Summary
Rett Syndrome (RTT) is caused by mutations of MECP2, a methyl CpG binding protein thought to act as a global transcriptional repressor. Here we show, using an isogenic human embryonic stem cell model of RTT, that MECP2 mutant neurons display key molecular and cellular features of this disorder. Unbiased global gene expression analyses demonstrate that MECP2 functions as global gene activator in neurons but not in neural precursors. Decreased transcription in neurons was coupled with a significant reduction in nascent protein synthesis and lack of MECP2 was manifested as a severe defect in the activity of the AKT/mTOR pathway. Lack of MECP2 also leads to impaired mitochondrial function in mutant neurons. Activation of AKT/mTOR signaling by exogenous growth factors or by depleting PTEN boosted protein synthesis and ameliorated disease phenotypes in mutant neurons. Our findings indicate a vital function for MECP2 in maintaining active gene transcription in human neuronal cells.
Keywords: Rett syndrome, human embryonic stem cells, AKT/mTOR, MECP2, gene expression, transcriptional activator
Introduction
Rett syndrome (RTT) is a monogenic X-linked neurodevelopmental disease that belongs to autism spectrum disorders (ASDs). It has been well established that mutations in the MECP2 gene account for the majority of RTT cases (Amir et al., 1999). We and others have generated mutant mice bearing loss-of-function alleles of Mecp2, and showed that they recapitulate many of the cellular and behavioral phenotypes of RTT patients (Chen et al., 2001; Guy et al., 2001). Recent work using these mouse models demonstrated that the disease pathology in MECP2 mutant mice could be reversed either via re-expression of the wild-type Mecp2 gene (Giacometti et al., 2007; Guy et al., 2007), or by exogenous growth factors (Chang et al., 2006; Tropea et al., 2009). These findings serve as important proof-of-principle evidence that RTT, and perhaps ASD in general, are treatable disorders. With the advent of induced pluripotent stem cell (iPSC) technology (Takahashi et al., 2007; Yu et al., 2007), it has become feasible to confirm conclusions from animal models in human cells by deriving patient-specific iPSCs for disease modeling and therapeutic investigation (Ananiev et al., 2011; Cheung et al., 2011; Marchetto et al., 2010). However, due to differences in genetic background and method of derivation, human embryonic stem cells (ESCs) and iPSCs display highly variable biological characteristics such as propensity to differentiate into specific lineages, complicating their use in disease modeling (Soldner and Jaenisch, 2012). This is of particular relevance to RTT, in which genetic background has been demonstrated to influence the severity of disease symptoms (Scala et al., 2007). To overcome this complication, isogenic experimental and control cells that differ exclusively at the disease-causing genetic alteration have been generated, allowing the study of disease-specific phenotypes under highly controlled conditions (Soldner et al, 2011).
It has been well established that MECP2 protein is abundantly present in neuronal cell types, and binds methylated-CpG sites throughout the neuronal genome (Skene et al., 2010). Such binding specificity and mode of distribution strongly suggested that MECP2 functions as a global transcriptional repressor (Nan et al., 1997). However, the role of MECP2 as a repressor received little support from in vivo studies as exhaustive gene expression analyses on pre-and post-symptomatic mutant mice provided little evidence of global transcriptional activation (Ben-Shachar et al., 2009; Chahrour et al., 2008; Jordan et al., 2007; Kriaucionis et al., 2006; Nuber et al., 2005; Tudor et al., 2002; Urdinguio et al., 2008).
In the present study, we used TALEN-mediated gene editing to generate human ESCs with a loss-of-function MECP2 allele. This strategy ensures that neuronal cells derived from the control and mutant ESCs differ only at the MECP2 gene. We found that although neural precursors (NPs) derived from mutant ESCs were largely normal as compared to their isogenic controls, an array of cellular and molecular abnormalities developed in differentiated MECP2 mutant neurons. To investigate the impact of MECP2 deletion on the neuronal transcriptome, we adopted a newly developed gene expression analysis method that took into consideration possible global shifts in transcriptional activities. We found a significant genome-wide transcriptional down-regulation in mutant neurons. This striking reduction of global transcription was echoed in significantly decreased protein synthesis levels. Pharmacological and genetic manipulations that boost protein synthesis ameliorated RTT-related disease phenotypes. These findings strongly support the notion that one of the key functions of MECP2 is to facilitate global transcription.
