Abstract
Nontypeable Haemophilus influenzae (NTHi) frequently colonize the human pharynx asymptomatically, and are an important cause of otitis media in children. Past studies have identified typeable H. influenzae as being clonal, but the population structure of NTHi has not been extensively characterized. The research presented here investigated the diversity and population structure in a well-characterized collection of NTHi isolated from the middle ears of children with otitis media or the pharynges of healthy children in three disparate geographic regions. Multilocus sequence typing identified 109 unique sequence types among 170 commensal and otitis media-associated NTHi isolates from Finland, Israel, and the US. The largest clonal complex contained only five sequence types, indicating a high level of genetic diversity. The eBURST v3, ClonalFrame 1.1, and structure 2.3.3 programs were used to further characterize diversity and population structure from the sequence typing data. Little clustering was apparent by either disease state (otitis media or commensalism) or geography in the ClonalFrame phylogeny. Population structure was clearly evident, with support for eight populations when all 170 isolates were analyzed. Interestingly, one population contained only commensal isolates, while two others consisted solely of otitis media isolates, suggesting associations between population structure and disease.
Keywords: Nontypeable Haemophilus influenzae, NTHi, Population structure, Otitis media, Multilocus sequence typing, MLST
1. Introduction
Haemophilus influenzae (Hi) are small, nonmotile, gram negative coccobacilli whose only natural hosts are humans (Kilian, 2005). The species can be divided into two major groups differentiated by the presence or absence of a polysaccharide capsule. Non-encapsulated (nontypeable, or NTHi) strains are frequent asymptomatic colonizers of the human pharynx, particularly in children, but can also cause a variety of respiratory infections, including otitis media, sinusitis, bronchitis, and pneumonia. The carriage rate of NTHi among healthy children varies between 25–81%; this wide distribution may be due to a number of factors, including proximity to other children (i.e. daycare centers), amount of antibiotic use, and exposure to secondhand smoke (Bou et al., 2000; Farjo et al., 2004; St Sauver et al., 2000). Furthermore, NTHi colonization is an active, dynamic process. A number of studies have shown that carriage is often marked by apparent rapid turnover of strains as well as simultaneous colonization with multiple NTHI strains (Dhooge et al., 2000; Murphy et al., 1999; Trottier et al., 1989). Previous data gathered in our laboratory support these findings (Farjo et al., 2004; LaCross et al., 2008; St Sauver et al., 2000).
While the population structure of H. influenzae has been investigated in previous studies, most of the literature dates back several decades and principally used typeable strains. In 1985, Musser and colleagues characterized 177 type b H. influenzae (Hib) isolates recovered from children with invasive disease by multilocus enzyme electrophoresis (MLEE) and determined that the sample exhibited significant clonality and major differences in the genetic structure of populations from the United States (US) and the Netherlands (Musser et al., 1985). Similarly, a study of over 2,200 typeable isolates from 30 countries was characterized by MLEE and, again, found that the population structure overall was clonal with strong patterns of geographic variation and a limited number of evolutionary lineages that largely corresponded to serotype (Musser et al., 1990; Musser et al., 1988a). For example, Hib isolates of electrophoretic type 100 comprised 4.5% of their Canadian sample, but this genotype was not found among isolates from the US. However, like many studies of clinically significant microorganisms, the vast majority of isolates were collected from cases of disease; of the 2,209 isolates in the study, nearly 90% were serotype b, and less than 5% of these were obtained from healthy carriers.
Interestingly, the population structure of H. influenzae appears to differ between typeable and non-typeable strains. In a 1985 study of 242 Hi disease isolates (65 nontypeable and 177 type b), Musser et al. found that each nontypeable (NTHi) isolate was of a unique electrophoretic type and that none of these were shared with a Hib isolate, indicating that the population of NTHi is extremely heterogeneous and distinct from that of typeable strains (Musser et al., 1986). Porras et al. used MLEE to characterize 135 Hi isolates from Sweden and the US, of which 81 were nontypeable. Seventy electrophoretic types were identified among the 81 NTHi isolates, and no electrophoretic type was shared between the geographic regions (Porras et al., 1986). While five electrophoretic types were found in both nontypeable and type b isolates, their version of MLEE assayed only six enzymes compared to the 15 used by Musser et al. (Musser et al., 1986), drastically reducing the discriminatory power of their method. More recently, Erwin et al. examined the population structure of all 656 Hi isolates in the MLST database circa 2006, including 322 NTHi isolates, based on a maximum-parsimony analysis (Erwin et al., 2008). However, as the authors noted, there may have been sampling bias, as nearly 90% of the NTHi in the MLST database were isolated from patients with symptomatic infections. Furthermore, NTHi submitted to the MLST database come from many different researchers and do not conform to any specific sampling scheme. Nevertheless, they were able to identify well defined phylogenetic groups of NTHi that differed in genetic content, though there was little apparent clustering by geography or clinical site of isolation.
Given the known genetic diversity of H. influenzae (Erwin et al., 2005; Erwin et al., 2008; LaCross et al., 2008; Musser et al., 1986), this study sought to explore the phylogenetic relationships and population structure of nontypeable strains. While phylogenies are used to infer evolutionary history, assessing population structure can reveal systematic differences in allele frequencies between subpopulations (which can be due to differences in ancestry). Furthermore, geography and disease status (i.e. commensal or otitis media-associated) were investigated as potential factors involved in the formation of population structure. A diverse collection of 170 commensal and otitis media-associated isolates from three disparate geographic regions were genotyped by MLST. Genetic diversity and phylogenetic relationships between the isolates were assessed using eBURST and the ClonalFrame program, while population structure was characterized using structure.
2. Materials and Methods
2.1. Bacterial Isolates
An initial set of 199 putative NTHi isolates was selected from existing collections representing three distinct geographic regions (Finland, Israel, and the US). Within each geographic region, half of the isolates had been collected from the middle ears of children with acute otitis media (hereafter designated ‘OM isolates’) and the remaining half had been collected from the throats or nasopharynges of healthy children (commensal isolates). This yielded six subgroups of isolates, the majority of which have been previously described in the literature: Finland OM (Kilpi et al., 2001); Finland commensal (Ukkonen et al., 2000); Israel OM (Leibovitz et al., 2003); Israel commensal (Greenberg et al., 2004); US OM (Krasan et al., 1999), as well as unpublished isolates from Dr. Stan Block and Dr. Alejandro Hoberman; and US commensal (Farjo et al., 2004; St Sauver et al., 2000). One hundred and twenty total isolates from Finland and Israel and 79 isolates from the US were randomly selected from within each subgroup for inclusion in the study. Only a single isolate was selected from each child, and all isolates were collected within an eight year period (1994 – 2002) from children under seven years of age (Table 1). The isolates were frozen in sterile skim milk at −80° C for storage.
Table 1.
Characteristics of H. influenzae isolate collections from Finland, Israel, and the US
| Isolation site | # in Initial Collectiona |
Typeable | Non- Hib |
Contam.c | Total Removed |
# in Final database |
Isolation Date |
Age in monthsd |
|---|---|---|---|---|---|---|---|---|
| Finland OM | 30 | 0 | 0 | 0 | 0 | 30 | 1994–96 | 2 – 24 |
| Finland Commensal | 30 | 4 | 0 | 0 | 4 | 26 | 1999 | 10 – 24 |
| Israel OM | 30 | 1 | 0 | 1 | 2 | 28 | 2000–01 | 6 – 48 |
| Israel Commensal | 30 | 3 | 2 | 3 | 8 | 22 | 2001–02 | 1 – 59 |
| US OM | 38 | 0 | 1 | 0 | 1 | 37 | 1996–2001 | 7 – 84 |
| US Commensal | 41 | 0 | 14 | 0 | 14 | 27 | 1998–2001 | < 36 |
| Total OM | 98 | 1 | 1 | 1 | 3 | 95 | 1994–2001 | 2 – 84 |
| Total Commensal | 101 | 7 | 16 | 3 | 26 | 75 | 1998–2002 | 1 – 59 |
Number of isolates randomly selected from the collection.
Number of selected isolates designated as non-H. influenzae.
Number of selected isolates with unresolvable contamination.
Age range in months of the children from whom the isolates were collected.
2.2. Preparation of Genomic DNA
The stored isolates were grown overnight on chocolate agar plates (BD Diagnostics, Sparks, MD) at 37°C with 5% CO2 in a humid environment. Genomic DNA was isolated using the Wizard genomic DNA purification kit (Promega, Madison, WI.) according to the manufacturer’s instructions and resuspended in 1× Tris-EDTA buffer (10 mM Tris-HCL and 1mM EDTA at pH 8). The majority of the DNA was kept at −20°C for storage, with a small aliquot stored at 4°C for use.
2.3. Isolate Speciation and Exclusion Criteria
Isolates were excluded from the final dataset if they met any of the following criteria: presence of the capsule locus genes, identification as non-Hi by phylogenetic clustering, or persistent and unresolvable contamination (e.g. superimposed peaks in the sequence chromatograms not resolvable by multiple re-isolations of genomic DNA from single colonies).
Typeable isolates were identified by the presence of a PCR amplification product reflecting the highly conserved bexA and bexB genes, which are required for transport of capsule components across the outer membrane, following the protocol of Davis et al. (Giebink, 1999). Among bexA and/or bexB positive isolates, the capsule type was identified by PCR of the type specific regions of the cap locus using the method of Falla et al. (Falla et al., 1994).
