Abstract
The paper presents state space models of the hemodynamic response (HR) of fNIRS to an impulse stimulus in three brain regions: motor cortex (MC), somatosensory cortex (SC), and visual cortex (VC). Nineteen healthy subjects were examined. For each cortex, three impulse HRs experimentally obtained were averaged. The averaged signal was converted to a state space equation by using the subspace method. The activation peak and the undershoot peak of the oxy-hemoglobin (HbO) in MC are noticeably higher than those in SC and VC. The time-to-peaks of the HbO in three brain regions are almost the same (about 6.76 76 ± 0.2 s). The time to undershoot peak in VC is the largest among three. The HbO decreases in the early stage (~0.46 s) in MC and VC, but it is not so in SC. These findings were well described with the developed state space equations. Another advantage of the proposed method is its easy applicability in generating the expected HR to arbitrary stimuli in an online (or real-time) imaging. Experimental results are demonstrated.
OCIS codes: (170.2655) Functional monitoring and imaging, (300.0300) Spectroscopy, (170.5380) Physiology, (100.2960) Image analysis
1. Introduction
This paper addresses a method for the reconstruction of a hemodynamic response (HR) for arbitrary stimuli in three different brain regions: the motor cortex (MC), the somatosensory cortex (SC), and the visual cortex (VC). In each cortex, an impulse HR function in state space form is first developed. Then, it is used to generate the expected HR for arbitrary stimuli. To that end, the state space method in dynamics and control is utilized. The coefficient matrices in the formulated state space equation are estimated by the subspace identification method. The main advantage of the state-space-based HR function lies in its generality (i.e., not restricted to a box-car-type stimulus). Also, it can describe the detailed characteristics of HR in terms of peak, undershoot, rise time, deactivation time, etc.
In the fMRI technique, the canonical HR function for a brain activity was modeled in the form of a linear combination of two gamma functions [1,2] or the Gaussian function [3]. The HR usually has been predicted by convolving the canonical HR function in fMRI with an experimental paradigm. The predicted HR and its derivatives have been utilized to establish the design matrix in the general linear model (GLM) [1,2,4], where the associated coefficients indicate the activity strengths of the signals [5]. Additionally, the blood-oxygen-level-dependent HR has been modeled as a balloon model [6,7], which was proved to be sufficient to account for nonlinearities in the event-related response [7]. In this model, a system of differential equations was utilized to describe the nonlinear relationship among deoxy-hemoglobin (HbR), blood volume, and total hemoglobin (HbT). Moreover, a dynamic causal model was utilized to build interconnections among selected brain regions [8,9] wherein the HR of each brain area was simulated by the balloon model and neuronal states.
In recent years, functional near-infrared spectroscopy (fNIRS) has been used as an alternative non-invasive brain imaging technique to fMRI [10–15]. fNIRS determines the concentration changes of oxy-hemoglobin (HbO) and HbR in tissues by measuring light-absorption variations at multi-wavelengths [16]. However, in most works related to GLM [4,10,17,18], the canonical HR function was borrowed from the fMRI literature [19]. Recently, a state space model has been utilized to reconstruct the HR to an impulse stimulus from the motor cortex prompted by right index-finger tapping, the model parameters having been estimated by the recursive least-square (RLS) method [20]. However, the coefficient matrices in the model, which determine the characteristics of the canonical HR function, were not well described.
In this study, the HRs (to impulse stimuli) for three brain regions are reconstructed as state space equations, wherein their coefficient matrices are estimated by the subspace identification method. The state-space-based HRs are compared one another, and their important characteristics (e.g., time-to-peak, activation peak, undershoot peak, deactivation time, etc.) are analyzed. It is shown that the HR functions are well-adjusted in terms of shape and peak amplitude in the main responses. Finally, the reconstructed HR of each brain region is validated across various experimental fNIRS data.
2. Methods
2.1. Subjects
Nineteen healthy volunteer subjects (17 males, 2 females; mean age: 27.74 years) were invited to participate in three experiments in relation to three brain regions: MC, SC, and VC, respectively. None of the subjects had any history of medical or neurological disease. To ensure direct contact of the optodes to the scalp, a small flexible plastic stick was used to separate the subjects’ hair. Additionally, all of the experiments were performed under dark and quiet conditions in order to reduce noise. All of the subjects were provided with clear instructions prior to the experiments. They were asked to sit comfortably on a chair and to relax without moving their body. If a subject experienced any discomfort, as manifested for example in body movement or sleepiness in the course of the experiment, he/she was asked to redo the test. Figure 1 illustrates the experimental paradigm, which entails 3 stimuli (the entire duration is 120 s). With regard to the MC, the subject was asked to perform right middle-finger tapping (RMFT); for the SC, the right index finger of the subject was poked by a nail; for the VC, the subject was asked to open his/her eyes and focus on a checkerboard display, which was shown on a 15-inch laptop screen for 3 s and then to relax during the rest period (with black background).
Fig. 1.

Experimental paradigm for obtaining impulse HR functions: For the motor cortex (MC), a right middle-finger tapping (RMFT) was repeated three times; for the somatosensory cortex (SC), the right index-finger poking using a nail was used, and for the visual cortex (VC), a 3-sec checkerboard display was employed.
2.2. Equipment and data conversion
A continuous-wave fNIRS instrument (DYNOT; NIRx Medical Technologies, USA) of dual wavelengths (i.e., 760 nm and 830 nm) was used, which was configured to have 1 emitter and 32 detectors at 70 Hz sampling rate [20]. In obtaining the optical density variations, the modified Beer-Lambert law (MBLL) was used [11,16,18,21–26].
| (1) |
where the superscript i indicates the i-th channel, ΔAi(k; λj) is the optical density variation at time k of the wavelength , a HbX(λj) denotes the extinction coefficient of HbX (i.e., HbO or HbR), is the concentration change of HbX, is the distance between the emitter and the detector at the i-th channel, and refers the differential path length factor. Then, solving for and , the concentration changes of HbX are obtained as follows.
| (2) |
In this work, the recorded optical density variation data were converted to the HbX concentration changes using the open-source software NIRS-SPM [18]. In this process, the fNIRS data were low-pass-filtered by a Gaussian filter of 1.5 s full-width at half-maximum and high-pass-filtered using a discrete-cosine transform with a 128 s cut-off period. This was done to remove noises related to the heart beat (~1 Hz) and respiration (~0.25 Hz).
2.3. Experimental design
Figure 2 shows the optodes configuration for MC and SC experiments, which includes 1 emitter and 10 detectors providing 10 channels. For MC, the emitter is positioned 2 cm toward the forehead from C3 (a reference point in the 10-20 international system). The detectors are set up around the emitter in the left MC. For SC, the optodes configuration (dotted red box) is designed similarly to the MC experiment. However, the emitter is positioned on top of C3, the red triangle in Fig. 2. The detectors are set up to measure the regions of interest in the left SC. Figure 3 depicts the optodes configuration for VC, which includes 1 emitter and 7 detectors providing 7 channels. The emitter is positioned at 1 cm apart from Oz to the right [27].