Results
TALEN-mediated targeting of the MeCP2 locus
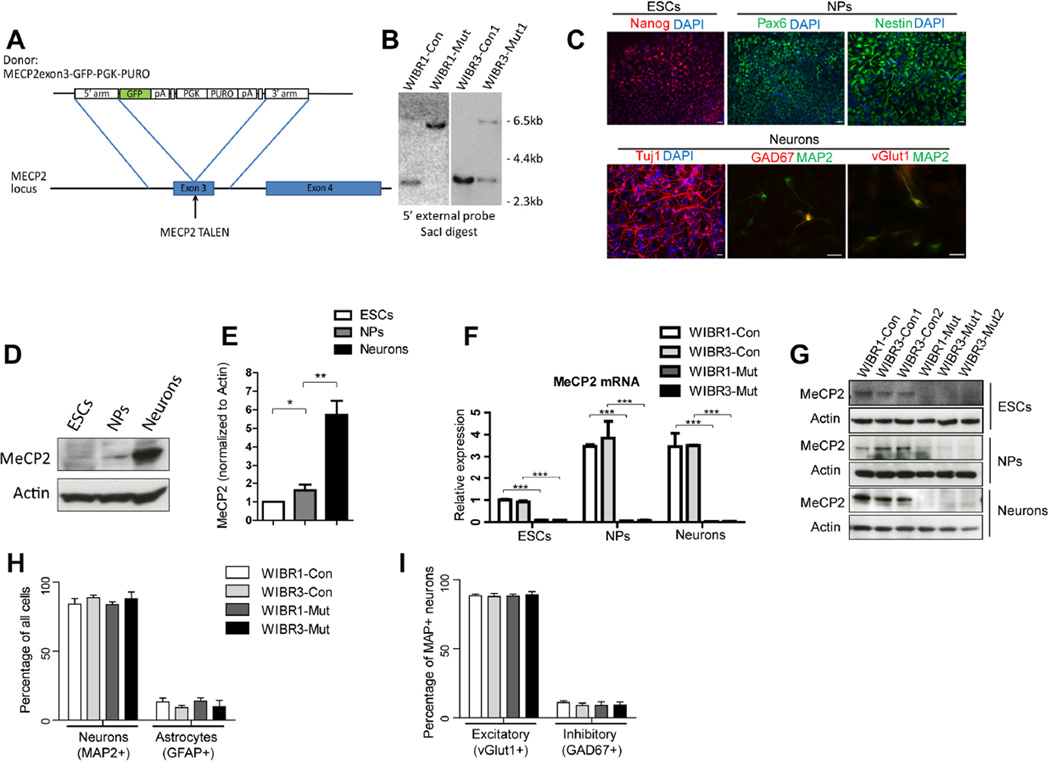
To generate a MECP2 loss-of-function allele, we designed TALENs to specifically target the third exon of the MECP2 gene, which encodes most of the methyl-CpG-binding domain (Figure 1A and S1A). A donor construct containing an in-frame eGFP-polyA sequence and a PGK-puro cassette flanked by two homology arms corresponding to the genomic sequence of the MECP2 gene was used for targeting (Figure 1A). This targeting strategy disrupts gene function and generates an endogenous reporter for MECP2 activity. We used a male (WIBR1) and a female ESC line (WIBR3) to generate MECP2 hemizygous mutant male and heterozygous mutant female clones (Figure 1B, S1B and S1C). The WIBR3 ESCs were maintained in the XaXi state, in which the targeted MECP2 allele resided on the active and the wild-type allele on the inactive X chromosome. For subsequent analyses, we used 1 pair of control and mutant WIBR1 ESCs (WIBR1-Con, WIBR1-Mut), and 2 sub-clones each of the control and mutant WIBR3 ESCs (WIBR3-Con1, WIBR3-Con2, WIBR3-Mut1, WIBR3-Mut2). In addition, similar to their isogenic wild-type controls, mutant ESCs maintained the expression of pluripotency marker expression (Oct4, Nanog and Sox2, Figure S1D) and formed teratomas when injected subcutaneously to NOD/SCID mice (Figure S1F–G).
Figure 1. Generation of isogenic pairs of control and MECP2 mutant human ESCs, NPs and neurons.
A) Schematic overview depicting the targeting strategy for the MECP2 locus.
B) Southern blot analysis of WIBR1, WIBR3 ESCs targeted with the MECP2exon3-GFP-PGK-PURO donor construct, using probes against the 5’ external sequence, showing targeted bands at 6.7kb, and wild-type band at 3.6kb.
