Abstract
PHF1 gene rearrangements have been recently described in around 50% of ossifying fibromyxoid tumors (OFMT) including benign and malignant cases, with a small subset showing EP400-PHF1 fusions. In the remaining cases no alternative gene fusions have been identified. PHF1-negative OFTs, especially if lacking S100 protein staining or peripheral ossification, are difficult to diagnose and distinguish from other soft tissue mimics. In seeking more comprehensive molecular characterization, we investigated a large cohort of 39 OFMT of various anatomic sites, immunoprofiles and grades of malignancy. Tumors were screened for PHF1 and EP400 rearrangements by FISH. RNA sequencing was performed in two index cases (OFMT1, OFMT3), negative for EP400-PHF1 fusions, followed by FusionSeq data analysis, a modular computational tool developed to discover gene fusions from paired-end RNA-seq data. Two novel fusions were identified ZC3H7B-BCOR in OFMT1 and MEAF6-PHF1 in OFMT3. After being validated by FISH and RT-PCR, these abnormalities were screened on the remaining cases. With these additional gene fusions, 33/39 (85%) of OFMTs demonstrated recurrent gene rearrangements, which can be used as molecular markers in challenging cases. The most common abnormality is PHF1 gene rearrangement (80%), being present in benign, atypical and malignant lesions, with fusion to EP400 in 44% of cases. ZC3H7B-BCOR and MEAF6-PHF1 fusions occurred predominantly in S100 protein-negative and malignant OFMT. As similar gene fusions were reported in endometrial stromal sarcomas, we screened for potential gene abnormalities in JAZF1 and EPC1 by FISH and found two additional cases with EPC1-PHF1 fusions.
Keywords: Ossifying fibromyxoid tumor, PHF1, EP400, BCOR, MEAF6
INTRODUCTION
OFMT is a rare soft tissue tumor of uncertain lineage, characterized histologically by multinodular growth and a distinctive shell of mature bone found at the periphery of the nodules. Microscopically, the tumors are composed of uniform round to oval cells embedded in a fibro-myxoid stroma. The tumors are often positive for S100 protein; however, the significance of this finding remains puzzling, since electron microscopic studies have failed to pinpoint a line of differentiation, excluding schwannian, melanocytic, chondroid or myoepithelial differentiation. In addition to its controversial histogenesis, the criteria for malignancy are not well defined, and atypical/malignant forms often deviate from the classic morphology, with lack of S100 protein expression or peripheral ossification. In the absence of objective molecular markers these atypical examples are difficult to distinguish from other look-alike soft tissue lesions.
The PHF1 gene, previously shown to be the 3′-partner of fusion genes in endometrial stromal tumors, has recently been implicated in the pathogenesis of about 50% of OFMTs, irrespective of whether they are diagnosed as typical, atypical, or malignant lesions (Gebre-Medhin et al., 2012; Graham et al., 2013). In only two tumors PHF1 was shown to fuse to EP400 (Gebre-Medhin et al., 2012; Endo et al., 2013), while in the remaining cases no alternative gene partners have been identified as yet. In this study we performed a detailed molecular analysis in a large cohort of OFMT lesions, covering a wide spectrum of clinical presentations and degree of malignancy. EP400-PHF1 negative tumors were investigated by RNA sequencing for novel translocation discovery and validated abnormalities were then screened in the remaining cases.
MATERIAL AND METHODS
The Pathology files of MSKCC and the personal consultations of the corresponding authors (CRA, CDF) were searched for cases of ossifying fibromyxoid tumor (OFMT), of any degree of malignancy. Pathologic diagnosis and immunohistochemical stains were re-reviewed in all cases. The histologic requirement for inclusion in the study was a predominantly classic morphologic appearance, the tumors being composed of relatively monotonous epithelioid, cuboidal or oval cells, arranged in cords or single files within a fibromyxoid stroma. Cases that displayed significant nuclear pleomorphism or conspicuous areas of spindling and fascicular growth were excluded. OFMT were classified as benign, for tumors with typical morphologic features and lacking cytologic atypia or increased mitotic activity. Tumors with increased cellularity but lacking increased mitotic activity, necrosis or nuclear pleomorphism were defined as atypical OFMTs. Malignant OFMTs showed increased cellularity, mitotic activity (>2MF/50HPFs) and/or nuclear pleomorphism or necrosis. The presence of ossification defined as a rim of lamellar bone was recorded in every case. Additional osteoid-like matrix deposition, if present, was separately recorded. Immunohistochemical stains, including S100 protein and desmin, were reviewed and results were correlated with degree of malignancy and fusion type (Table 1). The study was approved by the Institutional Review Board 02-060.
Table 1.
