Abstract
Aromatic amino acids often flank the transmembrane alpha helices of integral membrane proteins. By favoring locations within the membrane–water interface of the lipid bilayer, aromatic residues Trp, Tyr, and sometimes Phe may serve as anchors to help stabilize a transmembrane orientation. In this work, we compare the influence of interfacial Trp, Tyr, or Phe residues upon the properties of tilted helical transmembrane peptides. For such comparisons, it has been critical to start with no more than one interfacial aromatic residue near each end of a transmembrane helix, for example, that of GWALP23 (acetyl-GGALW5(LA)6LW19LAGA-[ethanol]amide). To this end, we have employed 2H-labeled alanines and solid-state NMR spectroscopy to investigate the consequences of moving or replacing W5 or W19 in GWALP23 with selected Tyr, Phe, or Trp residues at the same or proximate locations. We find that GWALP23 peptides having F5, Y5, or W5 exhibit essentially the same average tilt and similar dynamics in bilayer membranes of 1,2-dilauroylphosphatidylcholine (DLPC) or 1,2-dioleoylphosphatidylcholine (DOPC). When double Tyr anchors are present, in Y4,5GWALP23 the NMR observables are markedly more subject to dynamic averaging and at the same time are less responsive to the bilayer thickness. Decreased dynamics are nevertheless observed when ring hydrogen bonding is removed, such that F4,5GWALP23 exhibits a similar extent of low dynamic averaging as GWALP23 itself. When F5 is the sole aromatic group in the N-interfacial region, the dynamic averaging is (only) slightly more extensive than with W5, Y5, or Y4 alone or with F4,5, yet it is much less than that observed for Y4,5GWALP23. Interestingly, moving Y5 to Y4 or W19 to W18, while retaining only one hydrogen-bond-capable aromatic ring at each interface, maintains the low level of dynamic averaging but alters the helix azimuthal rotation. The rotation change is about 40° for Y4 regardless of whether the host lipid bilayer is DLPC or DOPC. The rotational change (Δρ) is more dramatic and more complex when W19 is moved to W18, as Δρ is about +90° in DLPC but about −60° in DOPC. Possible reasons for this curious lipid-dependent helix rotation could include not only the separation distances between flanking aromatic or hydrophobic residues but also the absolute location of the W19 indole ring. For the more usual cases, when the helix azimuthal rotation shows little dependence on the host bilayer identity, excepting W18GWALP23, the transmembrane helices adapt to different lipids primarily by changing the magnitude of their tilt. We conclude that, in the absence of other functional groups, interfacial aromatic residues determine the preferred orientations and dynamics of membrane-spanning peptides. The results furthermore suggest possibilities for rotational and dynamic control of membrane protein function.
Model peptides have proven to be useful for studying protein–lipid interactions, which in turn are important for the regulation of biological function. Model systems offer particular advantages for establishing general principles because specific changes in peptide sequence and structure can be made, and the direct effects on interaction with a surrounding lipid membrane can be analyzed. Several membrane proteins, including gramicidin channels, reveal the preferred location of tryptophan residues at the lipid–water interface.1−3 For model peptide design, a core leucine–alanine sequence4 will enhance sensitivity of the peptide to membrane thickness because of hydrophobic mismatch. Then, by altering the identities or positions of aromatic anchors that flank the core sequence, the effects of these placements on the orientations and dynamics of transmembrane helices can be investigated and inferences for membrane proteins can be deduced.
The early model WALP peptides (acetyl-GWWA(LA)nLWWA-[ethanol]amide), incorporating multiple Trp (W) anchors and the helical, hydrophobic repeating Leu–Ala core sequence, were important for helping to establish principles for the interfacial partitioning of Trp residues and the modulation of lipid phase behavior.4 Later, the four Trp residues were mutated to other aromatic or charged residues (Tyr, Phe, Lys, Arg, or His) to monitor the importance of the chemical and physical properties.5,6 The WALP family peptides adopt specific preferred tilted transmembrane orientations in lipid bilayer membranes,7,8 but, in addition to cone precession about the membrane normal,9 they also experience excessive dynamic averaging of solid-state NMR observables,10−13 caused by the presence of the four Trp residues.14 Importantly, the extent of dynamic averaging can be greatly reduced by decreasing the number of aromatic residues, as in GWALP23 (acetyl-GGALW(LA)6LWLAGA-[ethanol]amide),15 which possesses only two Trp residues and still maintains a preferred and well-defined tilted orientation12,14,16 with low dynamic averaging. The tryptophans in GWALP23 thereby flank a hydrophobic Leu–Ala core of the same length as that of WALP19.4,7 With fewer aromatic residues, it becomes easier to assess the roles of each of them. The results to date suggest that four Trp anchors are so dominating, and possibly competing,17 that they induce significant peptide dynamics, mainly because of rotational “slippage” about the helix axis.14,16
The favorable properties of GWALP23 have enabled this peptide to be employed as a highly suitable parent host framework for examining the influence of specific charged guest residues within the core hydrophobic sequence.18,19 It has furthermore been possible to examine the titration behavior of specific residues and the influence of ionization state upon helix orientation.20 Within the GWALP23 context, questions arose about the effects of changing the identity, number, and position of the aromatic residues. A peptide having a single Trp → Tyr replacement (Y5GWALP23) exhibits similar transmembrane orientation and dynamics as those of GWALP23,17 in three different lipids, such that the substitution of Tyr for Trp causes no fundamental change in the lipid–peptide interactions. However, when two Tyr residues are introduced to produce Y4,5GWALP23, the extent of dynamic averaging increases dramatically,17 reminiscent of the earlier WALP peptides.
With the Trp- and Tyr-containing members of the GWALP23 family as a backdrop, we have examined the influence of phenylalanine by altering the Tyr-containing models so as to have non-hydrogen-bonding Phe residues in F5GW19ALP23 and F4,5GW19ALP23 (acetyl-GGAF4F5(LA)6LW19LAGA-[ethanol]amide). We furthermore examined whether the dynamic behavior shown by Y4,5GW19ALP23 could be caused by Y4 alone, as opposed to the Y4,5 combination. Placing a Tyr residue at position 4 also provided a way of determining the effects of moving a single aromatic residue one position (100°) around the α helix by comparison with Y5GW19ALP23. A similar radial comparison between aromatic ring positions was made by moving Trp19 to position 18 to give GW5,18ALP23.
Materials and Methods
Peptides F5GWALP23, F4,5GWALP23, Y4GWALP23, and W18GWALP23 (Table 1) were synthesized on a model 433A synthesizer from Applied Biosystems by Life Technologies (Foster City, CA) using solid-phase methods, as described previously.17 Typically, two deuterated alanines of differing isotope abundances were incorporated into each synthesized peptide. Peptides were purified as described19,21 using an octyl silica column (Zorbax Rx-C8, 9.4 × 250 mm, 5 μm particle size; Agilent Technologies, Santa Clara, CA) and a gradient of 97–100% methanol (with 0.1% trifluoroacetic acid) over 28 min. Final peptide purity (>97%) was confirmed by reversed-phase HPLC, and peptide identity, by mass spectrometry (Figure S1 of the Supporting Information).
Table 1. Sequences of GWALP23-Like Peptides with Aromatic Substitutionsa.
| name | sequence |
|---|---|
| WALP23 | a-GWW3LALALALALALALALALWWA-e |
| GWALP23 | a-GGALW5LALALALALALALWLAGA-e |
| Y5GWALP23 | a-GGALY5LALALALALALALWLAGA-amide |
| Y4,5GWALP23 | a-GGAY4Y5LALALALALALALWLAGA-amide |
| F5GWALP23 | a-GGALF5LALALALALALALWLAGA-amide |
| F4,5GWALP23 | a-GGAF4F5LALALALALALALWLAGA-amide |
| Y4GWALP23 | a-GGAY4LLALALALALALALWLAGA-amide |
| W18GWALP23 | a-GGALW5LALALALALALAW18LLAGA-amide |
Abbreviations: a, acetyl; e, ethanolamide.
