Abstract
Macroautophagy (henceforth referred to as autophagy) is an in-bulk lysosomal degradative pathway that plays a crucial role in the maintenance of cellular homeostasis through the removal of damaged proteins and aged organelles. Following nutrient deprivation, a primary cellular response is the induction of autophagy that breaks down redundant cellular components and provides amino acids and additional precursor molecules for processes critical for cellular survival. In parallel, nutrient depletion leads to the mobilization of cellular lipid stores to supply free fatty acids for energy, thus pointing to regulatory and functional similarities between autophagy and lipid metabolism. The current chapter discusses the novel and mutually exclusive roles of autophagy in the regulation of lipid metabolism in the liver and of fat storage within the adipose tissue. Our studies in cultured hepatocytes and the murine liver have demonstrated that autophagy serves to degrade intracellular lipid stores through a process that we have termed “macrolipophagy” and that ablation of liver-specific autophagy leads to excessive hepatic lipid accumulation and the development of fatty liver. In contrast, preadipocytes in culture that lacked autophagy failed to differentiate into mature adipocytes and exhibited a reduction in fat storage that translated to decreased adipose tissue mass in an in vivo mouse model. These recent findings establish an association between autophagy and regulation of hepatic lipid metabolism and adipose tissue biology, thus providing new mechanistic insights into the regulation of these complex processes. These findings also highlight the possibility of novel therapeutic approaches, such as differential organ-specific regulation of autophagy to solve problems that arise from lipid over accumulation that occur in the metabolic syndrome and with aging.
1 An Introduction to Autophagy
Autophagy or “self-eating” is a conserved cellular process critical for maintaining cellular homeostasis (Cuervo 2008) through the targeting of altered cytosolic proteins, aged organelles and redundant cytosol, and even pathogenic organisms to the lysosomes for degradation. Besides this crucial role in quality control, autophagy has also been shown to be involved in a host of other functions such as growth and differentiation (Mizushima 2009), metabolic regulation (Singh et al. 2009a, b) and as an alternative energy source following nutrient deficiency. Three different forms of autophagy have been described in mammalian systems (Fig. 1), macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA) (Esclatine et al. 2009; Kon and Cuervo 2010). These three forms of autophagy co-exist in most cells at a given moment, although the induction of these discrete forms of autophagy differs temporally. For instance, under baseline cellular conditions all forms of autophagy functions at a basal level, however, following nutrient deficiency, macroautophagy is induced first and acquires maximal activation at 6–8 h of continuous stress (Esclatine et al. 2009). If the given stress persists beyond 6–8 h, macroautophagy gradually tapers off and induction of CMA occurs that attains maximal activation by 12–24 h (Kon and Cuervo 2010) and is maintained until the stress dissipates. Autophagy can also be qualitatively classified based upon the type of cargo that is being degraded, for instance, mitophagy mediates mitochondrial degradation (Tolkovsky 2009). Likewise, ribophagy (Beau et al. 2008), reticulophagy (Tasdemir et al. 2007), and pexophagy (Manjithaya et al. 2010) are required for the degradation of ribosomes, endoplasmic reticulum, and peroxisomes, respectively. Very recently, macroautophagy has also been shown to mediate the degradation of cellular lipid droplets under basal conditions as well as when macroautophagy is induced following lipogenic stimuli, by a process termed macrolipophagy (Singh et al. 2009a). This function of macroautophagy in the regulation of cellular lipid stores will be the focus of part of this chapter.