C) Immuno-staining for markers of ESC (Nanog), NPs (Pax6, Nestin) and neurons (Tuj1, MAP2) document the stepwise differentiation process. Both excitatory (vGlut1) and inhibitory (GAD67) neurons could be seen. Scale bar, 20um.
D and E) MeCP2 protein was specifically enriched in wild-type neurons compared to ESCs and NPs.
F) Quantitative RT-PCR analysis confirmed the absence of MECP2 mRNA in WIBR1-Mut and WIBR3-Mut ESCs, NPs and neurons.
G) Immuno-blot analyses for MECP2 confirmed complete ablation of wild-type MECP2 protein in WIBR1-Mut and WIBR3-Mut ESCs, NPs and neurons.
H) Quantification of neurons (MAP2+) and astrocytes (GFAP+) derived from control and MECP2 mutant NPs.
I) Quantification of excitatory neurons (MAP2+vGlut1+) and inhibitory neurons (MAP2+GAD67+) derived from control and MECP2 mutant NPs.
Results are mean +/− SEM. *p<0.05, **p<0.01, ***p<0.001. See also Figure S1.
Neural differentiation of MeCP2 mutant ESCs
Human ESCs and iPSCs can generate NPs, which can be induced to differentiate into different neural and glial subtypes (Soldner et al., 2009). To generate a homogenous population of NPs from control and MECP2 mutant ESCs, we utilized a SMAD-inhibition-based neural differentiation protocol (Chambers et al., 2009; Zhou et al., 2010). Adherent culture of control and mutant ESCs were differentiated side-by-side into neural rosettes, and further expanded as NPs in the presence of bFGF. These isogenic pairs of NPs were then cultured in medium that promotes terminal neural differentiation and maturation (Figure 1C). Using antibodies specific to antigens of these lineages, we demonstrate the generation of NPs (Pax6+, Nestin+) and neurons (TuJ1+, MAP2+) from pluripotent ESCs (Nanog+) (Figure 1C). Furthermore, using antibodies against vGlut1 and GAD67, we found that both excitatory and inhibitory neurons were present in the neuronal cultures (Figure 1C).
MECP2 is expressed in ESCs and NPs, but significantly more abundantly in neurons, as shown by immuno-blotting (Figure 1D–E). To confirm that the targeting strategy disrupted the MECP2 locus, we isolated mRNA from control and mutant ESCs, NPs and neurons. Quantitative RT-PCR revealed the absence of MECP2 expression in WIBR1-Mut and WIBR3-Mut cells (Figure 1F). Immuno-blotting confirmed the absence of MECP2 protein (Figure 1G). Furthermore, wild-type MECP2 protein was not detectable by immuno-staining (Figure S1E and not shown). The lack of wild-type MECP2 mRNA and protein was consistent with the hemizygous status of WIBR1-Mut ESCs, and the one X active (XaXi) state of WIBR3-Mut ESCs demonstrating loss of function in WIBR1-Mut and WIBR3-Mut cells.
Control and mutant NPs displayed similar morphology (Figure S1H) and expressed comparable levels of markers indicative of the cortical lineage (Figure S1I). Upon withdrawal of bFGF, both control and mutant NPs readily generated Doublecortin-positive immature neurons (Figure S1J). During the course of neuronal differentiation, predominantly excitatory neurons were generated (Figure 1H, I). Expression of genes characteristic for different cortical layers such as Calretinin (subplate), Reelin (layer I), FoxP2 (V/VI), CTIP2 (II to VI) and Cux2 (II) were similar in control and mutant cells (Figure S1K–P). These finding suggest that MECP2 deletion did not alter the differentiation potential of NPs.