Solid-state 2H NMR experiments, using mechanically aligned samples, were performed using methods that have been described previously.17 Mechanically aligned samples (1:60, peptide/lipid; 45% hydration, w/w) were prepared using DOPC, DMPC, or DLPC lipid from Avanti Polar Lipids (Alabaster, AL) and deuterium-depleted water from Cambridge Isotope Laboratories (Andover, MA). Bilayer alignment within each sample was confirmed using 31P NMR at 50 °C on a Bruker (Billerica, MA) Avance 300 spectrometer. Deuterium NMR spectra were recorded at 50 °C using both β = 0° (bilayer normal parallel to magnetic field) and β = 90° macroscopic sample orientations on a Bruker Avance 300 spectrometer utilizing a quadrupolar echo pulse sequence22 with 90 ms recycle delay, 3.2 μs pulse length, and 115 μs echo delay. Between 0.6 and 1.5 million scans were accumulated during each 2H NMR experiment. An exponential weighting function with 100 Hz line broadening was applied prior to Fourier transformation.
Helix orientations were analyzed by means of a semistatic “GALA” method based on three adjustable parameters: the average tilt τo of the helix axis, the average azimuthal rotation ρo about the helix axis, and a principal order parameter Szz, as described.7,17 An additional three-parameter modified Gaussian method is available, based on τo, ρo, a distribution width σρ, and a fixed στ.16 We also employed this modified Gaussian method, but στ was fixed at either 15° (DLPC) or 9° (DOPC; see Discussion) instead of the previously assumed value of 0° for στ. For the analysis of helix rotation, we analyzed some pairwise residue separation distances using a recently described procedure.23 Distances were compared to hydrophobic thicknesses of 20.9 Å for DLPC24 and 27.2 Å for DOPC,25 which are based on the location DC of the Gibbs dividing surface for the hydrocarbon region of the bilayer.25
Results
The designed peptides (Table 1 and Figure 1) were successfully synthesized and purified, as confirmed by analytical HPLC and MALDI-TOF mass spectrometry (Figure S1 of the Supporting Information). The repeating Leu–Ala sequence at the core of GWALP peptides favors folding into α-helical secondary structure within the hydrophobic region of the lipid bilayer. Indeed, the CD spectra for the new variants of GWALP23 (Figure 2) show a minimum near 208 nm and a broad shoulder near 222 nm, indicating that the secondary structure is indeed α-helical. The 31P NMR spectra for oriented samples of each peptide–lipid combination furthermore confirmed the presence of oriented lipid bilayers within samples that were aligned with the bilayer normal either parallel (β = 0°) or perpendicular (β = 90°) to the applied magnetic field. The spectra exhibit characteristic 31P resonances located close to −14.5 ppm for the β = 90° orientation and near +29 ppm when β = 0° (Figure S2 of the Supporting Information).
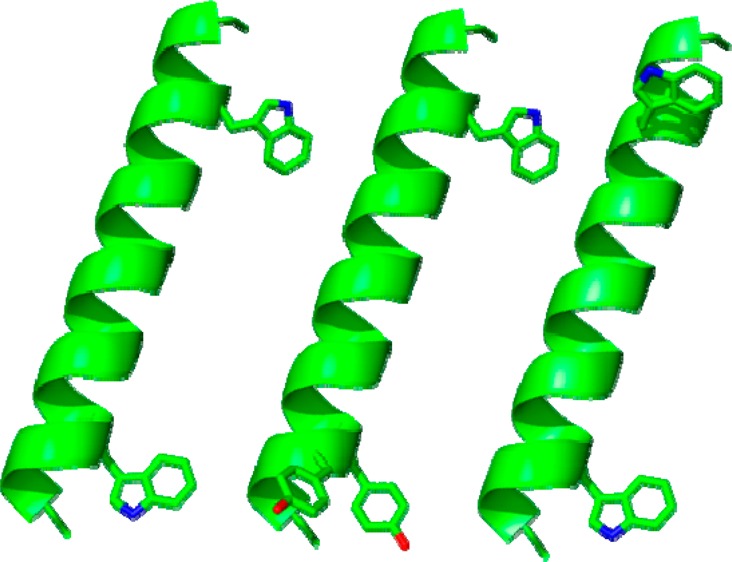
Figure 1.
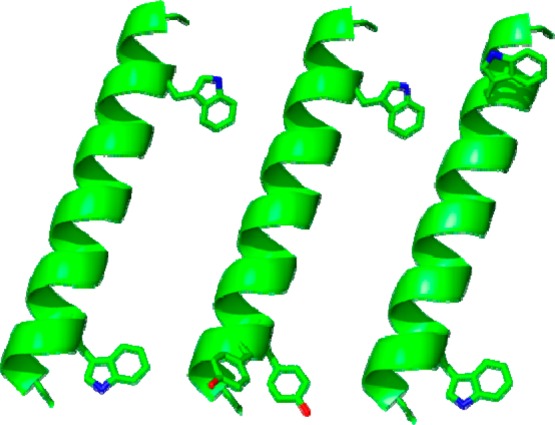
Representative models of GWALP23, Y4,5GWALP23, and W18GALP23 (left to right) showing the locations of aromatic side chains on a ribbon helix; drawn using PyMOL.30 The side-chain orientations are arbitrary.
Figure 2.
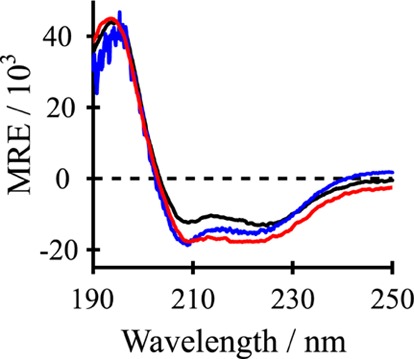
Circular dichroism (CD) spectra of F4,5GWALP23 (black), Y5GWALP23 (blue), and Y4GWALP23 (red) in DLPC (1:60 peptide/lipid). The y-axis units for mean residue ellipticity (MRE) are deg cm2 dmol–1.
Solid-state 2H NMR spectra from oriented samples of peptide and lipid enable the relative orientations and dynamic behavior of the folded peptide helices to be determined in lipid bilayer membranes. The aromatic residues on each end of the hydrophobic (LA)n core of GWALP peptides help to position the peptide termini at the respective membrane–water interfaces, at best in orientations that minimize hydrophobic mismatch between the lipid thickness and the peptide length. Deuterium-labeled (2H) alanine residues at various positions in the core Leu–Ala sequence of each peptide then allow characterization of the behavior of the peptides in aligned bilayers by means of solid-state 2H NMR spectroscopy. The 2H quadrupolar splitting magnitudes |Δνq| from the alanine CD3 (or C-D) groups serve to define a preferred tilted, dynamically averaged, orientation of the entire helix with respect to the bilayer normal in an applied magnetic field.7,8
On the basis of the solid-state 2H NMR spectra (Figures S3–S5 of the Supporting Information), we observed a wide range of quadrupolar splittings for F5 GWALP23, from 1 to 22 kHz in DLPC and 1–18 kHz in DOPC (Table 2), suggesting that the peptide helix tilts substantially in the bilayer membranes. (If the helix were not tilted, then all of the labeled alanines would give the same signal, namely, the same quadrupolar splitting, Δνq.) The wide ranges of 21 and 17 kHz are consistent with those seen previously for the W5 and Y5 analogues of GWALP23, suggesting similar global helix dynamics regardless of whether the aromatic residue is capable (W5 and Y5) or not (F5) of hydrogen bonding. Indeed, the ranges of Δνq values observed when W5 is present are 23 and 14 kHz in DLPC and DOPC, respectively. The corresponding ranges when Y5 is present are 21 and 13 kHz. These large and similar ranges of Δνq in all three of the X5GWALP23 peptides, where X is aromatic, suggest that all of these GWALP23 analogues are tilted to similar extents, and exhibit similar dynamics, in the lipid bilayer membranes.