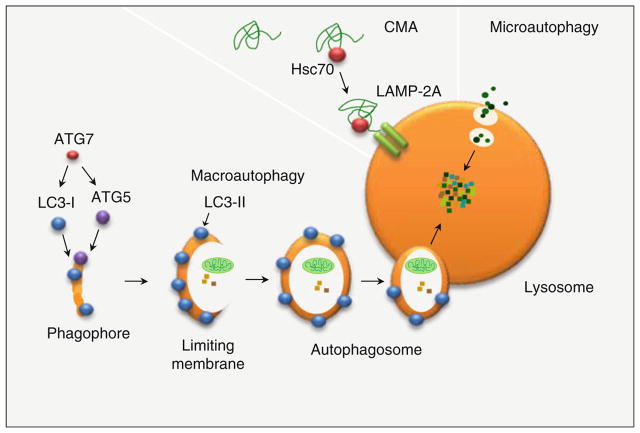
Fig. 1.
Types of autophagy in mammalian cells. Three types of autophagy have been described in mammalian cells, macroautophagy, an in-bulk degradative pathway that requires the de novo synthesis of a limiting membrane for sequestration and delivery of cargo to the lysosome. Microautophagy involves engulfment of cytosolic material by the lysosomal membrane itself. Chaperone-mediated autophagy targets specific proteins containing the recognition template, KFERQ motif to lysosomal membrane-associated translocation complex, LAMP-2A, for substrate internalization and degradation
1.1 Types of Autophagy
Macroautophagy is an in-bulk lysosomal degradative pathway that involves the initial de novo formation of a phagophore (He and Klionsky 2009), which then adds on additional membranes through yet unclear mechanisms and forms a limiting membrane. The limiting membrane sequesters portions of the cytosol destined for degradation and eventually seals upon itself to form a double-walled autophagosome (He and Klionsky 2009). The autophagosome lacks the acidic environment and the enzymes required for terminal digestion of the engulfed contents; thus, content degradation occurs via the fusion of the autophagosome with the lysosome that supplies the acidic environment as well as a battery of hydrolases. Microautophagy, a second form of autophagy, involves engulfment of cargo by the lysosomal membrane itself (Esclatine et al. 2009). Single-membrane vesicles that contain the cargo are pinched off the lysosomal membrane and rapidly degraded in the lysosomal lumen. A third form of autophagy, CMA, is highly specific for an estimate of 30% of the soluble cytosolic proteins (Kon and Cuervo 2010). In CMA, proteins that contain a recognition template, the KFERQ motif, are recognized by the cytosolic chaperone hsc70 and directed to the lysosome (Kon and Cuervo 2010). Upon the lysosomal surface, the protein–hsc70 complex is recognized by the lysosome-associated membrane protein (LAMP-2A) membrane receptor. The protein is then unfolded and internalized into the lysosome through the translocation complex comprised mainly of the LAMP-2A receptor (Cuervo and Dice 1996). The current chapter focuses on the role of macroautophagy in lipid metabolism; thus, the two additional forms of autophagy, microautophagy, and CMA will not be discussed any further.
1.2 Regulation of Macroautophagy
The induction of macroautophagy (henceforth termed autophagy) occurs following conditions such as cellular stress or starvation leading to the formation of autophagosomes. Autophagosome formation is a complex and highly regulated process that requires more than 30 autophagy-related proteins (ATG) that were identified by molecular dissection of the autophagic process through yeast genetic screens (He and Klionsky 2009). These ATG proteins form functional complexes that mediate individual steps of autophagy; initiation or induction, nucleation, membrane elongation, cargo recognition, and the fusion of autophagosomes with lysosomes.