MeCP2 mutant neurons show morphological and physiological defects
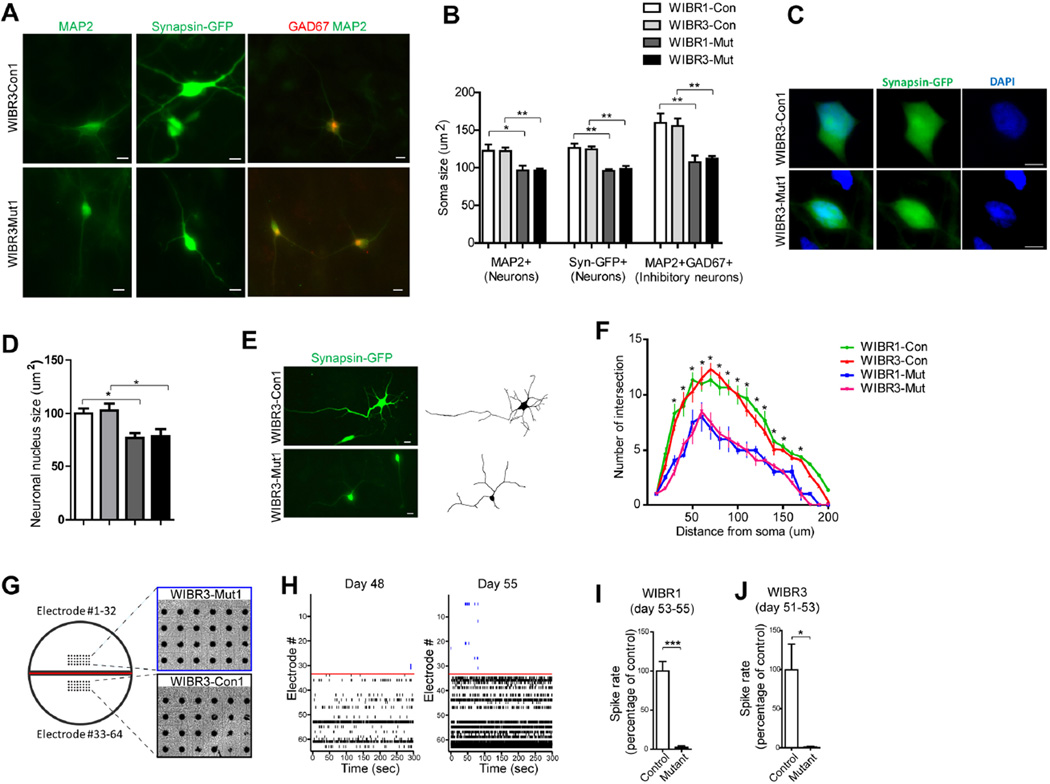
A prominent feature of RTT patients is microcephaly with the reduction in brain size correlating with the reduction in neuronal size (Akbarian, 2002; Chen et al., 2001). Consistent with these findings, soma and nuclei of mutant neurons expressing MAP2 or marked by a Synapsin-GFP virus (Kugler et al., 2003) displayed a significantly reduced size as compared to isogenic controls (Figure 2A–D). In contrast, mutant NPs displayed a similar soma and nuclear size as compared to controls (not shown). Because excitatory neurons comprise the majority of the mature neurons generated in our differentiation protocol (Figure 1I), the reduction of soma size mostly represents deficits in the excitatory neuron population. Recent studies suggest that cell-type specific deletion of MeCP2 in inhibitory neurons was sufficient to impair GABAergic function leading to severe behavior deficits (Chao et al., 2010). To investigate whether human inhibitory neurons require MECP2 for proper morphological development, we co-stained the neuronal culture with MAP2 and GAD67 and found a significant soma size reduction in mutant cells (Figure 2A and 2B). We also investigated whether MECP2 deletion affects neurite arborization of individual Synapsin-GFP labeled neurons. Sholl analysis revealed significantly reduced neurite complexity in MECP2 mutant neurons (Figure 2E and 2F). Utilizing a dual-chamber multi-electrode array culture system that allows for continuous monitoring the appearance of spontaneous activities, we plated control and mutant neurons side-by-side (Figure 2G), and found that action potential rates were significantly reduced in mutant neurons (Figure 2H to 2J). We conclude that MECP2 deletion impairs neuronal development and function.
Figure 2. MECP2 mutant human neurons show morphological and electrophysiological defects.
A) Representative images of Synapsin-GFP (middle panels), and immuno-staining with antibodies against MAP2 (left panels) show reduced soma size in WIBR3-Mut1 neurons. Co-staining for MAP2 and GAD67 (right panels) revealed reduced soma size in WIBR3-Mut1 inhibitory neurons. Scale bar, 10um.
B) Quantitative analysis of MAP2+, Synapsin-GFP+ neurons, and MAP2+GAD67+ inhibitory neurons demonstrated reduced soma size in MECP2 mutant neurons compared to their isogenic controls.
C) Representative images of Synapsin-GFP-positive control and MECP2 mutant neurons. Nuclei are highlighted with DAPI staining. Scale bar, 10um.
