Abstract
The serendipitous discovery of the spontaneous growth of protein crystals inside cells has opened the field of crystallography to chemically unmodified samples directly available from their natural environment. On the one hand, through in vivo crystallography, protocols for protein crystal preparation can be highly simplified, although the technique suffers from difficulties in sampling, particularly in the extraction of the crystals from the cells partly due to their small sizes. On the other hand, the extremely intense X-ray pulses emerging from X-ray free-electron laser (XFEL) sources, along with the appearance of serial femtosecond crystallography (SFX) is a milestone for radiation damage-free protein structural studies but requires micrometre-size crystals. The combination of SFX with in vivo crystallography has the potential to boost the applicability of these techniques, eventually bringing the field to the point where in vitro sample manipulations will no longer be required, and direct imaging of the crystals from within the cells will be achievable. To fully appreciate the diverse aspects of sample characterization, handling and analysis, SFX experiments at the Japanese SPring-8 angstrom compact free-electron laser were scheduled on various types of in vivo grown crystals. The first experiments have demonstrated the feasibility of the approach and suggest that future in vivo crystallography applications at XFELs will be another alternative to nano-crystallography.
Keywords: serial femtosecond crystallography, in vivo crystallography, X-ray free-electron laser
1. Introduction
The existence of structural biology lies on a fundamental fact: the knowledge of the sequence of a protein is not sufficient to determine the folding of the protein and how it will become functional. A detailed three-dimensional structure is required prior to inferring the function of proteins and their interactions with other biological entities. The study of diseases and their treatments is a fair example of such a requirement, whereby a boom in the developments of new medications resulting from structure-based drug-design approaches allowed engineering of hundreds of new molecules with potential pharmaceutical applications [1]. To fully appreciate and relate a protein structure to its biological function, the three-dimensional model produced should be accurate. Moreover, proteins are highly heterogeneous in nature; far from being rigid, they are dynamic entities, with post-translational modifications that often dictate their cellular localization and functional partners. Various techniques exist for studying the structures and the dynamics of macromolecules, among which protein crystallography, nuclear magnetic resonance, electron microscopy and atomic force microscopy are well documented. Nonetheless, the structure determination at atomic-resolution of biological macromolecules remains primarily dependent on synchrotron-base X-ray crystallography.
A detailed analysis of the Protein Data Bank (PDB, www.pdb.org) shows a constant increase in the number of coordinates deposited every year, predominantly originating from data collected at synchrotron X-ray sources (figure 1a). The unquestionable success of protein crystallography results from advances in the methods applied to the preparation of the samples, but also from various automations engineered all along the steps towards crystallization, data collection and structure determination. However, perusal of the molecular weight distribution of the proteins present in the PDB reveals a lognormal shape with its peak at 30 kDa (figure 1b), reflecting the difficulties encountered in protein crystallography of large-sized molecules. The accepted approaches to produce protein crystals, exclusively in vitro, rely on the necessary purification of the protein itself, which increases with the complexity of large molecules and molecular complexes. Eventually, a serious bottleneck of this technique lies in the need for large well-ordered protein crystals that can diffract to high resolution with limited X-ray dose. Big molecules rarely form such crystals, but rather generate nanometre-sized crystals, sensitive to radiation damage, with low-diffraction capabilities, and hence not exploitable at most third-generation synchrotron sources (low X-ray fluxes). As demonstrated previously [6], X-ray free-electron lasers (XFELs) opened new opportunities to overcome these drawbacks. Furthermore, using ultra-intense X-ray pulses from XFELs now makes it possible to collect high-quality structure factors from a flowing suspension of crystals of sub-micrometre size [3].
Figure 1.