Table 2. 2H NMR Quadrupolar Splittings (Δνq, in kHz) for Labeled Alanine CD3 Groups in F5GWALP23, F4,5GWALP23, and Y4GWALP23a.
| DLPC |
DMPC |
DOPC |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ala-d4 | F5 | F4,5 | Y4 | W5W18 | F5 | Y4 | F5 | F4,5 | Y4 | W5W18 |
| 7 | 17.8 | 23.7 | 15.8 | -- | -- | 17.6 | 16.2 | 16.2 | 11.8 | -- |
| 9 | 20.4 | 23.5 | 9.2 | 14.9 | -- | 6.4 | 2.8 | 0.8 | 1.5 | 6.6 |
| 11 | 21.9 | 25.7 | 18.8 | -- | -- | 17.9 | 17.6 | 18.6 | 16.4 | 15.4 |
| 13 | 13.8 | 19.6 | 9.2 | 9.0 | -- | 8.5 | 3.4 | 0.8 | 3.2 | 2.3 |
| 15 | 18.3 | 23.4 | 18.8 | 25.6 | -- | 18.4 | 17.6 | 18.6 | 18.4 | 5.6 |
| 17 | 1.0 | 1.8 | 1.3 | 21.6 | -- | 2.7 | 1.0 | 1.9 | 2.2 | 17.2 |
Sample orientation is β = 0°. Each value (in kHz) is the average of the magnitude observed at β = 0° and twice the magnitude observed for a β = 90° sample orientation. Values that are absent (--) were not recorded. The labeled alanines are identified, and the positions of the N-flanking aromatic amino acids are indicated as F5, F4,5, and Y4. The C-flanking W19 is present in all samples except for W5W18.
By using an α-helical geometry and a principal order parameter Szz to describe the helix motions, the sets of 2H-alanine quadrupolar splittings can be analyzed by a method known as “geometric analysis of labeled alanines” (GALA).10 The method finds the lowest RMSD values based on the helix tilt (τ), azimuthal rotation (ρ), and Szz as variables. Notably, the resulting tilt values for F5GWALP23 (about 7 and 21° in DOPC and DLPC, respectively) are nearly identical to those found previously8 for the W5 and Y5 peptides (Table 3). In the shorter DLPC lipids, the average tilt of the W19 peptides with a single N-flanking aromatic residue at position five is 21°, whereas in the longer DOPC lipids, the average tilt is 6°. Comparison of these τ values for DLPC and DOPC lipids suggests that each of the X5GWALP23 peptide responds to hydrophobic mismatch by tilting more in the thinner bilayers.
Table 3. Semistatic GALA Analysis of Transmembrane Orientations of Peptides of the GWALP23 Familya.
| DLPC |
DMPC |
DOPC |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| peptide | τ0 | ρ0 | Szz | RMSD (kHz) | τ0 | ρ0 | Szz | RMSD (kHz) | τ0 | ρ0 | Szz | RMSD (kHz) |
| bW5 | 21° | 305° | 0.71 | 0.7 | 9 | 311 | 0.88 | 1.0 | 6 | 323 | 0.87 | 0.6 |
| bY5 | 19 | 295 | 0.78 | 0.7 | 10 | 300 | 0.84 | 0.7 | 5 | 311 | 0.84 | 1.0 |
| F5 | 21 | 310 | 0.58 | 1.1 | 7 | 330 | 0.85 | 0.6 | ||||
| bY4,5 | 5 | 260 | 0.66 | 1.6 | 3 | 323 | 0.77 | 0.6 | 3 | 359 | 0.82 | 1.1 |
| F4,5 | 21 | 317 | 0.67 | 0.6 | 6 | 332 | 0.92 | 0.9 | ||||
| Y4 | 12 | 327 | 0.68 | 0.8 | 10 | 332 | 0.73 | 0.9 | 7 | 357 | 0.86 | 0.3 |
| W5W18 | 19 | 40 | 0.78 | 1.5 | 12 | 265 | 0.74 | 1.3 | ||||
The N-flanking aromatic residues are indicated by the abbreviation for each peptide. C-flanking W19 is also present in all samples except when the aromatic residues are W5 and W18.
Values from ref. (17).
It was surprising (see Discussion) to observe that the double-Phe derivative, F4,5GWALP23, with adjacent aromatic phenyl rings at positions 4 and 5, displays a wider range of 2H quadrupolar splittings than does F5GWALP23 itself. These results stand in stark contrast to the earlier comparison between Y4,5GWALP23 and Y5GWALP23.8 In the shortest lipid, DLPC, the range of Δνq for alanines in F4,5GWALP23 is about 24 kHz, from 1.8 to 25.7 kHz. In DOPC, the values cover a range of about 17 kHz, from 1.9 to 18.6 kHz (Table 2). These differences suggest that F4,5GWALP23 may be tilted to greater extent or may undergo less dynamic averaging than the single-anchored X5GWALP23 peptides. It is striking that the results for F4,5GWALP23 differ greatly from the previous characterization of Y4,5GWALP23.8 The larger quadrupolar splittings that are observed in 2H NMR spectra for the F4,5 peptide are greatly reduced in the spectra for Y4,5 (Figure 3). The ranges of the quadrupolar splitting magnitudes from alanines in Y4,5GWALP23 therefore span only 11 and 9 kHz in DLPC and DOPC, respectively (Table 2). By changing the identity of the aromatic anchors at positions 4 and 5 from Tyr to Phe (Y to F), thereby removing the hydrogen-bonding ability of the aromatic ring, the net effect is to double the kilohertz range that is spanned by the alanine quadrupolar splittings, a dramatic change! Furthermore, when comparing the change in Δνq range from DLPC to DOPC, one would expect a significant decrease due to less tilting necessary in the longer lipid. For F4,5GWALP23, the range indeed narrows from 24 to 17 kHz from DLPC to DOPC, but for Y4,5GWALP23, the range hardly changes. These results suggest that the Tyr (Y) anchors at positions 4 and 5 allow far less response to bilayer thickness than do the Phe (F) anchors, probably because the extent of dynamic averaging is large in both lipids when Y4 and Y5 are present together.
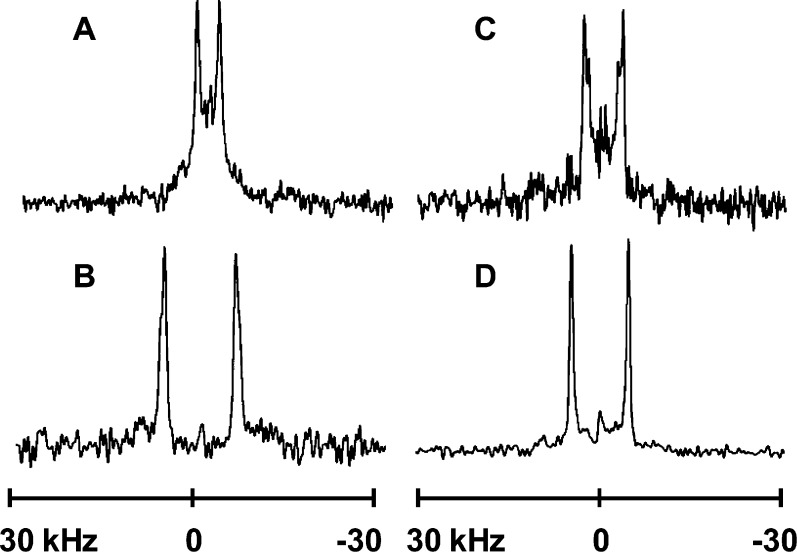
Figure 3.