Clinical and Pathologic Findings of OFMTs showing gene rearrangements
| OFMT | Age/ Sex | Location | Histology | S100 | Desmin | Fusion/Re- arrangement |
|---|---|---|---|---|---|---|
| 1* | 55/M | groin/thigh | Malignant, ossifying | Neg | Neg | ZC3H7B-BCOR |
| 2 | 76/M | Thigh | Malignant, ossifying | Neg | Neg | BCOR |
| 3* | 73/M | popliteal | Malignant, ossifying | Neg | Neg | MEAF6-PHF1 |
| 4 | 38/M | Shoulder | Malignant, ossifying | Neg | Neg | MEAF6-PHF1 |
| 5 | 66/F | Shoulder | Benign, ossifying | Neg | Pos | MEAF6-PHF1 |
| 6 | 56/F | Shoulder | Malignant, ossifying | Neg | Fpos | EP400-PHF1 |
| 7 | 24/F | Buttock | Benign, ossifying | Fpos | Fpos | EP400-PHF1 |
| 8 | 38/F | Thigh | Benign, ossifying | Pos | Neg | EP400-PHF1 |
| 9 | 48/M | Axilla | Benign, ossifying | Neg | Neg | EP400-PHF1 |
| 10 | 21/F | Thigh | Benign, non-ossifying | Pos | Neg | EP400-PHF1 |
| 11 | 73/M | Foot | Benign, non-ossifying | Pos | Neg | EP400-PHF1 |
| 12 | 46/F | skin, breast | Benign, non-ossifying | Pos | Pos | EP400-PHF1 |
| 13 | 23/F | Thigh | Benign, non-ossifying | Pos | Pos | EP400-PHF1 |
| 14 | 48/F | Neck | Benign, non-ossifying | Pos | Pos | EP400-PHF1 |
| 15 | 43/F | Buttock | Benign, non-ossifying | Pos | Pos | EP400-PHF1 |
| 16 | 65/F | Leg | Atypical, ossifying | N/A | N/A | EP400-PHF1 |
| 17 | 70/M | Neck | Atypical, ossifying | Pos | Pos | EP400-PHF1 |
| 18 | 69/F | supraclavicular | Atypical, ossifying | Pos | Pos | EP400-PHF1 |
| 19 | 59/F | Shoulder | Malignant, ossifying | Fpos | Pos | EP400-PHF1 |
| 20 | 69/F | chest wall | Malignant, ossifying | Neg | Pos | EP400-PHF1 |
| 21 | 41/F | Breast | Malignant, ossifying | Neg | Pos | EP400-PHF1 |
| 22 | 59/F | Paraspinal | Malignant, non-ossifying | Neg | Pos | EP400-PHF1 |
| 23 | 69/M | Thigh | Benign, non-ossifying | Neg | Pos | EPC1-PHF1 |
| 24 | 41/M | Forearm | Malignant, ossifying | Neg | Neg | EPC1-PHF1 |
| 25 | 49/F | supraclavicular | Benign, ossifying | N/A | N/A | PHF1 |
| 26 | 60/M | Finger | Benign, ossifying | Pos | Pos | PHF1 |
| 27 | 71/M | Hand | Benign, ossifying | Pos | Neg | PHF1 |
| 28 | 65/F | Tongue | Benign, ossifying | Pos | Pos | PHF1 |
| 29 | 32/M | Foot | Benign, non-ossifying | Pos | Fpos | PHF1 |
| 30 | 70/M | Cheek | Benign, non-ossifying | Pos | Pos | PHF1 |
| 31 | 55/F | Foot | Benign, non-ossifying | Pos | Pos | PHF1 |
| 32 | 72/F | Foot | Benign, non-ossifying | Pos | Pos | PHF1 |
| 33 | 59/M | Leg | Malignant, non-ossifying | Fpos | Neg | PHF1 |
index cases tested for RNAseq; F, female; M, male; Pos, positive; Neg, negative; Fpos, focally positive; N/A, not available.
RNA Sequencing
Total RNA was prepared for RNA sequencing in accordance with the standard Illumina mRNA sample preparation protocol (Illumina). Briefly, mRNA was isolated with oligo(dT) magnetic beads from total RNA (10 μg) extracted from case. The mRNA was fragmented by incubation at 94°C for 2.5 min in fragmentation buffer (Illumina). To reduce the inclusion of artifactual chimeric transcripts due to random priming of transcript fragments into the sequencing library because of inefficient A-tailing reactions that lead to self ligation of blunt-ended template molecules (Quail et al., 2008), an additional size-selection step (capturing 350–400 bp) was introduced prior to the adapter ligation step. The adaptor-ligated library was then enriched by PCR for 15 cycles and purified. The library was sized and quantified using DNA1000 kit (Agilent) on an Agilent 2100 Bioanalyzer according to the manufacturer’s instructions. Paired-end RNA-sequencing at read lengths of 50 or 51 bp was performed with the HiSeq 2500 (Illumina). Across the two samples a total of about 141 million paired-end reads were generated, corresponding to about 21 billion bases.
Analysis of RNA Sequencing Results with FusionSeq
All reads were independently aligned with STAR alignment software against the human genome reference sequence (hg19) and a splice junction library, simultaneously (Dobin et al., 2013). The mapped reads were converted into Mapped Read Format (Habegger et al., 2011) and analyzed with FusionSeq (Sboner et al., 2010) to identify potential fusion transcripts. FusionSeq is a computational method successfully applied to paired-end RNA-seq experiments for the identification of chimeric transcripts (Tanas et al., 2011; Pierron et al., 2012) (Mosquera et al., 2013). Briefly, paired-end reads mapped to different genes are first used to identify potential chimeric candidates. A cascade of filters, each taking into account different sources of noise in RNA-sequencing experiments, was then applied to remove spurious fusion transcript candidates. Once a confident list of fusion candidates was generated, they were ranked with several statistics to prioritize the experimental validation. In these cases, we used the DASPER score (Difference between the observed and Analytically calculated expected SPER): a higher DASPER score indicated a greater likelihood that the fusion candidate was authentic and did not occur randomly. See (Sboner et al., 2010) for further details about FusionSeq.
Fluorescence In Situ Hybridization (FISH)
FISH on interphase nuclei from paraffin-embedded 4-micron sections was performed applying custom probes using bacterial artificial chromosomes (BAC), covering and flanking genes that were identified as potential fusion partners in the RNA-seq experiment. BAC clones were chosen according to USCS genome browser (http://genome.uscs.edu), see Supplementary Table 1. The BAC clones were obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (CHORI) (Oakland, CA) (http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer’s instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution, as previously described (Antonescu et al., 2010). The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems). A positive score was interpreted when at least 20% of the nuclei showed a break-apart signal. Nuclei with incomplete set of signals were omitted from the score.
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
An aliquot of the RNA extracted above from frozen tissue (Trizol Reagent; Invitrogen; Grand Island, NY) was used to confirm the novel fusion transcript identified by FusionSeq. RNA quality was determined by Eukaryote Total RNA Nano Assay and cDNA quality was tested for PGK housekeeping gene (247 bp amplified product). Three microgram of total RNA was used for cDNA synthesis by SuperScript ® III First-Strand Synthesis Kit (Invitrogen, Carlsbad, CA). RT-PCR was performed using the Advantage-2 PCR kit (Clontech, Mountain View, CA) for 33 cycles at a 64.5°C annealing temperature, using the following primers: ZC3H7B-Ex10F: 5′ – CCTTCGGCTTGGTCATGGAC – 3′; BCOR-Ex7R: 5′ –GAGACTTTGCGTTTCCTGTCCAC– 3′; MEAF6-Ex4F: 5′ – CAGGAGTTCAGGACCAGCTC – 3′; PHF1-Ex3R: 5′ – CCTCAAACTGGACCAGACACAC – 3′. Amplified products were purified and sequenced by Sanger method.
