol Chem. 2007;282:426–435. doi: 10.1074/jbc.M607711200. [DOI] [PubMed] [Google Scholar]
- 18.Ren JG, Li Z, Crimmins DL, et al. Self-association of IQGAP1: characterization and functional sequelae. J Biol Chem. 2005;280:34548–34557. doi: 10.1074/jbc.M507321200. [DOI] [PubMed] [Google Scholar]
- 19.Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bisi S, Disanza A, Malinverno C, et al. Membrane and actin dynamics interplay at lamellipodia leading edge. Curr Opin Cell Biol. 2013;25:565–573. doi: 10.1016/j.ceb.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharya M, Su G, Su X, et al. IQGAP1 is necessary for pulmonary vascular barrier protection in murine acute lung injury and pneumonia. Am J Physiol Lung Cell Mol Physiol. 2012;303:L12–19. doi: 10.1152/ajplung.00375.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noritake J, Fukata M, Sato K, et al. Positive role of IQGAP1, an effector of Rac1, in actin-meshwork formation at sites of cell-cell contact. Mol Biol Cell. 2004;15:1065–1076. doi: 10.1091/mbc.E03-08-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hage B, Meinel K, Baum I, et al. Rac1 activation inhibits E-cadherin-mediated adherens junctions via binding to IQGAP1 in pancreatic carcinoma cells. Cell Commun Signal. 2009;7:23–35. doi: 10.1186/1478-811X-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fram S, King H, Sacks DB, et al. A PAK6-IQGAP1 complex promotes disassembly of cell-cell adhesions. Cell Mol Life Sci. 2013 doi: 10.1007/s00018-013-1528-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan Z, Zhang W, Tan W. A Labile Pool of IQGAP1 Disassembles Endothelial Adherens Junctions. Int J Mol Sci. 2013;14:13377–13390. doi: 10.3390/ijms140713377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews WJ, Bradley CA, Hamilton E, et al. A calcium-dependent interaction between calmodulin and the calponin homology domain of human IQGAP1. Mol Cell Biochem. 2012;371:217–223. doi: 10.1007/s11010-012-1438-0. [DOI] [PubMed] [Google Scholar]
- 27.Ren JG, Li Z, Sacks DB. IQGAP1 integrates Ca2+/calmodulin and B-Raf signaling. J Biol Chem. 2008;283:22972–22982. doi: 10.1074/jbc.M804626200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho YD, Joyal JL, Li Z, et al. IQGAP1 integrates Ca2+/calmodulin and Cdc42 signaling. J Biol Chem. 1999;274:464–470. doi: 10.1074/jbc.274.1.464. [DOI] [PubMed] [Google Scholar]
- 29.Psatha MI, Razi M, Koffer A, et al. Targeting of calcium:calmodulin signals to the cytoskeleton by IQGAP1. Cell Calcium. 2007;41:593–605. doi: 10.1016/j.ceca.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 30.Casteel DE, Turner S, Schwappacher R, et al. Rho isoform-specific interaction with IQGAP1 promotes breast cancer cell proliferation and migration. J Biol Chem. 2012;287:38367–38378. doi: 10.1074/jbc.M112.377499. [DOI] [PMC free article] [PubMed] [Google Scholar]