Fig. 2.

Optodes configuration for MC (black box) and SC (dotted red box): 1 emitter and 10 detectors → 10 channels.
Fig. 3.

Optodes configuration for VC: 1 emitter and 7 detectors → 7 channels.
2.4. Canonical HR
Even though both HbO and HbR in the fNIRS technique reflect the cerebral blood flow, HbO is more sensitive than HbR [5,28]. Therefore, in the present work, only HbO is utilized in further analyses. The t-values, p-values, and mean values of the averaged HbO (i.e., the averaged one of the three responses to three stimuli in Fig. 1) in each channel is analyzed in order to find the most active channel. Figure 4 depicts the shape of the canonical HR function to an impulse stimulus using the double-gamma function [2]. Its important characteristics include the peak amplitude (a 1), the time-to-peak (t 1), the magnitude of undershoot (a 2), and the time-to-undershoot (t 2), the mean value (m 1) in the activation period (i.e., 3 ~11 s, which is denoted by T 1), and the mean value (m 2) in the deactivation period (i.e., 15 ~28 s, which is denoted by T 2). In this work, these were utilized to evaluate the consistency between the HR measured from a brain region and the reconstructed one using the state-space model.
Fig. 4.

Characteristics of the canonical HR: a 1 stands for the peak amplitude of the main response, a 2 is the magnitude of undershoot, t 1 is the time-to-peak, t 2 is the time-to-undershoot, T 1 denotes the activation period, T 2 denotes the deactivation period, m 1 is the mean in T 1, and m 2 is the mean in T 2.
2.5. Regression model and statistical analysis
A stimulus will cause an increase in HbO [4], thus making the signal to rise. If there is no further stimulation, this response will return to the baseline. In practice, the acquired HbOs upon three stimuli might not be the same even in the same channel due to noises from the participants’ body movement or some unknown factors [29]. Therefore, the first step is to check whether the measured signal is eligible to be included for averaging or not. For this, the one-tail t-test is used, which is based on the estimated coefficient of the regression model as follows. The predicted HR to an impulse stimulus y M(k) is produced by convolving the impulse stimulus and the canonical HR function in Fig. 4, and is thereafter utilized as the reference signal.
| (3) |
where s(k) indicates the stimulus function and h(k) denotes the canonical HR function [2], which was chosen as the double-gamma function. The measured impulse response of each stimulus at the i-th channel is described in the following regression model.
| (4) |
where X(k) = [x 1(k) x 2(k) x 3(k) x 4(k)]T is the k-th sample vector of the regression model, the superscript T stands for the transpose operator, x 1(k) refers to y M(k) in Eq. (3). x 2(k) and x 3(k) are the first and second derivatives of x 1(k), respectively. x 4(k) indicates the null control regressor for compensating an offset value, is the coefficient vector representing the activity strengths, and vi(k) denotes the noise. The coefficient vector is estimated by the recursive least squares (RLS) algorithm as follows.
| (5) |
where ei(k) is the estimation error, is the estimated hemodynamic response, L(k) denotes the weighting vector for parameter updating, and P(k) represents the input covariance matrix. The initial values of are set to zero, and the initial covariance matrix P(0) are set to δI, where δ is a sufficiently large number (in this study, δ = 100). The first component of the estimate is utilized to calculate the t-value, as it is desired to test the null hypothesis (i.e., ). The t-value of the i-th channel at time k is computed as [5,30]
| (6) |
where c is a contrast vector for selecting the coefficient of interest, and denotes the residual sum-of-square given by
| (7) |
where r is the number of regression elements. The null hypothesis is accessed based on ti(k) of t-distribution with k - r degrees of freedom. If the impulse response is well fitted to the predicted HR, in Eq. (6) will become very small while is positive and steady. This causes the t-value to be large, positive, and steady in relation to the activation strength [25,31]. Therefore, two conditions (t is greater than 0 and p is less than 0.05) are utilized as the threshold criteria for accessing the activated HR in each stimulus.
The impulse HRs per channel per person are averaged over three responses in Fig. 1 (28 s × 3). In this, only those responses that its t-value and p-value satisfy the threshold criteria are included. Additionally, it is necessary to test whether m 2 of the HbO has returned to near the baseline. And then, to determine whether the obtained HbO was actually caused by the provided stimulus or not, m 1 is computed, see [4,27,32]. If m 1 in a channel is less than or equal to zero, the channel is concluded inactive and is discarded. If m 2 is larger than m 1, it is concluded de-active. The criteria to select the most active channel are to find the largest m 1 and the smallest standard deviation in T 2. To grasp an idea on the most active channel, the five variables (i.e., t, p, m 1, m 2, and SD) for all the channels and subjects are computed and three threshold criteria (i.e., t > 0, p < 0.05, m1 > 0) are checked. Those data not satisfying any criteria are excluded. The most active channel will be considered to represent the HR in the given brain region.
2.6. HR representation in state space form
To describe the impulse HR in state space form, the following discrete state space equation for a single-input single-output system is used.
| (8) |
where z(k) stands for the state vector of the model, u(k) represents the stimulus input or event, w(k) indicates the process noise assumed white, y(k) is the output of the reconstructed HR, v(k) denotes the white noise in the output equation. A ∈ ℜn × n, B ∈ ℜn × 1, C ∈ ℜ1 × n, and D ∈ ℜ1 × 1 are the matrices to be identified in the state-space modeling, and n indicates the order of the model. In the present study, the coefficient matrices (i.e., A, B, C, and D) are estimated with the Matlab function “moesp.m” available in [33]. The input arguments for this function (i.e., the block Hankel matrices) are generated as follows.
| (9) |
| (10) |
where q refers the number of block rows (i.e., q strictly greater than n) and N is the number of samples.
3. Paradigm design for verification
Figures 5(b) and 6(b) show the experimental paradigms for applying the developed method in Section 2. In this section, the DYNOT at a sampling frequency 1.81 Hz is utilized to record the data from three brain regions. The stimuli are right middle finger tapping (RMFT) in MC, poking stimulus in SC, and checkerboard in VC. The filtering and conversion processes of the measured signals are the same as done in Subsection 2.2.
Fig. 5.
Experiment for demonstrating the MC and SC models: (a) Optodes configuration (MC - black box; SC - dotted red box) and (b) the experimental paradigm including 5 taps/pokes at 30 and 90 s (1 tap/poke for 2 s); 10 taps/pokes at 60 and 120 s (1 tap/poke per s); and 1 tap (or poke) at 150, 170, and 190 s.
Fig. 6.

Experiment for demonstrating the VC model: (a) Optodes configuration and (b) experimental paradigm with 10 s checkerboard displays after the initial 30 s rest period.