D) Quantitative analyses of neuronal nucleus size demonstrated smaller nucleus in MECP2 mutant neurons.
E) Synapsin-GFP-labeling revealed the neurite morphology of control and mutant neurons. Scale bar, 10um.
F) Quantitative analyses of neurite morphology of Synapsin-GFP-positive neurons showed that MECP2 mutant neurons had reduced number of neurite intersections compared to controls.
G and H) Multi-electrode array analyses of isogenic control and MECP2 mutant neurons. Control and mutant neurons were plated on the same MED-64 arrays (G). Spike rasters depict spontaneous activities during a 5-minute recording (H). Note the gradual increase of activity in control neurons (electrode 33–64), and the largely absence of activity in mutant neurons (electrode 1–32).
(I and J) Reduced spike rate in both WIBR1-Mut and WIBR3-Mut neurons.
Results are mean +/− SEM. *p<0.05, **p<0.01.
MECP2 mutant neurons display global reductions in transcription
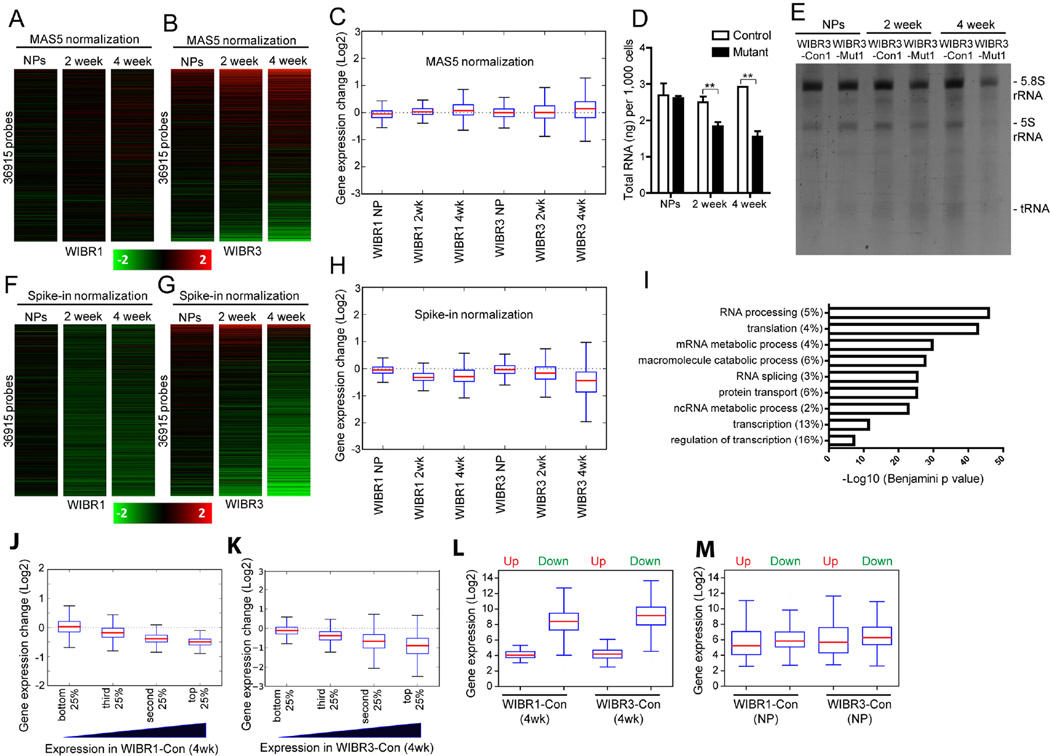
We next compared isogenic mutant and control cells to assess the effect of MECP2 deletion on global gene expression. Conventional global transcriptional analyses normalizing individual gene expression to total RNA (MAS5 for Affymetrix) indicated that the majority of genes were unchanged (Figure 3A and 3B). When the distribution of expression changes of all genes was examined, a minimal median change in mutant NPs and 2-week-differentiated mutant neurons and a small increase in 4-week-differentiated mutant neurons were found (Figure 3C). The minor gene expression changes in MECP2 mutant neurons, especially those found in the male WIBR1-Mut neurons, were consistent with previous findings in Mecp2 hemizygous mutant mice (Kriaucionis et al., 2006; Nuber et al., 2005; Tudor et al., 2002; Urdinguio et al., 2008). We found that the female WIBR3-Mut neurons displayed a higher level of differential expression compared to their isogenic controls, with a similar distribution of up- and down-regulated genes at 2-weeks, and slightly more up-regulated genes at 4-weeks of differentiation (Figure 3B).