To identify isolates misidentified in the original collection as non-Hi, a phylogeny based on six of the seven MLST loci (excluding fucK) was created in ClonalFrame using the conditions described below, with the exception that 200,000 total iterations (100,000 burnin iterations and 100,000 sampling iterations) were performed. Three non-Hi strains from the Pasteurellaceae family were included: H. haemolyticus strain HK386 (Norskov-Lauritsen et al., 2005), H. parainfluenzae strain T3T1 (GenBank ID FQ312002.1), and Pasteurella multocida strain Pm70 (May et al., 2001). Isolates that clustered with the non-Hi strains were excluded from further analysis.
In general, strains closely related to, but not strictly, Hi are negative for the MLST locus fucK, and this difference has been exploited to distinguish between the two groups (Norskov-Lauritsen, 2009; Norskov-Lauritsen et al., 2009). However, a recent paper by Ridderberg et al. has described at least one apparent H. influenzae strain in which the entire six gene fucose operon (containing fucK) is missing by PCR analysis, indicating that this absence is not a reliable indicator of species identity (Ridderberg et al., 2010). In the present study, the procedure described by Ridderberg was used to determine the presence or absence of the fucose operon in every isolate in which reliable amplification of fucK could not be achieved. Briefly, PCR was conducted with primers flanking the fucose operon using LongAmp Taq DNA polymerase (NEB, Ipswitch, MA), and the products were visualized by agarose gel electrophoresis stained with ethidium bromide. Isolates containing the operon yielded amplicons of approximately 10 kb, while isolates lacking the operon had amplicons of approximately 2 kb.
A total of 29 isolates met the exclusionary criteria, leaving 170 NTHi isolates in the final dataset.
2.4. MLST and eBURST
MLST was used to genotype the NTHi isolates following the protocol of Meats et al. (Meats et al., 2003). The eBURST v3 program was used to determine the relationships of the MLST sequences from the final dataset as described previously (Feil et al., 2004).
2.5. Phylogenetic Analysis
ClonalFrame 1.1 (Didelot and Falush, 2007) was used to construct phylogenies based on MLST gene sequences. Two independent ClonalFrame runs of 400,000 iterations each were performed on the final dataset of 170 isolates, using default values for all options. All seven MLST loci were used, with fucK negative isolates treated as having a gap at that locus. The first 200,000 iterations were considered the burnin period and were discarded, and the remaining iterations were sampled every 100 generations to produce 2,000 topologies in the posterior sample. Convergence of the Markov Chain Monte Carlo (MCMC) was assessed by the Gelman-Rubin test (Gelman and Rubin, 1992) as implemented by ClonalFrame. A Gelman-Rubin statistic above 1.2 indicates poor convergence (Didelot and Falush, 2008); the statistics for all parameters were below this value when convergence was compared between the two runs. An unrooted majority consensus tree was constructed from the posterior sample using SplitsTree 4.11.3 (Huson and Bryant, 2006).
2.6. Population Structure
The structure 2.3.3 program was used to identify the presence of population structure based on MLST gene sequences (Pritchard et al., 2000). Two datasets were used: the first contained all isolates, while the second consisted of only one example of each unique ST found in the sample. As some isolates were missing the fucK locus, only the remaining six MLST loci were used in both datasets. Twenty replicate runs of 100,000 burnin iterations and 100,000 sampling iterations were performed for each value of the number of populations K; all were based on the admixture model with correlated allele frequencies and independent values of the Dirichlet parameter α for each assumed population K. Convergence was assessed by visually examining the parameter traces. The number of assumed populations was increased until adding a population became uninformative (i.e. the individual membership proportions (Q) for the added population were very low and/or few individuals had a large portion of their ancestry from that population). The Greedy algorithm implemented in CLUMPP 1.1.2 (Jakobsson and Rosenberg, 2007) was used to identify potential distinct modes among the 20 replicate runs for each K value. To be within the same mode, two replicate runs at a given K must have had a symmetric similarity coefficient (SSC) ≥ 0.9. The estimated individual membership proportions were then averaged among all runs within the same mode for a given value of K. Plots of structure results were produced using distruct 1.1 (Rosenberg, 2004). This method of analysis is similar to those described in earlier studies (Kopelman et al., 2009; Verdu et al., 2009; Wang et al., 2007).
3. Results
3.1. Isolate Characteristics
Eight bexA and bexB positive isolates (one type a, two type e, and five type f) were identified and removed from the final dataset. The advantage of using both bexA and bexB over traditional slide agglutination techniques using type-specific antisera or methods detecting bexA alone is that bexB PCR will detect rare strains that are bexA negative but bexB positive, which renders them phenotypically nontypeable but genetically far closer to typeable strains. A positive result for either gene indicates the presence of the cap locus and thus that the isolate is at least genetically typeable. Additionally, four isolates with unresolvable superimposed peaks in the MLST loci sequence chromatograms and six isolates identified as non-Hi in our laboratory (McCrea et al., 2008) were excluded from further analysis.
ClonalFrame 1.1 (Didelot and Falush, 2007) analysis using six of the seven MLST loci (excluding fucK) from the remaining 181 isolates expressed as an unrooted majority consensus tree is shown in Figure 1. Eleven of the 181 isolates clustered with H. haemolyticus strain HK386 and were excluded from further analyses, leaving a final dataset of 170 NTHi isolates In total, 29 of the original 199 isolates (three OM, 26 commensal) were either typeable, non-H. influenzae, or persistently contaminated and were excluded from further analysis (Table 1).
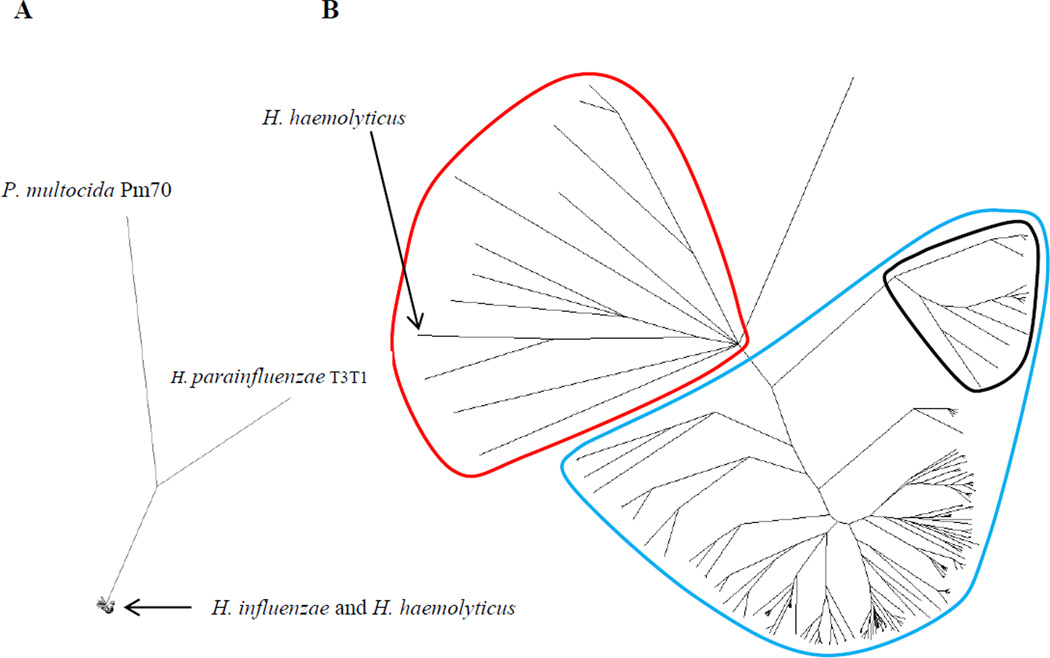
Figure 1.
Unrooted majority consensus tree of 181 Haemophilus isolates. H. haemolyticus strain HK386 (Norskov-Lauritsen et al., 2005), H. parainfluenzae strain T3T1 (GenBank ID FQ312002.1), and Pasteurella multocida strain Pm70 (May et al., 2001) were included as outgroups. A. Zoomed out view showing the relative positions of the four species. B. Zoomed in view illustrating the 11 isolates (circled in red) that cluster with H. haemolyticus HK386 compared with the remaining 170 NTHi isolates (circled in blue), as well as 14 fucK negative commensal isolates positioned between the two groups (circled in black).
3.2. MLST
The 170 NTHi isolates in the final dataset genotyped by MLST, yielded a total of 109 STs, 45 of which were found only in OM isolates, 51 only in commensal isolates, and 13 in both OM and commensal isolates. Of 53 STs previously undescribed in the MLST database, 20 were found only in OM isolates and 33 only in commensal isolates. The majority of STs (81 of 109) were found only once, and only ten of the remaining STs were found three or more times. Thirteen STs were comprised of both commensal and OM isolates. Fifteen isolates could not be amplified with the fucK primers, and were found to be missing the entire fucose operon after following the protocol of Ridderberg et al. (Ridderberg et al., 2010). Because the MLST database is currently unable to assign a sequence type to isolates missing one of the seven loci, all isolates missing fucK have been assigned placeholder STs starting at 10,000. General characteristics of the MLST genotyping are detailed in Table 2, and the specific ST assigned to each isolate is listed in Table S1.
Table 2.