2H NMR spectra for labeled alanines in selected X4,5 peptides in DLPC and DOPC: (A) Y4,5GWALP23 in DLPC, (B) F4,5GWALP23 in DLPC, (C) Y4,5GWALP23 in DOPC, and (D) F4,5GWALP23 in DOPC. In each peptide, Ala-15 is 100% deuterated and Ala-11 is 50% deuterated. β = 90° sample orientation; 50 °C.
The results of tilt analysis for F4,5GWALP23 also are strikingly different from the previous results for Y4,5GWALP23.8 The tilt value for F4,5GWALP23 in DLPC (∼21°) is much larger than the “apparent” tilt of the Y4,5 analogue (∼5°) (Table 3). The actual tilt of Y4,5GWALP23 in DLPC is nevertheless obscured by the extensive dynamics of this peptide.8 Remarkably, F4,5GWALP23 does not suffer from the high dynamics and gives quadrupolar wave fits for tilt and azimuthal rotation similar to those for F5GWALP23 in DLPC and DOPC (Figure 4). In DLPC, the wave amplitude actually is larger for F4,5GWALP23 than for F5GWALP23 (Figure 4A), suggesting somewhat lower dynamic averaging for F4,5GWALP23, and the wave amplitudes for both F4,5GWALP23 and F5GWALP23 are notably very much larger than that for Y4,5GWALP23 in DLPC.17 In DOPC, these three peptides have rather similar small tilt angles (Figure 4B and Table 3), yet the dynamic averaging remains much larger for Y4,5GWALP23 (see Discussion).
Figure 4.
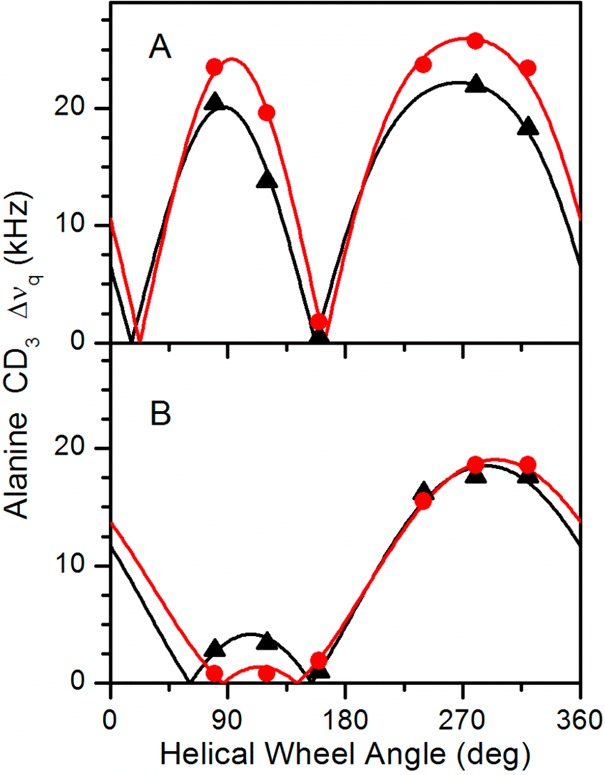
Quadrupolar wave plots for F5GWALP23 (black, triangles) and F4,5GWALP23 (red, circles) in (A) DLPC and (B) DOPC.
For the case of moving the tyrosine residue from position 5 to position 4 in the sequence, the helix tilt and dynamics remain unchanged, yet a phase change is seen in the quadrupolar wave plot (Figure 5), indicating a change in the helix azimuthal rotation (ρo) or direction of the tilt. Semistatic GALA analysis of Y4GWALP23 indicates that the rotation of the peptide changes by about 31° in DLPC and 46° in DOPC (Table 3) when Y5 is moved radially by 100° to position 4. A top view or helical wheel illustrates these differences in the direction of tilt for Y4GWALP23 and Y5GWALP23 (Figure 5B). Importantly, the extensive dynamics observed for double-Tyr derivative Y4,5GWALP23 are not caused by either Y4 or Y5 alone, but rather by the presence of the two tyrosines together.
Figure 5.
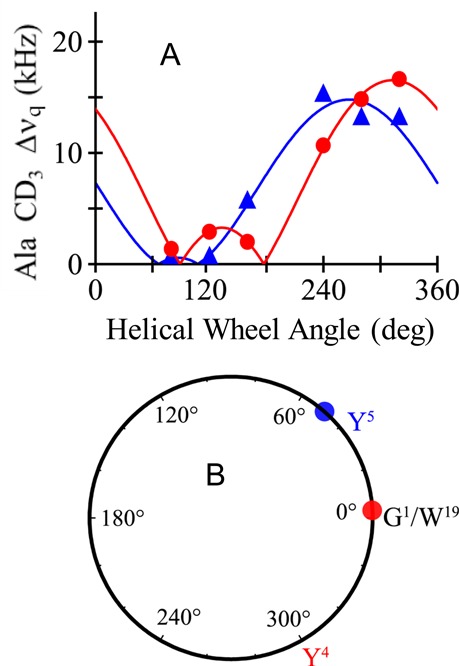
(A) Quadrupolar wave plots for Y4GWALP23 (red, circles) and Y5GWALP23 (blue, triangles) in DOPC. (B) Helical wheel diagram to illustrate the relative azimuthal rotation ρ for Y4GWALP23 (red circle) and Y5GWALP23 (blue circle) in DOPC, offset by ∼50°. The labels Y4 and Y5 represent the respective radial locations of the tyrosines, which differ by 100° on the helical wheel.
Because of the change in azimuthal rotation that accompanies the moving of Y5 to Y4, we decided to investigate the consequence of moving W19 to position 18 in GWALP23. The results (Figure 6) indicate a larger rotational shift of almost 100° for the peptide helix in DLPC when W19 in GWALP23 is moved by 100° to W18. This large shift suggests a dominant role for W19 in determining the azimuthal rotation preference of GWALP23 in lipid bilayer membranes. Yet, a similar comparison between GWALP23 and W18GWALP23 in DOPC brings a surprise (Table 3). Namely, the preferred azimuthal rotation of the W18GWALP23 helix seems to depend upon the lipid environment. Within the realm of transmembrane peptides that exhibit low dynamic averaging, this result for W18GWALP23 stands in contrast to the essentially constant rotational preferences of other GWALP23 family peptides in bilayers of differing thickness.12 Namely, for the transmembrane helices of GWALP23, R2,22GWALP23, and K2,22GWALP23,12 as well as R14GWALP23,14,18 Y5GWALP23,17 Y19GWALP23,26 Y4GWALP23, F5GWALP23, and F4,5GWALP23 (this work), the most probable azimuthal rotation ρo about the helix axis does not vary appreciably when each particular helix is moved from DLPC to DMPC or DOPC. For cases of excess dynamics, namely, W2,22GWALP2312 and Y4,5GWALP23,17 which also incorporate extra aromatic residues, σρ is large and ρo is not well defined from lipid to lipid. In this respect, W18GWALP23 stands alone with low dynamic averaging and yet has a value of ρo that changes by about 135° from DLPC to DOPC (Table 3). Even if one considers a Gaussian analysis (see Discussion), the value of ρo still varies widely from DLPC to DOPC. We therefore sought to consider possible reasons for this lipid-dependent variation in helix azimuthal rotation.
Figure 6.
