Abstract
Multiple Sclerosis (MS) is an autoimmune disease that affects the central nervous system. One approved treatment for relapsing forms of MS is YEAK, a random copolymer of the amino acids Tyr, Glu, Ala and Lys. YFAK, a second-generation copolymer composed of Tyr, Phe, Ala and Lys, is more successful in treating experimental autoimmune encephalitis (EAE), a mouse model of MS. Although originally designed and optimized based on the autoantigen myelin basic protein (MBP) and the MBP-derived peptide MBP85-99 presented to the MS-associated Class II MHC molecule HLA-DR2, YEAK and YFAK also stimulate cytokine and chemokine production in APCs that lack Class II MHC products. How YEAK and YFAK copolymers interact with APCs remains enigmatic. We used biotinylated YFAK to affinity-purify YFAK-interacting proteins from RAW264.7 cells and tested APCs from mice deficient in several of the newly identified interactors for their capacity to secrete CCL22 in response to YEAK and YFAK. We propose that initial contact of YFAK with cells is mediated mainly by electrostatic interactions, and find that interaction of YFAK with host proteins is strongly dependent on ionic strength. Cells deficient in enzymes involved in sulfation of proteins and proteoglycans showed strongly reduced binding of biotinylated YFAK. Last, cells stimulated with YFAK in the presence of heparin, structurally similar to heparan sulfates, failed to produce CCL22. We conclude that charge-dependent interactions of copolymers that alleviate MS/EAE are critical for their effects exerted on APCs and may well be the main initial mediators of these therapeutically active copolymers.
Introduction
Multiple Sclerosis (MS) is a chronic inflammatory autoimmune disease that affects the central nervous system (CNS). CD4+ T cells that recognize antigens of the myelin sheath are held responsible for the inflammation and demyelination that characterize MS. The random amino acid copolymers Copaxone® (Copolymer-1, glatiramer acetate, poly(Y, E, A, K)n, called YEAK) and poly(Y, F, A, K)n (hereafter referred to as YFAK) ameliorate experimental autoimmune encephalomyelitis (1-4) and other autoimmune diseases (5). Notwithstanding its heterogeneous composition, YEAK is one of the few compounds approved for clinical treatment of MS. YFAK and YEAK may suppress inflammation via regulatory and TH2-polarized CD4+ T cells that secrete IL-10, (1, 2, 6-8), via IL-10 producing B cells (9-11), or by modulating cytokine and chemokine secretion by dendritic cells and macrophages (12-14). Bone marrow-derived myeloid cells (15-17) and RAW264.7 cells exposed to YFAK and YEAK secrete CCL22, a chemoattractant for regulatory and TH2 helper T-cells. Whereas the cellular targets cells have been defined especially for YEAK (15, 16), not much is known about the molecular target(s) and mechanism(s) of action of these copolymers. They have been variously proposed to be presented via Class II MHC products or to engage TLRs, but Class II MHC-deficient antigen presenting cells still respond to copolymers (12, 16, 17). Here we sought to identify molecular target(s) by searching for proteins and other macromolecules that interact with biotinylated versions of YFAK and YEAK. We identify gp96 as the major interaction partner of YFAK and YEAK in RAW264.7 cells, confirmed by analysis of gp96-deficient cell lines and primary antigen presenting cells. Although gp96 is a strong interactor of the copolymers, binding of YFAK to gp96 cannot account for all of its biological effects. We show that copolymers bind to sulfated proteins and/or sulfated glycosaminoglycans, most likely through electrostatic interactions.
Material and Methods
Copolymers and modification of copolymers
YEAK was obtained from Hanna Pharmaceuticals (Wilmington, DE). YFAK was synthesized as described in (12). The random copolymers were biotinylated with a twofold molar excess (assuming an average MW for the copolymers of 5500 Da (YFAK) and 7000 Da (YEAK)) of Sulfo ChromaLink Biotin reagent (Solulink, Inc., San Diego) in 50mM sodium phosphate pH=7.6, 75mM sodium chloride and 37.5% (v/v) acetonitrile for 14 hours at room temperature. Copolymers were resolved from hydrolyzed/unreacted biotin reagent on a silica-based TSK-Gel G2000 SW size exclusion column (Tosoh Bioscience LLC, King of Prussia, PA). Labeling efficiency was determined spectroscopically to be approximately one biotin molecule per four YFAK molecules and one biotin molecule per six YEAK molecules. We aimed for a low degree of substitution with biotin to minimize structural alterations in the already quite heterogeneous copolymers. .
Cells and cell lines
RAW264.7 cells were obtained from ATCC (TIB-71). B70Z/30 NF-kB-GFP and E4.126 cells (18) were a gift from Brian Seed (Harvard Medical School). Murine bone marrow-derived dendritic cells (BMDCs) were generated by culturing bone marrow cells in RPMI supplemented with 10% inactivated fetal calf serum, 0.0005% (v/v) β-mercaptoethanol, 0.1mM non-essential amino acids, 1mM pyruvate (Gibco), 1ng/ml IL-4 (R&D Systems) and 1ng/ml GM-CSF (R&D Systems) for seven days.
Radiolabeling and affinity purification experiments
For biosynthetic labeling, cells were fed overnight (~14-18 hours) with 50μl [35S]methionine/cysteine (1175Ci/mmol, Perkin Elmer Life Sciences) per ml DMEM supplemented with 10% IFS. Cells were washed once in PBS, detached in PBS containing 5mM EDTA, and washed once again in PBS. Cells were lysed in 12.5mM HEPES pH 7.76, 50mM NaCl (or as otherwise indicated in the text), 5mM CaCl2, 2.5mM MgCl2, 1% w/v digitonin and ‘complete protease inhibitor’ cocktail tablets (Roche) for one hour. Lysates were centrifuged for 20 minutes at 12.000xg and equal amounts of radioactive lysate (based on trichloroacetic acid-precipitable cpm) of soluble fractions (‘cleared lysates’) were used for affinity purification experiments. YFAK-biotin or YEAK-biotin were added to the lysates for three hours and captured with High Capacity NeutrAvidin beads (Pierce Biotechnology, Inc.) for one hour. Beads were washed four times in lysis buffer with 0.1% digitonin.. Polypeptides were either eluted in reducing SDS sample buffer or in Glycoprotein Denaturing Buffer (New England Biolabs) for subsequent treatment with EndoH or PNGaseF (both obtained from New England Biolabs). EndoH and PNGaseF digestions were performed according to the manufacturer's instructions.