General characteristics of the MLST genotyping
| Isolates |
fucK (−) Isolates |
fucK (−) STs |
Total STs |
New STs | |
|---|---|---|---|---|---|
| Finland OM | 30 | 0 | 0 | 21 | 3 |
| Finland Commensal | 26 | 0 | 0 | 24 | 6 |
| Finland Total | 56 | 0 | 0 | 45 | 9 |
| Israel OM | 28 | 0 | 0 | 22 | 13 |
| Israel Commensal | 22 | 2 | 2 | 21 | 13 |
| Israel Total | 50 | 2 | 2 | 43 | 26 |
| US OM | 37 | 1 | 1 | 25 | 4 |
| US Commensal | 27 | 12 | 9 | 24 | 14 |
| US Total | 64 | 13 | 10 | 49 | 18 |
| Total OM | 95 | 1 | 1 | 45 | 20 |
| Total Commensal | 75 | 14 | 11 | 51 | 33 |
| Total found in botha | NA | NA | 0 | 13 | 0 |
| Overall Total | 170 | 15 | 12 | 109 | 53 |
Listing of STs containing both OM and commensal isolates.
3.3. eBURST Analysis
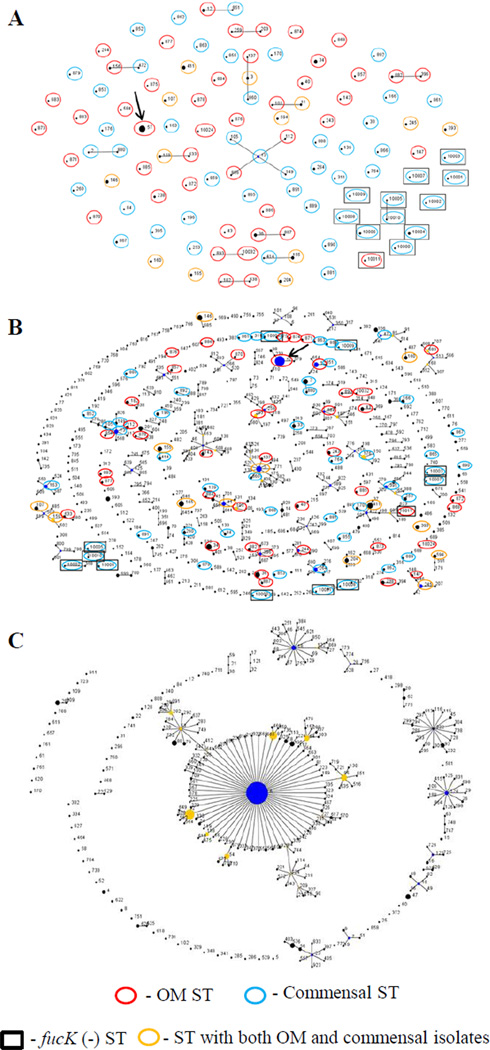
The MLST data from the final dataset of 170 NTHi isolates was analyzed with eBURST v3 (Figure 2A). Most STs are not closely enough related to another ST in the sample to form a complex and are thus unconnected, and little clustering is evident by disease (OM/commensal). The largest clonal complex consists of only five STs, and contains both OM and commensal STs. Only ten STs contain three or more isolates, and in eight of these STs the isolates are from at least two of the geographic regions. By far the most common ST is ST57, which represents 18 of the 170 NTHi isolates in the final dataset, and 28 of all the NTHi isolates in the MLST database. Intriguingly, all 18 ST57 isolates in the final dataset were collected from the middle ears of children with otitis media, and were similarly distributed among the three geographic regions (five, six, and seven isolates from Finland, Israel, and the US, respectively).
Figure 2.
eBURST analyses of H. influenzae isolates. The most conservative definition of a clonal complex was used, by which STs are included in the group only if they share alleles at a minimum of six of the seven loci with at least one other ST in that group. The size of the circles is proportional to the abundance of the corresponding STs in the data set, and the relative placement of unconnected STs is random. The key at the bottom identifies aspects of the STs identified in this study. The thick black arrows point to ST57. A. Analysis of the 109 STs found in the 170 NTHi isolates of the final dataset. B. Analysis of all 537 NTHi STs found in the MLST database (accessed 03-31-11) and the 12 fucK negative STs from part A. The largest clonal complex consists of 19 STs. C. eBURST analysis of all 281 typeable STs in the MLST database (accessed 06-28-11). The central clonal complex consists of 126 type b STs.
This high level of diversity remains consistent when all 537 NTHi STs (836 isolates) in the MLST database (accessed March 31st, 2011), as well as the 12 fucK negative STs (15 isolates), were analyzed (Figure 2B). The 537 STs include all 97 fucK positive STs identified in this study. Again, little clustering is evident, and the few complexes are relatively small (the largest being composed of 19 STs). This suggests that the high diversity observed in the collection of 170 isolates from three geographic areas is not merely an artifact of incomplete sampling, but may instead represent the true diversity of NTHi.
The high diversity of NTHi strains can be contrasted with that of typeable strains (see the eBURST plot of all typeable STs in the MLST database accessed June 28th, 2011, Figure 2C). Nearly half (126, or 45%) of the 281 STs among typeable strains comprises a single clonal complex, with many of the rest forming smaller groups. Type b STs predominate, making up 2/3 of all typeable STs in the database (187 of 281). This includes the central clonal complex observed in Figure 2C, in which all 126 STs are type b.
3.4. Phylogenetic analysis
ClonalFrame was used to infer the phylogenic relationships between the 170 NTHi isolates in the final dataset. Figure 3A shows this tree color coded by disease status (OM/commensal), while Figure 3B show the same tree color coded by geographic region (Finland/Israel/US). Only limited clustering is apparent by disease status, as in the eBURST analyses, and geographic region appears to play a similarly small role. The majority of clades are composed of both OM and commensal isolates from multiple geographic regions, suggesting that neither of these factors impose an overwhelming constraint on the phylogenic relationships of the isolates in this sample.
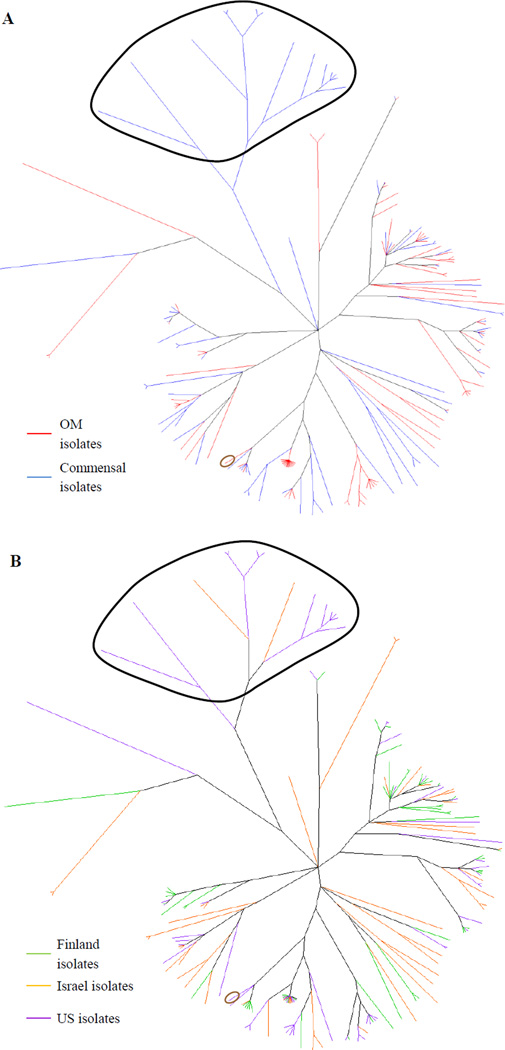
Figure 3.
Unrooted majority rule consensus tree constructed from the MLST data from 170 NTHi isolates. Commensal fucK negative isolates are circled in black, while the OM fucK isolate is circled in brown. A. Branches connecting only to OM isolates are colored red, while branches connecting only to commensal isolates are colored blue. B. Branches connecting only to Finland isolates are colored green, branches connecting only to Israel isolates are colored orange, and branches connecting only to US isolates are colored purple.
One exception to this lack of clustering are the 14 commensal fucK negative isolates circled in black in Figure 3, which form a distinct cluster that is fairly distant from the majority of the tree, in contrast to the OM fucK negative isolate (circled in brown), which falls in the midst of other NTHi isolates in the core of the tree. This group can also be seen in Figure 1B circled in black, where the isolates occupy a position between H. haemolyticus and the rest of the NTHi.
ClonalFrame was also used to investigate the relative rates and contributions of recombination and mutation in the sample in the form of two ratios: ρ/θ and r/m. ρ/θ is the ratio of the recombination rate to the mutation rate, and is therefore a measure of how often recombination events occur relative to mutations. However, as a single recombination event could potentially introduce many more nucleotide changes than a mutation, the ratio of probabilities that a given site is altered via recombination or mutation, r/m, is perhaps more informative. In essence, it is a direct measure of the importance of recombination relative to mutation in the diversification of the sample. These measures for this sample, as well as δ (the average tract length of a recombination event), are reported in Table 3.
Table 3.
Ratios of the rate and effect of recombination versus mutation inferred by ClonalFrame
Ratio of the rates of recombination versus mutation.
Ratio of the effects of recombination versus mutation.
Average tract length of a recombination event.