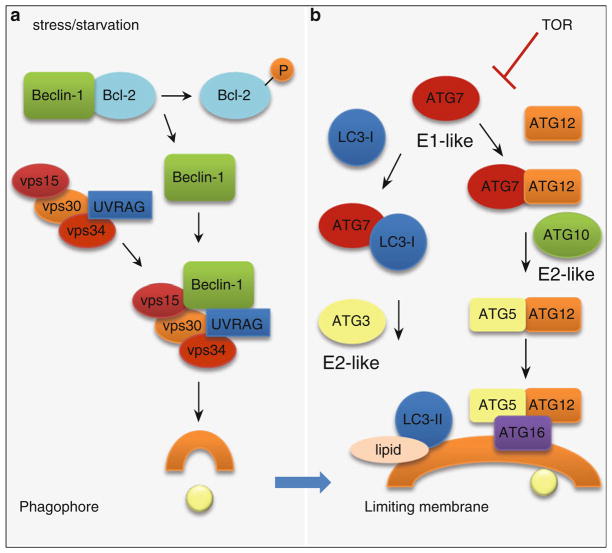
The induction of autophagy requires the activation of the autophagy initiation complex or the class III phosphatidylinositol-3-kinase (PI3K) (Fig. 2a), which occurs through the release of Beclin-1 (ATG6 in the yeast) from the Bcl-2–Beclin-1 complex through starvation or stress-induced phosphorylation of Bcl-2 (Wei et al. 2008). Beclin-1 is then recruited along with additional molecules, vps15, vps30, vps34, ATG14L, and UV radiation-resistance associated gene protein (UVRAG) to form the active class III PI3K complex (Petiot et al. 2000; Tassa et al. 2003). The activation of class III PI3K allows the mobilization of this induction complex to the site of autophagosome formation. A process called nucleation involves the mobilization of this initiation complex to the site of the limiting membrane formation (He and Klionsky 2009). In addition, lipid phosphorylation by the PI3K complex is critical for the recruitment of additional ATG molecules to the phagophore leading to elongation of the limiting membrane. The formation of the limiting membrane requires two parallel conjugation cascades, the microtubule-associated protein 1 light chain 3 (LC3) or ATG8 and the ATG5–12 conjugation cascades (Fig. 2b) that are similar to the ubiquitin conjugation system involving discrete ligases for activation and enzymatic conjugation of substrates (He and Klionsky 2009). These conjugation events occur on the surface of the limiting membrane and are crucial for membrane elongation. The ubiquitin-like ligase ATG7 is required for conjugation of ATG5 and ATG12 to form the ATG5–12 complex. Independently, cleavage and activation of cytosolic LC3-I occurs by the proteolytic cleavage of a cysteinyl residue by the protease ATG4. Activated LC3-I acquires a phosphatidylethanolamine residue that then forms the membrane-associated LC3-II (Kabeya et al. 2000) through a reaction that requires ATG7, in addition to the E2-like activity of ATG3. The molecular mechanisms of the later steps that mediate sealing of the limiting membrane to form autophagosomes and fusion events that form autophagolysosomes are yet unclear. However, it is thought that additional ATGs, specific SNARE, and Rab proteins as well as cytoskeletal elements may be involved in these fusion events (He and Klionsky 2009).
Fig. 2.
Autophagy induction complex (a) and (b) conjugation cascades that lead to autophagosome formation. The induction of autophagy requires release of Beclin-1 from Bcl-2 and its binding to vps15, vps30, vps34, and UVRAG to form the active Class III PI3K complex. Lipid phosphorylation by active class III PI3K recruits the two conjugation cascades to the site of autophagosome formation. The LC3 and ATG5–12 conjugation cascades require the ubiquitin-like ligase activity of ATG7. Products of these two cascades, LC3-II and the ATG5–12–16 conjugate, form structural components of the limiting membrane
Autophagy is centrally regulated by the upstream serine/threonine kinase mTOR (mammalian target of rapamycin) (Jung et al. 2009). Under conditions of nutrient deprivation or following rapamycin treatment, inhibition of mTOR occurs leading to activation of Unc-51-like kinase-1 and -2 (ULK) that phosphorylates and activates ATG13 and FIP200 (focal adhesion kinase family-interacting protein of 200 kDa), thus, leading to the induction of autophagy (Jung et al. 2009). Nutritional excess, in particular, amino acids activate mTOR that phosphorylates and inactivates the ULK–ATG13–FIP200 complex resulting in the switching off of autophagy (Jung et al. 2009). Another crucial regulator of starvation-induced autophagy is c-Jun N-terminal kinase that mediates the phosphorylation of Bcl-2 to release Beclin-1 in response to nutrient deprivation (Wei et al. 2008).