Long-Range PCR
Genomic DNA was extracted from frozen tissue using the Phenol/Chloroform assay and quality was confirmed by electrophoresis. 0.5 μg genomic DNA was amplified with the LongRange PCR Kit (QIAGEN, Germantown, MD) in order to assess the intronic breaks. The following primer sets were used to investigate both derivatives for each gene fusion: derivative 22: ZC3H7B-Ex10F: 5′ – CCTTCGGCTTGGTCATGGAC – 3′; BCOR-Ex7R: 5′ –GAGACTTTGCGTTTCCTGTCCAC– 3′; derivative x: BCOR-Ex6F: 5′ – CGACTGGGAAAGGTTGAAAGG – 3′; ZC3H7B-Ex12R: 5′ – GATGAGCAAGGCAGTGTTGGG – 3′; derivative 1: MEAF6-Ex5F: 5′ – CTCAGGGAGTCACCACAGCAG – 3′ ; PHF1-Ex2R: 5′ - CCAAAGTGAGGAGGCACCAG – 3′; derivative 6: PHF1-Ex1F: 5′ – CTTTGGCTGCTGCGTCATAC – 3′; MEAF6-In6R: 5′ – GGTCTCAAAAAGGCATACTGGTG – 3′.
RESULTS
Pathologic Features
Fifty OFMTs were selected for the study based on their typical morphologic appearance; however, eleven cases were subsequently excluded due to FISH failure secondary to prior decalcification. Thus the study group was composed of thirty-nine tumors, showing classic histologic features and adequate tissue for FISH. There were 22 females and 17 males, with a mean age at diagnosis of 54 years-old (range 21–76). The tumor location included: lower extremities, 17 (thigh, 8; foot, 6; buttock/hip, 3); upper extremity, 7 (shoulder, 4; forearm, hand/finger); trunk, 9 (back/paraspinal, 3; axilla, 3; breast, 2; chest wall, 1) and head & neck, 6. Most tumors showed a multinodular growth pattern, surrounded by an incomplete fibrous pseudocapsule, which was variably ossified or hyalinized (Figs. 1A, B, D, E). Despite this rather well-defined appearance at low power, small microscopic foci were commonly present outside the fibrous capsule. Additionally, two of the superficial cases involving skin showed a diffuse, infiltrative growth up to the epidermis. In some tumors thick collagenous bands separated the tumor into large compartments (Fig. 1G).
Figure 1.
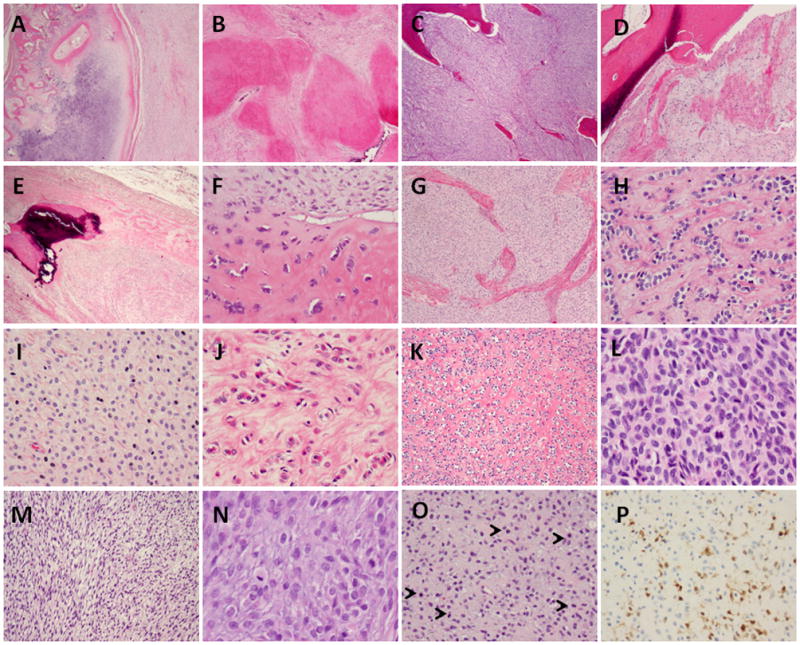
Morphologic spectrum of OFMT harboring different fusion transcript types. Low power reveals a thick peripheral rim of lamellar bone accompanied by an incomplete cartilaginous cap (A), other areas showed densely hyalinized nodules, lacking mineralization (B), while central areas showed the cellular component admixed with lamellar bone (C) (A–C, OFMT1, ZC3H7B-BCOR fusion); (D) Peripheral ossification in OFMT3 showing MEAF6-PHF1; (E) benign OFMT showing partly ossified partly hyalinized pseudocapsule (OFMT7, EP400-PHF1 fusion); (F) a subset of malignant OFMT showed lesional cells embedded within osteoid matrix, reminiscent of osteosarcoma (OFMT3, MEAF6-PHF1); or (G) thick fibrous bands separating the tumor into broad compartments (OFMT14, EP400-PHF1); (H) Benign OFMT with classic cord-like arrangement separated by a dense collagenous stroma (OFMT26, PHF1 rearranged); (I) epithelioid phenotype with more abundant eosinophilic cytoplasm (OFMT15, EP400-PHF1); or (J) a distinctive rhabdoid appearance (OFMT7, EP400-PHF1); (K) malignant OFMT showing a biphasic appearance composed of a benign hypocellular component associated with a conspicuous fibrous stroma, in abrupt transition to a (L) malignant cellular component with high mitotic activity (OFMT21, EP400-PHF1); (M) rare cases of malignant OFMT showed a more spindled/fusiform appearance, arranged in short intersecting fascicles (OFMT20, EP400-PHF1); (N) Most malignant OFMT showed increased cellularity and mitotic activity (OFMT1, ZC3H7B-BCOR); (O) with a loose extracellular stroma and focally very high mitotic activity (5 mitoses, highlighted with arrows) (OFMT33, PHF1 rearranged). (P) The pattern of S100 protein reactivity in malignant OFMT was typically focal (OFMT33, PHF1 rearranged).
Twenty-one cases were classified as benign, three were atypical and fifteen were malignant (Tables 1 and 2). Benign OFMT showed uniform cytomorphology, ranging from delicate, small cells with scant cytoplasm, ill-defined cell borders and uniform ovoid nuclei with fine chromatin (Fig. 1H), to more epithelioid cells with better defined cell borders and moderate amount of cytoplasm, ranging from amphophilic (Fig. 1I) to more densely eosinophilic, reminiscent of rhabdoid phenotype in two cases (Fig. 1J). Eight of the benign OFMTs showed peripheral ossification, while the remaining 13 did not. Atypical OFMT showed increased cellularity and a loose myxoid stroma, but was not accompanied by increased mitotic activity. All 3 atypical OFMTs showed areas of ossification.