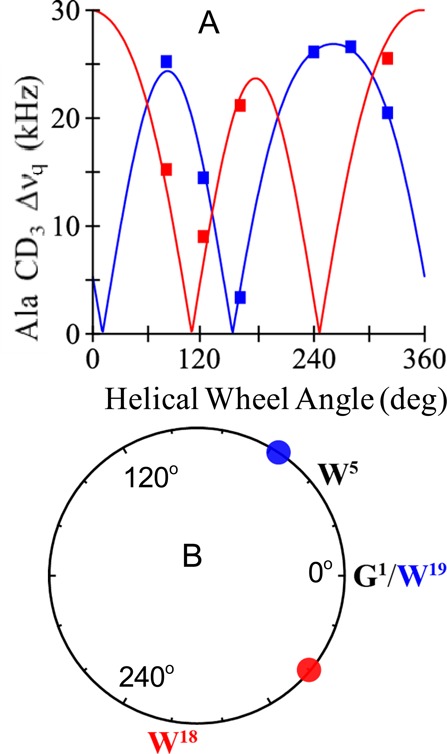
(A) Quadrupolar wave plots for W18GWALP23 (red) and GWALP23 itself (blue) in DLPC. (B) Helical wheel diagram to illustrate the relative azimuthal rotation ρ for W18GWALP23 (red circle) in relation to (W19)GWALP23 (blue circle) in DLPC, offset by ∼100°. The labels W18 and W19 represent the respective radial locations of the tryptophans, which differ by 100° on the helical wheel.
Recently,23 a method was presented for correlating helix rotational preference with flanking residue positions on a tilted transmembrane helix. We have applied this method to W18GWALP23 (Figure 7) as well as to the parent GWALP23 helix (Figure 8). These peptides are sequence isomers that differ only with respect a substitution of W18L19 in place of L18W19; yet, their rotational preferences are remarkably different. A flanking residue analysis suggests that the rotation of W18GWALP23 is governed not by the W18-to-W5 distance (along the bilayer normal) but perhaps by the L20-to-L4 distance (Figure 7). Indeed, the tilted W18GWALP23 helix rotates about its axis to give approximately the minimum L20-to-L4 distance in DLPC (Figure 7A) and approximately the maximum L20-to-L4 distance in DOPC (Figure 7B). Notably, the tilt angle for W18GWALP23 appears to be essentially the same in DOPC as in DLPC (Table 3, Figure 7), with the main adaption seeming to involve the helix rotation. We will revisit these issues using a modified Gaussian analysis (see Discussion).
Figure 7.
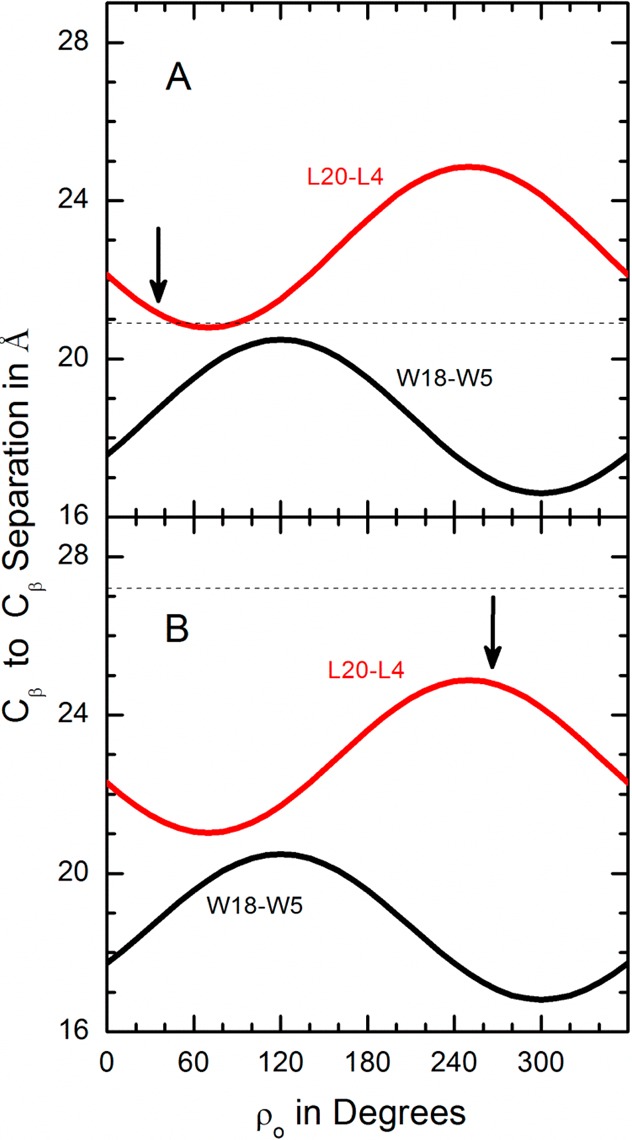
Aromatic (W18–W5) and hydrophobic (L20–L4) residue Cβ separation distances in angstroms along the bilayer normal as functions of rotation of W18GWALP23 about its tilted helix axis (A) when τo = 18° in DLPC or (B) when τo = 17° in DOPC. The preferred ρo values are shown by the arrows. The respective bilayer thicknesses are indicated by the dashed segments.
Figure 8.
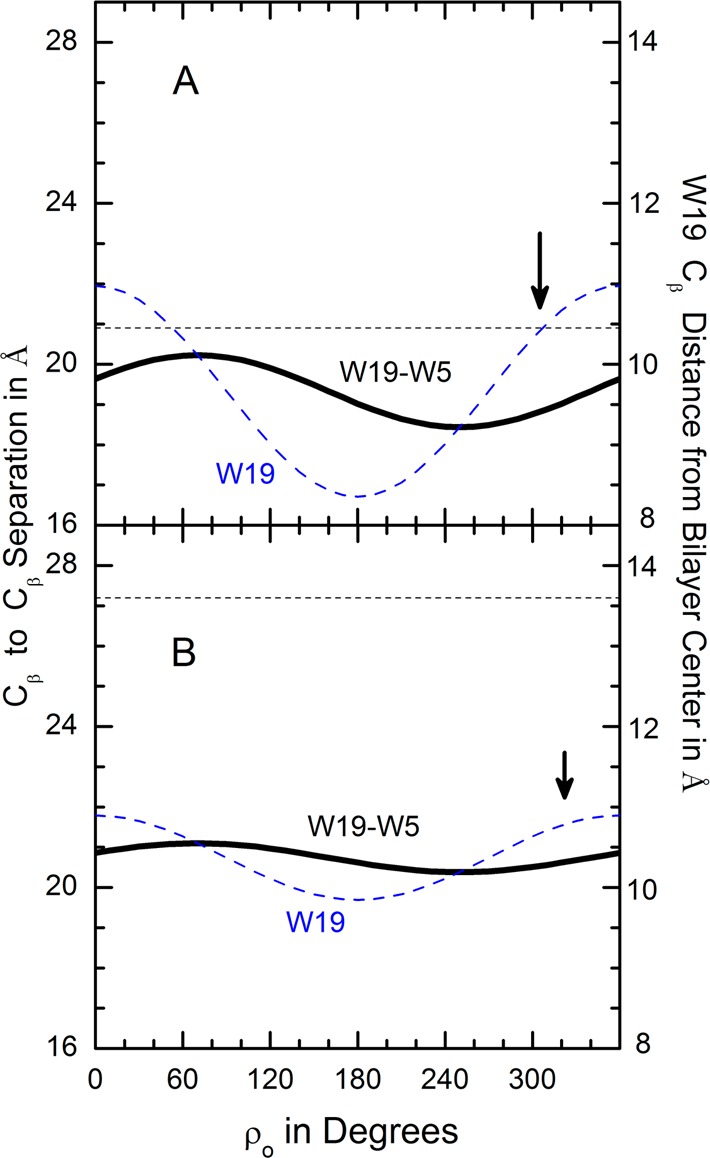
Aromatic residue Cβ separation distance along the bilayer normal and W19 Cβ distance from bilayer center as functions of rotation of GWALP23 about its tilted helix axis (A) when τo = 23° in DLPC or (B) when τo = 9° in DOPC. The preferred ρo values are shown by the arrows. The respective bilayer thicknesses are indicated by the dashed segments.