Figure 3. Reduced total RNA and global transcriptional down-regulation in MECP2 mutant neurons.
A and B) Heatmaps showing the fold change of expression in mutant versus control WIBR1 (A) or WIBR3 (B) cells using a standard Affymetrix microarray normalization method (MAS5).
C) Boxplots of transcriptional changes between control and mutant cells, normalized by standard Affymetrix microarray normalization method (MAS5).
D) Two- and four-week differentiated MECP2 mutant neurons have reduced total RNA levels compared to their isogenic controls. Note the comparable levels of total RNA in control and mutant NPs.
E) TBE urea gel of ethidium bromide-stained total RNA extracted from equivalent numbers of control and MECP2 mutant cells. Bands correspond to 5.8S rRNA, 5S rRNA and tRNA.
F and G) Heatmap showing the fold change of expression in mutant versus control WIBR1 (F) or WIBR3 (G) cells using RNA spike-in standards.
H) Boxplots of transcriptional changes between mutant and control cells, normalized by RNA spike-in standards.
I) Gene ontology analysis of genes commonly down-regulated in 4-week-old mutant neurons. Graph shows the top GO terms of biological processes significantly affected in MECP2 mutant neurons.
J and K) Boxplots of fold transcriptional changes of all genes, sub-divided into 4 equal groups based on their expression levels in 4-week-differentiated WIBR1-Con (J) and WIBR3-Con (K) neurons.
L and M) Boxplots of gene expression distributions in 4-week-differentiated control neurons (L) or control NPs (M), based on whether they were up- or down-regulated in the isogenic mutant neurons.
Results are mean +/− SEM. *p<0.05, **p<0.01. See also Figure S2.
The validity of conventional gene expression analyses, which usually are based on the assumption that total RNA content per cell remains constant, has recently been questioned. It was discovered that c-Myc over-expression results in a significant increase in nuclear size and an amplified per-cell expression of more than 90% of all genes. However, when the expression data were normalized using the traditional analysis method, i.e. normalization to total input RNA, this genome-wide gene expression change was not detected (Lin et al., 2012; Loven et al., 2012). These observations are highly relevant to MECP2 mutant cells, which display a significantly reduced soma and nuclear size (Figure 2A to 2D).
To investigate whether the reduced soma and nuclear size of MECP2 mutant cells affected per cell RNA levels, we isolated total RNA from equal numbers of NPs and differentiating neurons. Figure 3D shows that mutant neurons at 2 and 4 weeks of differentiation have 27% and 47% less total RNA, respectively, as compared to their isogenic controls. A significant reduction of differently sized rRNAs was found in differentiating mutant neurons consistent with ribosomal RNA (rRNA) and transfer RNA (tRNA) constituting the majority of total RNA (Figure 3E). In contrast, NPs of either genotype had similar levels of total RNA and rRNA (Figure 3D and 3E).
To assess how normalization to input cell number affects the interpretation of global transcriptional analyses, we prepared RNA from an identical number of control and mutant cells and added synthetic RNA spike-in standards as surrogates for cell numbers (Loven et al., 2012). After normalizing gene expression to RNA spike-ins, an average of 50% of all genes were down-regulated more than 1.2-fold in mutant neurons at 2-weeks of differentiation (Figure 3F and 3G; 62.3% in WIBR1-Mut; 37.9% in WIBR3-Mut). At 4-weeks of differentiation, the percentage of down-regulated genes in MECP2 mutant neurons was 60% (Figure 3F and 3G; 54.1% in WIBR1-Mut; 64.6% in WIBR3-Mut). In contrast, only about 7% of all genes were up-regulated more than 1.2-fold at 2- and 4-weeks of differentiation (2.4% and 5.8% in WIBR1-Mut; 12.6% and 7.4% in WBIR3-Mut). At the NP stage, expression levels were more evenly distributed, with 14% down-regulated and 8% up-regulated 1.2-fold or more (13.5% down and 5.8% up in WIBR1-Mut; 15.4% up and 10.5% down in WIBR3-Mut).