Large-scale affinity purification experiments and mass-spectrometry analysis
Approximately 2×108 RAW264.7 cells per condition were lysed in digitonin lysis buffer as described above. Cleared lysates were incubated with approximately 250μg biotinylated YFAK or YEAK for at least four hours. Copolymers with interacting molecules were retrieved with 250μl (bed volume) NeutrAvidin beads. After extensive washing in digitonin wash buffer (0.1% digitonin) beads were eluted by boiling in reducing SDS sample buffer containing 10μM biotin.. Samples were resolved on 10% SDS-polyacrylamide gels. Gels were stained with colloidal Coomassie (19), slices of individual lanes were excised, proteins were digested with trypsin, and analysed by tandem mass-spectrometry.
GO analysis
GO analyses were based on the list of 222 proteins (Section (A) of Supplemental Table 1) that were recovered in association with biotinylated YFAK from RAW264.7 lysates in both of two independent experiments. GO term enrichments for the different GO categories and p values plotted in the right panels of figure 2 were determined with the GeneGO MetaCore program (https://portal.genego.com/). To calculate the counts of enriched GO terms depicted in the left panels of figure 2, we performed GO analysis on a reduced set of GO categories (GO-slim) on mouse gene models available through the GO tools at http://go.princeton.edu.
FIGURE 2.
Biotinylated copolymers bind to a diverse set of proteins in RAW264.7 cells. (A) Top panel: UV traces of the chromatographically separated biotinylation reaction mixture for YFAK. Bottom panel: UV traces of pooled fractions of the first peak of the biotinylation reaction mixture for YFAK (purified biotinylated YFAK). (B) Straptavidin blot of unlabelled YFAK (-) and purified biotinylated YFAK from bottom panel in figure 2A (+). (C) FACS blots of RAW264.7 cells stained with biotinylated FYAK. Left panel: Grey shade: SA-PE staining; dotted line: Staining with hydrolyzed biotinylating reagent and SA-PE; solid line: CD11-PE staining. Right panel. Grey shade: SA-PE staining; thin solid line: 0.01μg/ml biotinylated YFAK and SA-PE; dotted line: 0.1μg/ml biotinylated YFAK and SA-PE; dashed line: 1μg/ml biotinylated YFAK and SA-PE; bold solid line: 10μg/ml biotinylated YFAK and SA-PE. (D) Autoradiograph of co-precipitating proteins with biotinylated YFAK or YEAK from 35S-Met/Cys-labelled RAW264.7 cell lysate. (E) Autoradiograph of PNGaseF-treated co-precipitating proteins with biotinylated YFAK from 35S-Met/Cys-labelled RAW264.7 cell lysate. Asterisk indicates gp96.
Immunoblotting and antibodies
RAW264.7 cells were lysed in digitonin lysis buffer for one hour. Cell debris and nuclei were pelleted and discarded. YFAK-biotin was added to cleared cell lysates and incubated for another hour. YFAK-biotin was recovered by NeutrAvidin beads and gp96 was recovered with a polyclonal anti-gp96 rabbit serum and protein-G beads. Recovered proteins were separated by Tris-Glycine (for detection of gp96) or Tris-Tricine (to detect YFAK-biotin) SDS-PAGE. Proteins were transferred to PVDF membranes and (immuno)blotted with standard methods. YFAK-biotin was detected with Streptavidin-HRP (GE Healthcare), gp96 was detected with the same polyclonal rabbit antibody that was used for immunoprecipitation.
FACS analysis
Cells were detached in PBS containing 5mM EDTA, washed in cold PBS containing 2% (w/v) BSA (FACS buffer) and incubated with the indicated concentrations of biotinylated copolymers for 30 minutes on ice. Cells were washed with FACS buffer and incubated on ice with 0.5μg/ml Streptavidin-PE (Invitrogen) for another 30 minutes. After washing with FACS buffer, cells were analyzed with a FACSCalibur flow cytometer (BD Biosciences). FACS data were analyzed using Flowjo software (Tree star, OR)
ELISA
ELISA assays were done as described (12). In short, 500.000 cells were stimulated in 250μl medium containing different concentrations of stimulant. For experiments shown in Fig. 7, 500.000 RAW267.4 cells or 100.000 BMDCs were stimulated for 4 hours in serum-free medium. LPS (from E.coli 055:B5) was obtained from Sigma-Aldrich. CCL22 ELISA kits were purchased from R&D Systems and used according to the manufacturer's protocol.
FIGURE 7.
Heparin treatment reduces CCL22 secretion in response to YFAK. (A) CCL22-ELISA of supernatant of RAW267.4 cells stimulated for 4 hours with 50μg/ml YFAK in presence (black bars) or absence (white bars) of 100μg/ml heparin in serum-free medium. (B) CCL22-ELISA of supernatant of BMDCs stimulated for 4 hours with 50μg/ml YFAK in absence (white bars) or presence (black bars) of 12.5, 25, or 50μg/ml heparin (titration indicated by black wedges below graph) in serum-free medium. n.d.: not detected.
Mice
Wild-type C57BL/6 mice were obtained from the Jackson Laboratory. Animals were housed at the Whitehead Institute for Biomedical Research and maintained according to protocols approved by the Massachusetts Institute of Technology Committee on Animal Care. Bones from gp96−/− animal bones were provided by Zihai Li (Medical University of South Carolina), bones from CD91−/− and CD91−/−LDLr−/− deficient animals were provided by Dudley Strickland (University of Maryland). Bones from MyD88−/−TRIF−/− deficient animals were provided by Kate Fitzgerald (University of Massachusetts, Worcester) and from App−/− animals by Tracy Young-Pearse, Harvard Medical School).
Results
Copolymers elicit CCL22 secretion in RAW264.7 cells and in mouse BMDCs
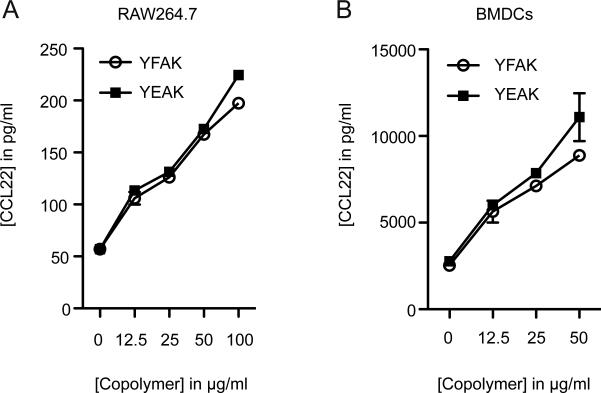
The copolymer YFAK was designed to optimize the YEAK copolymer for treatment of EAE. The immunosuppressive effect of YEAK has been ascribed to its binding to Class II MHC molecules, which would then interfere with the interaction of MHC Class II/autoantigenic peptide complexes and autoreactive TCRs (3). Glutamic acid was replaced by phenylalanine to improve binding to Class II MHC molecules, based on the known binding motifs of the autoantigenic peptide MBP85-99 to HLA-DR2 (DRB1*1501) (3). Only recently attention was drawn to Class II MHC-independent effects of the YFAK and YEAK copolymers on innate immune cells, such as monocytes, macrophages and dendritic cells (12, 15-17).We confirmed the earlier observation that exposure to YFAK and YEAK stimulate RAW264.7 cells (Fig. 1A) and BMDCs (Fig. 1B) to secrete the TH2 and Treg chemoattractant CCL22. Attempts to identify an innate immune receptor using various knock-out and mutant cells as well as blocking antibodies had been unsuccessful (17).