However, the rotation of GWALP23 (Figure 8) seems not to correlate with either the W19-to-W5 distance or the L20-to-L4 distance. Indeed, we surmise that W19 itself may play a special role. Being near the C-terminal, the indole side chain of W19 must adopt non-standard torsion angles27,28 in order to aim its NH bond toward the aqueous solution. Furthermore, the helix properties are more sensitive to substitution of W19 by Tyr than to substitution of W5 by Tyr,26 again suggesting a special role for the W19 indole. Within this context, the tilted GWALP23 helix in DLPC is rotated so as to provide an (perhaps fortuitous) exact match between the W19 Cβ distance from the bilayer center and half of the DLPC bilayer’s hydrophobic thickness (Figure 8A). In the thicker DOPC, the GWALP23 helix is less tilted such that the W19 Cβ position, with respect to the bilayer center, varies less steeply with rotation (Figure 8B). There is a detectable, yet minor, change in ρo, so as to move the W19 side chain outward, but only slightly, with the main adaption being the smaller helix tilt in the thicker lipid bilayer. In contrast, W18GWALP23 adapts to the changing lipid environment more by changing its helix rotation than by changing its tilt (Figure 7).
Discussion
Recent reports have compared the influence of Tyr versus Trp in the N-flanking position 517 and the C-flanking position 19 (26) in membrane-spanning GWALP23 peptides. The main findings were that either Tyr or Trp at each location will support a similar tilted transmembrane helix orientation, with only about a 10 relative azimuthal rotation about the helix axis when Tyr is substituted for Trp. Furthermore, the extent of dynamic averaging of solid-state NMR observables remains low as long as only one interfacial aromatic residue is present at each end of the membrane-spanning GWALP23 helix (with no extra aromatic residues potentially competing for lipid headgroup interactions).
In the present work, we compare the results for Trp/Tyr with the consequences of Phe substitutions at positions 4 and 5 in GWALP23. Additionally, we examine the consequences of moving a single aromatic side chain, either Y5 or W19, 100° around the helix axis by interchanging either Y5 and L4 or W19 and L18. Key findings are (a) an unexpectedly low extent of dynamic averaging for F4,5GWALP23 and (b) a preference of azimuthal rotation of W18GWALP23 that depends upon the identity (thickness) of the host lipid bilayer membrane. We will discuss first the results with the tyrosine-to-phenylalanine substitutions. Because of the differences in dynamics, we will also present the outcomes for a modified Gaussian treatment of the helix rotational dynamic averaging. Then, we will discuss the changes in rotational preference when aromatic and leucine residue locations are switched on the N-flanking side or the C-flanking side of the core helix.
Changes Involving Y5 → F5 or Y4,Y5 → F4,F5
A significant question concerns the hydrogen-bonding property of the OH group on the Y5 ring, which could interact favorably, albeit transiently (under conditions of dynamic hydrogen-bond exchange), with lipid heads groups and interfacial water molecules. The potential for hydrogen bonding is nevertheless absent when the F5 phenyl ring is substituted for Y5. With F5GWALP23, we find that hydrogen bonding by the N-flanking aromatic ring is not necessarily needed to define a preferred, stable transmembrane orientation, with limited dynamic averaging, for the core (LA)6L transmembrane helix. In this context, the indole ring of W19 seems to have special importance for defining the orientation and low dynamics (see below) and should not be overlooked or underestimated. Notably, F5GWALP23 exhibits a similar transmembrane orientation as that of Y5GWALP23 and W5GWALP23 (Figure 4 and Table 3), with low dynamic averaging. Furthermore, F5GWALP23, as well as its Y5 and W5 cousins, adapts to changes in the lipid bilayer thickness mainly by changing its tilt (Figure 4 and Table 3), with little change of the helix azimuthal rotation. The slightly smaller amplitude of the quadrupolar wave for F5GWALP23 in DLPC (Figure 4) nevertheless suggests increased dynamic averaging, which indeed is borne out by the lower estimate of 0.58 for Szz for F5GWALP23 in DLPC (Table 3). This order parameter is notably low compared to those observed for gramicidin A29 and Y5GWALP23 or GWALP23 itself (Table 3). Moreover, we note the uniformly lower estimates for Szz from the semistatic GALA analysis of each peptide helix in DLPC compared to DOPC (Table 3).
The change from Y4,5GWALP23 to F4,5GWALP23 brings big changes to the properties of the transmembrane helix, mainly to reduce the very extensive dynamic averaging that occurs when Y4 and Y5 are present together. It has been noted that Y4,5GWALP23 exhibits dramatically more dynamic averaging than is the case when only one tyrosine is present in Y5GWALP23.17 What happens when the hydrogen-bonding ability is removed from the Y4 and Y5 aromatic rings? Remarkably, the dynamic averaging is seen to diminish when F4 and F5 are present (compare Figure 4 to Figure 6B in ref (17)). We surmise that the phenyl rings of F4 and F5 are favorably placed if they are just “in the neighborhood” of the interfacial region, without need of any specific interactions with the lipid head groups. Phenol rings Y4 and Y5, on the other hand, may compete, perhaps alternately, for direct hydrogen-bonding interactions with lipid head groups. Such competition, if not able to optimize simultaneously a Y4 interaction and a Y5 interaction, could lead to increased helix rotational dynamics of the type suggested previously.11,13 Importantly, neither Y4 nor Y5 alone causes the extensive dynamic averaging (see below); rather, it is the pair of tyrosines together that leads to the high level of dynamics, as has been noted also for pairs of tryptophans.12
Modified Gaussian Treatment of the Dynamics
We sought to compare a semistatic treatment with a modified Gaussian treatment of the dynamic properties of each of the transmembrane peptides. The semistatic GALA treatment7,8 employs three adjustable parameters: tilt, τo, and azimuthal rotation, ρo, to describe helix orientation, together with a principal order parameter, Szz, to provide a highly abbreviated estimate of overall dynamics. A Gaussian treatment offers advantages of describing estimates for helix “wobble” στ and rotational “slippage” σρ, but it does so at the expense of requiring four adjustable parameters instead of three.13 As an alternative, a modified Gaussian treatment has been suggested,16 in which στ is held constant and dynamic differences are embodied in an adjustable σρ. We have employed this modified procedure16 but differing in that we set στ to a small finite value instead of to zero while maintaining Szz of 0.88 as an estimate of internal motion of the peptide.14
Notably, the results of such a modified Gaussian analysis (Table 4) agree substantially with the conclusions from a semistatic GALA analysis (Table 3). Both methods indicate that each peptide (except possibly W18GWALP23; see below) adapts from DOPC to DLPC by increasing its tilt. The methods in all cases give the same preferred values of ρo in each lipid and are in agreement concerning the small variations when the identity of aromatic residue 5 is changed. Additionally, the same trends for in ρo are seen with either method when a peptide helix is moved from DOPC to DLPC. Generally, the change of lipid involves only a small Δρo, excepting the case of W18GWALP23 (see below) and the highly dynamic Y4,5GWALP23, noted previously.17
Table 4. Modified Gaussian Analysis of Orientations and Dynamics for Peptides of the GWALP23 Familya,b.
| DLPC |
DOPC |
|||||||
|---|---|---|---|---|---|---|---|---|
| peptide | τ0 | ρ0 | σρ | RMSD (kHz) | τ0 | ρ0 | σρ | RMSD (kHz) |
| W5 | 23 | 304 | 33 | 0.7 | 9 | 321 | 48 | 0.7 |
| Y5 | 23 | 295 | 32 | 0.0 | 6 | 313 | 34 | 1.0 |
| F5 | 17 | 314 | 21 | 1.7c | 9 | 329 | 40 | 0.6 |
| Y4,5 | 14 | 259 | >90d | 1.7 | 6 | 344 | 72 | 0.9e |
| F4,5 | 18 | 314 | 0f | 0.7 | 10 | 329 | 54 | 1.6 |
| Y4 | 11 | 328 | 22 | 0.9 | 11 | 353 | 56 | 0.4 |
| W5W18 | 18 | 35 | 8f | 1.4 | 17 | 264 | 50 | 1.2 |
The N-flanking aromatic residues are indicated by the abbreviation for each peptide. C-flanking W19 is also present in all samples except when the aromatic residues are W5 and W18.