A similar global down-regulation was seen when a higher threshold (1.5-fold) was used, which shows that mutant neurons on average had ~10 times the number of down-regulated genes compared to up-regulated ones. By examining the distribution of expression changes of all genes, we saw a median 25% percent reduction in 2-week differentiated mutant neurons, and 37% reduction in 4-week differentiated mutant neurons (Figure 3H, 32.2% and 29.6% in WIBR1-Mut; 17.1% and 43.7% in WIBR3-Mut) whereas average gene expression in mutant NPs was not significantly changed. Gene ontology analysis revealed that genes involved in the biological processes of transcription and translation were enriched amongst the most significantly down-regulated genes (Figure 3I). Through this series of analyses, we found that female mutant WIBR3 cells displayed more differential gene expression changes than male WIBR1 mutant cells when compared to the isogenic controls (Figure 3F to 3H), possibly reflecting the inherent stability differences between cultured male and female pluripotent stem cells (Anguera et al., 2012; Mekhoubad et al., 2012; Zvetkova et al., 2005). Indeed, WIBR3-derived female NPs and neurons had increased expression levels of X-linked genes as compared to WIBR1-derived male cells (Figure S2A–B). Importantly, these unbiased global gene expression analyses revealed a consistent genome-wide transcriptional reduction in both WIBR1 and WIBR3 mutant neurons.
MECP2 mutant neurons display preferential reductions of active transcription
To further quantify expression changes, we divided all genes based on their expression levels (in control neurons) into 4 equal groups. Figure 3J and 3K show that higher expression was correlated with a more dramatic extent of down-regulation in 4-week-old mutant neurons. The most highly expressed genes (top 25%) showed a median 70% reduction in expression, whereas the lowest expressed genes (bottom 25%) showed only a median 4% reduction. A similar trend was seen in 2-week-old neurons (Figure S2C–D), but not in NPs (Figure S2E–F). In addition to analyzing the expression of all genes, we examined genes that were significantly up-or down-regulated in 4-week-old mutant neurons of both WIBR1 and WIBR3 genetic background (85 and 7265 commonly up and down-regulated genes). Figure 3L (and Figure S2G) shows that genes that were down-regulated in the absence of MECP2 were expressed at a substantially higher level, as compared to genes that were up-regulated. Notably, when the expression level of genes that were significantly up-or down-regulated in mutant NPs was examined, no such correlation was seen (Figure 3M). The selective group of up-regulated genes in 4-week-old mutant neurons, albeit numerically few, was less active in wild-type neurons and may represent instances where MECP2 binding exerted repressive effects. Indeed, some of the up-regulated genes (CALCR, Nanog, UGT2B10) were predominantly expressed outside of the nervous system, and may be mis-expressed due to the lack of MECP2-mediated repression (Figure S2H–J). Our results demonstrate that active genes were more susceptible to down-regulation upon MECP2 deletion strongly indicating a transcriptional activator function for MECP2.
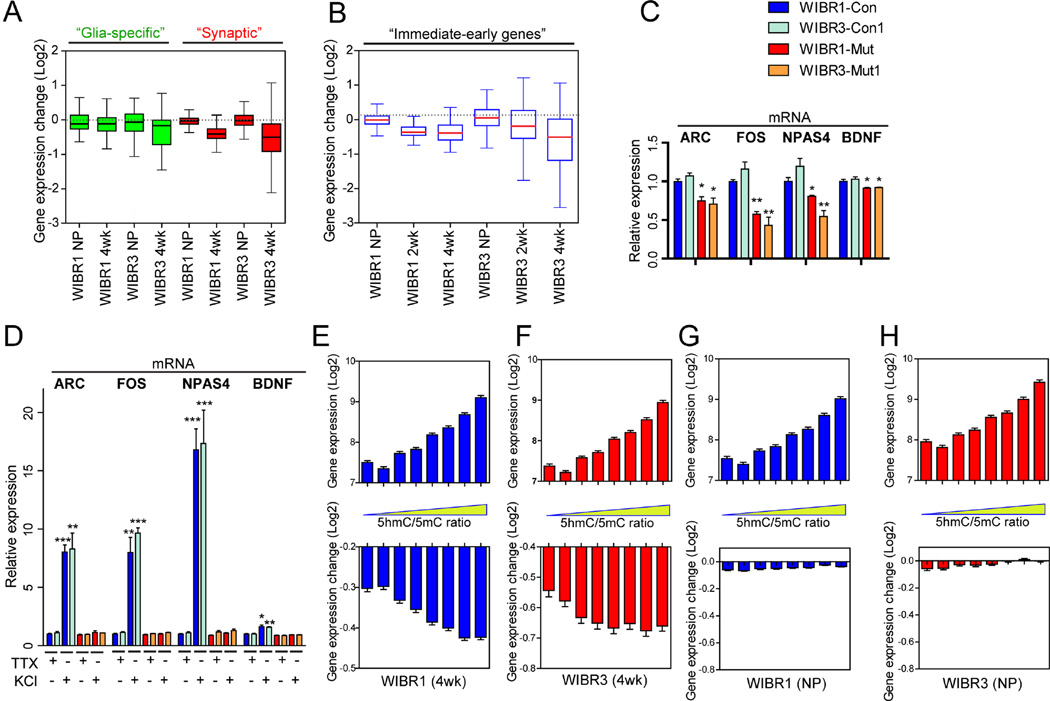
We compared the specificity of transcriptional reduction of genes involved in nervous system development (Figure S3A, human.brain-map.org) and in synapse formation (Pirooznia et al., 2012). Figures 4A shows that both groups of genes were collectively down-regulated in MECP2 mutant neurons. In contrast, the expression levels of a group of genes known to be specifically expressed in astrocytes and oligodendrocytes were largely unaffected in mutant cells (Figure 4A) (Cahoy et al., 2008).