95% credibility regions.
The ratio of the rates of recombination and mutation (ρ/θ) is one, indicating that the two processes occur at approximately the same rate. However, the ratio of the probabilities that a given nucleotide is changed by recombination or mutation (r/m) is 5.05. This indicates that despite both processes occurring at the same rate, recombination introduces over five times more nucleotide substitutions than do point mutations. The higher rate and impact of recombination than mutation in this sample of NTHi is consistent with data reported previously (Cody et al., 2003; LaCross et al., 2008; Perez-Losada et al., 2006; Vos and Didelot, 2009).
3.5. Population Structure
The structure 2.3.3 program, a model-based clustering method that has been extensively used to infer population structure in humans (Jakobsson et al., 2008; Kopelman et al., 2009; Verdu et al., 2009; Wang et al., 2007), animals (Riehle et al., 2011; Rosenberg et al., 2001), plants (Caniato et al., 2011; Jacobs et al., 2011), and bacteria (den Bakker et al., 2008; Falush et al., 2003b; Sheppard et al., 2010; Sheppard et al., 2008), was used to assess population structure in the sample. Two datasets were utilized, one including only a single example of each of the 109 unique STs found previously (unique STs dataset), and the other including all 170 NTHi isolates (all isolates dataset). For both datasets, 20 independent runs were performed, and the CLUMPP program was used to assess multimodality among the runs. This is an essential step during the analysis and interpretation of structure results, as the algorithm it implements can identify non-symmetric modes (or clustering solutions with high posterior probabilities), particularly in complex datasets with large values of K (i.e. large numbers of populations). The current implementation of structure typically does not cross between these modes, which can lead to different runs producing very different answers (Pritchard et al., 2010). Within each distinct mode for a given K, the estimated lnP(D) (log probability of observing the data) and Q (individual membership proportions) were averaged.
Multimodality was indeed apparent within the two datasets for many values of K, from a high of 12 distinct modes found among the 20 runs at K=6 in the all isolates dataset to a low of a single mode at K=2 in the unique STs dataset. A summary of this information is presented in Table S2. The mode that maximized the lnP(D) at each value of K was chosen for further analysis. The number of assumed populations was increased and those data assessed until adding another population became uninformative, identified by the individual membership proportions Q for that population being on average very low and few individuals having a large portion of their ancestry from that population. For the unique STs dataset, this occurred at K = 7, where the average Q was 4.5% and only two STs had greater than 65% of their ancestry from that population. In the all isolates dataset, up to eight populations were well supported; if K was increased to nine, the average Q for the added population was 2.3% and only two isolates had more than 65% of their ancestry from that population. The individual membership proportions Q from the eight well-supported populations for all 170 isolates are given in Table S1.
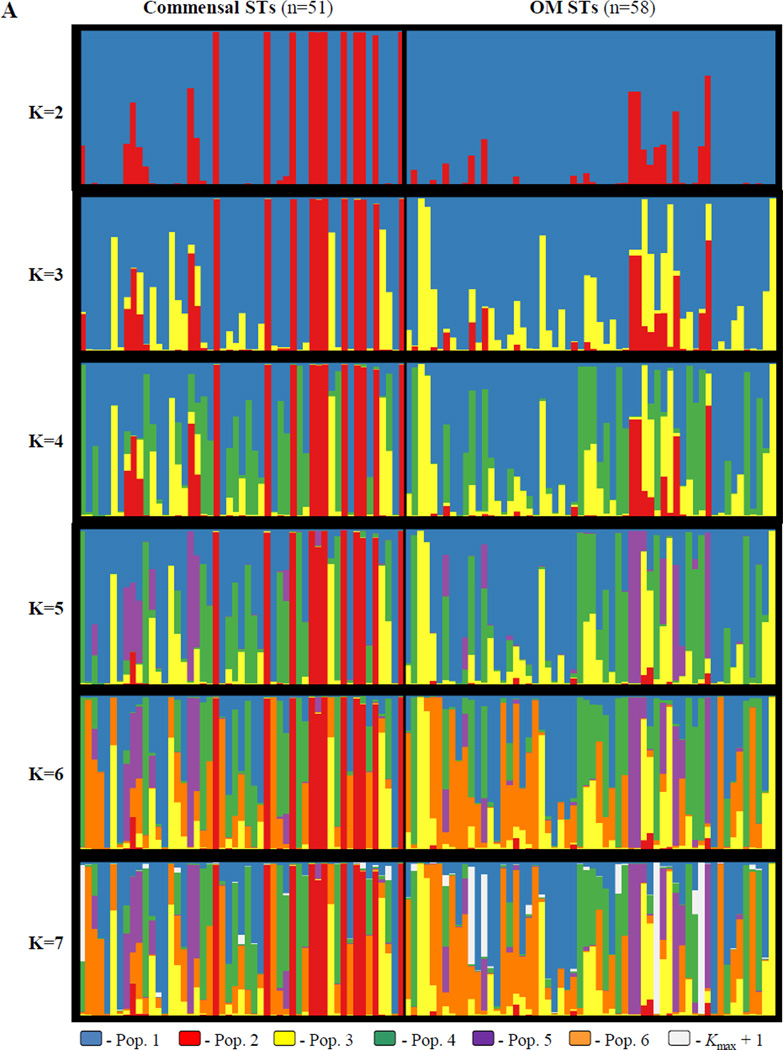
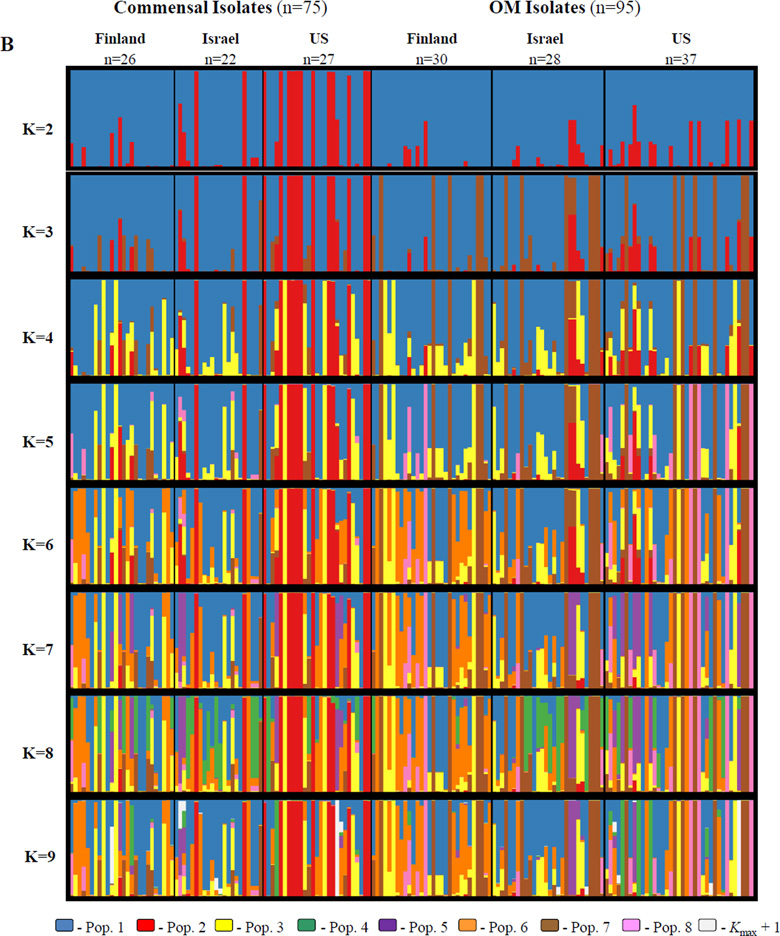
Figure 4A illustrates the clustering solutions inferred by structure from the unique STs dataset that maximize lnP(D) for each value of K from 2 to 7. Commensal STs were those STs containing only isolates collected from healthy children in this sample (n = 51), while OM STs contained at least one isolate collected from a case of otitis media (n = 58). The lack of support for adding a population past K = 6 can be seen in the bottom panel, in which almost none of the of the STs trace their ancestry back to the added population (seen in white) and the clustering solution is otherwise nearly identical to that at K = 6. Figure 4B displays the same information for the all isolates dataset, with K ranging from 2 to 9. Once again, the lack of support for additional populations past K = 8 can be seen in the bottom panel, in which extremely few isolates of the sample have membership in the K = 9 population (seen in white). The clustering solution at K = 9 differs from the K = 8 solution in that far fewer of the isolates have significant membership in the K = 8 population (colored green). However, the lack of significant ancestry in the K = 9 population, combined with a plateau in the lnP(D) at K = 8 and K = 9 (−10,127.2 and −10,083.9, respectively, while K = 7 is −10,846.6), indicates that K = 8 is the best choice for this sample. These choices are reinforced by the considerable amount of information gained at K = 6 for the unique STs dataset and at K = 8 for the all isolates dataset, where the added populations (orange in Figure 4A, green in Figure 4B) comprise a significant portion of the sample’s ancestry.
Figure 4.
Population structure inferred by structure for A: the 109 unique commensal and OM NTHi sequence types; and B: all 170 commensal and OM NTHi isolates. The number of predefined populations (K) is indicated to the left of each plot. Each isolate is represented by a vertical line partitioned into K colored components according to the estimated individual membership proportion in each population (Q). The average of all replicate runs within the mode with the highest likelihood at each K is shown. Population color coding is consistent throughout the figure. Kmax + 1 refers to the first K that is no longer informative (i.e. A: K=7 and B: K=9).