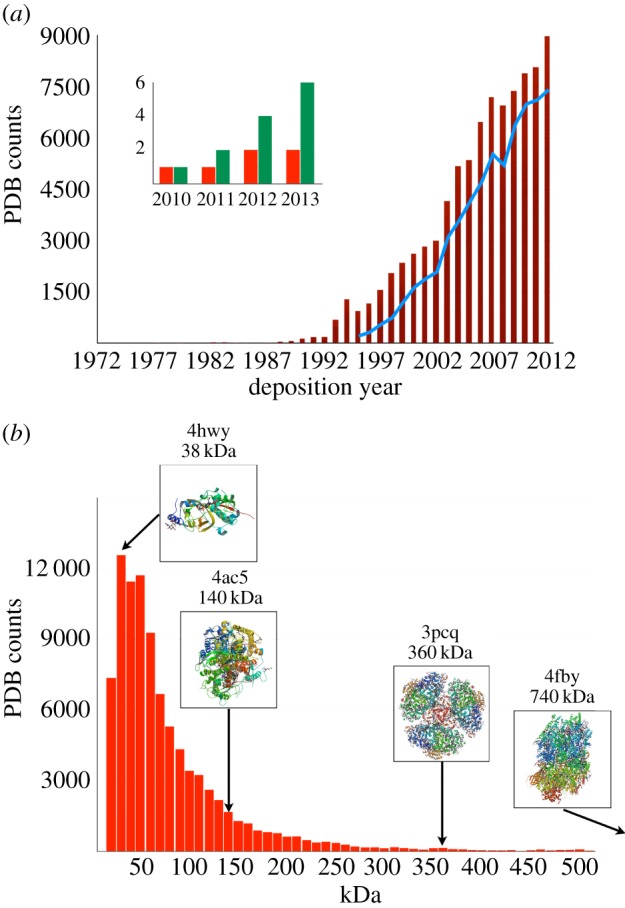
(a) Evolution of the PDB in terms of the total number of coordinates deposited each year (dark-red bar chart) and the total number of coordinates originating from data collected at synchrotron X-ray sources (blue line graph). The inset represents the total number of coordinates originating from data collected at XFELs (green bar chart, right) with the corresponding number of coordinates deposited each year (red bar chart, left). (b) Molecular weight distribution of the coordinates in the PDB (red bar chart). Within the insets are presented the structures of each molecule for which XFEL data were used (PDB accession numbers 4ac5 [2], 3pcq [3], 4fby [4] and 4hwy [5]). (Online version in colour.)
The realization of serial femtosecond crystallography (SFX) at XFELs has been coupled with the earlier serendipitous discovery of protein microcrystals that spontaneously form within cells, such as during the infection of insect cells with naturally occurring viral particles [7–10]. In a previous report, the applicability of SFX on in vivo grown crystals was demonstrated through the study of cathepsin B [5], during which it was discovered that the protein structure includes post-translational modifications that would not be seen in conventional expression systems. With these latest SFX results, the number of reported observations of microcrystals has increased, forcing nano-crystallography to appear as a general technique that could replace months or years of crystallization trials. In the present report, we are questioning whether in vivo grown crystals when associated with SFX could be a solution for solving the structure of systems that have not been amenable to conventional crystallography such as macromolecular complexes and chemically untreated proteins still bearing their post-translational modifications in general.
2. Material and methods
(a). Characterization of the in vivo grown crystals
Brief characteristics of the in vivo expression systems and the protein crystals grown within used in this study are listed in table 1. Further details are to be reported in separate communications. For every protein sample studied, the size of the crystals was sufficient for microscopic visualization. A typical crystal of a human neuraminidase [12] grown in vivo is shown in figure 2. Various methods were applied for identification and characterization of the crystals, including immunoblotting (figure 2), X-ray diffraction studies and rigorous extraction and analysis of the crystal content. Attempts to image the samples through the second-order nonlinear imaging of chiral crystal system were not successful, possibly due to the weak signal emitting from these crystals lying in a crowded environment.
Table 1.