Table 2.
Clinical and Pathologic Findings in Translocation-Negative OFMT (n=6)
| OFMT | Age/ Sex | Location | Histology | S100 | Desmin |
|---|---|---|---|---|---|
| 34 | 53/M | back | Benign, non-ossifying | Pos | Pos |
| 35 | 34/F | axilla | Benign, non-ossifying | Neg | Pos |
| 36 | 71/F | hip | Malignant, ossifying | Fpos | Pos |
| 37 | 24/M | back | Malignant, ossifying | Neg | Pos |
| 38 | 51/M | axilla | Malignant, ossifying | Fpos | Neg |
| 39 | 70/F | thigh | Malignant, ossifying | Neg | Pos |
F, female; M, male; Pos, positive; Neg, negative; Fpos, focally positive.
Despite their increased cellularity and brisk mitotic activity (Figs. 1K–O), most malignant OFMTs did not show highly pleomorphic components or large areas of spindling/fascicular growth. Instead, the tumors had a more compact/solid architecture, composed of packed oval to short fusiform cells separated by a loose intervening myxoid stroma (Fig. 1M). The myxoid quality of the extracellular matrix predominated in the malignant examples compared to the more densely fibrous or fibromyxoid stroma in the benign cases, where tumors showed a more rigid, cord-like epithelioid morphology. Most malignant OFMTs showed a peripheral shell of lamellar bone (Figs 1A,D); except for two cases were no ossification was noted. A subset of malignant OFMT had, in addition to the lamellar bone areas, focal osteoid-like matrix deposition, surrounding individual tumor cells, reminiscent of an extraskeletal osteosarcoma (Fig. 1F). Only two tumors showed large areas of necrosis.
Within the entire cohort, immunohistochemical stains for S100 protein was positive in 60% and desmin in 70% of cases. Most malignant OFMTs were negative for S100 protein, with only 4 cases showing focal (Fig. 1P) or more diffuse staining. The reverse was true in the benign and atypical OFMT, most showing reactivity for S100 protein, with only 4 tumors being negative. Desmin positivity was seen in half of the malignant OFMT, but in most (71%) of benign and atypical lesions.
FusionSeq Identifies Novel Fusion Involving ZC3H7B-BCOR
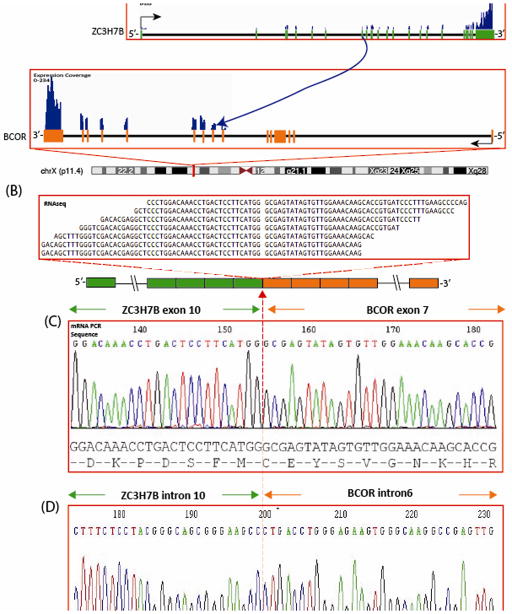
FusionSeq identified a ZC3H7B-BCOR fusion as the top candidate in OFMT1, a malignant OFMT (Figs. 2A,B). Alignment of the reads suggested a fusion of ZC3H7B exon 10 with exon 7 of BCOR, fusion transcript sequence, which was then confirmed by RT-PCR (Fig. 2C). Furthermore, FISH analysis using a fusion-assay showed rearrangements in both ZC3H7B and BCOR genes (Fig. 4). Long range DNA PCR, showed the fusion of intron 10 (732bp) of ZC3H7B with 1306 bp of intron 6 of BCOR (Fig. 2D). Remaining PHF1-negative cases were tested for BCOR gene abnormalities by FISH and one additional case was identified, OFMT2 (Table 1); however this lacked a break-apart signal in ZC3H7B. Interestingly, both cases were classified as malignant but with typical ossification, occurred in the thigh of males aged 55 and 76 respectively, and were negative for both S100 protein and desmin (Figs. 1A–C, N).
Figure 2.
ZC3H7B-BCOR gene fusion in malignant ossifying fibromyxoid tumor (OFMT1). (A) Schematic representation of the ZC3H7B-BCOR fusion indicating the loci that are joint together; ZC3H7B exon 10 being fused to BCOR exon 7; (B) RNA reads covering the fusion junction were isolated independent to FusionSeq analysis work flow, supporting the ZC3H7B-BCOR fusion candidate; (C) Experimental validation of the fusion by RT-PCR shows the junction sequence between exon 10 of ZC3H7B and exon 7 of BCOR; (D) Long range DNA PCR showing the fusion of intron 10 of ZC3H7B to the intron 6 of BCOR.
Figure 4.