FIGURE 1.
RAW264.7 cells and BMDCs secrete CCL22 after stimulation with random copolymers. (A) RAW264.7 (shown are means and ±SEM of triplicates from one experiments) or (B) BMDCs were stimulated with different concentrations of YFAK or YEAK for 24 hours (shown are average values and ±SEM of three independent experiments for YFAK and mean and ±SEM of one experiment for YEAK). CCL22 concentrations were measured by ELISA.
Biotinylation of YFAK and YEAK
To obtain insight into the molecular mechanism of action of YFAK and YEAK on antigen presenting cells, we sought to identify interacting partners of the copolymer. We prepared biotinylated versions of either copolymer by chemical modification at lysine residues, using a reagent that includes a biotin portion, a (PEG)3 linker, a bis-aryl hydrazone chromophore and an amine-reactive succinimidyl ester. Unreacted and hydrolyzed labeling reagents were separated from the copolymers by size-exclusion chromatography (shown for YFAK in Fig. 2A). The presence of the aryl hydrazone chromophore in the copolymer peaks indicated successful biotinylation. After electrophoretic separation by Tris-Tricine SDS-PAGE and transfer to PVDF membranes, the biotinylated copolymers were readily detected with streptavidin-HRP (shown for YFAK-biotin in Fig. 2B), confirming successful conjugation. Consistent with their inherent heterogeneity, both copolymers presented themselves as a diffuse smear over a range of apparent molecular weights. To avoid significant alteration of the immunological properties of the copolymers we established conditions that resulted in substoichiometric modification. Capitalizing on the bis-aryl hydrazone chromophore's absorption at λ=354nm, labeling efficiency was determined to be approximately 0.31 or 0.17 moles of biotin per mole of YFAK or YEAK, respectively.
Copolymers bind to the surface of RAW264.7
We next examined the ability of the biotinylated copolymers to bind to intact cells. RAW264.7 cells were incubated for 30 minutes at 4°C with different concentrations of YFAK-biotin (0.01, 0.1, 1 and 10μg/ml) and then stained with PE-conjugated streptavidin (SA-PE). We observed concentration-dependent binding of YFAK-biotin to the cell surface, with maximal binding at 10μg/ml amongst the concentrations tested here (Fig. 2C, right panel). Binding was specific, since neither SA-PE alone nor hydrolyzed ChromaLink reagent plus SA-PE gave any signal above background (Fig. 2C, left panel).
Affinity purification using biotinylated copolymers
Given that YFAK can stimulate RAW264.7 cells to secrete CCL22 and that YFAK-biotin binds to RAW264.7 cells, we next sought to identify proteins capable of binding to YFAK-biotin.. We prepared digitonin extracts of 35S-Met/Cys labeled RAW264.7 cells. YFAK-biotin was added to the lysate and subsequently recovered with NeutrAvidin-conjugated beads. As shown in Figure 2D, neither NeutrAvidin beads alone nor non-biotinylated YFAK recovered significant amounts of protein from the cell lysates. In contrast, both biotinylated YFAK and YEAK retrieved a diverse set of co-precipitating proteins. The overall pattern of recovered proteins was similar for both copolymers, although some unique bands were present for each condition.
We reasoned that APCs might detect YFAK by means of (a) cell surface receptor(s) that would acquire N-linked sugar modification(s) while passing through the secretory pathway. To distinguish proteins with N-linked glycan(s) from proteins without them, we treated the samples with the N-glycosidase PNGaseF. Indeed, some of the interaction partners of YFAK were sensitive to PNGaseF (Fig. 2E). The most intense signal under the conditions used was a glycoprotein that migrated slightly below the 100kDa marker (Fig. 2E, asterisk).
Next, we analyzed the polypeptides recovered in complex with YFAK-biotin and YEAK-biotin by mass spectrometry to establish their identity. We performed two independent experiments with YFAK-biotin from RAW264.7 cell lysates. For subsequent analysis we created a list of proteins recovered in both experiments (Supplemental Table 1). Furthermore, proteins for which the coverage, number of unique peptides or number of total peptides recovered in the control sample (NeutrAvidin beads only) was 20% or higher than in the experimental sample (YFAK-biotin added) were deleted from the list. Applying these criteria, we obtained a list of 222 interactors of YFAK-biotin for which we performed a Gene Ontology (GO) term enrichment analysis for the three GO categories ‘cellular component’, ‘biological process’ and ‘molecular function’ (Fig. 3 and Supplemental Table 1). The most significantly enriched molecular function was ‘RNA binding’ (p value 2.6*10-29), followed by ‘aminoacyl-tRNA ligase activity’, ‘ligase activity, forming aminoacyl-tRNA and related compounds’ and ‘ligase activity,forming carbon-oxygen bounds’ (p values 1.3*10-20, respectively). In terms of absolute numbers,’ion binding’ and again ‘RNA binding’ were the most abundant GO terms in the ‘molecular function’ category.
FIGURE 3.
Gene ontology (GO) term enrichment analysis of FYAK-interacting proteins. Top 10 GO terms were ranked according to numbers of counts (left panels) or p values (right panels) of GO enrichment analyses for the (A) ‘cellular component’, (B) ‘biological process’ or (C) ‘molecular function’ category of FYAK-associated proteins. See Material and Methods sections for description of GO term enrichment analysis. The full list of GO terms is available in Supplementary Table 1.