2 Autophagy and Lipid Metabolism
An established role of autophagy is the degradation of cellular organelles, redundant cytosol and proteins. However, only in these recent years, some reports have demonstrated a novel relationship between autophagy and cellular lipid metabolism, thus unfolding a new area of research.
2.1 Autophagy and Regulation of Intracellular Lipid Stores
A recently reported function of autophagy within hepatocytes is the degradation of intracellular lipid stores (Singh et al. 2009a). Although, the lipolytic function of lysosomes had been known earlier, the mechanism of delivery of lipids into the lysosome was unclear. As mentioned in the earlier sections, the lysosomes contain numerous hydrolases and lipases that function in an acidic environment (pH < 5.2) to breakdown the delivered cargo.
In this recent report (Singh et al. 2009a), it has been demonstrated that the delivery of cellular lipid droplets to the lysosomes occur through the sequestration of lipid droplets by autophagosomes and the subsequent fusion of these autophagosomes with lysosomes leading to degradation of lipid droplets (Fig. 3). Studies in cultured hepatocytes that lacked autophagy by pharmacological inhibition with 3-methyladenine or by using RNA interference against ATG5 and ATG7 have revealed that inhibition of autophagy leads to increased hepatocellular triglyceride (TG) accumulation (Singh et al. 2009a) when compared to controls. Increased TG accumulation occurs under both basal condition and when hepatocytes are provided a lipogenic stimulus, such as treatment with physiological concentrations of oleic acid or following culture in a lipogenic medium, methionine, and choline-deficient medium. Electron microscopic studies demonstrated that inhibition of autophagy in hepatocytes and liver leads to marked increase in the number and size of lipid droplets demonstrating that accumulation of lipids occur in the form of lipid droplets (Singh et al. 2009a). Interestingly, increased hepatocellular TG stores results from decreased lipolysis of lipid stores due to decreased delivery of lipid cargo into the lysosomes, and not from increased hepatocellular TG synthesis or a reduction in secretion in the form of VLDL. Immunofluorescence co-localization experiments between a neutral lipid dye (bodipy 493/503) and the autophagosomal marker (LC3) or the lysosomal marker (LAMP1) have revealed co-localization of cellular lipids with components of the autophagosomal and lysosomal system under conditions that activate autophagy (Singh et al. 2009a), such as treatment with rapamycin (inhibitor of TOR) or provision of a lipid stimulus. Furthermore, pharmacological or genetic ablation of autophagy decreased the observed bodipy-LC3 and bodipy-LAMP1 co-localizations.
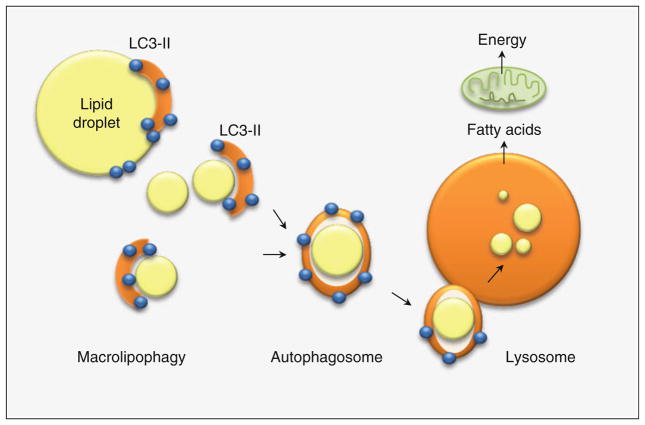
Fig. 3.