List of in vivo grown crystals. Cockroach milk protein (C.m.p.) is a cockroach protein involved in food storage [11]; hNeu1 is a human neuraminidase [12]. Type 1 and 2 refer to the viscosity of the extracted sample solution before injection, with type 1 samples being less viscous than type 2. The [buffer] line corresponds to the buffer of the sample after extraction from the cells; cell lysate/PBS stands for the overall cell-lysate together with its lysis buffer immediately after treatment of the cells. The [volume] line corresponds to the total volume of injected sample. Cathepsin B was added as a reference for comparison and was not used in these experiments.
| spheroid | C.m.p. | hNeu1 | hNeu1 | CatB | |
|---|---|---|---|---|---|
| type | 1 | 1 | 2 | 2 | 1 |
| host | LD652 cells | cockroach | CHO cells | HEK293FT cells | Sf9 cells |
| molecular weight (kDa) | 114.9 | 17.8 | 45.5 | 45.5 | 37.2 |
| buffer | tris | water | cell lysate/PBS | cell lysate/PBS | PBS |
| crystal size (µm3) | 4 × 4 × 4 | 15 × 15 × 5 | 1 × 1 × 1 | 1 × 1 × 1 | 0.5 × 0.5 × 3 |
| hit rate | — | approximately 20% | n.d. | approximately 0.2% | — |
| volume (ml) | — | 5 | 45 | 15 | — |
| PDB | — | — | — | — | 4hwy |
| reference | — | [11] | [12] | [12] | [5] |
Figure 2.
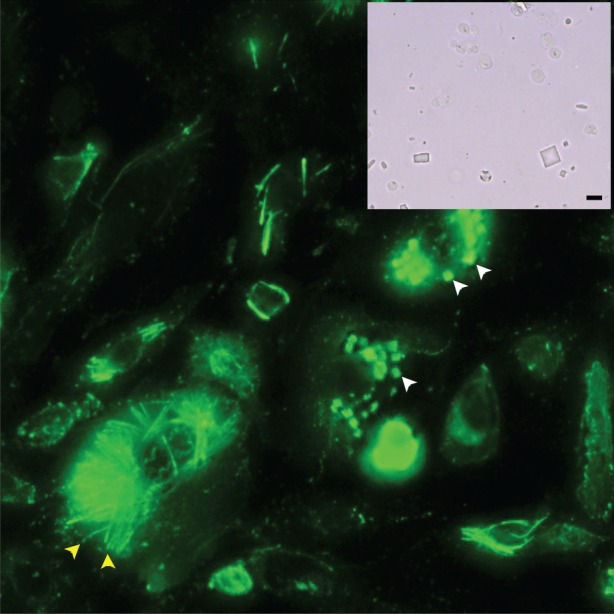
In vivo grown crystals enclosed inside mammalian CHO cells (a) and after extraction from human HEK293FT cells (inlet). The crystals vary in size and can reach dimensions of 15 × 15 × 3 μm3. White (top right) and yellow (bottom left) arrows point towards square- and needle-like crystals, respectively. The scale bar on the inlet picture represents 10 μm. (Online version in colour.)
(b). Crystal injection, data collection and analysis
Crystals extracted from their host cells were kept in solutions at 293 K on a rotary device after being filtered through 20 μm filtering units. An HPLC system (LC-20AD type, Shimadzu Scientific Instruments) was used for injecting the crystals into a sample chamber filled with helium in a liquid jet of approximately 20 μm width [13]. The experiments were performed at the SPring-8 angstrom compact free-electron laser (SACLA) beamline 3 experimental hutch 3 [14,15]. Diffraction data were recorded on an octal multi-port charged-coupled device (2048 × 2048 pixels) detector [13]. The distance of the detector to the interaction region was physically set at approximately 50 mm and was further refined virtually through powder diffraction pattern fitting. The final calculated distance was approximately 52 mm. Diffraction experiments were performed at an energy of 7 keV with single pulses of 10 fs and 100 µJ on average. The X-rays were focused to the interaction point by Kirkpatrick–Baez mirrors to a focal point of 1 μm2 (full width at half maximum) [16]. Images were recorded at 20 Hz. Diffraction pictures were screened for the identification of Bragg diffraction spots using in-house software after background removal. False positives were removed from the pool of possible protein diffraction pictures by visual inspection.