With two exceptions (populations 7 and 8 from the all isolates dataset), the structure analyses inferred the same populations for both datasets. When considering those individuals (whether STs or isolates) with a large proportion of their ancestry from one population, they are clustered together in both analyses, though they are not necessarily inferred in the same order. This trend holds for the vast majority of STs and isolates with a large percentage of their ancestry from one population. Some differences are seen in assignment of STs and isolates with a more admixed heritage, but overall the similarity of clustering between the two analyses is very high. Henceforth, the populations inferred by structure will be referred to by the labels presented in Figure 4. Both figures are color coded identically, so the red colored population 2 (for example) refers to the same genetic cluster in both datasets.
One major difference between the analysis in Figure 4A and 4B is that analysis of the all isolates dataset Figure 4B identified two additional populations, labeled population 7 (brown) and population 8 (pink). Population 7 is comprised of 18 OM isolates of ST57, while population 8 consists of five OM isolates of ST34. This may be a situation similar to those mentioned by the authors of structure, in which having multiple family members (or in this case, multiple isolates with the same ST) can lead to an overestimation of K, though there tends to be little effect of the assignment of individuals to populations for a given K (Falush et al., 2003a; Pritchard et al., 2010). Indeed, as mentioned above, with the exception of populations 7 and 8, the overall clustering solutions between the two analyses are nearly identical. However, these two clusters, though perhaps not true populations in the usual sense, are of interest as they represent large groups of identical genotypes associated solely with otitis media, despite being found in different times, places, and people.
The complexity of the structure plots in Figures 4A and 4B at the chosen levels of K (six and eight, respectively) make discerning trends in population structure by either geographic area or disease difficult. The most readily apparent features are isolates and STs with a very high proportion of their ancestry from population 2 (in red), which seem to be found exclusively among commensal STs in Figure 4A, and with two exceptions only among US commensal isolates in Figure 4B. Population 2 corresponds exactly to the 14 fucK negative isolates (11 STs) identified during MLST genotyping. As fucK sequences were not used for the analysis of population structure, identification of fucK negative strains as a distinct cluster is not biased due to their deletion at that locus. The phylogenetic analysis presented in Figure 3, which placed these isolates in a distinct clade apart from the remaining NTHi, reinforces this clustering solution.
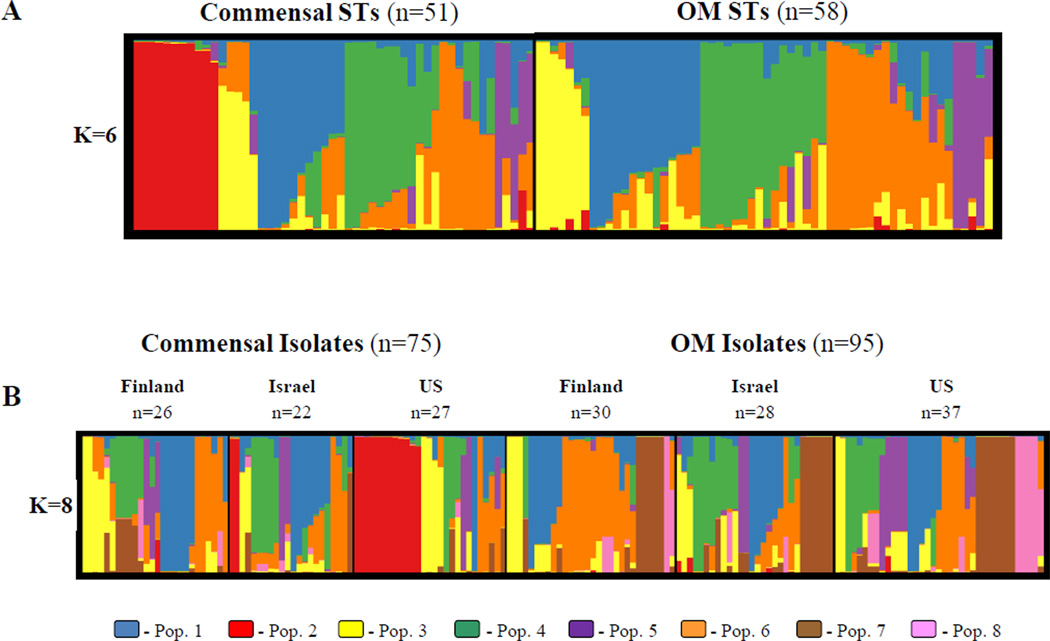
Rearrangement of the population structure plots presents a clearer picture, as shown in Figure 5. Panel A presents the K = 6 plot from Figure 4A while panel B presents the K = 8 plot from Figure 4B; both plots have been sorted by individual membership proportion in each K. From this figure, the clustering of population 2 (in red) only among commensal STs and isolates is even more obvious. However, most other populations are fairly evenly distributed between disease states and geographic areas. One exception may be population 6 (in orange), which appears to be larger among OM STs in this sample. In terms of STs with a high proportion of their ancestry from that population, both commensal and OM groups have five STs with a Q greater than 75%, but the OM group has an additional eight STs with a Q greater than 60% compared to only two from the commensal group. Panel B offers a more nuanced picture, showing that the greater abundance of population 6 STs in the OM group can be traced to the Finland OM group, which has seven isolates with a Q greater than 75%. The groups with the next highest number of isolates with a population 6 Q greater than 75%, the Finland commensal and US OM collections, have only three apiece.
Figure 5.
Inferred population structure from Figures 4A and 4B sorted by individual membership proportion. The number of predefined populations (K) is indicated to the left of each plot. Color coding is consistent with Figure 4. A. Unique STs dataset. B. All isolates dataset.
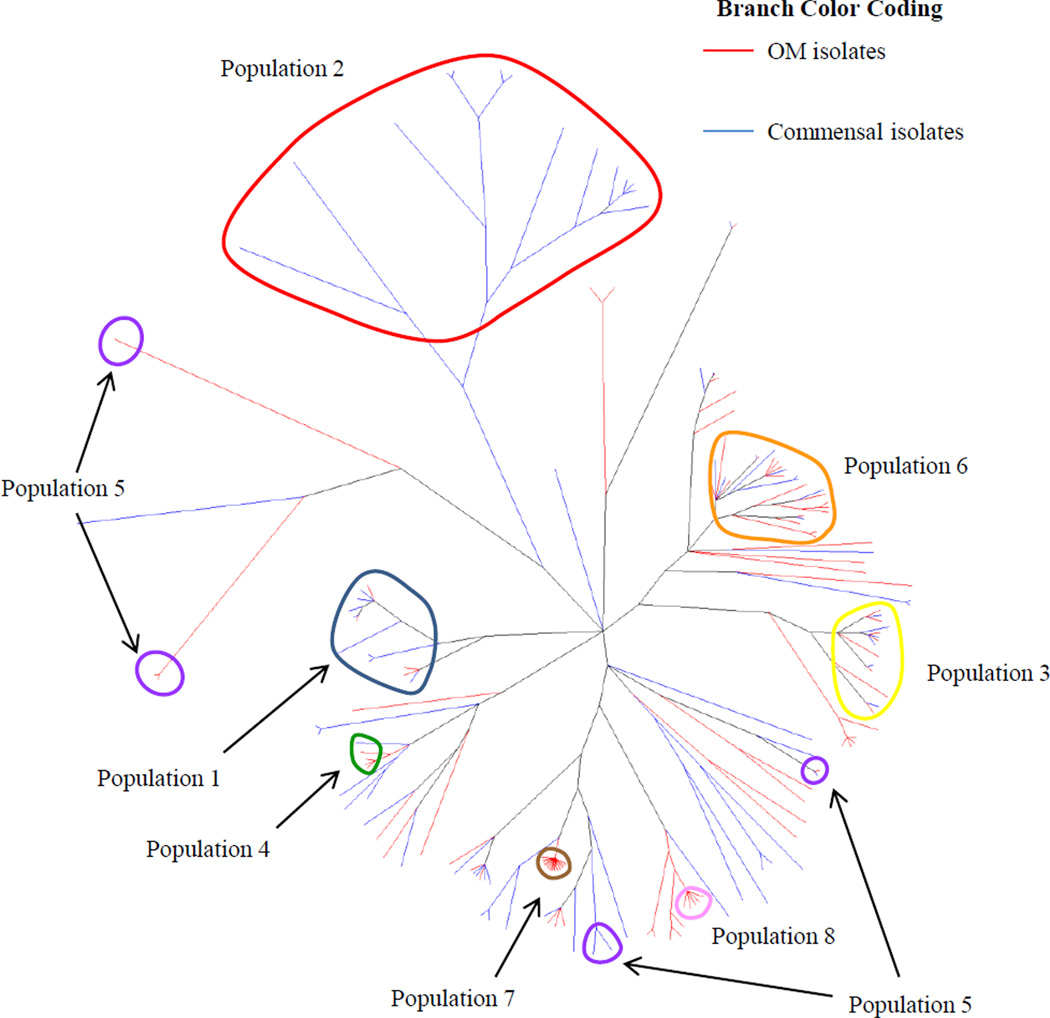
The populations inferred by structure can also be mapped onto the phylogeny estimated by ClonalFrame, as shown in Figure 6. Seven of the eight populations identified in the all isolates dataset correspond to monophyletic groups, indicating that both programs arrived at similar conclusions regarding the ancestry of those isolates. However, population 5 (in purple) is polyphyletic and mapped onto portions of three separate clades. This could denote a greater uncertainty or difficulty in estimating the ancestry of these isolates. Alternatively, it could simply be an illustration of differing results from using the different methods implemented by the two programs.