The process of macrolipophagy occurs by the de novo formation of a limiting membrane that sequesters cytosolic lipid droplets and delivers the lipid cargo to the lysosomes for degradation
In mammals, induction of autophagy occurs in response to starvation and consequently, lipid droplet components such as structural proteins TIP47 and adipophilin (ADRP) could be detected within isolated autophagic vacuoles and lysosomes from starved mice liver (Singh et al. 2009a). Conversely, immunoblotting demonstrated increased autophagosome-associated LC3-II levels within lipid droplets fractions isolated from livers of starved rodents (Singh et al. 2009a). Additionally, immunoelectronmicrographs demonstrated increased presence of LC3-positive membranes on lipid droplets in liver sections from starved animals. Thus, these in vivo studies have demonstrated a functional interaction between cellular lipid stores and the autophagic machinery that effectively allows the targeting of a lipid cargo to the lysosomes for degradation. A final confirmation of the process of “macrolipophagy” was obtained through studies in genetically modified mice that lacked autophagy within the liver (ATG7 conditional knockouts) (Singh et al. 2009a). These animals exhibit markedly enlarged livers that are lipid laden as demonstrated biochemically by increased TG and cholesterol levels (Singh et al. 2009a). In addition, Oil Red O staining of autophagy-deficient livers revealed marked increase in the number and size of lipid droplets when compared to control littermates (Singh et al. 2009a). Interestingly, the differences observed between controls and autophagy-ablated mice increased further when mice were fed a diet that provided 60% of calories in the form of fat. Although the lipophagic function of autophagy was initially demonstrated in cultured hepatocytes, in the liver, and in fibroblasts in culture (Singh et al. 2009a), it is now clear that macrolipophagy also regulates lipid stores in neurons (Martinez-Vicente et al. 2010).
Despite these exciting advances, a number of questions need to be addressed. It is still unclear how autophagosomes sequester lipid droplets given the enormous size of these lipid stores. An interesting possibility is that unconjugated LC3-I, which localizes on lipid droplets in basal condition, could acquire a phosphatidylethanol-amine to form LC3-II that then generates a limiting membrane in situ sequestering the lipid droplet. Another integral question is to explore the possibilities of crosstalks between lysosomal lipases that degrade sequestered lipids and cytosolic neutral lipases. It is conceivable that smaller lipid droplets are engulfed completely by the autophagic apparatus, whereas larger lipid droplets are perhaps broken down into smaller droplets by autophagy that promotes further sequestration as well as increase the net lipid droplet surface area for efficient cytosolic neutral lipase activity.
2.2 Intracellular Lipids and Regulation of Autophagy
In contrast to the induction of autophagy observed following acute lipogenic stimulus, sustained lipogenic challenge or even acutely elevated abnormal lipid levels lead to compromised macrolipophagic function. Studies in cultured hepatocytes and mice fed a high-fat diet for prolonged periods have revealed that in these model systems there is a failure to mobilize intracellular lipid stores by macroautophagy (Singh et al. 2009a). This was reflected by electron microscopic evidence of decreased LC3-positive membranes on lipid droplets, as well as reduced areas of degeneration on lipid droplets following chronic lipogenic stimulus. Interestingly, a recent study by Koga et al. demonstrates that this autophagic defect following high-fat diet feeding is not limited to degradation of lipid cargo, but also alters degradation of all forms of autophagic cargo, including proteins, highlighting the fact that the primary defect lies in the autophagic apparatus (Koga et al. 2010). In this remarkable study, a dissection of the individual steps that mediate autophagy has demonstrated that high-fat diet feeding has no effect on induction or limiting membrane/autophagosome formation. In fact, the defect occurs at the level of autophagosome–lysosome fusion (Koga et al. 2010). The authors put forth an attractive possibility that alterations in membrane lipid composition following chronic high-fat diet feeding could potentially lead to decreased autophagosomal and lysosomal fusion. A crucial membrane lipid, cholesterol, plays an important role in membrane structure and function including membrane fusion events. Pharmacological agents that alter the concentration of autophagosomal and lysosomal membrane cholesterols resulted in altered membrane fusion events (Koga et al. 2010).