Analysis followed Strandberg et al.,16 but στ was assigned a finite value instead of 0° (see Methods). Except as noted, στ was set to 15° in DLPC or 9° in DOPC.
The value of στ was 20° because no satisfactory solution was found when στ = 15°.
The value of σρ remains > 90° even if στ is set to 20°.
The value of στ was 13° because no satisfactory solution was found when στ = 9°.
The value of σρ is perhaps artificially low because of the choice of στ. Because of the limited data set, further solutions were not explored.
The respective outcomes for fitting the dynamics also are similar. The semistatic analysis (Table 3) leads to estimates for Szz that are higher in DOPC than in DLPC, seemingly because the relatively smaller tilt angles in DOPC do not demand a downgrade of Szz during the fitting process. For the modified Gaussian analysis, we set στ to 15° in DLPC (where τo is larger) and to 9° in DOPC (where τo is smaller). (When presented with a binary choice of either 9 or 15°, the analytical software selected 15° for each peptide in DLPC but 9° for each peptide in DOPC; the values subsequently were fixed.) With these constraints, the resulting fitted values for σρ are similar between DLPC and DOPC (Table 4), with somewhat larger values of σρ emerging for the fits in DOPC (perhaps reflecting the somewhat smaller choice for στ). For the highly dynamic Y4,5GWALP23, σρ is seen to be very large in both DLPC and DOPC, thereby confirming earlier conclusions based on semistatic analysis.17 With a caveat that the helix behavior should be examined in more than one lipid, similar conclusions about the extent of dynamic averaging (whether limited, moderate, or high) for each transmembrane helix emerge from the semistatic GALA and modified Gaussian analyses. It is useful and productive to apply and compare both methods.
Changes in Rotational Preference when Y5 Is Moved or W18 Is Moved
We observed sizable changes in the preference for helix azimuthal rotation when a key aromatic ring, the side chain of either residue 5 or residue 19, has been moved by 100° around the helix axis (Tables 3 and 4). In this regard, the Trp residue, W19, seems once again to have a special importance, as the changes to the rotational preference are larger and lipid-dependent when this tryptophan is moved.
When Y4 is moved to Y5, the preferred ρo changes by about 30–35° in DLPC and by about 40–45° in DOPC. Conspicuously, the deduced values of Δρo are similar between the semistatic and modified Gaussian methods of analysis (Tables 3 and 4). The rotational change furthermore is slightly less than half of the 100° radial shift that accompanies the move of Tyr from position 5 to position 4. The results suggest that a compromise may be imposed by the interplay of W19 (fixed) and either Y4 or Y5 (variable), with the influence of W19 on the rotational preference nevertheless dominating to a small extent over that of Y4 or Y5 (because Δρo < 50° in both lipids). We recall also that the identity of residue 5 exerts a smaller influence on the azimuthal rotation.17 Interestingly, ρo decreases ∼10° when W5 is changed to Y5, but it increases ∼10° when W5 is changed to F5; the molecular reasons that may underlie these small but opposite values of Δρo are elusive at this time.
The movement of W19 to position 18 has major influence on the helix azimuthal rotation. Interestingly, an interchange of W19 and L18 leads to a change in ρo of +90° in DLPC but −60° in DOPC in spite of the fact that σρ remains low in DLPC and moderate in DOPC (Tables 3 and 4). These results once again highlight the major importance of W19, or more generally of the C-flanking aromatic residue, for the helix azimuthal rotation. Moreover, the semistatic and modified Gaussian methods agree on not only the absolute magnitudes but also the extent of change in both lipids (Tables 3 and 4). Why does the preferred rotation of W18GWALP23, but not of the other peptides that experience low dynamic averaging, depend upon the host lipid bilayer? While the possible answers to this question remain incomplete at this time, some clues may emerge from consideration of Figures 7 and 8. With W18 and W5 separated by 140° on the helical wheel (Figures 1 and 6), the transmembrane helix azimuthal rotation seems not to correlate with the W5-to-W18 separation distance along the bilayer normal (Figure 7) but rather with the L4-to-L20 distance, which could better describe the longitudinal hydrophobic length of the helix. Indeed, W18GWALP23 seemingly rotates so as to minimize the L4-to-L20 transmembrane distance in DLPC and to maximize the L4-to-L20 transmembrane distance in DOPC (Figure 7). While more examples would be needed to establish a causal relationship instead of a mere correlation, this principle could help to explain the lipid-dependent helix rotation for the special case of the W18GWALP23 helix.
The particular importance of W19 for the transmembrane orientation of GWALP23 is evident in Figure 8. Notably, in GWALP23, the two Trp residues are on the same side of a helical wheel, with a radial displacement of only 40° (Figures 1 and 5). Rather than any dependence on the W5-to-W19 longitudinal separation, GWALP23 rotates in DLPC so that the distance from Cβ of W19 to the bilayer center matches the hydrophobic thickness of a DLPC monolayer (Figure 8A). From DLPC to DOPC, the main adjustment is that GWALP23 is less tilted in DOPC. The rather minimal rotational adjustment serves to move W19 slightly farther away from the DOPC bilayer center (Figure 8B).
Concluding Remarks
For the model membrane-spanning helix of acetyl-GGALW(LA)6LWLAGA-amide (GWALP23), a well-defined transmembrane orientation with only limited dynamic averaging, other than long-axis precession about the bilayer normal,9 is retained following the single aromatic residue substitution of F5 or Y5 in place of W5. By contrast, all known model transmembrane peptide helices having more than two interfacial Trp or Tyr residues characterized to date exhibit extensive dynamic averaging of the solid-state NMR observables. Remarkably, nevertheless, the peptide F4,5GW19ALP23 fits into the category of low dynamics and not high dynamics, perhaps because of the absence of hydrogen-bonding ability of the phenyl rings. Y5 confers similar properties as W5. When the single Y5 is moved to Y4, within the context where W19 is held constant to flank the other end of the core helix, there ensues a change of about 40° in the preferred helix azimuthal rotation regardless of whether the host lipid membrane is DLPC or DOPC. By contrast, when the single W19 is moved to W18, within the context where W5 is held constant to flank the other end of the core helix, the preferred helix azimuthal rotation is seen to vary greatly, by ∼130°, between DLPC and DOPC. The results suggest that a dynamic interplay between lipid membrane thickness and protein helix rotation may regulate aspects of biological function.
Acknowledgments
We thank Vitaly Vostrikov for software for semistatic GALA and modified Gaussian methods for analysis of helix orientations and dynamics.