3.1 Tapping experiment
Two healthy volunteers (1 female, 1 male; mean age: 27.5 years) were invited to participate in the RMFT-based MC experiment. Figure 5(a) illustrates the channel configuration (black box), where 8 emitters and 8 detectors are employed. The location of C3 coincides with the small blue circle. The subjects were asked to perform five taps at 30 s and 90 s (1 tap per 2 s), 10 taps at 60 s and 120 s (1 tap per second), one tap at 150s, 170 s, and 190 s (1 tap per second), and do nothing during the rest period.
3.2 Poking experiment
In the poking stimulus experiment, the optodes configuration (red dotted box) and experimental paradigm were the same like the MC experiment. However, the optodes were moved so that the location of C3 coincided with the red triangle; see Fig. 5(a). Three healthy male volunteers (mean age: 29.67 years) were invited to participate. They were asked to sit comfortably on a chair and to close their eyes during the experiment. Five pokes at 30 s and 90 s (1 poke per 2 s), 10 pokes at 60 s and 120 s (1 poke per second), and one poke at 150s, 170 s, and 190 s (1 poke per second) were administered to the right index finger of the participant.
3.3 Checkerboard experiment
For VC, two healthy volunteers (1 female, 1 male; mean age: 29.5 years) were asked to look at the 15-inch laptop screen showing a checkerboard for 10 s. They were instructed to sit comfortably on a chair and not to close their eyes during the entire 180 s experiment duration. After the initial 30 s rest period (with black background), the checkerboard was displayed five times at 30 s, 60 s, 90 s, 120 s, and 150 s. Figure 6(a) illustrates the channel configuration.
4. Results and discussion
4.1 Averaging
Figures 7(a) and 7(b) depict the HbOs (blue solid curves) in Ch. 7 and Ch. 8, respectively, which were produced by three middle-finger taps. Each red thick bars represents a tap and the black dashed curve represents the predicted HR obtained by convolving the red impulses with the canonical HR function in [2]. Figures 7(c) and 7(d) illustrate the averaged HbOs obtained from Figs. 7(a) and 7(b), respectively (i.e., the averaged value for 28 s time span). In Fig. 7(a), it is seen that the rises/peaks of HbO are consistent with the given stimuli (and with the predicted HR as well). However, in Fig. 7(b), only the second response is comparable to the predicted HR. This reveals that the same stimulus may not result in the same response even in the same channel. Some previous works [19,34] also discussed about the HR’s variances in time, amplitude, and shape from region to region and subject to subject. Therefore, it is necessary to examine whether the obtained HR is permissible to averaging. The one-tail t-test was utilized to test the null hypothesis. The response at each stimulus was estimated based on the predicted HR using Eq. (4). The t-values were computed using Eq. (6) with c = [1 0 0 0]T. If the response satisfied the threshold criteria (i.e., t > 0, p < 0.05), it was included for averaging. Table 1 shows the entire t-values of 19 subjects who participated in the MC experiment. The same approach was applied to the analyses of SC and VC as well.
Fig. 7.
Examples of HRs (blue solid curves) upon middle-finger taps (thick red impulses) in MC (Subject 10): (a) active channel (Ch. 7); (b) de-active channel (Ch. 8); (c) the averaged HR from (a); (d) the averaged HR from (b).
Table 1. Examples of t-values in MC (Ch. 7, Ch. 8).
| (scale: 10−5 µM) | ||||||
|---|---|---|---|---|---|---|
|
|
Channel 7 |
Channel 8 |
||||
| Subjects | Stim.1 | Stim.2 | Stim.3 | Stim.1 | Stim.2 | Stim.3 |
| 1 | 97.01 | 73.51 | −10.73* | 62.84 | 153.69 | 66.69 |
| 2 | 17.98 | −61.27* | 60.99 | 52.50 | 6.48 | −24.71* |
| 3 | −37.98* | −7.15* | −56.55* | 44.81 | 31.00 | 22.35 |
| 4 | 119.66 | −21.56 | 23.18 | 39.41 | 36.13 | 96.72 |
| 5 | −42.09* | 121.34 | 81.41 | −3.56* | 76.52 | 54.70 |
| 6 | 92.51 | −33.23* | 69.46 | 100.88 | −35.71* | 63.48 |
| 7 | 13.91 | 14.80 | 125.47 | 77.20 | 47.33 | −36.81* |
| 8 | 122.40 | 29.32 | −154.80* | −34.06 | 41.50 | 139.14 |
| 9 | 96.55 | 29.90 | 122.64 | 112.66 | 83.71 | 43.22 |
| 10 | 91.20 | 113.47 | 11.97 | −60.88* | 142.07 | −115.27* |
| 11 | 4.37 | 103.80 | 21.63 | 18.31 | 67.21 | 13.68 |
| 12 | 26.92 | −77.02* | −12.73* | −56.18* | 52.87 | 130.17 |
| 13 | −1.35* | 40.60 | 46.96 | 155.21 | 24.16 | −34.48* |
| 14 | 160.48 | 127.81 | 58.35 | 14.99 | −80.76* | −76.22* |
| 15 | 135.46 | 97.58 | 42.34 | 80.07 | −119.26* | 104.85 |
| 16 | 2.89 | −1.46* | 17.18 | −27.81* | −7.65* | −84.61* |
| 17 | −9.22* | −31.13* | −14.13* | −4.06* | 86.18 | 52.67 |
| 18 | 29.88 | 41.32 | 130.12 | −62.35* | 34.93 | −35.99* |
| 19 | 31.68 | −8.34* | 47.48 | −28.30* | −71.52* | −5.38* |
Note: The asterisk indicates the case that p > 0.05.
4.2 HR to impulse stimulus in three brain regions: MC, SC, and VC
Table 2 shows m 1 (see Fig. 4) of the averaged HR obtained in Subsection 4.1. Table 3 depicts m 2. The most distinctive HR that shows the largest m 1 and the smallest standard deviation of m 2 is considered as the representative impulse HR in that brain region. In order to demonstrate the difference between m 1 and m 2, the two-sample t-test (i.e., the ttest2.m function in Matlab, the Mathworks, Inc.) was utilized. The null hypothesis was that m 1 and m 2 were the same. Table 4 shows that p < 0.05 is found from all the channels except Ch. 9. This result reveals that m 1 and m 2 are significantly different.