This dynamic cross-talk between autophagy and liver lipids has direct bearing to nonalcoholic fatty liver disease (NAFLD), the hepatic manifestation of the metabolic syndrome. The first step in the pathogenesis of NAFLD is the development of a fatty liver or hepatic steatosis that then predisposes to a “second hit,” which is development of oxidative stress and inflammation leading to steatohepatitis and liver injury (Day and James 1998). In the human, it is conceivable that prolonged consumption of processed diets rich in fat, such as the western diet, can impair autophagy through the effects of lipids on autophagosome–lysosome fusion perpetuating a vicious circle of further hepatic fat accumulation. In addition, it remains to be explored whether compromised autophagy in the setting of fatty liver predisposes to the second hit that then leads to the development of steatohepatitis. In other words, is autophagy a central process that not only confers protection against steatosis but also blocks the development of steatohepatitis and end-stage liver disease in the setting of steatosis?
3 Autophagy and Adipose Tissue
The lipophagic function of autophagy demonstrated in the liver raised yet another critical question. Does autophagy play a similar role in the major fat-storing tissue in mammals, the adipose tissue?
3.1 Autophagy and White Adipose Tissue
As discussed in the previous section of this chapter, macrolipophagy has been demonstrated to be one of the mechanisms that mediate mobilization of intracellular lipid stores in hepatocytes, fibroblasts, and neurons. However, the function of autophagy in the dedicated fat-storing cell, the adipocyte, appears to be paradoxical to what has been observed in the aforementioned cell types. Recent studies by two independent groups have demonstrated a novel role of autophagy in the regulation of adipocyte differentiation and fat storage (Singh et al. 2009b; Zhang et al. 2009). In a study by Singh et al., inhibition of autophagy by RNA interference against ATG proteins in 3T3-L1 preadipocytes blocked the differentiation of preadipocytes into mature adipocytes (Singh et al. 2009b). This inhibition of differentiation of preadipocytes was associated with reduced expression of key adipogenic factors, CEBP-α and PPAR-γ. In addition, differentiation failure was associated with reduced TG storage, and at the molecular level by reduced levels of terminal differentiation markers, such as fatty acid binding protein-4 (FABP-4/aP-2), stearoyl-CoA desaturase, fatty acid synthase, and glucose transporter-4 in the autophagy-deficient adipocytes (Singh et al. 2009b).
White adipose tissue (WAT)-specific inhibition of autophagy in vivo by ablation of ATG7 not only reduced adipose tissue mass and differentiation but also imparted a remarkable brown adipose tissue (BAT)-like phenotype (Singh et al. 2009b). Reflective of this WAT to BAT transdifferentiation in the autophagy-deficient adipose tissue was histological evidence of smaller adipocytes with rounded nuclei that contained smaller, numerous multiloculated lipid droplets as opposed to control adipocytes with flattened nuclei containing a single large lipid droplet. In addition, morphometric analyses demonstrated increased mitochondrial content within the autophagy-deficient WAT (Singh et al. 2009b). To conclusively claim that inhibition of autophagy results in transdifferentiation of WAT into BAT-like tissue, it is imperative to demonstrate the expression of BAT-specific markers within the WAT. Indeed, immunoblotting demonstrated increased molecular markers of BAT; the PPAR-γ transcriptional coactivator (PGC)-1α is crucial for brown adipogenesis and mitochondrial biogenesis, as well as presence of the specific BAT marker uncoupling protein-1 (UCP-1) (Singh et al. 2009b). One might argue that increased mitochondrial content could perhaps be a result of reduced mitophagy and not from increased biogenesis? However, if such a scenario were to be true, one would observe accumulation of predominantly dysfunctional mitochondria that would contribute to reduced oxidative function and cellular toxicity. Evidence against this possibility has been the demonstration of increased β-oxidation rates in adipose tissues of autophagy-ablated animals, regardless of whether mice were fed regular chow or high-fat diet. The physiological consequence of this dramatic alteration in WAT morphology including the acquisition of BAT-like properties is a remarkably lean mouse (Singh et al. 2009b) and maintained insulin sensitivity despite high-fat diet feeding (Singh et al. 2009b).