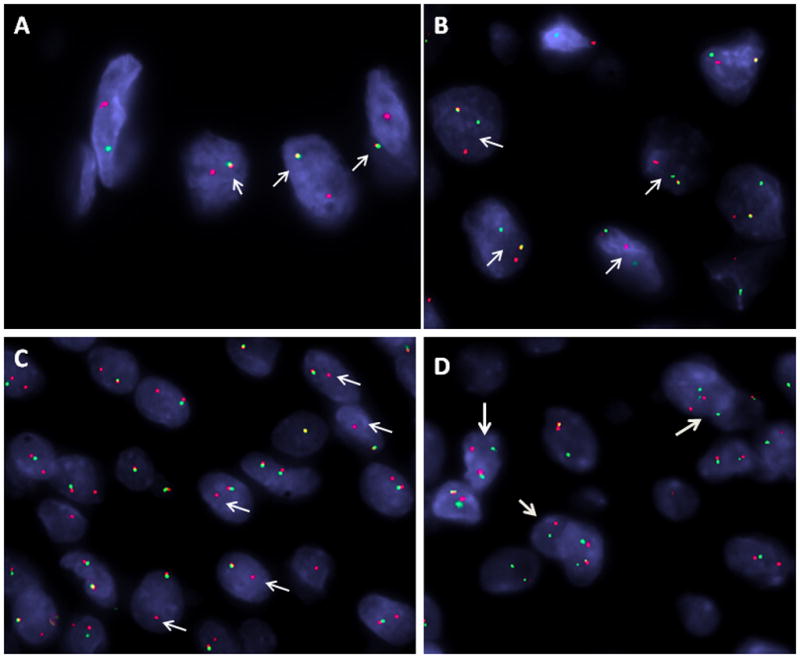
FISH validation of OFMT-related gene rearrangements. (A) Fusion assay with BCOR (green, telomeric) and ZC3H7B (red, centromeric) showing one yellow fused signal (OFMT1, male patient, only one BCOR allele on Xp11); (B) Break-apart assay showing a split MEAF6 signal (OFMT4; red centromeric, green telomeric); (C) Unbalanced EP400 gene rearrangement, showing break-apart signal with deletion of telomeric (green) part (OFMT14; red, centromeric); (D) PHF1 break apart signal (OFMT14; red centromeric, green, telomeric).
Novel MEAF6-PHF1 Fusions in a subset of S100 protein negative OFMTs
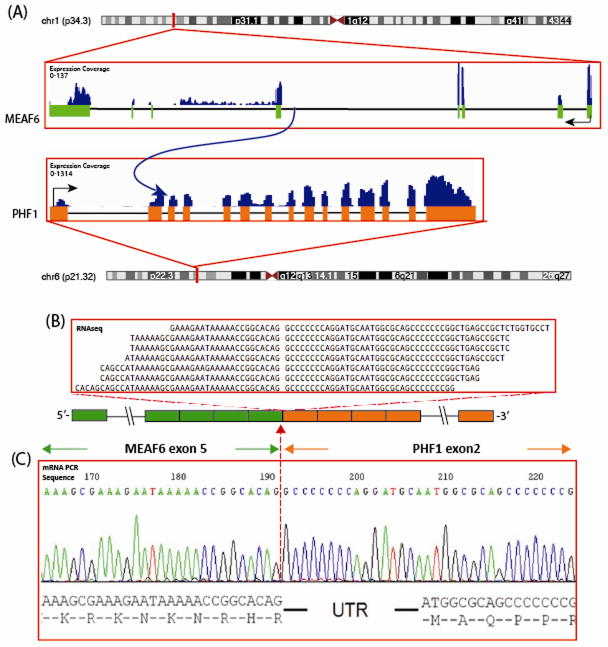
FusionSeq identified in the 2nd index case, OFMT3, a MEAF6-PHF1 as the top candidate. Alignment of the reads suggested a fusion of MEAF6 exon 5 with exon 2 of PHF1, which was confirmed by RT-PCR (Fig. 3). Exon 1 of PHF1 and the first 16 bp of exon 2 are untranslated. FISH confirmed the presence of PHF1 gene rearrangement, while showed only one copy of the MEAF6, in keeping with an unbalanced translocation. Long Range DNA PCR also failed to identify the intronic breakpoint, using a variety of different primer strategies. The remaining PHF1-rearranged cases, lacking an EP400 abnormality, were screened by FISH for potential MEAF6 break-apart signals. Two additional cases were positive for a MEAF6-PHF1 fusion (Fig 4, Table 1). The three MEAF6-PHF1-positive tumors showed a peripheral rim of lamellar bone but lacked S100 protein reactivity. One of them showed expression of desmin. Two of them occurred in the shoulder, and the third in the popliteal fossa. Two of them occurred in males and were classified as malignant, while the last one occurred in a woman and was benign. The index case OFMT3 showed, in addition to the mature shell of bone, focal areas of osteoid-like matrix deposition, surrounding individual tumor cells, reminiscent of an osteosarcoma (Fig. 1F).
Figure 3.
MEAF6-PHF1 gene fusion in malignant ossifying fibromyxoid tumor (OFMT3). (A) Schematic representation of the MEAF6-PHF1 fusion indicating the loci that are joint together; MEAF6 exon 5 being fused to PHF1 exon 2; (B) RNA reads covering the fusion junction were isolated independent to FusionSeq analysis work flow, supporting this fusion candidate; (C) Experimental validation of the fusion by RT-PCR shows the junction sequence between exon 5 of MEAF6 to exon 2 of PHF1.
EP400-PHF1 is the most common recurrent fusion in OFMT
PHF1 gene rearrangements were identified in 31/39 cases (80%). The most common fusion partner for PHF1 was EP400, present in 17 (55%) cases (Figs. 1G, I–L, 4). Of these, 11 (69%) cases were positive for S100 protein (with two cases being only focal) and twelve (75%) showed reactivity for desmin. Ten cases showed typical peripheral ossification and were variably distributed within extremity, trunk and head and neck. Nine were classified as benign (53%), while the remaining were divided among atypical or malignant.
OFMT share EPC1-PHF1 fusions with endometrial stromal sarcomas (ESS)
As EPC1 and JAZF1 have been described as additional gene partners involved in PHF1 fusions in ESS, we hypothesized that similar gene fusions may be implicated in OFMT with PHF1 gene rearrangements. Thus, 5 PHF1-rearranged OFMT cases lacking a fusion partner were tested by FISH for abnormalities in JAZF1 and EPC1. Two of the 5 cases showed EPC1 breakapart with an unbalanced telomeric deletion (Suplem Fig. 1, Table 1), while no JAZF1 gene abnormalities were seen in any of the cases. Both EPC1-PHF1 positive OFMT tumors were negative for S100 protein and one showed desmin reactivity.
PHF1-rearranged OFMT, lacking an identifiable fusion partner, is more often benign and S100 protein positive
Nine tumors were positive for PHF1 break-apart by FISH, but lacked abnormalities in EP400, MEAF6 and EPC1. No differences in anatomic location or morphology were noted between the different PHF1-rearranged genetic subsets (Figs. 1H, O, P). All except one was benign and all 8 tumors tested were S100 protein positive. Six (75%) tumors showed desmin reactivity. Six tumors were located in the extremity, while three cases were in the head and neck.