Table 2. Mean value during the activation period (m 1): MC.
| (scale: 10−5 µM) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Subs. | Ch.1 | Ch.2 | Ch.3 | Ch.4 | Ch.5 | Ch.6 | Ch.7 | Ch.8 | Ch.9 | Ch.10 |
| 1 | 16.11 | −3.93 | −0.60 | 6.07 | 4.21 | 8.22 | 3.82 | 4.03 | 4.30 | −2.00 |
| 2 | −5.23 | 7.13 | −1.94 | −5.22 | 4.81 | 6.51 | 5.10 | 5.18 | 7.09 | 4.33 |
| 3 | 0.21 | 3.24 | −3.48 | 1.14 | −2.31 | 1.45 | 0.00 | −0.01 | 8.73 | 6.23 |
| 4 | 4.77 | 2.12 | 3.39 | −0.43 | −5.02 | 7.56 | −0.22 | 2.11 | −10.94 | −1.83 |
| 5 | 1.67 | −4.33 | 3.36 | 2.58 | −0.14 | −0.85 | 0.20 | 4.18 | 3.30 | 5.50 |
| 6 | 5.87 | −5.42 | 1.35 | 3.68 | −1.54 | −8.92 | 6.67 | −4.54 | 8.32 | 6.78 |
| 7 | −4.02 | −3.08 | 1.37 | 5.62 | 6.72 | −2.35 | 4.66 | −1.82 | −1.65 | 5.81 |
| 8 | −1.63 | 2.04 | 3.81 | 0.73 | 1.04 | −0.63 | 5.01 | 1.83 | 10.77 | −1.72 |
| 9 | −2.92 | −2.61 | −0.25 | −4.20 | 6.91 | 2.61 | 2.14 | 2.13 | 3.67 | 5.15 |
| 10 | −6.08 | −11.16 | 1.90 | 4.18 | 3.56 | 18.37 | 11.24 | 4.80 | 0.51 | −3.51 |
| 11 | 4.99 | 3.18 | 2.72 | 1.18 | 5.29 | −0.07 | 7.04 | 1.30 | 1.74 | 4.41 |
| 12 | 2.47 | 1.73 | −4.20 | −7.16 | −0.39 | −9.93 | 1.68 | 3.08 | 9.23 | −1.43 |
| 13 | 0.82 | 0.03 | −2.79 | 3.43 | −2.70 | −9.52 | 6.70 | 2.80 | 2.15 | 0.11 |
| 14 | 1.59 | 0.00 | 1.49 | 1.22 | −2.37 | 2.24 | 5.05 | −6.17 | 0.00 | 2.03 |
| 15 | 4.29 | 2.17 | 0.00 | −4.52 | 4.68 | 2.19 | 3.48 | −5.53 | 7.09 | 3.42 |
| 16 | 7.41 | 4.82 | 7.23 | −0.62 | −1.04 | 0.00 | −3.34 | 0.00 | 12.40 | 16.33 |
| 17 | 3.05 | 4.77 | −3.84 | 0.00 | 4.08 | 4.27 | 0.00 | 3.16 | 4.05 | 5.25 |
| 18 | 2.85 | −2.91 | −1.34 | −1.16 | 1.39 | −3.78 | 5.67 | −5.39 | −3.08 | −6.10 |
| 19 | −0.05 | 0.47 | −2.52 | 0.21 | 6.41 | 4.31 | 2.25 | 0.00 | −1.74 | 1.79 |
| Avg* | 1.90 | -0.09 | 0.30 | 0.35 | 1.77 | 1.14 | 3.53 | 0.59 | 3.47 | 2.66 |
| SD** | 5.10 | 4.39 | 3.07 | 3.64 | 3.69 | 6.77 | 3.41 | 3.65 | 5.65 | 5.02 |
* The Avg row indicates the average value over 19 subjects.
** SD denotes standard deviation.
Table 3. Mean value during the deactivation period (m 2): MC.
| (scale: 10−5 µM) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Subs. | Ch.1 | Ch.2 | Ch.3 | Ch.4 | Ch.5 | Ch.6 | Ch.7 | Ch.8 | Ch.9 | Ch.10 |
| 1 | 6.71 | −7.11 | −3.27 | −1.39 | 1.69 | 4.39 | 1.11 | −0.69 | 4.69 | −4.67 |
| 2 | −9.72 | 1.31 | −2.67 | −6.17 | 4.83 | 0.96 | −0.76 | 0.67 | −5.48 | 1.77 |
| 3 | −4.76 | 1.11 | −11.34 | −4.13 | −3.80 | −1.62 | 0.00 | −2.01 | 2.04 | 3.77 |
| 4 | 0.54 | −5.33 | 1.99 | −3.32 | −8.76 | 0.04 | −5.06 | −1.83 | −9.07 | −4.98 |
| 5 | −2.94 | −7.18 | 0.82 | 1.76 | −4.28 | −9.85 | −5.03 | −0.68 | −3.08 | 1.31 |
| 6 | −9.53 | −2.59 | −5.22 | 1.06 | −1.40 | −11.33 | 4.15 | −10.01 | 2.79 | −1.16 |
| 7 | −12.73 | −6.12 | −0.97 | 0.63 | −1.74 | −5.91 | 0.05 | −7.13 | −5.44 | 1.66 |
| 8 | −1.20 | −0.95 | 0.37 | −1.57 | −2.46 | −1.76 | 1.20 | −2.95 | 3.28 | −4.38 |
| 9 | −0.45 | −1.49 | −1.31 | −3.18 | 4.83 | −3.08 | −1.78 | −3.12 | −2.40 | −5.96 |
| 10 | −5.51 | −11.29 | −3.45 | 1.76 | −2.56 | 3.70 | 2.29 | −4.08 | −7.18 | −8.89 |
| 11 | −0.25 | 1.02 | −2.77 | −1.88 | 1.19 | −7.57 | 1.64 | −0.13 | −2.82 | 0.57 |
| 12 | −3.05 | −5.75 | −6.66 | −11.21 | −2.37 | −11.32 | 0.07 | −3.97 | 11.57 | −3.98 |
| 13 | −5.67 | −5.58 | −6.01 | −2.74 | −3.74 | −14.56 | 3.26 | −2.52 | −0.08 | −4.42 |
| 14 | 0.61 | 0.00 | −0.49 | 2.14 | −1.37 | 3.19 | −0.72 | −6.25 | 0.00 | −2.71 |
| 15 | −1.36 | −1.56 | 0.00 | −6.87 | 4.79 | −7.83 | −1.25 | −11.09 | 2.34 | −2.85 |
| 16 | 6.65 | −0.99 | 3.15 | −4.89 | −3.61 | 0.00 | −2.47 | 0.00 | 15.67 | 10.05 |
| 17 | −0.69 | −1.16 | −8.64 | 0.00 | 1.87 | −0.24 | 0.00 | −2.81 | −4.61 | −0.14 |
| 18 | 3.66 | −5.95 | −5.58 | −2.46 | −1.54 | −8.33 | 1.26 | −9.32 | −9.31 | −10.0 |
| 19 | −11.55 | −12.30 | −4.08 | −3.24 | 3.69 | 2.48 | −2.76 | 0.00 | −0.81 | 2.02 |
| Avg* | -2.70 | -3.79 | -2.95 | -2.40 | -0.78 | -3.61 | -0.25 | -3.58 | -0.42 | -1.74 |
| SD** | 5.54 | 4.03 | 3.71 | 3.40 | 3.68 | 5.80 | 2.46 | 3.58 | 6.46 | 4.73 |
* The Avg row indicates the average value over 19 subjects.