Despite these developments, it remains to be determined how absence of WAT-specific autophagy regulates adipose tissue differentiation and modulates the switch from WAT to a BAT-like phenotype. Recent studies have identified a number of proteins that regulate adipocyte biology (Lefterova and Lazar 2009). It could be possible that inhibition of autophagy blocks degradation of these critical adipocyte proteins that regulate adipogenesis and differentiation. Conversely, inhibition of autophagy could accumulate factors that impart BAT-like characteristics. An attractive candidate is PRD1-BF1-RIZ1 homologous domain containing 16 (PRDM-16), a transcriptional regulator that has recently been shown to promote a BAT-like phenotype through its interaction with PPAR-γ and transcriptional upregulation of BAT-specific genes (Seale et al. 2008). A second possibility is a putative function of autophagy on adipocyte proliferation through effects on a subpopulation of adipose stromal vascular cells that are lineage − CD29+, CD34+, Sca-1+, and CD24+ and have been identified recently as WAT progenitor cells (Rodeheffer et al. 2008). However, since no differences have been observed in the relative percentage of these progenitor cells between control and autophagy-deficient fat pads (Singh et al. 2009b), a possibility exists that qualitative differences in progenitors may contribute to reduced adipose mass in WAT-specific autophagy null animals. It may also be possible that inhibition of autophagy perhaps alters the adipocyte metabolic milieu that exerts an indirect effect on fat storage. Despite all these possibilities, one cardinal question remains: how can one explain the differential roles of autophagy in the liver and adipose tissue? A plausible explanation for the divergent roles of autophagy in the liver and adipose tissue is the presence of an efficient lipolytic mechanism in the adipose tissue, i.e., the presence of the adipose TG lipase and hormone-sensitive lipase that act in tandem to mobilize adipose fat stores and liberate free fatty acids. Thus, a lipolytic function of autophagy in the adipose tissue akin to what has been described in the liver would prove to be redundant. Moreover, it can be envisioned that the mutually exclusive roles of autophagy in the liver and the adipose tissue could appear to be complementing each other to protect against fat storage in organs not functionally suited to store fat, such as liver and heart. In other words, the function of autophagy to promote adipose mass and differentiation allows efficient sequestration of excessive fat away from circulation, thus sparing the metabolic insult of fat accumulation in organs such as liver and heart.
3.2 Autophagy and Brown Adipose Tissue
The fact that inhibition of autophagy within WAT promotes the dramatic remodeling of WAT into a BAT-like phenotype does make one wonder: what would be the outcome of regulating autophagy within the interscapular BAT per se? The study by Singh et al. used a transgenic Cre mouse, the expression of which was driven by the aP2 promoter. The expression of aP2-driven Cre recombinase occurs in both WAT and BAT allowing the deletion of floxed ATG7 and hence autophagy in both adipose tissue compartments (Singh et al. 2009b). Interestingly, the inhibition of autophagy induced morphological and functional changes in BAT as well. This was suggested by the findings of a modest yet significant increase in the interscapular BAT mass in the autophagy null animals (Singh et al. 2009b). Histological sections of interscapular BAT from regular chow- and high-fat diet-fed autophagy null mice were remarkable for a reduction in the number of lipid droplets. Autophagy-deficient BATs demonstrated modest increases in UCP-1 and PGC-1α, as well as increased levels of mitochondrial markers cytochrome oxidase and cytochrome c, regardless of whether the mice were fed regular chow or a high-fat diet (Singh et al. 2009b). This was associated with a twofold increase in oxidative capacity of the autophagy-ablated BAT (Singh et al. 2009b). Thus, changes in BAT in animals lacking autophagy were consistent with those found in WAT but less significant because they occurred in the background of a fat depot already composed of brown adipocytes. A striking question is: why such a similar change, as observed in autophagy-null WAT, is occurring in autophagy-deficient BAT that is functionally and developmentally discrete from WAT? Is the fundamental mechanism increased retention of PRDM16 in both WAT and BAT that drives BAT gene expression? Only future studies will delineate the complex roles of autophagy in the regulation of adipocyte differentiation and fat storage and metabolism.