Figure 6.
Consensus tree from Figure 3 with populations inferred by structure circled. Isolates with a high proportion of their ancestry from a given population were considered to be members of that population and thus included in that circle. Population colors are coded as in Figures 4 and 5.
4. Discussion
Advances in molecular biology and bioinformatics have greatly aided the investigation of bacterial population structure. Falush et al. were able to infer ancestral populations of H. pylori whose spread could be mapped to historical human migrations (Falush et al., 2003b). Recently, Sheppard and colleagues expanded on their work characterizing the convergence of Campylobacter jejuni and C. coli (Sheppard et al., 2008) and found that despite the high diversity of the two species, strong structuring of the populations by host source was apparent, which was stronger than the structuring by geography (Sheppard et al., 2010). Budroni et al. investigated 20 full genome sequences for N. meningitidis and found evidence for a population structured into phylogenetic clades, despite high rates of detectable recombination throughout the bacterial genome (Budroni et al., 2011). Intriguingly, they identified 22 restriction modification systems whose distribution coincided with the clades, suggesting that the observed population structure may in part be due to a differential barrier to gene flow generated by the restriction systems.
The population structure of nontypeable H. influenzae has been less well characterized. Much of the research in this field relied upon MLEE and concentrated on serotypeable isolates, including only a nominal collection of NTHi at best. However, these investigators observed that while the population of typeable Hi, and type b in particular, is clonal, the population of NTHi is large, heterogeneous, and distinct from that of type b isolates (Musser et al., 1986; Musser et al., 1985; Musser et al., 1990; Musser et al., 1988a, b; Porras et al., 1986). More recently, PFGE has been used to again find a clonal population structure in Hib isolates from Australian Aborigines and non-Aborigines (Moor et al., 1999) and to identify a lack of change in the population of Hib genotypes causing vaccine failures in the United Kingdom as compared to genotypes found in the pre-vaccine era (Aracil et al., 2006). Erwin and colleagues used existing MLST data to identify well defined phylogenetic groups of NTHi that differed in genetic content but not geographic or clinical site of isolation (Erwin et al., 2008).
The research presented here investigated the diversity and population structure of a collection of both commensal and otitis media-associated putative NTHi. One striking aspect from Table 1 is that all but one of the isolates identified as non-H. influenzae were in the commensal groups, with the overwhelming majority (14 of 16) in the US commensal subgroup. This is likely due to differences in study design and research priorities. Commensal isolates from Finland (Ukkonen et al., 2000) and Israel (Greenberg et al., 2004) were both collected in health care settings (e.g. hospitals, primary clinics, etc.) with a limited number of isolates obtained from a given child. In general, the investigators conducting these studies were more interested in whether any NTHi was present in the sample, rather than its potential diversity. In contrast, commensal isolates from the US (Farjo et al., 2004; St Sauver et al., 2000) were obtained from healthy children at a number of daycare centers in Michigan, and up to 30 isolates per child were collected. Investigating the diversity of the NTHi present among those children was one of the goals of the studies. Thus, the US commensal subgroup of isolates may represent a more complete picture of the flora inhabiting the naso- and oropharynges of a healthy child, including examples of what we are now able to distinguish as something closely related to, but separate from, nontypeable H. influenzae sensu stricto.
MLST was used to genotype 170 NTHi isolates from Finland, Israel, and the US, identifying 109 unique STs, of which 53 were previously undescribed. Like the study by Musser et al. (Musser et al., 1986) using MLEE to compare NTHi to Hib isolates (and unlike the similar study by Porras et al. (Porras et al., 1986)), no ST found among the NTHi isolates was shared by any of the eight typeable isolates identified in this study. This discrepancy has several potential causes, such as a lack of discriminatory power caused by Porras’ use of only a small number of MLEE loci (six versus the 15 used by Musser) or their potential misidentification of typeable strains as nontypeable, including capsule deficient strains with a typeable genetic background (Giebink, 1999). In the present study, nearly three quarters of the STs (81 of 109) were identified only once, and thirteen STs were comprised of both commensal and OM isolates, which is not unexpected. The naso- and oropharynges are the reservoir of NTHi, from which emerge strains able to travel up the Eustachian tubes to the middle ear spaces and initiate otitis media. Thus, one would anticipate isolating genotypes otherwise implicated in disease from the naso- and oropharynges of currently healthy children.
Interestingly, 15 isolates comprising 12 STs were found to have a deletion of the entire fucose operon, which includes the MLST locus fucK. This phenomenon has been previously described in two isolates by Ridderberg et al., who found that one isolate clustered with confirmed Hi in their phylogenetic tree, while the other isolate occupied a more intermediary position between Hi strains and ‘variant’ strains (including non-hemolytic H. haemolyticus) (Ridderberg et al., 2010). The phylogeny constructed in ClonalFrame (Figure 3) presents a similar picture, with the single OM fucK negative isolate falling in the core of the tree and the remaining 14 commensal fucK negative isolates forming a more distant clade. In Figure 1B, this group (circled in black), while still among the NTHi portion of the tree, occupies an intermediate position between NTHi and H. haemolyticus. This supports the position that distinct divisions between closely related bacterial species frequently offer an inadequate summation of the true relationships among the bacteria (Smith et al., 2000). This group seems, however, to deserve classification as NTHi because, apart from the phylogenies presented here, six of the 14 isolates have been tested for the presence of the iga and lgtC loci by microarray, and all six were positive (data not shown). Reactivity against probes for these genes has been shown to be a robust discriminatory marker for distinguishing H. influenzae from H. haemolyticus (McCrea et al., 2008), and is part of our laboratory’s criteria for true NTHi.
Analysis with the eBURST revealed that few STs were related closely enough to form clonal complexes, with no significant clustering by either disease or geographic area. To be a member of a clonal complex, a ST had to be identical to at least one other ST in the complex at a minimum of six of the seven MLST loci. The largest group consisted of only five STs, and contained both commensal and OM isolates from multiple geographic areas. Analysis of all 537 NTHi STs in the MLST database confirmed this high level of diversity, with the largest group composed of only 19 STs. These findings are similar to those reported previously (LaCross et al., 2008), and show that the considerable expansion of the database in the years since that analysis and those presented by Erwin et al. (Erwin et al., 2008) have not provided reason to alter our understanding of NTHi as a very diverse organism. Further credence is provided by the more consistent sampling scheme employed in this study compared to the MLST database as a whole. While the 170 isolates came from multiple studies with various original goals, care was taken to match the isolates, in an approximate manner, on time, geography, and age of the host children, and only one isolate was selected per child. This last criterion is an important point, as it means that while multiple isolates with the same MLST genotype were identified, they do not constitute clones in the traditional molecular biologic sense, as they were collected from different individuals at different times, and often from very different parts of the world.
The phylogenetic analysis performed in ClonalFrame further demonstrates the high diversity of NTHi. Some distinct clades were identified, but similar to the results reported by Erwin et al. (Erwin et al., 2008) and those apparent from the eBURST analyses, the clustering does not seem to be predicated on either site of isolation or geographic location. With the exception of the commensal fucK negative isolates, the clades are composed of both commensal and OM isolates from multiple geographic regions.
ClonalFrame was also used to estimate the relative rates and contributions of recombination and mutation. While the relative rates of recombination and mutation were approximately equal, recombination was estimated to introduce over five times more nucleotide substitutions than mutation (r/m = 5.05), higher than that found by Vos and Didelot using the same method (r/m = 3.7) (Vos and Didelot, 2009). However, the authors combined both typeable and nontypeable isolates in their analysis. Perez-Losada et al. found that NTHi have higher rates of recombination at four of the seven MLST loci than do typeable isolates (Perez-Losada et al., 2006), which may account for the lower r/m ratio found by Vos and Didelot. Perez-Losada et al. also reported a measure of recombination relative to mutation (the per allele ratio of recombination to mutation Γ/ΓWf = 2.51), but as they utilized a completely different analytic technique, comparing values between the studies is difficult. The higher influence of recombination on the evolution NTHi relative to mutation supports the theory that recombination predominates in the evolution of the species.
Despite the high genetic diversity of NTHi, significant population structure was apparent in this sample. As can be seen in Figures 4 and 5, numerous isolates and STs trace the majority of their ancestry to a single population, in spite of the presence of admixed genotypes with significant ancestry from multiple populations. However, in concordance with the eBURST and ClonalFrame analyses, there is no large scale structuring by either site of isolation or geography. Most populations are present in roughly equal proportions in all disease and geographic region subgroups.
Population 2 (in red), comprised of the commensal fucK negative isolates, is an exception, with all but two isolates and STs coming from the US. This cluster may represent a divergent population of NTHi with a significantly reduced ability to cause otitis media. While the majority of this population was collected from a single geographic region, it is possible that broader sampling techniques such as those used to collect the US commensal isolates (Farjo et al., 2004; St Sauver et al., 2000) would reveal additional members of this population in other regions. Population 6 (in orange) is another cluster that appears to be differentially distributed, such that isolates with a high percentage of their ancestry from population 6 are more common among the OM isolates, particularly so among the Finland OM subgroup. This may represent a case of combined geographic and disease population structuring, and would be an interesting target for further study.