** SD denotes standard deviation.
Table 4. p-value between m 1 and m 2 for MC.
| Ch.1 | Ch.2 | Ch.3 | Ch.4 | Ch.5 | Ch.6 | Ch.7 | Ch.8 | Ch.9 | Ch.10 | |
|---|---|---|---|---|---|---|---|---|---|---|
| p | 0.0115 | 0.0105 | 0.0056 | 0.0209 | 0.0402 | 0.0259 | 0.0004 | 0.0011 | 0.0561 | 0.0086 |
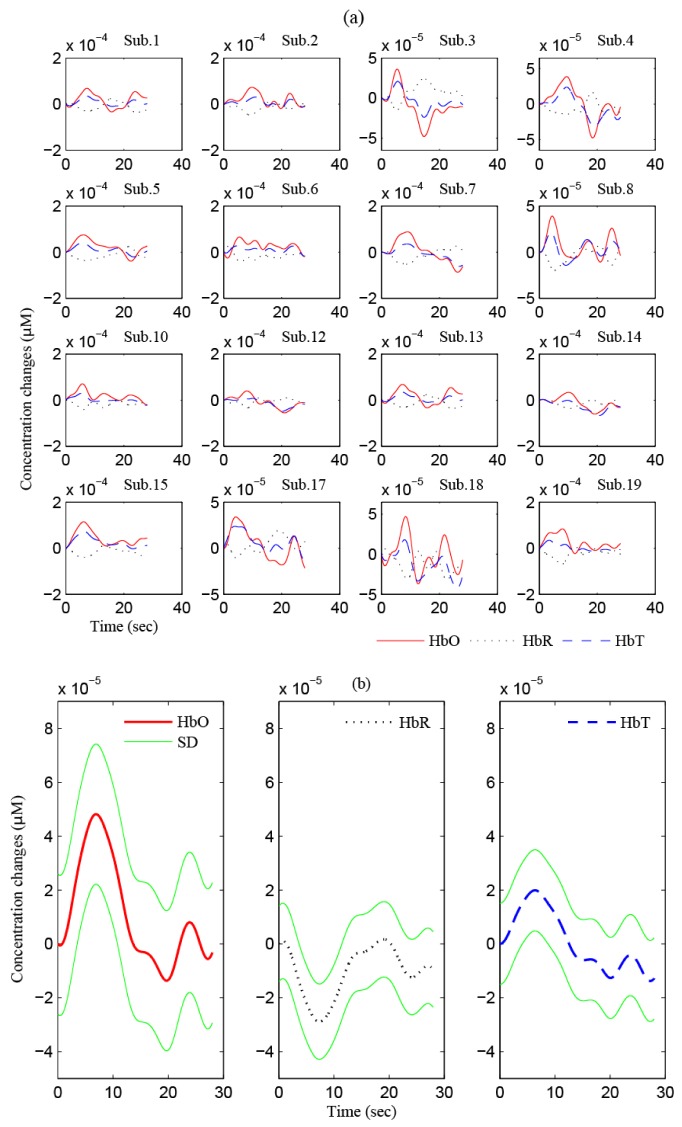
From the second last row in Table 2 and the last row in Table 3, the largest m 1 (i.e., 3.53 × 10−5 μM) and the smallest standard deviation (i.e., 2.46 × 10−5 μM) in T 2 are found from Ch. 7. Moreover, the m 1 in this channel is significantly higher than the m 2 (−0.25 × 10−5 μM). Therefore, Ch. 7 is considered as the most active channel in MC. If the mean value from some subjects (Ch. 7) is less than or equal to zero, the subject is supposed to be excluded from further analysis: In the MC experiment, four subjects (3, 4, 16, and 17) were excluded. The HRs in Ch. 7 for all other 15 subjects are shown in Fig. 8(a) . They all show an increased HR in T 1 followed by a return to the baseline. Finally, Fig. 8(b) depicts the averaged HbO, HbR and HbT over the 15 subjects with their standard deviations (solid green curves).
Fig. 8.
HRs in MC (Ch. 7): (a) The averaged HR over three stimuli in Fig. 1, (b) the averaged HR over 15 subjects in (a) and its standard deviation.
In the SC case, even though tables like Tables 2 and 3 are omitted due to space limitation, it is mentioned that Sub. 7 was excluded because of the lost data in some channels. The largest m 1 (1.91 × 10−5 µM) and the smallest standard deviation (2.59 × 10−5 μM) in T 2 over 18 subjects were found in Ch. 4. Therefore, Ch. 4 is identified as the most active channel in SC. Figure 9(a) shows the averaged HR over three stimuli, in which four subjects (1, 9, 17, and 19) were excluded additionally since their m 1 values were less than zero. Figure 9(b) depicts the averaged HbO, HbR, and HbT over the 14 subjects together with its standard deviation (green solid curves).
Fig. 9.
HRs in SC (Ch. 4): (a) The averaged HR over three stimuli in Fig. 1, (b) the averaged HR over 14 subjects in (a) and its standard deviation.
Finally, in the case of VC, Subs. 11 and 16 were excluded as the acquired data in some channels were lost. The largest m 1 and the smallest standard deviation in T 2 of the averaged HR across 17 subjects were found in Ch. 7, which were 3.54 × 10−5 μM and 2.02 × 10−5 μM, respectively. Therefore, Ch. 7 was regarded as the most active channel in VC. Subject 9 was excluded owing to m 1 < 0. Figure 10(a) shows the HRs of individual 16 subjects. Figure 10(b) illustrates the averaged HbO, HbR, and HbT over 16 subjects and its standard deviation (green solid curves). To summarize the entire development above, Fig. 11 compares the entire impulse HbO (solid line), HbR (dotted line), and HbT (dashed line) in three brain regions (MC, SC, VC).
Fig. 10.
HRs in VC (Ch. 7): (a) The averaged HR over three stimuli in Fig. 1, (b) the averaged HR over 16 subjects in (a) and its standard deviation.
Fig. 11.
Comparison of the impulse HbO (solid line), HbR (dotted line), and HbT (dashed line): (a) MC (Ch. 7), (b) SC (Ch. 4), and (c) VC (Ch. 7).
Additionally, Fig. 12 compares only the impulse HbOs in three brain regions. As can be seen, the HbO increases upon the stimulus and then falls to the baseline after making an undershoot peak. This result is consistent with the relevant previous works [26,27,32,34–36]. The amplitude of the main response and the undershoot peak of the MC are significantly higher than those of the SC and the VC. This result is quite consistent with the previous research [37]. But, the time-to-peaks (i.e., t 1) of three HbOs did not differ much, and they are about 6.76 ± 0.2 s. It is noted that the HbO decreased in the early stage (~0.46 s) in both MC and VC, whereas it was not so in the case of SC. This might be related to the initial dip in the cases of MC and VC [27,38].
Fig. 12.

Comparison of the impulse HbOs in three brain regions.
In addition, to compare the HbOs among the three brain regions, the ttest2.m function in Matlab was utilized again. The obtained p-values of the HbO pairs of MC vs. SC, MC vs. VC, and SC vs. VC were 6.3907 × 10−10, 2.7076 × 10−4, and 0.0267, respectively. Since p < 0.05 in all three cases, it is concluded that the HbOs from MC, SC, and VC are different from each other.