3. Results and discussion
(a). Sample viscosity and injector requirements
In the present experiments, the viscosity of the sample solutions to be injected differed depending on the procedure adopted to extract the crystals (table 1). While low-viscosity samples (hereafter referred as type 1) are kept in water after extensive purification and isolation of the crystals, the high-viscosity samples (hereafter referred as type 2) remain largely in their native environment, due to the more simplistic sample preparation procedure. The only two purification steps of these crystals consist in opening the cell membranes by cell lysis, followed by filtering to remove large sized particles. This procedure was chosen over a refined purification approach mostly as a consequence of the instability of the crystals once outside of their natural environment. To minimize the risks of injector clogging from such viscous samples, a nozzle with an inner diameter of 75 μm was chosen, resulting in a jet of approximately 20 μm after gas focusing. The samples were injected at 0.5 ml min−1, and no clogging of the nozzle occurred even after injecting up to 45 ml of sample. However, the clear advantage of using wider jets for avoiding clogging was counterbalanced by an increase of the background on the diffraction pictures with a direct influence on the quality of the data collected.
(b). Diffraction from in vivo crystals: proof of principle
The hit rate in SFX experiments is considered as the mean number of indexed patterns per recorded diffraction picture. It greatly varies, depending on numerous parameters among which are the relationship of the sample crystal size and concentration to the volume of sample jet occupied by crystals, the beam stability and fluence, and the detector sensitivity. During the present experiments, the most successful sample (cockroach milk protein [11]) showed a hit rate reaching 20%, whereas more difficult samples only led to approximately 0.2% hit rate (200 indexed patterns out of 75 750 recorded pictures). The only evident difference between the two jetting conditions was the dilution effect for the type 2 samples. Indeed, the likelihood of having a crystal at the X-ray interaction region depends on the concentration of crystal in the liquid jet.
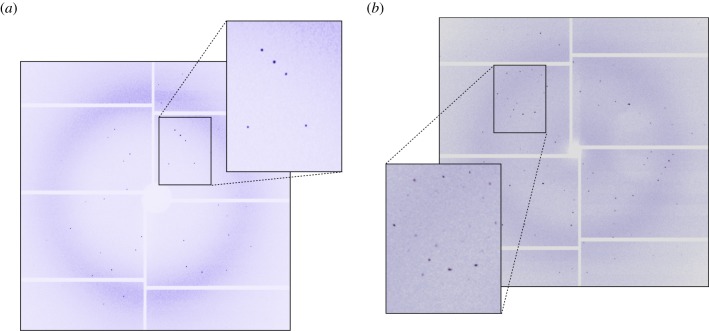
As an indicator of crystal quality, the diffracting power of the in vivo grown crystals could be considered as a good estimate. While the human neuraminidase crystals diffracted at a visible resolution of 3 Å (figure 3a), the diffraction limit of the crystals from cockroaches reached 1.6 Å, up to the edges of the detector set-up at the minimum available sample-to-detector distance (figure 3b). These results, combined with the reported structures of cathepsin B [5], clearly indicate that in vivo grown crystals have a strong potential for structural studies at XFEL sources.
Figure 3.
Typical diffraction picture for the in vivo crystals of the mammalian neuraminidase hNeu1 (a) and the cockroach milk protein (b). hNeu1 crystals diffracted to 3.0 Å resolution, and cockroach milk protein crystals to 1.6 Å. The contrast of the picture was adapted to optimize visualization of the diffraction spots. (Online version in colour.)
4. Conclusion
In the present short communication, we clearly confirmed that the implementation of in vivo crystallography at XFEL sources opens a new window to structural biology. One repetitive issue that develops when working on such systems questions the possibility of crystallizing any kind of protein using this method. Answering this question is beyond the scope of this article, but we could easily assume that by providing the right conditions, it should be feasible to engineer such a platform for ‘natural’ crystallization. As shown in this report, diffraction data can be recorded from in vivo grown crystals, and whether the crystals are produced from insects, insect cells, mammalian cells or virus-infected cells does not affect the quality of the crystals and recorded data. Improvements in the extraction procedure of the in vivo crystals will be necessary for making in vivo crystallography the method of choice for structural biologists.