The major differences between the structure analyses of the two datasets lie in populations 7 and 8 (brown and pink, respectively). Both populations are composed of multiple isolates of the same MLST genotype, 18 ST57 strains among population 7 and 5 ST34 strains among population 8. The identification of these groups of identical genotypes as populations by structure may be an artifact that reflects a violation of a model assumption, but has apparently had little detrimental effect on the proper clustering of other isolates and proved helpful in distinguishing these STs as being of interest. The sole sequence type in population 7, ST57, was by far the most commonly identified genotype in this sample, occurring over three times more frequently than the next most common ST (ST34, containing five isolates). Additionally, every isolate of both ST57 and ST34 was recovered from the middle ear of a child with otitis media, and was found in roughly equal proportions among the three geographic regions. Given that each of the ST57 isolates was also collected from a different child, this suggests a strong association between that particular genotype and otitis media, with little association with geography. A similar trend for ST34 is seen, but the smaller number of isolates precludes firm hypotheses. When analyzed as part of the unique STs dataset, both ST57 and ST34 were significantly admixed, tracing approximately 60% of their ancestry to population 4 and 30% to population 5, which precludes a recombination defect, at least one distant in evolution, as an explanation for conservation of the MLST alleles that form the basis for the structure analysis. As it was not found among any of the commensal isolates, despite the naso- and oropharynges being the reservoir for NTHi, ST57 may represent a rare genotype with increased virulence for disease in the middle ear (thus, the OM collections would be ‘enriched’ for this virulent genotype). This genotype, along with ST34, has the potential to be a useful target for research into the mechanisms of NTHi virulence.
Supplementary Material
Highlights.
We investigated population structure among nontypeable Haemophilus influenzae.
MLST was used to type 170 commensal and disease associated isolates.
Two frequently isolated sequence types were only associated with disease.
There was clear evidence for population structure, despite high genetic diversity.
The populations were not structured by disease or geography.
Acknowledgements
This work was supported with funding from the Molecular Mechanisms of Microbial Pathogenesis Training Program (NCL) (AI007528), and the National Institutes of Health (JRG) (R01-DC05840 and R01-AI125630).
We wish to thank Drs. Noah Rosenberg and Paul Verdu for their advice on the analysis of population structure, and Mayuri Patel and Emefah Loccoh for their excellent technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aracil B, Slack M, Perez-Vazquez M, Roman F, Ramsay M, Campos J. Molecular epidemiology of Haemophilus influenzae type b causing vaccine failures in the United Kingdom. J Clin Microbiol. 2006;44:1645–1649. doi: 10.1128/JCM.44.5.1645-1649.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou R, Dominguez A, Fontanals D, Sanfeliu I, Pons I, Renau J, Pineda V, Lobera E, Latorre C, Majo M, Salleras L. Prevalence of Haemophilus influenzae pharyngeal carriers in the school population of Catalonia. Working Group on invasive disease caused by Haemophilus influenzae. Eur J Epidemiol. 2000;16:521–526. doi: 10.1023/a:1007632122570. [DOI] [PubMed] [Google Scholar]
- Budroni S, Siena E, Hotopp JC, Seib KL, Serruto D, Nofroni C, Comanducci M, Riley DR, Daugherty SC, Angiuoli SV, Covacci A, Pizza M, Rappuoli R, Moxon ER, Tettelin H, Medini D. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc Natl Acad Sci U S A. 2011;108:4494–4499. doi: 10.1073/pnas.1019751108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caniato FF, Guimaraes CT, Hamblin M, Billot C, Rami JF, Hufnagel B, Kochian LV, Liu J, Garcia AA, Hash CT, Ramu P, Mitchell S, Kresovich S, Oliveira AC, de Avellar G, Borem A, Glaszmann JC, Schaffert RE, Magalhaes JV. The Relationship between Population Structure and Aluminum Tolerance in Cultivated Sorghum. PLoS ONE. 2011;6:e20830. doi: 10.1371/journal.pone.0020830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody AJ, Field D, Feil EJ, Stringer S, Deadman ME, Tsolaki AG, Gratz B, Bouchet V, Goldstein R, Hood DW, Moxon ER. High rates of recombination in otitis media isolates of non-typeable Haemophilus influenzae. Infect Genet Evol. 2003;3:57–66. doi: 10.1016/s1567-1348(02)00152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Bakker HC, Didelot X, Fortes ED, Nightingale KK, Wiedmann M. Lineage specific recombination rates and microevolution in Listeria monocytogenes. BMC evolutionary biology. 2008;8:277. doi: 10.1186/1471-2148-8-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhooge I, Vaneechoutte M, Claeys G, Verschraegen G, Van Cauwenberge P. Turnover of Haemophilus influenzae isolates in otitis-prone children. Int J Pediatr Otorhinolaryngol. 2000;54:7–12. doi: 10.1016/s0165-5876(00)00321-9. [DOI] [PubMed] [Google Scholar]
- Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Falush D. ClonalFrame User Guide. 2008 [Google Scholar]
- Erwin AL, Nelson KL, Mhlanga-Mutangadura T, Bonthuis PJ, Geelhood JL, Morlin G, Unrath WC, Campos J, Crook DW, Farley MM, Henderson FW, Jacobs RF, Muhlemann K, Satola SW, van Alphen L, Golomb M, Smith AL. Characterization of genetic and phenotypic diversity of invasive nontypeable Haemophilus influenzae. Infect Immun. 2005;73:5853–5863. doi: 10.1128/IAI.73.9.5853-5863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin AL, Sandstedt SA, Bonthuis PJ, Geelhood JL, Nelson KL, Unrath WC, Diggle MA, Theodore MJ, Pleatman CR, Mothershed EA, Sacchi CT, Mayer LW, Gilsdorf JR, Smith AL. Analysis of genetic relatedness of Haemophilus influenzae isolates by multilocus sequence typing. J Bacteriol. 2008;190:1473–1483. doi: 10.1128/JB.01207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falla TJ, Crook DW, Brophy LN, Maskell D, Kroll JS, Moxon ER. PCR for capsular typing of Haemophilus influenzae. J Clin Microbiol. 1994;32:2382–2386. doi: 10.1128/jcm.32.10.2382-2386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003a;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, Blaser MJ, Graham DY, Vacher S, Perez-Perez GI, Yamaoka Y, Megraud F, Otto K, Reichard U, Katzowitsch E, Wang X, Achtman M, Suerbaum S. Traces of human migrations in Helicobacter pylori populations. Science. 2003b;299:1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- Farjo RS, Foxman B, Patel MJ, Zhang L, Pettigrew MM, McCoy SI, Marrs CF, Gilsdorf JR. Diversity and sharing of Haemophilus influenzae strains colonizing healthy children attending day-care centers. Pediatr Infect Dis J. 2004;23:41–46. doi: 10.1097/01.inf.0000106981.89572.d1. [DOI] [PubMed] [Google Scholar]
- Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman A, Rubin DB. Inference from iterative simulation using multiple sequences. Stat Sci. 1992;7:457–511. [Google Scholar]
- Giebink GS. Otitis media: the chinchilla model. Microb Drug Resist. 1999;5:57–72. doi: 10.1089/mdr.1999.5.57. [DOI] [PubMed] [Google Scholar]
- Greenberg D, Broides A, Blancovich I, Peled N, Givon-Lavi N, Dagan R. Relative importance of nasopharyngeal versus oropharyngeal sampling for isolation of Streptococcus pneumoniae and Haemophilus influenzae from healthy and sick individuals varies with age. J Clin Microbiol. 2004;42:4604–4609. doi: 10.1128/JCM.42.10.4604-4609.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Jacobs MM, Smulders MJ, van den Berg RG, Vosman B. What's in a name; genetic structure in Solanum section Petota studied using population-genetic tools. BMC Evol Biol. 2011;11:42. doi: 10.1186/1471-2148-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, Bras JM, Schymick JC, Hernandez DG, Traynor BJ, Simon-Sanchez J, Matarin M, Britton A, van de Leemput J, Rafferty I, Bucan M, Cann HM, Hardy JA, Rosenberg NA, Singleton AB. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Kilian M. Genus Haemophilus. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey's Manual of Systematic Bacteriology. 2 ed. New York: Springer-Verlag; 2005. pp. 883–904. [Google Scholar]
- Kilpi T, Herva E, Kaijalainen T, Syrjanen R, Takala AK. Bacteriology of acute otitis media in a cohort of Finnish children followed for the first two years of life. Pediatr Infect Dis J. 2001;20:654–662. doi: 10.1097/00006454-200107000-00004. [DOI] [PubMed] [Google Scholar]