4.3 State space models
In this section, the A, B, C, and D matrices of the averaged HbOs in MC, SC, and VC are derived by the method discussed in Subsection 2.6. The system order in Eq. (8) is set to n = 6 (this was determined by trial and error). The number of rows in the Hankel matrices is q = 187 (for MC), q = 119 (for SC), and q = 795 (for VC) (q was found by trial and error as well). A systematic method for determining n and q is left as a future work. The state space model of the impulse HbO in MC is given as follows.
| (11) |
where a scale-up factor of 102 has been used. It is noted that t 1, t 2, and a 2 are related to the system matrix A. a 1 is influenced by the input and output matrices B and C. The direct transmission term D is close to zero. Figure 13(a) compares the reconstructed HbO from Eq. (11) (red solid line) and the corresponding experimental result (dashed blue line). As can be seen, they are very close.
Fig. 13.

Comparison between the experimental HbOs (blue dashed curves) and the reconstructed ones (red solid curves): (a) MC, (b) SC, and (c) VC.
Similarly, the coefficient matrices for the HbO concentration change in SC (the dotted line in Fig. 12) are obtained as follows. They are slightly different from Eq. (11).
| (12) |
Figure 13(b) illustrates the reconstructed impulse HbO in SC (red solid line), which fits to the experimentally obtained HbO (blue dashed line) very well.
Finally, the coefficient matrices for the HbO concentration changes (the dashed line in Fig. 12) to the 3 s checkerboard display in VC are obtained as follows.
| (13) |
It is noted that the elements in the left-lower corner in the system matrix A of MC and SC are zero, while it is not so in the case of VC. Figure 13(c) compares the reconstructed HbO in VC (red solid line) and the experimentally obtained one (blue dashed line). The characteristic parameters in Fig. 4 are tabulated in Table 5 . It is seen that t 1, a 1, and m 1 values between experiment and reconstruction are close, but there exist some difference in t 2 and a 2. This is due to that the signals in the deactivation period are low and fluctuating.
Table 5. Comparison of HR parameters between experiment and reconstruction: MC, SC, and VC.
| Characteristics of HR | MC | SC | VC | |
|---|---|---|---|---|
| t 1 (s) | Experiment | 6.72 | 6.58 | 6.98 |
| Reconstructed | 6.57 | 5.38 | 7.03 | |
| t 2 (s) | Experiment | 9.99 | 8.01 | 12.70 |
| Reconstructed | 10.57 | 12.43 | 11.42 | |
| a 1 (μM × 10−5) | Experiment | 5.37 | 3.93 | 4.82 |
| Reconstructed | 5.37 | 3.93 | 4.86 | |
| a 2 (μM × 10−5) | Experiment | −0.77 | −0.16 | −1.37 |
| Reconstructed | −0.30 | −0.74 | −0.97 | |
| m 1 (μM × 10−5) | Experiment | 4.14 | 3.03 | 3.84 |
| Reconstructed | 4.01 | 3.20 | 3.90 | |
5. Application to arbitrary stimuli
In this section, the developed method is applied to arbitrary stimuli. The blue pulses and boxes in Fig. 14(a) -14(c) indicate the given stimuli. Like the previous cases, tapping for MC, poking for SC, and checkerboard for VC are used. Figure 14 compares the desired HRs generated by two methods: one by Eq. (3) and another by the method in Subsection 4.3. The red dashed curves are generated by Eq. (3). The black solid curves are generated through Eqs. (11)-(13), respectively. It is noted that the red dashed curves in Figs. 14(a) and 14(b) are the same (since it was generated by Eq. (3)), but there exist a noticeable difference between black solid curves.
Fig. 14.
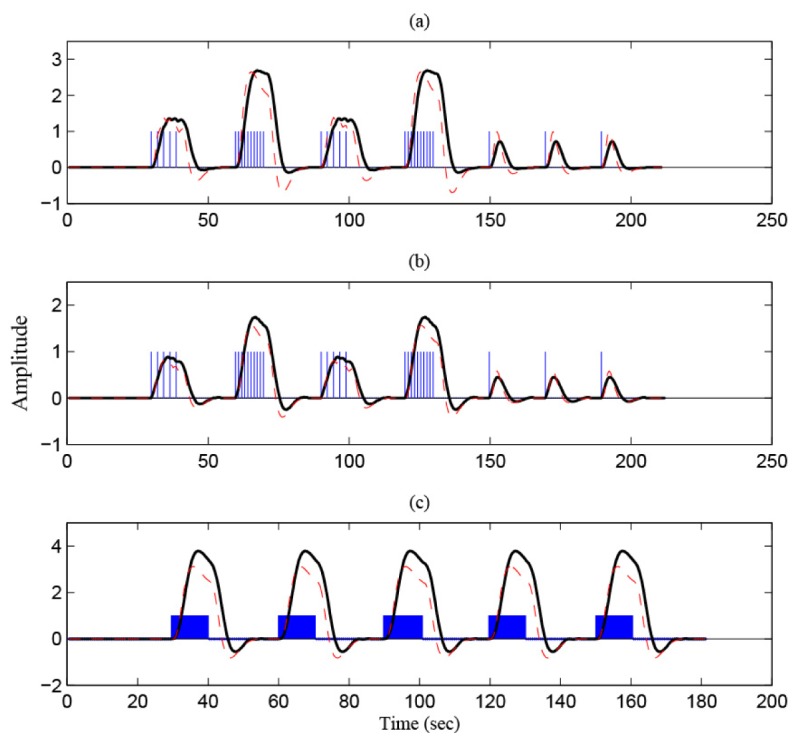
Comparison of the predicted HRs (the red dashed curves are through Eq. (3) and the black solid curves are through the state space method): (a) MC, (b) SC, and (c) VC.
To illustrate the effectiveness of the developed method, the black curves in Fig. 14 are used as x 1(k) in the regression model in Eq. (4). The RLS method in Eq. (5) is applied to identify the HR to the arbitrary stimuli in blue color (which is done online) and to check the activity strength (i.e., ) of all the channels online.
5.1. Tapping experiment
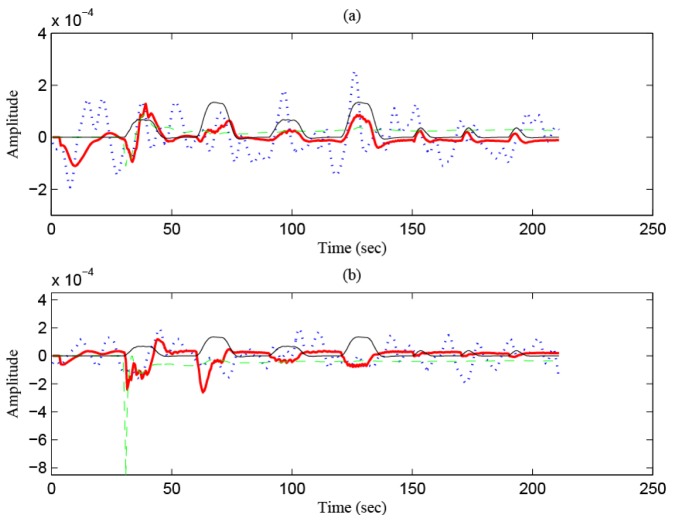
The red thick solid curves in Figs. 15(a) and 15(b) show the identified HR in an active channel (Ch. 12) and that in a de-active channel (Ch. 11), respectively (Subject 2). The dotted curves are the measured data. The black solid curve is the predicted HR (the same curve in Fig. 14). The green dashed curves indicate the activity strengths of the identified HRs. The active channel shows a positive activity strength, , whereas the de-active channel shows a negative strength, .