Copolymers interact with gp96
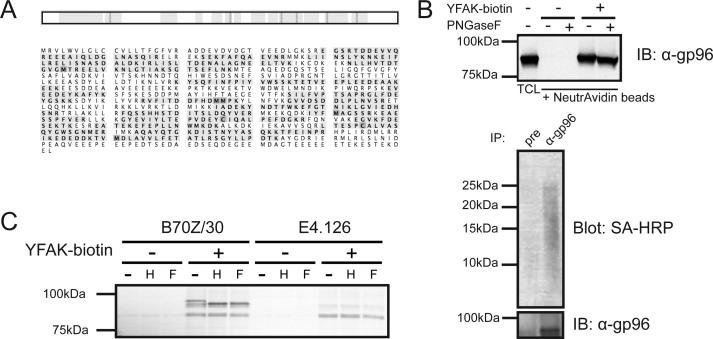
Amongst the interacting proteins identified here, one prominent hit consistently identified in the two independent affinity purification experiments and with known involvement in immune signaling was gp96 (also known as endoplasmin, GRP94, heat shock protein 90 beta or ERp99). Fig. 4A shows the sequence coverage from one such experiment: 45 unique gp96-derived peptides were recovered, which corresponds to a sequence coverage of 49%. Coprecipitated gp96 was detected also by immunoblot after retrieval of YFAK-biotin bound materials and vice versa (Fig. 4B), confirming the interaction of YFAK with gp96. PNGaseF sensitivity of gp96 (fig. 4B) is consistent with previous data, showing that gp96 is a glycosylated protein with five potential N-glycosylation sites (20, 21), not all of which are used. We confirmed these results by a similar affinity purification experiment using YFAK-biotin on lysates from the gp96-deficient B-cell line E4.125 (18) and its parental gp96-proficient line B70Z/3 (Fig. 4C). Only the latter yielded a positive signal for gp96.
FIGURE 4.
YFAK interacts with gp96. (A) Exemplary sequence coverage of gp96 co-precipitated with biotinylated FYAK from RAW264.7 cell lysate. Top: Schematic map of gp96 (white bar) and recovered peptides (yellow, overlaps in green); bottom: Amino acid sequence of gp96 with recovered peptides (yellow, overlaps in green). (B) Top: Anti-gp96 immunoblot of co-precipitates with YFAK-biotin from RAW264.7 cell lysate; bottom: Streptavidin and anti-gp96 blot of immunoprecipitates with an anti-gp96 rabbit serum or pre-immune serum from the same rabbit (‘pre’). (C) Autoradiograph of co-precipitating proteins with biotinylated YFAK from 35S-Met/Cys-labelled B70Z/30 (gp96-proficient) and E4.126 (gp96-deficient) cell lysate. H: EndoH treated, F: PNGaseF treated.
CCL22 secretion in cells deficient for interactors of YFAK and YEAK
Next, we tested whether the absence of some of the proteins identified as interactors of YFAK-biotin has consequences for CCL22 production in antigen presenting cells (APCs). We generated BMDCs from gp96-deficient bone marrow and stimulated cells with different concentrations of YFAK, YEAK or the TLR4 agonist LPS. As expected, wild-type BMDCs secreted elevated amounts of CCL22 in response to LPS (Fig. 5A, lower panel) (12). In contrast, gp96−/− cells did not secrete additional CCL22 upon LPS stimulation, consistent with gp96's role as an essential chaperone for most TLRs, including TLR4 (Fig. 5A, lower panel) (22). However, upon stimulation with either YFAK or YEAK we saw no reduction in CCL22 secretion by gp96−/− cells (Fig. 5A, upper panel). We conclude that gp96 is not necessary for BMDCs to release CCL22 in response to YFAK or YEAK. Beyond that, it is unlikely that client proteins of gp96 are required for detection and downstream signaling events triggered by YFAK or YEAK. Substrates of gp96 include all TLRs except TLR3 (23), most of the integrin pairs found on cells of the hematopoetic system (exceptions are α5β1, α6β1, and αIIbβ3) (24), and several other surface molecules, including four members of the extended LDL receptor family and CD180/Ly86 (25).
FIGURE 5.
CCL22 secretion upon YFAK and YEAK stimulation is not altered in APCs from gp96-deficient, MyD88−/−TRIF−/− or LRP-deficient mice. (A) Gp96-deficient BMDCs (one experiment, n=1), (B) MyD88−/−TRIF−/− BMDCs (DKO), (mean values ±SEM of two independent experiments, n=1 per genotype and experiment) and (C) LRP-deficient BMDMs (mean values ±SEM of two mice per genotype, one experiment) were stimulated for 24 hours with 12.5, 25, 50 or 100μg/ml of YFAK or YEAK (upper panels) and CCL22 in the culture supernatants was measured by ELISA. Gp96-deficient and MyD88/TRIF-double deficient cells were stimulated with 1, 10, 100 or 1000ng/ml LPS as well (lower panel in (A) and (B).
To test whether any of the TLRs is necessary for copolymer-dependent CCL22 secretion, we stimulated MyD88−/−TRIF−/− double knockout BMDCs. As predicted, MyD88−/−TRIF−/−double knockout BMDCs were unresponsive to LPS (Fig. 5B, lower panel). However, CCL22 production evoked by YFAK or YEAK was unaffected (Fig. 5B, upper panel), ruling out a role for toll-like receptor, IL-1 receptor or IL18 receptor signaling (and other MyD88 and TRIF-dependent pathways (26, 27)). This result also excludes the possibility of CCL22 secretion due to contamination of YFAK or YEAK with microbial components (e.g. LPS), that signal through TLRs.
Another interactor identified in the affinity purification experiments with biotinylated copolymers was CD91 (also known as LRP1). Several structurally diverse ligands have been identified for CD91, including various heat shock proteins such as gp96 (28, 29), calreticulin and HSP90 (29), but also P. aeroginosa exotoxin A (30), RAP, alpha 2-macroglobulin (31), lipoprotein lipase and others (32). Some immunomodulatory heat shock proteins, such as gp96, modulate helper T cell polarization through CD91 (33). Heparan sulfate is important for binding of gp96 to CD91 (34). YFAK binds to both CD91 and some of its ligands (e.g. gp96 and HSP90, Supplemental Table 1) and, as will be shown below, binding of YFAK to cells is mediated mainly by heparan sulfate. Thus, we asked whether YFAK or YEAK might also signal through CD91 to facilitate CCL22 secretion by APCs. We stimulated CD91-deficient BMDMs from CD91fl/fl LysM-Cre mice with YFAK or YEAK. However, we saw no difference in the amounts of CCL22 secreted (Fig. 5C). Since the functions of CD91 and the LDL receptor partly overlap (35), we also examined BMDMs from LDLr−/−CD91fl/fl LysM-Cre mice. CCL22 secretion by these double knock-out cells was unaffected as well (data not shown). We conclude that neither LDLr nor CD91 are essential for CCL22 secretion in response to YFAK and YEAK.
We further identified the amyloid beta precursor protein (App) as an interactor of YFAK. App is a cell surface receptor best known as the precursor for different amyloid beta species implicated in Alzheimer's disease. Although App is expressed on myeloid cells, not much is known about the function of App in these cells. App comprises an intracellular domain presumed capable of signal transduction. Thus, we asked whether App is involved in the signaling cascade(s) triggered by YFAK or YEAK. We stimulated BMDCs from App−/− mice and measured CCL22 in the cell culture supernatant. Again, we saw no difference in secretion of CCL22 between BMDCs from wild-type control mice and App-KO mice (data not shown), excluding an essential role for App in this signaling pathway.