4 Conclusion
These exciting developments present autophagy as an organ-specific therapeutic target, which can be potentially manipulated to maintain energy homeostasis. In the liver, it is beneficial to upregulate autophagy that increases disposal of hepatic fat. In contrast, inhibition of adipose-specific autophagy enhances tissue oxidative capacity by imparting BAT-like properties that in turn promotes a lean insulin-sensitive phenotype. In addition, the effect of modulation of autophagy on the quantity and functioning of existing brown fat may have important implications for the development of novel therapeutic options for conditions that stem from increased adipose expansion such as the metabolic syndrome.
Acknowledgments
This work was supported by National Institutes of Health grants from the National Institute of Diabetes and Digestive and Kidney Diseases to Dr. Ana Maria Cuervo (AMC) and Dr. Mark Czaja and from the National Institute of Aging and a Glenn Award to AMC; RS is supported by a NIH NIDDK K01 (DK087776-01) grant. The author thanks Dr. Ana Maria Cuervo for thought-provoking discussions and Dr. Susmita Kaushik for the critical reading of this manuscript.
References
- Beau I, Esclatine A, Codogno P. Lost to translation: when autophagy targets mature ribosomes. Trends Cell Biol. 2008;18:311–314. doi: 10.1016/j.tcb.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- Day CP, James OF. Steatohepatitis: a tale of two “hits”? Gastroenterology. 1998;114:842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- Esclatine A, Chaumorcel M, Codogno P. Macroautophagy signaling and regulation. Curr Top Microbiol Immunol. 2009;335:33–70. doi: 10.1007/978-3-642-00302-8_2. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon M, Cuervo AM. Chaperone-mediated autophagy in health and disease. FEBS Lett. 2010;584:1399–1404. doi: 10.1016/j.febslet.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Lazar MA. New developments in adipogenesis. Trends Endocrinol Metab. 2009;20:107–114. doi: 10.1016/j.tem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- Manjithaya R, Nazarko TY, Farre JC, Subramani S. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 2010;584:1367–1373. doi: 10.1016/j.febslet.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Talloczy Z, Wong E, Tang G, Koga H, Kaushik S, de Vries R, Arias E, Harris S, Sulzer D, Cuervo AM. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat Neurosci. 2010;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Physiological functions of autophagy. Curr Top Microbiol Immunol. 2009;335:71–84. doi: 10.1007/978-3-642-00302-8_3. [DOI] [PubMed] [Google Scholar]
- Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009a;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J Clin Invest. 2009b;119:3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir E, Maiuri MC, Tajeddine N, Vitale I, Criollo A, Vicencio JM, Hickman JA, Geneste O, Kroemer G. Cell cycle-dependent induction of autophagy, mitophagy and reticulophagy. Cell Cycle. 2007;6:2263–2267. doi: 10.4161/cc.6.18.4681. [DOI] [PubMed] [Google Scholar]
- Tassa A, Roux MP, Attaix D, Bechet DM. Class III phosphoinositide 3-kinase–Beclin1 complex mediates the amino acid-dependent regulation of autophagy in C2C12 myotubes. Biochem J. 2003;376:577–586. doi: 10.1042/BJ20030826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkovsky AM. Mitophagy. Biochim Biophys Acta. 2009;1793:1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc Natl Acad Sci USA. 2009;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]