Fig. 15.
Identified HRs in MC (Subject 2): (a) Active channel (Ch. 12), (b) de-active channel (Ch. 11).
5.2. Poking experiment
Figures 16(a) and 16(b), respectively, illustrate the identified HRs (red thick curve) in an active channel (Ch. 13) with and that in a de-active channel (Ch. 23) with . The green dashed curves depict the activity strengths. It is seen that the identified HR is congruent well with the expected HR in the active channel.
Fig. 16.
Identified HRs in SC (Subject 1): (a) Active channel (Ch. 13), (b) de-active channel (Ch. 23).
5.3. Checkerboard experiment
Figures 17(a) and 17(b) describe the estimated HRs in Ch. 1 (active) and Ch. 3 (de-active). The identified HR in Ch. 1 is in good agreement with the experimental HR (blue dotted curve) and the predicted HR (black solid curve).
Fig. 17.
Identified HRs in VC (Subject 1): (a) Active channel (Ch. 1), (b) de-active channel (Ch. 3).
6. Conclusions
In this study, the state space method was utilized to reconstruct the HR to an impulse stimulus in three brain areas. Through the careful investigation, it was found that the HbOs in MC, SC, and VC are statistically different. Even the time-to-peak in the activation period of the HbO was similar (about 6.76 ± 0.2 s), all other parameters differed noticeably: The peak amplitude of the main response and the undershoot peak in MC were noticeably higher than those in SC and VC. Also, the time to undershoot peak in VC was the largest among three. These differences were able to be fully incorporated in the state space equations of the impulse HRs in MC, SC, and VC, while the canonical HR function in the form of double-gamma function cannot. This is the main advantage of the state space approach. Another important advantage of the proposed method lies in its easy applicability in generating the expected HR to arbitrary stimuli, as demonstrated in Section 4. It is remarked that the more state space models of brain activities are identified, the broader and varied brain functions can be estimated.
Acknowledgment
This work was supported by the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology, Korea (grant no. MEST-2012-R1A2A2A01046411).
References and links
- 1.Friston K. J., Fletcher P., Josephs O., Holmes A., Rugg M. D., Turner R., “Event-related fMRI: characterizing differential responses,” Neuroimage 7(1), 30–40 (1998). 10.1006/nimg.1997.0306 [DOI] [PubMed] [Google Scholar]
- 2.Lindquist M. A., Meng Loh J., Atlas L. Y., Wager T. D., “Modeling the hemodynamic response function in fMRI: efficiency, bias and mis-modeling,” Neuroimage 45(1Suppl), S187–S198 (2009). 10.1016/j.neuroimage.2008.10.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen H. F., Yao D. Z., Liu Z. X., “A comparison of Gamma and Gaussian dynamic convolution models of the fMRI BOLD response,” Magn. Reson. Imaging 23(1), 83–88 (2005). 10.1016/j.mri.2004.11.002 [DOI] [PubMed] [Google Scholar]
- 4.Ciftçi K., Sankur B., Kahya Y. P., Akin A., “Constraining the general linear model for sensible hemodynamic response function waveforms,” Med. Biol. Eng. Comput. 46(8), 779–787 (2008). 10.1007/s11517-008-0347-6 [DOI] [PubMed] [Google Scholar]
- 5.Hu X. S., Hong K. S., Ge S. S., Jeong M. Y., “Kalman estimator- and general linear model-based on-line brain activation mapping by near-infrared spectroscopy,” Biomed. Eng. Online 9(1), 82 (2010). 10.1186/1475-925X-9-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buxton R. B., Wong E. C., Frank L. R., “Dynamics of blood flow and oxygenation changes during brain activation: the balloon model,” Magn. Reson. Med. 39(6), 855–864 (1998). 10.1002/mrm.1910390602 [DOI] [PubMed] [Google Scholar]
- 7.Friston K. J., Mechelli A., Turner R., Price C. J., “Nonlinear responses in fMRI: the balloon model, volterra kernels, and other hemodynamics,” Neuroimage 12(4), 466–477 (2000). 10.1006/nimg.2000.0630 [DOI] [PubMed] [Google Scholar]
- 8.Friston K. J., Harrison L., Penny W., “Dynamic causal modelling,” Neuroimage 19(4), 1273–1302 (2003). 10.1016/S1053-8119(03)00202-7 [DOI] [PubMed] [Google Scholar]
- 9.Stephan K. E., Weiskopf N., Drysdale P. M., Robinson P. A., Friston K. J., “Comparing hemodynamic models with DCM,” Neuroimage 38(3), 387–401 (2007). 10.1016/j.neuroimage.2007.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen-Adad J., Chapuisat S., Doyon J., Rossignol S., Lina J. M., Benali H., Lesage F., “Activation detection in diffuse optical imaging by means of the general linear model,” Med. Image Anal. 11(6), 616–629 (2007). 10.1016/j.media.2007.06.002 [DOI] [PubMed] [Google Scholar]
- 11.Kamran M. A., Hong K. S., “Linear parameter-varying model and adaptive filtering technique for detecting neuronal activities: an fNIRS study,” J. Neural Eng. 10(5), 056002 (2013). 10.1088/1741-2560/10/5/056002 [DOI] [PubMed] [Google Scholar]
- 12.Naseer N., Hong M. J., Hong K. S., “Online binary decision decoding using functional near-infrared spectroscopy for the development of brain-computer interface,” Exp. Brain Res. 232(2), 555–564 (2014). 10.1007/s00221-013-3764-1 [DOI] [PubMed] [Google Scholar]
- 13.Hu X. S., Hong K. S., Ge S. S., “Recognition of stimulus-evoked neuronal optical response by identifying chaos levels of near-infrared spectroscopy time series,” Neurosci. Lett. 504(2), 115–120 (2011). 10.1016/j.neulet.2011.09.011 [DOI] [PubMed] [Google Scholar]
- 14.Shimokawa T., Kosaka T., Yamashita O., Hiroe N., Amita T., Inoue Y., Sato M. A., “Extended hierarchical Bayesian diffuse optical tomography for removing scalp artifact,” Biomed. Opt. Express 4(11), 2411–2432 (2013). 10.1364/BOE.4.002411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Re R., Contini D., Turola M., Spinelli L., Zucchelli L., Caffini M., Cubeddu R., Torricelli A., “Multi-channel medical device for time domain functional near infrared spectroscopy based on wavelength space multiplexing,” Biomed. Opt. Express 4(10), 2231–2246 (2013). 10.1364/BOE.4.002231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cope M., Delpy D. T., “System for long-term measurement of cerebral blood and tissue oxygenation on newborn infants by near infra-red transillumination,” Med. Biol. Eng. Comput. 26(3), 289–294 (1988). 10.1007/BF02447083 [DOI] [PubMed] [Google Scholar]