Fifteen percent of OFMT are fusion-negative
There were 6 (15%) tumors that were negative for all FISH probes tested and no frozen tissue was available for further RNA sequencing (Table 2). Four of these lesions occurred in the trunk, two each in the back and axilla, while the remaining two lesions occurred in the hip and thigh area. All except one case was desmin positive, while three showed S100 protein positivity (focally in two cases). Two of them were benign, while the others were classified as malignant. There were no discernible differences in their morphologic appearance when compared to fusion-positive tumors; four of them showed the classic peripheral rim of lamellar bone. All tumors in this group were tested for FUS gene abnormalities to exclude the possibility of an unusual low grade fibromyxoid sarcoma, the closest diagnostic mimic to non-ossifying OFMT.
DISCUSSION
Ossifying fibromyxoid tumor (OFMT) is a rare and enigmatic soft tissue tumor, of uncertain histogenesis and until recently of unknown genetic abnormalities. It was initially defined by Enzinger as a subcutaneous tumor with a peripheral or septal shell of bone, lobulated growth, and small, bland cells arranged in cords and nests within a fibromyxoid stroma (Enzinger et al., 1989). Most OFMT are benign, however, reports have documented both cyto-architecturally and clinically atypical OFMT, including cases with histologic features of malignancy (generally including some combination of high cellularity, nuclear atypia, and elevated mitotic activity) and confirmed metastatic disease (Kilpatrick et al., 1995; Folpe and Weiss, 2003). The diagnosis of malignant OFMT remains challenging and somewhat controversial, since it has not always been defined or accepted as the cellular and mitotically active counterpart of conventional OFMT. As such, atypical variants were reported with minimal or no conventional OFMT component and/or lack of bone formation, and a lower rate of S100 protein expression (Folpe and Weiss, 2003). S100 protein was reported as being almost universally expressed in typical, benign OFMT (Miettinen et al., 2008), while malignant OFMT cases have a significantly lower rate of S100 reactivity (Folpe and Weiss, 2003). This immunophenotypic difference has been proposed by some as evidence that malignant lesions are fundamentally distinct from conventional OFMTs (Miettinen et al., 2008), while others suggested that loss of S100 protein may be related to malignant transformation (Folpe and Weiss, 2003). As Miettinen et al. pointed out, the lack of objective diagnostic criteria in some of the reported atypical/malignant cases questions their subclassification as OFMT (Miettinen et al., 2008). Thus the identification of an unifying genetic marker, such as a recurrent tumor-specific translocation, has long been needed for the apparently wide morphologic spectrum of OFMT, which could then help in defining the “outer limits” of this entity and resolve the nature of biologically malignant tumors with some OFMT-like features (Miettinen et al., 2008).
Gebre-Medhin et al (Gebre-Medhin et al., 2012) recently identified the presence of PHF1 gene rearrangements on 6p21 in more than half of the OFMT lesions tested, with higher incidence in benign lesions (4/4) compared to malignant OFTs (1/6). In one of their tumors PHF1 was fused to EP400 on 12q24.3 (Gebre-Medhin et al., 2012). An additional OFMT case carrying a t(6;12)(p21;q24.3) resulting in a EP400-PHF1 fusion has subsequently been reported in a 71 year-old female with a soft tissue mass in the palm (Endo et al., 2013). Furthermore, a very recent study screening a larger cohort of 41 OFMTs confirmed the incidence of PHF1 gene rearrangements by FISH at nearly 50%, including roughly similar percentages of typical, atypical, and malignant tumors (Graham et al., 2013). The recurrent PHF1 abnormalities identified in both typical and malignant OFMTs suggest a shared pathogenesis for these lesions and suggest the utility of FISH testing for PHF1 gene rearrangements when diagnosing morphologically challenging cases. This is further supported by a gene expression profiling study, which showed extensive overlap between typical OFMT and the cases classified as malignant, in keeping with a single pathologic entity (Graham et al., 2011).
The PHF1 protein interacts with the polycomb-repressive complex 2 (PRC2), which, in turn, regulates the expression of a variety of developmental genes. PHF1 encodes the PHD finger protein 1 (PHF1), which is involved in chromatin structure regulation, forming Polycomb repressive complex 2 (PRC2) with EZH1, EZH2, SUZ12, regulating histone H3 at lysine 27 (H3K27) methylation.
RNA sequencing, followed by FusionSeq data analysis, in one of the index cases (OFMT1) showed the presence of a t(X;22)(p11;q13) translocation resulting in a ZC3H7B-BCOR fusion. A similar translocation has recently been identified in two cases of endometrial stromal sarcoma (Panagopoulos et al., 2013). Interestingly of the three patients with available karyotypes in the study of Gebre-Medhin et al, two cases that were positive for PHF1 gene rearrangements but negative for EP400 by FISH, showed an Xp11 locus abnormality, either involving a standard translocation with 6p, or in a three-way exchange with chromosomes 6 and 7 (Gebre-Medhin et al., 2012). Based on our findings, these cases with Xp11 locus rearrangements would suggest BCOR gene fusions. BCOR gene rearrangements have recently been described in a subset of small blue round cell tumors, as the 5′ fusion partner to CCNB3 (encoding the testis-specific cyclin B3, which induces oncogenic activation of the CCNB3 protein (Pierron et al., 2012). In contrast, in the OFMT setting, BCOR, encoding for BCL6 co-repressor, is the 3′ partner in the fusion with ZC3H7B, resulting in BCOR mRNA expression, suggesting a different mechanism of oncogenesis. Additionally a BCOR-RARA fusion has been reported in a variant of acute promyelocytic leukemia, resulting in a dominant-negative manner on RARA transcriptional activation (Yamamoto et al., 2010).