YFAK binds to heparan sulfate proteoglycans
Since lysine residues are a key component of YFAK, we reasoned that the observed interactions might be mediated by electrostatic interactions. To examine this possibility we first tested if binding of YFAK-biotin to target proteins is disrupted by raising of ionic strength. Recovery of most if not all of the proteins gradually decreased with increasing salt concentrations in the lysis buffer (Fig. 6A), consistent with the notion that YFAK binds through electrostatic interactions. Does YFAK also bind to negatively charged heparan sulfate proteoglycans (HSPGs)? To test this, we performed cell binding assays with three recently described knock-out cell lines that lack functional B3GAT3, B4GALT7 or SLC35B2 (36). These enzymes are involved in sulfation of heparan sulfate precursors and other macromolecules. Although the parental Hap1 cell line bound readily detectable amounts of YFAK-biotin (Fig. 6B, top panel), binding to the KO lines with the above mentioned gene disruptions was much reduced (Fig. 6B, left column). YFAK binding was restored in cells in which the function of the disrupted genes had been reconstituted by transduction with an intact cDNA version of the disrupted gene (Fig. 6B, right panels), but not in cells reconstituted with cDNAs that encoded catalytically inactive mutants (Fig. 6B, middle panels). We conclude that YFAK binds to sulfated molecules on the cell surface via electrostatic interactions. We have not been able to demonstrate CCL22 production by the parental Hap1 line or its mutant derivatives under any condition of stimulation.
FIGURE 6.
Biotinylated YFAK interacts with heparan sulfate and other sulfated molecules. (A) Autoradiograph of co-precipitating proteins from cell lysates of 35S-Met/Cys-labelled RAW264.7 cells with gradually increasing sodium chloride concentrations present in the lysis and wash buffers. (B) FACS plots of different KO cell lines deficient in enzymes involved in sulfation of proteins and glysaminoglycans stained with YFAK. Left columns: KO cells transduced with empty vector; middle columns: KO cells transduced with HA-tagged enzymatically-inactive mutants of the deleted genes; right columns: KO cells transduced with HA-tagged enzymatically-active mutants of the deleted genes. Grey filled: SA-PE alone; no filling: YFAK-biotin (10μg/ml) and SA-PE.
To determine whether the interaction of YFAK with HSPGs has functional consequences, we stimulated RAW264.7 cells or BMDCs with YFAK in the presence or absence of heparin, a highly sulfated glycosaminoglycan structurally similar to but distinct from HSPGs (Fig. 7). We reasoned that if binding of YFAK to HSPGs is important to prompt APCs to eventually secrete CCL22, then preventing this interaction with a competitor such as heparin for HSPGs to bind to YFAK should reduce CCL22 production. Indeed, coincubation with YFAK and heparin abolished or markedly decreased CCL22 secretion in RAW264.7 cells (Fig. 7A) or BMDCs (Fig. 7B), respectively.
Discussion
We designed experiments to explore the molecular details of how random amino acid copolymers that alleviate EAE in vivo interact with cells of the innate immune system, notably macrophages and dendritic cells. By making use of biotinylated versions of these copolymers, we used an affinity purification strategy to recover interacting proteins. In this manner we identified proteins that interact with YEAK and YFAK, and defined sulfated glycosaminoglycans as a major cell surface structure necessary for interaction of YFAK with cells.
We generated biotinylated copolymers to affinity purify interacting proteins from RAW264.7 cell lysates. We found overlapping sets of proteins to interact with either copolymer. Gene ontology (GO) enrichment analysis for the ‘biological process’, ‘molecular function’ and ‘cellular component’ categories revealed diverse functions assigned to the hits. The most significantly enriched biological processes were ‘cellular macromolecule metabolic process’ and ‘gene expression’ and the most significantly enriched molecular function was ‘RNA binding’. Although not further investigated here, it is possible that the copolymers exert their effects by interfering with cellular events involved in generation or processing of (maybe specific) mRNAs and/or events involved in translation. It is unlikely that the copolymers gain access to the cytosol to exert their effect, given their size and charge, unless one were to postulate an endosomal escape route sufficiently efficient to allow delivery of the biologically active component to cytosolic targets. Regardless, the list presented here might serve as a resource for further studies on downstream cellular targets of the amino acid copolymers investigated here.
We were primarily interested in molecules displayed at the cell surface as possible copolymer targets, as this could identify (a) cell surface receptor(s) that is/are capable of activating APCs in a manner that would program helper T cells to a tolerogenic and/or anti-inflammatory response (as described for YEAK in (16)). First we focused on gp96 as the major interactor identified. Gp96 has been assigned an endogenous immunomodulatory function. Despite its C-terminal ER retention motif (KDEL), gp96 is found on the cell surface, a feature that appears to be phylogenetically conserved (37), although the cell biological mechanisms that underlie such surface display remain obscure. Gp96 participates in both adaptive and innate immune responses by eliciting Class I MHC-restricted CD8+ T cells specific for the antigenic peptides it chaperones (38-40), or by directly activating cells of the innate immune system to secrete cytokines (41, 42). Not only gp96, but also other immunogenic heat shock proteins dictate helper T cell responses by stimulating APCs in a CD91-dependent manner (33). Furthermore it was proposed that gp96 functions as a TH2-specific costimulatory molecule (43). Here we identify gp96 as a strong interactor of YEAK and YFAK. We confirmed this association by reciprocal precipitation, followed by immunoblotting in RAW264.7. We used a gp96-deficient cell line to further validate these results. However, gp96 is not essential for CCL22 production as shown by the analysis of gp96-deficient BMDCs. Secretion of CCL22 in response to copolymers occurs also in the absence of gp96. This finding implies that other proteins reliant on gp96 chaperone activity for their native fold (reviewed in (44)), are also dispensable for CCL22 induction by YEAK or YFAK. These include most integrin receptor pairs except α5β1, α6β1, and αIIbβ3 (24), at least four members (LDLR, LRP6, Sorl1 and LRP8) of the LDL receptor family (25), and all TLRs except TLR3 (45). Moreover, the interaction we observed for YFAK and LRP (CD91) is not required for CCL22 induction in BMDMs, again as verified by experiments in a CD91−/− background.
Experiments with MyD88/TRIF double-knockout BMDCs showed that neither of these two proteins is essential for CCL22 secretion stimulated by YEAK or YFAK. This result excludes any non-redundant role of MyD88, TRIF, any of the TLRs, IL-18R, IL-1R, and potentially other as-yet-unidentified MyD88 and/or TRIF-dependent pathways. Many TLRs and CLRs have also been excluded by a recent study (17).