- 17.Saager R. B., Telleri N. L., Berger A. J., “Two-detector corrected near infrared spectroscopy (C-NIRS) detects hemodynamic activation responses more robustly than single-detector NIRS,” Neuroimage 55(4), 1679–1685 (2011). 10.1016/j.neuroimage.2011.01.043 [DOI] [PubMed] [Google Scholar]
- 18.Ye J. C., Tak S., Jang K. E., Jung J., Jang J., “NIRS-SPM: statistical parametric mapping for near-infrared spectroscopy,” Neuroimage 44(2), 428–447 (2009). 10.1016/j.neuroimage.2008.08.036 [DOI] [PubMed] [Google Scholar]
- 19.Hassanpour M. S., White B. R., Eggebrecht A. T., Ferradal S. L., Snyder A. Z., Culver J. P., “Statistical analysis of high density diffuse optical tomography,” Neuroimage 85(Pt 1), 104–116 (2014). 10.1016/j.neuroimage.2013.05.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aqil M., Hong K. S., Jeong M. Y., Ge S. S., “Detection of event-related hemodynamic response to neuroactivation by dynamic modeling of brain activity,” Neuroimage 63(1), 553–568 (2012). 10.1016/j.neuroimage.2012.07.006 [DOI] [PubMed] [Google Scholar]
- 21.Hu X. S., Hong K. S., Ge S. S., “fNIRS-based online deception decoding,” J. Neural Eng. 9(2), 026012 (2012). 10.1088/1741-2560/9/2/026012 [DOI] [PubMed] [Google Scholar]
- 22.Schelkanova I., Toronov V., “Independent component analysis of broadband near-infrared spectroscopy data acquired on adult human head,” Biomed. Opt. Express 3(1), 64–74 (2012). 10.1364/BOE.3.000064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan Z., “Combining independent component analysis and Granger causality to investigate brain network dynamics with fNIRS measurements,” Biomed. Opt. Express 4(11), 2629–2643 (2013). 10.1364/BOE.4.002629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naseer N., Hong K. S., “Classification of functional near-infrared spectroscopy signals corresponding to the right- and left-wrist motor imagery for development of a brain-computer interface,” Neurosci. Lett. 553, 84–89 (2013). 10.1016/j.neulet.2013.08.021 [DOI] [PubMed] [Google Scholar]
- 25.Santosa H., Hong M. J., Kim S. P., Hong K. S., “Noise reduction in functional near-infrared spectroscopy signals by independent component analysis,” Rev. Sci. Instrum. 84(7), 073106 (2013). 10.1063/1.4812785 [DOI] [PubMed] [Google Scholar]
- 26.Villringer A., Chance B., “Non-invasive optical spectroscopy and imaging of human brain function,” Trends Neurosci. 20(10), 435–442 (1997). 10.1016/S0166-2236(97)01132-6 [DOI] [PubMed] [Google Scholar]
- 27.Jasdzewski G., Strangman G., Wagner J., Kwong K. K., Poldrack R. A., Boas D. A., “Differences in the hemodynamic response to event-related motor and visual paradigms as measured by near-infrared spectroscopy,” Neuroimage 20(1), 479–488 (2003). 10.1016/S1053-8119(03)00311-2 [DOI] [PubMed] [Google Scholar]
- 28.Hoshi Y., Kobayashi N., Tamura M., “Interpretation of near-infrared spectroscopy signals: a study with a newly developed perfused rat brain model,” J. Appl. Physiol. 90(5), 1657–1662 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Brigadoi S., Ceccherini L., Cutini S., Scarpa F., Scatturin P., Selb J., Gagnon L., Boas D. A., Cooper R. J., “Motion artifacts in functional near-infrared spectroscopy: a comparison of motion correction techniques applied to real cognitive data,” Neuroimage 85(Pt 1), 181–191 (2014). 10.1016/j.neuroimage.2013.04.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barker J. W., Aarabi A., Huppert T. J., “Autoregressive model based algorithm for correcting motion and serially correlated errors in fNIRS,” Biomed. Opt. Express 4(8), 1366–1379 (2013). 10.1364/BOE.4.001366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu X. S., Hong K. S., Ge S. S., “Reduction of trial-to-trial variability in functional near-infrared spectroscopy signals by accounting for resting-state functional connectivity,” J. Biomed. Opt. 18(1), 017003 (2013). 10.1117/1.JBO.18.1.017003 [DOI] [PubMed] [Google Scholar]
- 32.Franceschini M. A., Fantini S., Thompson J. H., Culver J. P., Boas D. A., “Hemodynamic evoked response of the sensorimotor cortex measured noninvasively with near-infrared optical imaging,” Psychophysiology 40(4), 548–560 (2003). 10.1111/1469-8986.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.T. Katayama, Subspace methods for system identification, E. D. Sontag, M. Thoma, A. Isidori, J. H. vanSchuppen ed. (Springer-Verlag London Limited, 2005). [Google Scholar]
- 34.Miezin F. M., Maccotta L., Ollinger J. M., Petersen S. E., Buckner R. L., “Characterizing the hemodynamic response: effects of presentation rate, sampling procedure, and the possibility of ordering brain activity based on relative timing,” Neuroimage 11(6), 735–759 (2000). 10.1006/nimg.2000.0568 [DOI] [PubMed] [Google Scholar]
- 35.Bhutta M. R., Hong K. S., Kim B. M., Hong M. J., Kim Y. H., Lee S. H., “Note: three wavelengths near-infrared spectroscopy system for compensating the light absorbance by water,” Rev. Sci. Instrum. 85(2), 026111 (2014). 10.1063/1.4865124 [DOI] [PubMed] [Google Scholar]
- 36.Khan M. J., Hong M. J., Hong K. S., “Decoding of four movement directions using hybrid NIRS-EEG brain-computer interface,” Front. Hum. Neurosci. 8, 244 (2014). 10.3389/fnhum.2014.00244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baraldi P., Manginelli A. A., Maieron M., Liberati D., Porro C. A., “An ARX model-based approach to trial by trial identification of fMRI-BOLD responses,” Neuroimage 37(1), 189–201 (2007). 10.1016/j.neuroimage.2007.02.045 [DOI] [PubMed] [Google Scholar]
- 38.Yacoub E., Shmuel A., Pfeuffer J., Van De Moortele P. F., Adriany G., Ugurbil K., Hu X. P., “Investigation of the initial dip in fMRI at 7 Tesla,” NMR Biomed. 14(7-8), 408–412 (2001). 10.1002/nbm.715 [DOI] [PubMed] [Google Scholar]