PHF1 rearrangements have previously been associated with endometrial stromal sarcoma (ESS), in that context being fused with either JAZF1 or EPC1 (Micci et al., 2006). The JAZF1-PHF1 and EPC1-PHF1 fusions account for a minority (9%) of ESS cases and have not been detected in benign endometrial stromal nodules (Chiang et al., 2011). The PHF1-chromosomal rearrangements are highly complex in both tumor types (Micci et al., 2006; Gebre-Medhin et al., 2012), and, due to the transcriptional orientation of the genes, neither JAZF1-PHF1 nor EPC1-PHF1 fusions can arise through a simple translocation (Micci et al., 2006). As seen in our OFMT3, PHF1-rearranged ESS showed a similar breakpoint within intron 1, thus the entire PHF1 coding region is translated in the chimeric protein, including its tudor, PHD zinc finger and MTF2 domains (Panagopoulos et al., 2012). Furthermore, similar with ESS pathogenesis, it appears that translocation genes involved in acetylation (MEAF6, EPC1) and methylation (PHF1) have a role in the neoplastic development of OFMT. EPC1 encodes a member of the polycomb group (PcG) family, which is a component of the NuA4 histone acetyltransferase complex and can act as both a transcriptional activator and repressor. Potential mistargeted acetylation or methylation most likely results in loosening of the heterochromatin and induction of aberrant gene expression. In light of our present findings, the recent case report of a cardiac ossifying sarcoma of non-endometrial stromal origin most likely represents a malignant OFMT, showing the JAZF1-PHF1 (Schoolmeester et al., 2013).
Furthermore, MEAF6-PHF1fusion has been described in one ESS from a 43 year-old female with classic morphology (Panagopoulos et al., 2012). MEAF6 is ubiquitously expressed and encodes a protein which is part of the histone acetyltransferase multisubunit complexes of the MYST family. The MYST histone acetyltransferases are highly conserved in eukaryotes and carry out a significant proportion of all nuclear acetylation, playing a critical role in gene-specific transcriptional regulation, DNA damage and repair (Cai et al., 2003; Avvakumov and Cote, 2007).
The presence of identical chromosomal translocations in ESS and OFMT, two pathologic entities with no morphologic or immunophenotypic overlap, is quite intriguing, although shared abnormalities have been described in a variety of other seemingly unrelated tumors, including EWSR1-CREB1 in angiomatoid fibrous histiocytoma and clear cell sarcoma (Antonescu et al., 2006; Antonescu et al., 2007), ETV6-NTRK3 in infantile fibrosarcoma, leukemia and secretory breast carcinoma (Knezevich et al., 1998; Tognon et al., 2002), and ALK-gene rearrangements in inflammatory myofibroblastic tumor, lung carcinoma and lymphoma (Morris et al., 1994; Lawrence, 2000; Soda et al., 2007).
In summary, our study identified three novel fusions ZC3H7B-BCOR, MEAF6-PHF1 and EPC1-PHF1 in OFMTs. With these additional gene fusions, the majority (85%) of OFMTs with classic morphologic appearances demonstrated recurrent gene rearrangements, regardless of the degree of malignancy, presence of ossification or immunoprofile, which can therefore be used as molecular markers in challenging cases. The most common abnormality is PHF1 gene rearrangement (80%), being present in benign, atypical and malignant lesions, with fusion to EP400 in 44% of cases. ZC3H7B-BCOR, MEAF6-PHF1 and EPC1-PHF1 fusions occurred predominantly in S100 protein-negative and malignant OFMT. Similar gene fusions have been reported in endometrial stromal sarcoma, a tumor seemingly unrelated to OFMT.
Supplementary Material
Supplementary Figure 1. FISH showing unbalanced rearrangement for EPC1 gene, with break-apart and loss of the centromeric (green) signal of EPC1 (OFMT23).
Supplementary Table 1. Custom BAC probes used for FISH analysis
Acknowledgments
We thank Agnes Viale and the Genomic Core Lab for the excellent expertise with RNA sequencing and Milagros Soto for editorial assistance.
Supported in part by: P01CA47179 (CRA, SS), P50 CA 140146-01 (CRA, SS).
Footnotes
Conflict of interest: none
References
- Antonescu CR, Dal Cin P, Nafa K, Teot LA, Surti U, Fletcher CD, Ladanyi M. EWSR1-CREB1 is the predominant gene fusion in angiomatoid fibrous histiocytoma. Genes Chromosomes Cancer. 2007;46:1051–1060. doi: 10.1002/gcc.20491. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Nafa K, Segal NH, Dal Cin P, Ladanyi M. EWS-CREB1: a recurrent variant fusion in clear cell sarcoma--association with gastrointestinal location and absence of melanocytic differentiation. Clin Cancer Res. 2006;12:5356–5362. doi: 10.1158/1078-0432.CCR-05-2811. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Zhang L, Chang NE, Pawel BR, Travis W, Katabi N, Edelman M, Rosenberg AE, Nielsen GP, Dal Cin P, Fletcher CD. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278:42733–42736. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- Chiang S, Ali R, Melnyk N, McAlpine JN, Huntsman DG, Gilks CB, Lee CH, Oliva E. Frequency of known gene rearrangements in endometrial stromal tumors. Am J Surg Pathol. 2011;35:1364–1372. doi: 10.1097/PAS.0b013e3182262743. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M, Kohashi K, Yamamoto H, Ishii T, Yoshida T, Matsunobu T, Iwamoto Y, Oda Y. Ossifying fibromyxoid tumor presenting EP400-PHF1 fusion gene. Hum Pathol. 2013;44:2603–2608. doi: 10.1016/j.humpath.2013.04.003. [DOI] [PubMed] [Google Scholar]
- Enzinger FM, Weiss SW, Liang CY. Ossifying fibromyxoid tumor of soft parts. A clinicopathological analysis of 59 cases. Am J Surg Pathol. 1989;13:817–827. doi: 10.1097/00000478-198910000-00001. [DOI] [PubMed] [Google Scholar]
- Folpe AL, Weiss SW. Ossifying fibromyxoid tumor of soft parts: a clinicopathologic study of 70 cases with emphasis on atypical and malignant variants. Am J Surg Pathol. 2003;27:421–431. doi: 10.1097/00000478-200304000-00001. [DOI] [PubMed] [Google Scholar]