Given the physicochemical properties and chemical heterogeneity of the copolymers investigated here, and in view of the negative results summarized above, we hypothesized that interactions of copolymers with their targets might be mediated by electrostatic and polar interactions and therefore be of a less specific nature. YFAK is composed of tyrosine, phenylalanine, alanine and lysine in a molar ratio of Y:F:A:K 1 : 1.2 : 23.5 : 6.0 (amino acid input mixture used for copolymer synthesis), which results in a strong net positive charge. YEAK (Y:E:A:K 1 : 1.5 : 4.5 : 3.6) is also positively charged, although not as strongly as YFAK, because glutamic acid residues contribute negative charges and will partly compensate for the charged lysine residues. In our affinity purification experiments with biotinylated copolymers in radiolabeled cell lysates we found that the amount of recovered proteins correlated inversely with the sodium chloride concentration in the buffers used, arguing in favor of our hypothesis. The interaction of streptavidin with biotinylated DNA is not affected by high sodium chloride concentration (46). We reasoned that the positively charged copolymers might interact with negatively charged structures on the cell-surface, such as sulfated proteoglycans and sialylated proteins or lipids. Indeed, YFAK did not bind to cells lacking functional SLC35B2 (3’-phosphoadenosine 5’-phosphosulfate transporter I (PAPSTI)) and bound much more weakly to cells lacking B3GAT3 (glucuronosyltransferase I), or B4GALT7 (galactosyltransferase I). B3GAT3- and B4GALT-deficient cells entirely lack heparan sulfates, but are able to produce other sulfated protein species. Cells deficient for PAPSTI retain approximately 5-10% of heparan sulfate levels compared to wild-type cells, but produce hardly any proteins with other sulfate modifications (36). Thus, YFAK predominantly binds to cells through electrostatic interactions. HSPGs account for the majority of these interactions, since binding to B3GAT3- and B4GALT-null cells is greatly reduced. However, because binding to B3GAT3- and B4GALT-null cells is less completely abolished while binding to SLC35B2 (PAPS transporter)-deficient cells is almost completely absent, YFAK also binds to other sulfated molecules (obvious candidates are sulfated tyrosines or chondroitin sulfate). Charge-based interactions of YFAK with target cells is functionally relevant, because cells stimulated with YFAK in the presence of the competitor heparin do not produce CCL22.
The exact role of this interaction remains to be defined. (Co)receptor(s) for YFAK might be sulfated themselves. Once bound to the cell surface, YFAK might trigger a conformational change of and/or cross-link sulfated proteoglycans and/or associated receptors, upon which cells get activated and eventually secrete e.g. CCL22. The identification of such (a) receptor(s) will help to understand how the class of immunomodulating copolymers investigated here is sensed by a cell and which ‘downstream’ signaling events occur. HSPGs might also serve as scavengers that capture YFAK and deliver it to a cellular compartment that is appropriately equipped to sense YFAK and transmit a signal to elicit CCL22 production. Interestingly, YFAK and YEAK are similar in nature to cationic cell-penetrating peptides (CPPs). Although the mechanism of translocation of CPPs to the cytosol is still a matter of debate, there is agreement that they interact in a first step with cellular heparan sulfates and negatively charged components of the phospholipids bilayer (reviewed in (47)). By analogy, YFAK and YEAK might follow a similar path, gain access to the cytosol/nucleus an/or other organelles and act there on their target molecule(s). Possibly these are components of the translational machinery, the most enriched ‘biological process’ amongst the interaction partners identified in this study.
In summary we identify cellular interaction partners of YFAK and YEAK. For some of the identified receptors, we tested functional relevance for CCL22 production in BMDCs or BMDMs. Although none of the tested interactors proved to be essential or play a non-redundant role for CCL22 production, the list of interactors presented here provides a basis for further investigations on the molecular mechanism of YFAK and YEAK.
Supplementary Material
Acknowledgments
We thank Prat Thiru and Bingbing Yuan from the BaRC at the Whitehead Institute for help with analysis of the mass-spectrometry data. We are grateful to Zihai Li (Medical University of South Carolina), Dudley Strickland (University of Maryland), Kate Fitzgerald and Tracy Young-Pearse (Harvard Medical School) for providing bones from KO mice.
This work was supported by the NIH grant RO1 AI087879 to H.L.P. and the NIH grant AI049524 to J.S..
Abbreviations used in this article
- App
amyloid precursor protein
- B3GAT3
beta-1,3-glucuronyltransferase 3
- B4GALT7
xylosylprotein beta 1,4-galactosyltransferase
- BMDC
bone marrow-derived dendritic cell
- BMDM
bone marrow-derived macrophage
- CLR
C-type lectin receptor
- CNS
central nervous system
- CPP
cell-penetrating peptide
- EAE
experimental autoimmune encephalitis
- EndoH
endoglycosidase H
- GO
gene ontology
- HSPG
heparan sulfate proteoglycan
- KO
knock-out
- LDL
low-density lipoprotein
- MBP
myelin basic protein
- MS
multiple sclerosis
- PEG
polyethylene glycol
- PNGaseF
Peptide-N-Glycosidase F
- SA
streptavidin
- SLC35B2
solute carrier family 35, member B2
- TRIF
TIR-domain-containing adaptor-inducing interferon-β
References
- 1.Illes Z, Stern JN, Reddy J, Waldner H, Mycko MP, Brosnan CF, Ellmerich S, Altmann DM, Santambrogio L, Strominger JL, Kuchroo VK. Modified amino acid copolymers suppress myelin basic protein 85-99-induced encephalomyelitis in humanized mice through different effects on T cells. Proc Natl Acad Sci U S A. 2004;101:11749–11754. doi: 10.1073/pnas.0403833101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stern JN, Illes Z, Reddy J, Keskin DB, Sheu E, Fridkis-Hareli M, Nishimura H, Brosnan CF, Santambrogio L, Kuchroo VK, Strominger JL. Amelioration of proteolipid protein 139-151-induced encephalomyelitis in SJL mice by modified amino acid copolymers and their mechanisms. Proc Natl Acad Sci U S A. 2004;101:11743–11748. doi: 10.1073/pnas.0403832101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fridkis-Hareli M, Santambrogio L, Stern JN, Fugger L, Brosnan C, Strominger JL. Novel synthetic amino acid copolymers that inhibit autoantigen-specific T cell responses and suppress experimental autoimmune encephalomyelitis. J Clin Invest. 2002;109:1635–1643. doi: 10.1172/JCI15402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teitelbaum D, Meshorer A, Hirshfeld T, Arnon R, Sela M. Suppression of experimental allergic encephalomyelitis by a synthetic polypeptide. Eur J Immunol. 1971;1:242–248. doi: 10.1002/eji.1830010406. [DOI] [PubMed] [Google Scholar]