Acknowledgements
The experiments were performed at the beamline 3 experimental hutch 3 of SACLA with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) (Proposal No. 2013A8039). We additionally thank all the crew at the beamline for constant help and understanding.
Funding statement
This work was supported by the X-ray Free-Electron Laser Priority Strategy Program (MEXT, Japan). F.C. is supported by an ARC Future Fellowship.
References
- 1.Anderson A. 2003. The process of structure-based drug design. Chem. Biol. 10, 787–797. ( 10.1016/j.chembiol.2003.09.002) [DOI] [PubMed] [Google Scholar]
- 2.Johansson LC, et al. 2012. Lipidic phase membrane protein serial femtosecond crystallography. Nat. Methods 9, 263–265. ( 10.1038/nmet.1867) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman H, et al. 2011. Femtosecond X-ray protein nanocrystallography. Nature 470, 73–77. ( 10.1038/nature09750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kern J, et al. 2012. Room temperature femtosecond X-ray diffraction of photosystem II microcrystals. Proc. Natl Acad. Sci. USA 109, 9721–9726. ( 10.1073/pnas.1204598109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Redecke L, et al. 2013. Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser. Science 339, 227–230. ( 10.1126/science.1229663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman H, et al. 2004. Femtosecond diffractive imaging with a soft-X-ray free-electron laser. Nat. Phys. 2, 839–843. ( 10.1038/nphys461) [DOI] [Google Scholar]
- 7.Hasegawa H, et al. 2011. In vivo crystallization of human IgG in the endoplasmic reticulum of engineered Chinese hamster ovary (CHO) cells. J. Biol. Chem. 286, 19 917–19 931. ( 10.1074/jbc.M110.204362) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coulibaly F, Chiu E, Ikeda K, Gutmann S, Haebel PW, Schulze-Briese C, Mori H, Metcalf P. 2007. The molecular organization of cypovirus polyhedral. Nature 446, 97–101. ( 10.1038/nature05628) [DOI] [PubMed] [Google Scholar]
- 9.Coulibaly F, et al. 2009. The atomic structure of baculovirus polyhedra reveals the independent emergence of infectious crystals in DNA and sRNA viruses. Proc. Natl Acad. Sci. USA 106, 22 205–22 210. ( 10.1073/pnas.0910686106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan G, Maldonado F, Zhang Y, Kincaid R, Ellisman MH, Gastinel LB. 1996. In vivo calcineurin crystals formed using the baculovirus expression system. Microsc. Res. Tech. 34, 77–86. () [DOI] [PubMed] [Google Scholar]
- 11.Williford A, Stay B, Bhattacharya D. 2004. Evolution of a novel function: nutritive milk in the viviparous cockroach, Diploptera punctate. Evol. Dev. 6, 67–77. ( 10.1111/j.1525-142X.2004.04012.x) [DOI] [PubMed] [Google Scholar]
- 12.Seyrantepe V, Poupetova H, Froissart R, Zabot M-T, Maire I, Pshezhetsky AV. 2003. Molecular pathology of NEU1 gene in sialidosis. Hum. Mutat. 22, 343–352. ( 10.1002/humu.10268) [DOI] [PubMed] [Google Scholar]
- 13.Song C, et al. 2013. Multiple application X-ray imaging chamber for single-shot diffraction experiments with femtosecond X-ray laser pulses. J. Appl. Crystallogr. 47, 188–197. ( 10.1107/S1600576713029944) [DOI] [Google Scholar]
- 14.Ishikawa T, et al. 2012. A compact X-ray free-electron laser emitting in the sub-angstrom region. Nat. Photonics 6, 540–544. ( 10.1038/nphoton.2012.141) [DOI] [Google Scholar]
- 15.Tono K, et al. 2013. Beamline for X-ray free electron laser at SACLA. J. Phys. 425, 072006 ( 10.1088/1742-6596/425/7/072006) [DOI] [Google Scholar]
- 16.Yumoto H, et al. 2013. Focusing of X-ray free-electron laser pulses with reflective optics. Nat. Photonics 7, 43–47. ( 10.1038/nphoton.2012.306) [DOI] [Google Scholar]