Glossary
Abbreviations Used
- CD
circular dichroism
- DLPC
1,2-dilauroylphosphatidylcholine
- DMPC
1,2-dimyristoylphosphatidylcholine
- DOPC
1,2-dioleoylphosphatidylcholine
- Fmoc
fluorenylmethoxycarbonyl
- GALA
geometric analysis of labeled alanines
- GWALP23
acetyl-GGALW(LA)6LWLAGA-[ethanol]amide
- MtBE
methyl-t-butyl ether
- RMSD
root mean squared deviation
- TFA
trifluoroacetic acid
- WWALP23
acetyl-GWALW(LA)6LWLAWA-[ethanol]amide
Supporting Information Available
2H NMR, 31P NMR, and mass spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported in part by NSF MCB grant 1327611 and by the Arkansas Biosciences Institute. The peptide, NMR, and mass spectrometry facilities are supported by NIH grants GM103429 and GM103450.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- O’Connell A. M.; Koeppe R. E. II; Andersen O. S. (1990) Kinetics of gramicidin channel formation in lipid bilayers: transmembrane monomer association. Science 250, 1256–1259. [DOI] [PubMed] [Google Scholar]
- Schiffer M.; Chang C. H.; Stevens F. J. (1992) The functions of tryptophan residues in membrane proteins. Protein Eng. 5, 213–214. [DOI] [PubMed] [Google Scholar]
- Landolt-Marticorena C.; Williams K. A.; Deber C. M.; Reithmeier R. A. (1993) Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J. Mol. Biol. 229, 602–608. [DOI] [PubMed] [Google Scholar]
- Killian J. A.; Salemink I.; de Planque M. R.; Lindblom G.; Koeppe R. E. II; Greathouse D. V. (1996) Induction of nonbilayer structures in diacylphosphatidylcholine model membranes by transmembrane alpha-helical peptides: importance of hydrophobic mismatch and proposed role of tryptophans. Biochemistry 35, 1037–1045. [DOI] [PubMed] [Google Scholar]
- de Planque M. R.; Kruijtzer J. A.; Liskamp R. M.; Marsh D.; Greathouse D. V.; Koeppe R. E. II; de Kruijff B.; Killian J. A. (1999) Different membrane anchoring positions of tryptophan and lysine in synthetic transmembrane alpha-helical peptides. J. Biol. Chem. 274, 20839–20846. [DOI] [PubMed] [Google Scholar]
- de Planque M. R.; Boots J. W.; Rijkers D. T.; Liskamp R. M.; Greathouse D. V.; Killian J. A. (2002) The effects of hydrophobic mismatch between phosphatidylcholine bilayers and transmembrane alpha-helical peptides depend on the nature of interfacially exposed aromatic and charged residues. Biochemistry 41, 8396–8404. [DOI] [PubMed] [Google Scholar]
- van der Wel P. C.; Strandberg E.; Killian J. A.; Koeppe R. E. II. (2002) Geometry and intrinsic tilt of a tryptophan-anchored transmembrane alpha-helix determined by 2H NMR. Biophys. J. 83, 1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandberg E.; Ozdirekcan S.; Rijkers D. T.; van der Wel P. C.; Koeppe R. E. II; Liskamp R. M.; Killian J. A. (2004) Tilt angles of transmembrane model peptides in oriented and non-oriented lipid bilayers as determined by 2H solid-state NMR. Biophys. J. 86, 3709–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; Im W. (2008) Transmembrane helix tilting: Insights from calculating the potential of mean force. Phys. Rev. Lett. 100, 018103. [DOI] [PubMed] [Google Scholar]
- Özdirekcan S.; Etchebest C.; Killian J. A.; Fuchs P. F. J. (2007) On the orientation of a designed transmembrane peptide: toward the right tilt angle?. J. Am. Chem. Soc. 129, 15174–15181. [DOI] [PubMed] [Google Scholar]
- Esteban-Martín S.; Salgado J. (2007) The dynamic orientation of membrane-bound peptides: bridging simulations and experiments. Biophys. J. 93, 4278–4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostrikov V. V.; Daily A. E.; Greathouse D. V.; Koeppe R. E. II. (2010) Charged or aromatic anchor residue dependence of transmembrane peptide tilt. J. Biol. Chem. 285, 31723–31730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandberg E.; Esteban-Martin S.; Salgado J.; Ulrich A. S. (2009) Orientation and dynamics of peptides in membranes calculated from 2H-NMR data. Biophys. J. 96, 3223–3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostrikov V. V.; Grant C. V.; Opella S. J.; Koeppe R. E. II. (2011) On the combined analysis of 2H and 15N/1H solid-state NMR data for determination of transmembrane peptide orientation and dynamics. Biophys. J. 101, 2939–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostrikov V. V.; Grant C. V.; Daily A. E.; Opella S. J.; Koeppe R. E. II. (2008) Comparison of “polarization inversion with spin exchange at magic angle” and “geometric analysis of labeled alanines” methods for transmembrane helix alignment. J. Am. Chem. Soc. 130, 12584–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandberg E.; Esteban-Martin S.; Ulrich A. S.; Salgado J. (2012) Hydrophobic mismatch of mobile transmembrane helices: Merging theory and experiments. Biochim. Biophys. Acta 1818, 1242–1249. [DOI] [PubMed] [Google Scholar]
- Gleason N. J.; Vostrikov V. V.; Greathouse D. V.; Grant C. V.; Opella S. J.; Koeppe R. E. II (2012) Tyrosine replacing tryptophan as an anchor in GWALP peptides. Biochemistry 51, 2044–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostrikov V. V.; Hall B. A.; Greathouse D. V.; Koeppe R. E. II; Sansom M. S. P. (2010) Changes in transmembrane helix alignment by arginine residues revealed by solid-state NMR experiments and coarse-grained MD simulations. J. Am. Chem. Soc. 132, 5803–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostrikov V. V.; Hall B. A.; Sansom M. S. P.; Koeppe R. E. II. (2012) Accommodation of a central arginine in a transmembrane peptide by changing the placement of anchor residues. J. Phys. Chem. B 116, 12980–12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason N. J.; Vostrikov V. V.; Greathouse D. V.; Koeppe R. E. II. (2013) Buried lysine, but not arginine, titrates and alters transmembrane helix tilt. Proc. Natl. Acad. Sci. U.S.A. 110, 1692–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel D. P.; Cherezov V.; Greathouse D. V.; Koeppe R. E. II; Killian J. A.; Caffrey M. (2006) Transmembrane peptides stabilize inverted cubic phases in a biphasic length-dependent manner: Implications for protein-induced membrane fusion. Biophys. J. 90, 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J. H.; Jeffrey K. R.; Bloom M.; Valic M. I.; Higgs T. P. (1976) Quadrupolar echo deuteron magnetic resonance spectroscopy in ordered hydrocarbon chains. Chem. Phys. Lett. 42, 390–394. [Google Scholar]
- Sánchez-Munoz O. L.; Strandberg E.; Esteban-Martín E.; Grage S. L.; Ulrich A. S.; Salgado J. (2013) Canonical azimuthal rotations and flanking residues constrain the orientation of transmembrane helices. Biophys. J. 104, 1508–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kučerka N.; Liu Y.; Chu N.; Petrache H. I.; Tristram-Nagle S.; Nagle J. F. (2005) Structure of fully hydrated fluid phase DMPC and DLPC lipid bilayers using X-ray scattering from oriented multilamellar arrays and from unilamellar vesicles. Biophys. J. 88, 2626–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Nagle J. F. (2004) Diffuse scattering provides material parameters and electron density profiles of biomembranes. Phys. Rev. E 69, 040901 (R). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason N. J.; Greathouse D. V.; Grant C. V.; Opella S. J.; Koeppe R. E. II. (2013) Single tryptophan and tyrosine comparisons in the N-terminal and C-terminal interface regions of transmembrane GWALP peptides. J. Phys. Chem. B 117, 13786–13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Wel P. C.; Reed N. D.; Greathouse D. V.; Koeppe R. E. II. (2007) Orientation and motion of tryptophan interfacial anchors in membrane-spanning peptides. Biochemistry 46, 7514–7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostrikov V. V.; Koeppe R. E. II. (2011) Response of GWALP transmembrane peptides to changes in the tryptophan anchor positions. Biochemistry 50, 7522–7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Separovic F.; Pax R.; Cornell B. (1993) NMR order parameter analysis of a peptide plane aligned in a lyotropic liquid crystal. Mol. Phys. 78, 357–369. [Google Scholar]
- DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.