- 5.Yin H, Vistica BP, Chan CC, Strominger JL, Gery I. Inhibition of experimental autoimmune uveitis by amino acid copolymers. J Neuroimmunol. 2009;215:43–48. doi: 10.1016/j.jneuroim.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aharoni R, Teitelbaum D, Arnon R. T suppressor hybridomas and interleukin-2-dependent lines induced by copolymer 1 or by spinal cord homogenate down-regulate experimental allergic encephalomyelitis. Eur J Immunol. 1993;23:17–25. doi: 10.1002/eji.1830230105. [DOI] [PubMed] [Google Scholar]
- 7.Aharoni R, Teitelbaum D, Sela M, Arnon R. Copolymer 1 induces T cells of the T helper type 2 that crossreact with myelin basic protein and suppress experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 1997;94:10821–10826. doi: 10.1073/pnas.94.20.10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aharoni R, Teitelbaum D, Sela M, Arnon R. Bystander suppression of experimental autoimmune encephalomyelitis by T cell lines and clones of the Th2 type induced by copolymer 1. J Neuroimmunol. 1998;91:135–146. doi: 10.1016/s0165-5728(98)00166-0. [DOI] [PubMed] [Google Scholar]
- 9.Begum-Haque S, Christy M, Ochoa-Reparaz J, Nowak EC, Mielcarz D, Haque A, Kasper LH. Augmentation of regulatory B cell activity in experimental allergic encephalomyelitis by glatiramer acetate. J Neuroimmunol. 232:136–144. doi: 10.1016/j.jneuroim.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Begum-Haque S, Sharma A, Christy M, Lentini T, Ochoa-Reparaz J, Fayed IF, Mielcarz D, Haque A, Kasper LH. Increased expression of B cell- associated regulatory cytokines by glatiramer acetate in mice with experimental autoimmune encephalomyelitis. J Neuroimmunol. 219:47–53. doi: 10.1016/j.jneuroim.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 11.Kala M, Rhodes SN, Piao WH, Shi FD, Campagnolo DI, Vollmer TL. B cells from glatiramer acetate-treated mice suppress experimental autoimmune encephalomyelitis. Exp Neurol. 221:136–145. doi: 10.1016/j.expneurol.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 12.Kovalchin J, Krieger J, Genova M, Kawamoto N, Augustyniak M, Collins K, Bloom T, Masci A, Hittinger T, Dufour I, Strominger JL, Zanelli E. Macrophage-specific chemokines induced via innate immunity by amino acid copolymers and their role in EAE. PLoS One. 6:e26274. doi: 10.1371/journal.pone.0026274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim HJ, Ifergan I, Antel JP, Seguin R, Duddy M, Lapierre Y, Jalili F, Bar-Or A. Type 2 monocyte and microglia differentiation mediated by glatiramer acetate therapy in patients with multiple sclerosis. J Immunol. 2004;172:7144–7153. doi: 10.4049/jimmunol.172.11.7144. [DOI] [PubMed] [Google Scholar]
- 14.Vieira PL, Heystek HC, Wormmeester J, Wierenga EA, Kapsenberg ML. Glatiramer acetate (copolymer-1, copaxone) promotes Th2 cell development and increased IL-10 production through modulation of dendritic cells. J Immunol. 2003;170:4483–4488. doi: 10.4049/jimmunol.170.9.4483. [DOI] [PubMed] [Google Scholar]
- 15.Toker A, Slaney CY, Backstrom BT, Harper JL. Glatiramer acetate treatment directly targets CD11b(+)Ly6G(−) monocytes and enhances the suppression of autoreactive T cells in experimental autoimmune encephalomyelitis. Scand J Immunol. 74:235–243. doi: 10.1111/j.1365-3083.2011.02575.x. [DOI] [PubMed] [Google Scholar]
- 16.Weber MS, Prod'homme T, Youssef S, Dunn SE, Rundle CD, Lee L, Patarroyo JC, Stuve O, Sobel RA, Steinman L, Zamvil SS. Type II monocytes modulate T cell-mediated central nervous system autoimmune disease. Nat Med. 2007;13:935–943. doi: 10.1038/nm1620. [DOI] [PubMed] [Google Scholar]
- 17.Kawamoto N, Ohnishi H, Kondo N, Strominger JL. The role of dendritic cells in the generation of CD4+ CD25HI Foxp3+ T cells induced by amino acid copolymers. Int Immunol. 25:53–65. doi: 10.1093/intimm/dxs087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Randow F, Seed B. Endoplasmic reticulum chaperone gp96 is required for innate immunity but not cell viability. Nat Cell Biol. 2001;3:891–896. doi: 10.1038/ncb1001-891. [DOI] [PubMed] [Google Scholar]
- 19.Dyballa N, Metzger S. Fast and sensitive colloidal coomassie G-250 staining for proteins in polyacrylamide gels. J Vis Exp. 2009 doi: 10.3791/1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altmeyer A, Maki RG, Feldweg AM, Heike M, Protopopov VP, Masur SK, Srivastava PK. Tumor-specific cell surface expression of the-KDEL containing, endoplasmic reticular heat shock protein gp96. Int J Cancer. 1996;69:340–349. doi: 10.1002/(SICI)1097-0215(19960822)69:4<340::AID-IJC18>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 21.Maki RG, Old LJ, Srivastava PK. Human homologue of murine tumor rejection antigen gp96: 5′-regulatory and coding regions and relationship to stress-induced proteins. Proc Natl Acad Sci U S A. 1990;87:5658–5662. doi: 10.1073/pnas.87.15.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee CC, Avalos AM, Ploegh HL. Accessory molecules for Toll-like receptors and their function. Nat Rev Immunol. 12:168–179. doi: 10.1038/nri3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y, Liu B, Dai J, Srivastava PK, Zammit DJ, Lefrancois L, Li Z. Heat shock protein gp96 is a master chaperone for toll-like receptors and is important in the innate function of macrophages. Immunity. 2007;26:215–226. doi: 10.1016/j.immuni.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staron M, Yang Y, Liu B, Li J, Shen Y, Zuniga-Pflucker JC, Aguila HL, Goldschneider I, Li Z. gp96, an endoplasmic reticulum master chaperone for integrins and Toll-like receptors, selectively regulates early T and B lymphopoiesis. Blood. 