- Kopelman NM, Stone L, Wang C, Gefel D, Feldman MW, Hillel J, Rosenberg NA. Genomic microsatellites identify shared Jewish ancestry intermediate between Middle Eastern and European populations. BMC Genet. 2009;10:80. doi: 10.1186/1471-2156-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasan GP, Cutter D, Block SL, St Geme JW., 3rd Adhesin expression in matched nasopharyngeal and middle ear isolates of nontypeable Haemophilus influenzae from children with acute otitis media. Infect Immun. 1999;67:449–454. doi: 10.1128/iai.67.1.449-454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCross NC, Marrs CF, Patel M, Sandstedt SA, Gilsdorf JR. High Genetic Diversity of Nontypeable Haemophilus influenzae Among Two Children Attending a Daycare Center. J Clin Microbiol. 2008;46:3817–3821. doi: 10.1128/JCM.00940-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibovitz E, Piglansky L, Raiz S, Greenberg D, Hamed KA, Ledeine JM, Press J, Leiberman A, Echols RM, Pierce PF, Jacobs MR, Dagan R. Bacteriologic and clinical efficacy of oral gatifloxacin for the treatment of recurrent/nonresponsive acute otitis media: an open label, noncomparative, double tympanocentesis study. Pediatr Infect Dis J. 2003;22:943–949. doi: 10.1097/01.inf.0000095468.89866.14. [DOI] [PubMed] [Google Scholar]
- May BJ, Zhang Q, Li LL, Paustian ML, Whittam TS, Kapur V. Complete genomic sequence of Pasteurella multocida, Pm70. Proc Natl Acad Sci U S A. 2001;98:3460–3465. doi: 10.1073/pnas.051634598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrea KW, Xie J, LaCross N, Patel M, Mukundan D, Murphy TF, Marrs CF, Gilsdorf JR. Relationships of nontypeable Haemophilus influenzae strains to hemolytic and nonhemolytic Haemophilus haemolyticus strains. J Clin Microbiol. 2008;46:406–416. doi: 10.1128/JCM.01832-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meats E, Feil EJ, Stringer S, Cody AJ, Goldstein R, Kroll JS, Popovic T, Spratt BG. Characterization of encapsulated and noncapsulated Haemophilus influenzae and determination of phylogenetic relationships by multilocus sequence typing. J Clin Microbiol. 2003;41:1623–1636. doi: 10.1128/JCM.41.4.1623-1636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor PE, Collignon PC, Gilbert GL. Pulsed-field gel electrophoresis used to investigate genetic diversity of Haemophilus influenzae type b isolates in Australia shows differences between Aboriginal and non-Aboriginal isolates. J Clin Microbiol. 1999;37:1524–1531. doi: 10.1128/jcm.37.5.1524-1531.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TF, Sethi S, Klingman KL, Brueggemann AB, Doern GV. Simultaneous respiratory tract colonization by multiple strains of nontypeable Haemophilus influenzae in chronic obstructive pulmonary disease: implications for antibiotic therapy. J Infect Dis. 1999;180:404–409. doi: 10.1086/314870. [DOI] [PubMed] [Google Scholar]
- Musser JM, Barenkamp SJ, Granoff DM, Selander RK. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect Immun. 1986;52:183–191. doi: 10.1128/iai.52.1.183-191.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser JM, Granoff DM, Pattison PE, Selander RK. A population genetic framework for the study of invasive diseases caused by serotype b strains of Haemophilus influenzae. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:5078–5082. doi: 10.1073/pnas.82.15.5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser JM, Kroll JS, Granoff DM, Moxon ER, Brodeur BR, Campos J, Dabernat H, Frederiksen W, Hamel J, Hammond G, Hoiby EA, Jonsdottir KE, Kabeer M, Kallings I, Koornhof HJ, Law B, Li KI, Montgomery J, Pattison PE, Piffaretti J, Takala AK, Thong ML, Wall RA, Ward JI, Selander RK. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev Infect Dis. 1990;12:75–111. doi: 10.1093/clinids/12.1.75. [DOI] [PubMed] [Google Scholar]
- Musser JM, Kroll JS, Moxon ER, Selander RK. Clonal population structure of encapsulated Haemophilus influenzae. Infect Immun. 1988a;56:1837–1845. doi: 10.1128/iai.56.8.1837-1845.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser JM, Kroll JS, Moxon ER, Selander RK. Evolutionary genetics of the encapsulated strains of Haemophilus influenzae. Proc Natl Acad Sci U S A. 1988b;85:7758–7762. doi: 10.1073/pnas.85.20.7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norskov-Lauritsen N. Detection of cryptic genospecies misidentified as Haemophilus influenzae in routine clinical samples by assessment of marker genes fucK, hap, sodC. J Clin Microbiol. 2009;47:2590–2592. doi: 10.1128/JCM.00013-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norskov-Lauritsen N, Bruun B, Kilian M. Multilocus sequence phylogenetic study of the genus Haemophilus with description of Haemophilus pittmaniae sp. nov. Int J Syst Evol Microbiol. 2005;55:449–456. doi: 10.1099/ijs.0.63325-0. [DOI] [PubMed] [Google Scholar]
- Norskov-Lauritsen N, Overballe MD, Kilian M. Delineation of the species Haemophilus influenzae by phenotype, multilocus sequence phylogeny, and detection of marker genes. J Bacteriol. 2009;191:822–831. doi: 10.1128/JB.00782-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Losada M, Browne EB, Madsen A, Wirth T, Viscidi RP, Crandall KA. Population genetics of microbial pathogens estimated from multilocus sequence typing (MLST) data. Infect Genet Evol. 2006;6:97–112. doi: 10.1016/j.meegid.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porras O, Caugant DA, Gray B, Lagergard T, Levin BR, Svanborg-Eden C. Difference in structure between type b and nontypable Haemophilus influenzae populations. Infect Immun. 1986;53:79–89. doi: 10.1128/iai.53.1.79-89.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Wen X, Falush D. structure 2.3 documentation. 2010 [Google Scholar]
- Ridderberg W, Fenger MG, Norskov-Lauritsen N. Haemophilus influenzae may be untypable by the multilocus sequence typing scheme due to a complete deletion of the fucose operon. J Med Microbiol. 2010;59:740–742. doi: 10.1099/jmm.0.018424-0. [DOI] [PubMed] [Google Scholar]
- Riehle MM, Guelbeogo WM, Gneme A, Eiglmeier K, Holm I, Bischoff E, Garnier T, Snyder GM, Li X, Markianos K, Sagnon N, Vernick KD. A cryptic subgroup of Anopheles gambiae is highly susceptible to human malaria parasites. Science. 2011;331:596–598. doi: 10.1126/science.1196759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004;4:2. [Google Scholar]
- Rosenberg NA, Burke T, Elo K, Feldman MW, Freidlin PJ, Groenen MA, Hillel J, Maki-Tanila A, Tixier-Boichard M, Vignal A, Wimmers K, Weigend S. Empirical evaluation of genetic clustering methods using multilocus genotypes from 20 chicken breeds. Genetics. 2001;159:699–713. doi: 10.1093/genetics/159.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, Colles F, Richardson J, Cody AJ, Elson R, Lawson A, Brick G, Meldrum R, Little CL, Owen RJ, Maiden MC, McCarthy ND. Host association of Campylobacter genotypes transcends geographic variation. Appl Environ Microbiol. 2010;76:5269–5277. doi: 10.1128/AEM.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard SK, McCarthy ND, Falush D, Maiden MC. Convergence of Campylobacter species: implications for bacterial evolution. Science. 2008;320:237–239. doi: 10.1126/science.1155532. [DOI] [PubMed] [Google Scholar]
- Smith JM, Feil EJ, Smith NH. Population structure and evolutionary dynamics of pathogenic bacteria. Bioessays. 2000;22:1115–1122. doi: 10.1002/1521-1878(200012)22:12<1115::AID-BIES9>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- St Sauver J, Marrs CF, Foxman B, Somsel P, Madera R, Gilsdorf JR. Risk factors for otitis media and carriage of multiple strains of Haemophilus influenzae and Streptococcus pneumoniae. Emerg Infect Dis. 2000;6:622–630. doi: 10.3201/eid0606.000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trottier S, Stenberg K, Svanborg-Eden C. Turnover of nontypable Haemophilus influenzae in the nasopharynges of healthy children. J Clin Microbiol. 1989;27:2175–2179. doi: 10.1128/jcm.27.10.2175-2179.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukkonen P, Varis K, Jernfors M, Herva E, Jokinen J, Ruokokoski E, Zopf D, Kilpi T. Treatment of acute otitis media with an antiadhesive oligosaccharide: a randomised, double-blind, placebo-controlled trial. Lancet. 2000;356:1398–1402. doi: 10.1016/S0140-6736(00)02843-9. [DOI] [PubMed] [Google Scholar]
- Verdu P, Austerlitz F, Estoup A, Vitalis R, Georges M, Thery S, Froment A, Le Bomin S, Gessain A, Hombert JM, Van der Veen L, Quintana-Murci L, Bahuchet S, Heyer E. Origins and genetic diversity of pygmy hunter-gatherers from Western Central Africa. Curr Biol. 2009;19:312–318. doi: 10.1016/j.cub.2008.12.049. [DOI] [PubMed] [Google Scholar]
- Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- Wang S, Lewis CM, Jakobsson M, Ramachandran S, Ray N, Bedoya G, Rojas W, Parra MV, Molina JA, Gallo C, Mazzotti G, Poletti G, Hill K, Hurtado AM, Labuda D, Klitz W, Barrantes R, Bortolini MC, Salzano FM, Petzl-Erler ML, Tsuneto LT, Llop E, Rothhammer F, Excoffier L, Feldman MW, Rosenberg NA, Ruiz-Linares A. Genetic variation and population structure in native Americans. PLoS Genet. 2007;3:e185. doi: 10.1371/journal.pgen.0030185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.