- Gebre-Medhin S, Nord KH, Moller E, Mandahl N, Magnusson L, Nilsson J, Jo VY, Vult von Steyern F, Brosjo O, Larsson O, Domanski HA, Sciot R, Debiec-Rychter M, Fletcher CD, Mertens F. Recurrent rearrangement of the PHF1 gene in ossifying fibromyxoid tumors. Am J Pathol. 2012;181:1069–1077. doi: 10.1016/j.ajpath.2012.05.030. [DOI] [PubMed] [Google Scholar]
- Graham RP, Dry S, Li X, Binder S, Bahrami A, Raimondi SC, Dogan A, Chakraborty S, Souchek JJ, Folpe AL. Ossifying fibromyxoid tumor of soft parts: a clinicopathologic, proteomic, and genomic study. Am J Surg Pathol. 2011;35:1615–1625. doi: 10.1097/PAS.0b013e3182284a3f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RP, Weiss SW, Sukov WR, Goldblum JR, Billings SD, Dotlic S, Folpe AL. PHF1 Rearrangements in Ossifying Fibromyxoid Tumors of Soft Parts: A Fluorescence In Situ Hybridization Study of 41 Cases With Emphasis on the Malignant Variant. Am J Surg Pathol. 2013;37:1751–1755. doi: 10.1097/PAS.0b013e31829644b4. [DOI] [PubMed] [Google Scholar]
- Habegger L, Sboner A, Gianoulis TA, Rozowsky J, Agarwal A, Snyder M, Gerstein M. RSEQtools: a modular framework to analyze RNA-Seq data using compact, anonymized data summaries. Bioinformatics. 2011;27:281–283. doi: 10.1093/bioinformatics/btq643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick SE, Ward WG, Mozes M, Miettinen M, Fukunaga M, Fletcher CD. Atypical and malignant variants of ossifying fibromyxoid tumor. Clinicopathologic analysis of six cases. Am J Surg Pathol. 1995;19:1039–1046. doi: 10.1097/00000478-199509000-00007. [DOI] [PubMed] [Google Scholar]
- Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PH. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet. 1998;18:184–187. doi: 10.1038/ng0298-184. [DOI] [PubMed] [Google Scholar]
- Lawrence B, Perez-Atayde A, Hibbard MK, Rubin BP, Dal Cin P, Pinkus JL, Pinkus GS, Xiao S, Yi ES, Fletcher CD, Fletcher JA. TPM3-ALK and TPM4-ALK oncogenes in inflammatory myofibroblastic tumors. Am J Pathol. 2000;157:377–384. doi: 10.1016/S0002-9440(10)64550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micci F, Panagopoulos I, Bjerkehagen B, Heim S. Consistent rearrangement of chromosomal band 6p21 with generation of fusion genes JAZF1/PHF1 and EPC1/PHF1 in endometrial stromal sarcoma. Cancer Res. 2006;66:107–112. doi: 10.1158/0008-5472.CAN-05-2485. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Finnell V, Fetsch JF. Ossifying fibromyxoid tumor of soft parts--a clinicopathologic and immunohistochemical study of 104 cases with long-term follow-up and a critical review of the literature. Am J Surg Pathol. 2008;32:996–1005. doi: 10.1097/PAS.0b013e318160736a. [DOI] [PubMed] [Google Scholar]
- Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- Mosquera JM, Sboner A, Zhang L, Kitabayashi N, Chen CL, Sung YS, Wexler LH, Laquaglia MP, Edelman M, Sreekantaiah C, Rubin MA, Antonescu CR. Recurrent NCOA2 gene rearrangements in congenital/infantile spindle cell rhabdomyosarcoma. Genes Chromosomes Cancer. 2013;52:538–550. doi: 10.1002/gcc.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagopoulos I, Micci F, Thorsen J, Gorunova L, Eibak AM, Bjerkehagen B, Davidson B, Heim S. Novel fusion of MYST/Esa1-associated factor 6 and PHF1 in endometrial stromal sarcoma. PLoS One. 2012;7:e39354. doi: 10.1371/journal.pone.0039354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagopoulos I, Thorsen J, Gorunova L, Haugom L, Bjerkehagen B, Davidson B, Heim S, Micci F. Fusion of the ZC3H7B and BCOR genes in endometrial stromal sarcomas carrying an X;22-translocation. Genes Chromosomes Cancer. 2013;52:610–618. doi: 10.1002/gcc.22057. [DOI] [PubMed] [Google Scholar]
- Pierron G, Tirode F, Lucchesi C, Reynaud S, Ballet S, Cohen-Gogo S, Perrin V, Coindre JM, Delattre O. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet. 2012;44:461–466. doi: 10.1038/ng.1107. [DOI] [PubMed] [Google Scholar]
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sboner A, Habegger L, Pflueger D, Terry S, Chen DZ, Rozowsky JS, Tewari AK, Kitabayashi N, Moss BJ, Chee MS, Demichelis F, Rubin MA, Gerstein MB. FusionSeq: a modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data. Genome Biol. 2010;11:R104. doi: 10.1186/gb-2010-11-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoolmeester JK, Sukov WR, Maleszewski JJ, Bedroske PP, Folpe AL, Hodge JC. JAZF1 rearrangement in a mesenchymal tumor of nonendometrial stromal origin: report of an unusual ossifying sarcoma of the heart demonstrating JAZF1/PHF1 fusion. Am J Surg Pathol. 2013;37:938–942. doi: 10.1097/PAS.0b013e318282da9d. [DOI] [PubMed] [Google Scholar]
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, Bando M, Ohno S, Ishikawa Y, Aburatani H, Niki T, Sohara Y, Sugiyama Y, Mano H. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- Tanas MR, Sboner A, Oliveira AM, Erickson-Johnson MR, Hespelt J, Hanwright PJ, Flanagan J, Luo Y, Fenwick K, Natrajan R, Mitsopoulos C, Zvelebil M, Hoch BL, Weiss SW, Debiec-Rychter M, Sciot R, West RB, Lazar AJ, Ashworth A, Reis-Filho JS, Lord CJ, Gerstein MB, Rubin MA, Rubin BP. Identification of a disease-defining gene fusion in epithelioid hemangioendothelioma. Sci Transl Med. 2011;3:98ra82. doi: 10.1126/scitranslmed.3002409. [DOI] [PubMed] [Google Scholar]
- Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, Becker L, Carneiro F, MacPherson N, Horsman D, Poremba C, Sorensen PH. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–376. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Tsuzuki S, Tsuzuki M, Handa K, Inaguma Y, Emi N. BCOR as a novel fusion partner of retinoic acid receptor alpha in a t(X;17)(p11;q12) variant of acute promyelocytic leukemia. Blood. 2010;116:4274–4283. doi: 10.1182/blood-2010-01-264432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. FISH showing unbalanced rearrangement for EPC1 gene, with break-apart and loss of the centromeric (green) signal of EPC1 (OFMT23).
Supplementary Table 1. Custom BAC probes used for FISH analysis