115:2380–2390. doi: 10.1182/blood-2009-07-233031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weekes MP, Antrobus R, Talbot S, Hor S, Simecek N, Smith DL, Bloor S, Randow F, Lehner PJ. Proteomic plasma membrane profiling reveals an essential role for gp96 in the cell surface expression of LDLR family members, including the LDL receptor and LRP6. J Proteome Res. 11:1475–1484. doi: 10.1021/pr201135e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seubert A, Calabro S, Santini L, Galli B, Genovese A, Valentini S, Aprea S, Colaprico A, D'Oro U, Giuliani MM, Pallaoro M, Pizza M, O'Hagan DT, Wack A, Rappuoli R, De Gregorio E. Adjuvanticity of the oil-in-water emulsion MF59 is independent of Nlrp3 inflammasome but requires the adaptor protein MyD88. Proc Natl Acad Sci U S A. 108:11169–11174. doi: 10.1073/pnas.1107941108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He B, Santamaria R, Xu W, Cols M, Chen K, Puga I, Shan M, Xiong H, Bussel JB, Chiu A, Puel A, Reichenbach J, Marodi L, Doffinger R, Vasconcelos J, Issekutz A, Krause J, Davies G, Li X, Grimbacher B, Plebani A, Meffre E, Picard C, Cunningham-Rundles C, Casanova JL, Cerutti A. The transmembrane activator TACI triggers immunoglobulin class switching by activating B cells through the adaptor MyD88. Nat Immunol. 11:836–845. doi: 10.1038/ni.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Binder RJ, Han DK, Srivastava PK. CD91: a receptor for heat shock protein gp96. Nat Immunol. 2000;1:151–155. doi: 10.1038/77835. [DOI] [PubMed] [Google Scholar]
- 29.Basu S, Binder RJ, Ramalingam T, Srivastava PK. CD91 is a common receptor for heat shock proteins gp96, hsp90, hsp70, and calreticulin. Immunity. 2001;14:303–313. doi: 10.1016/s1074-7613(01)00111-x. [DOI] [PubMed] [Google Scholar]
- 30.Willnow TE, Herz J. Genetic deficiency in low density lipoprotein receptor-related protein confers cellular resistance to Pseudomonas exotoxin A. Evidence that this protein is required for uptake and degradation of multiple ligands. J Cell Sci. 1994;107(Pt 3):719–726. [PubMed] [Google Scholar]
- 31.Ashcom JD, Tiller SE, Dickerson K, Cravens JL, Argraves WS, Strickland DK. The human alpha 2-macroglobulin receptor: identification of a 420-kD cell surface glycoprotein specific for the activated conformation of alpha 2-macroglobulin. J Cell Biol. 1990;110:1041–1048. doi: 10.1083/jcb.110.4.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herz J, Strickland DK. LRP: a multifunctional scavenger and signaling receptor. J Clin Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pawaria S, Binder RJ. CD91-dependent programming of T-helper cell responses following heat shock protein immunization. Nat Commun. 2:521. doi: 10.1038/ncomms1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jockheck-Clark AR, Bowers EV, Totonchy MB, Neubauer J, Pizzo SV, Nicchitta CV. Re-examination of CD91 function in GRP94 (glycoprotein 96) surface binding, uptake, and peptide cross-presentation. J Immunol. 185:6819–6830. doi: 10.4049/jimmunol.1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bovenschen N, Mertens K, Hu L, Havekes LM, van Vlijmen BJ. LDL receptor cooperates with LDL receptor-related protein in regulating plasma levels of coagulation factor VIII in vivo. Blood. 2005;106:906–912. doi: 10.1182/blood-2004-11-4230. [DOI] [PubMed] [Google Scholar]
- 36.Rosmarin DM, Carette JE, Olive AJ, Starnbach MN, Brummelkamp TR, Ploegh HL. Attachment of Chlamydia trachomatis L2 to host cells requires sulfation. Proc Natl Acad Sci U S A. 109:10059–10064. doi: 10.1073/pnas.1120244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robert J, Menoret A, Cohen N. Cell surface expression of the endoplasmic reticular heat shock protein gp96 is phylogenetically conserved. J Immunol. 1999;163:4133–4139. [PubMed] [Google Scholar]
- 38.Blachere NE, Li Z, Chandawarkar RY, Suto R, Jaikaria NS, Basu S, Udono H, Srivastava PK. Heat shock protein-peptide complexes, reconstituted in vitro, elicit peptide-specific cytotoxic T lymphocyte response and tumor immunity. J Exp Med. 1997;186:1315–1322. doi: 10.1084/jem.186.8.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Udono H, Levey DL, Srivastava PK. Cellular requirements for tumor-specific immunity elicited by heat shock proteins: tumor rejection antigen gp96 primes CD8+ T cells in vivo. Proc Natl Acad Sci U S A. 1994;91:3077–3081. doi: 10.1073/pnas.91.8.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suto R, Srivastava PK. A mechanism for the specific immunogenicity of heat shock protein-chaperoned peptides. Science. 1995;269:1585–1588. doi: 10.1126/science.7545313. [DOI] [PubMed] [Google Scholar]
- 41.Singh-Jasuja H, Scherer HU, Hilf N, Arnold-Schild D, Rammensee HG, Toes RE, Schild H. The heat shock protein gp96 induces maturation of dendritic cells and down-regulation of its receptor. Eur J Immunol. 2000;30:2211–2215. doi: 10.1002/1521-4141(2000)30:8<2211::AID-IMMU2211>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 42.Binder RJ, Anderson KM, Basu S, Srivastava PK. Cutting edge: heat shock protein gp96 induces maturation and migration of CD11c+ cells in vivo. J Immunol. 2000;165:6029–6035. doi: 10.4049/jimmunol.165.11.6029. [DOI] [PubMed] [Google Scholar]
- 43.Banerjee PP, Vinay DS, Mathew A, Raje M, Parekh V, Prasad DV, Kumar A, Mitra D, Mishra GC. Evidence that glycoprotein 96 (B2), a stress protein, functions as a Th2-specific costimulatory molecule. J Immunol. 2002;169:3507–3518. doi: 10.4049/jimmunol.169.7.3507. [DOI] [PubMed] [Google Scholar]
- 44.Eletto D, Dersh D, Argon Y. GRP94 in ER quality control and stress responses. Semin Cell Dev Biol. 21:479–485. doi: 10.1016/j.semcdb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu B, Yang Y, Qiu Z, Staron M, Hong F, Li Y, Wu S, Hao B, Bona R, Han D, Li Z. Folding of Toll-like receptors by the HSP90 paralogue gp96 requires a substrate-specific cochaperone. Nat Commun. 1:79. doi: 10.1038/ncomms1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holmberg A, Blomstergren A, Nord O, Lukacs M, Lundeberg J, Uhlen M. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis. 2005;26:501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
- 47.Madani F, Lindberg S, Langel U, Futaki S, Graslund A. Mechanisms of cellular uptake of cell-penetrating peptides. J Biophys. 2011:414729. doi: 10.1155/2011/414729. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.