Figure 4. Preferential reduction of active transcription in MECP2 mutant neurons.
A and B) Boxplots of transcriptional changes between mutant and control cells, in groups of genes identified as glia-specific (A, left), synaptic (A, right) and immediate-early genes (B).
C) Quantitative RT-PCR analysis confirmed the basal down-regulation of immediate-early genes (Arc, Fos, NPAS4 and BDNF) in WIBR1-Mut and WIBR3-Mut neurons.
D) Quantitative RT-PCR analysis revealed the lack of activity-induced up-regulation of immediate-early genes (Arc, Fos, NPAS4 and BDNF) in WIBR1-Mut and WIBR3-Mut neurons.
E to H) Correlation of wild-type gene expression levels (top panels) and gene expression changes in the absence of MECP2 (lower panels) to the 5hmC/5mC ratios from mouse brain. Note that higher 5hmC/5mC ratio correlated with higher expression levels and increased probability of down-regulation in 4-week-old mutant neurons, but not mutant NPs.
Results are mean +/− SEM. *p<0.05, **p<0.01, ***p<0.001. See also Figure S3.
Recent reports have suggested a role for MECP2 in regulating the expression of immediate early genes, a distinct group of genes found to be critically dependent on transcriptional regulation (Kron et al., 2012; Su et al., 2012; Tullai et al., 2007; Yasui et al., 2007). When comparing the expression of a group of immediate early genes known to be activated by neuronal activity (Bateup et al., 2013), these genes were expressed at relatively high levels in control neurons (Figure S3B) and collectively down-regulated in mutant neurons (Figure 4B). Quantitative RT-PCR further confirmed the down-regulation of some of these key immediate early genes, such as Arc, Fos, NPAS4 and BDNF (Figure 4C). This coincides with mutant neurons displaying reduced network activities (Figure 2H-2G) thereby correlates transcription reduction with physiological impairment. To further investigate whether MECP2 plays a direct role in regulating immediate early gene expression, we treated control and mutant neurons with tetrodotoxin, followed by de-polarization with KCl. Quantitative RT-PCR revealed significant increases in the Arc, Fos, NPAS4 and BDNF expression in control neurons, while mutant neurons failed to display such induction (Figure 4D).
A recent study in mice showed that MECP2 binds to both 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC), with the latter enriched within active genes (Mellen et al., 2012). This observation is consistent with our finding that binding of MECP2 facilitates active transcription, and predicts that genes with higher 5hmC/5mC ratios would be more likely to be down-regulated in the absence of MECP2. We extracted the ranking of 5hmC/5mC ratios from wild-type mouse neurons, and correlated the expression of these genes in 4-week-old human neurons. Figure 4E–4F shows that genes with higher 5hmC/5mC ratios were more highly expressed in control human neurons (top panels), and were more likely down-regulated in MECP2 human mutant neurons (lower panels). In contrast, gene expression changes seen in MECP2 mutant NPs were not correlated to the 5hmC/5mC ratio (Figure 4G–4H) suggesting that this mode of MECP2-mediated differential gene expression is specific for differentiated neurons. Our findings suggest that MECP2 exerts bimodal regulation of gene expression, dictated by its binding to 5hmC or 5mC.
MECP2 mutant neurons display impaired global translation and AKT/mTOR activity
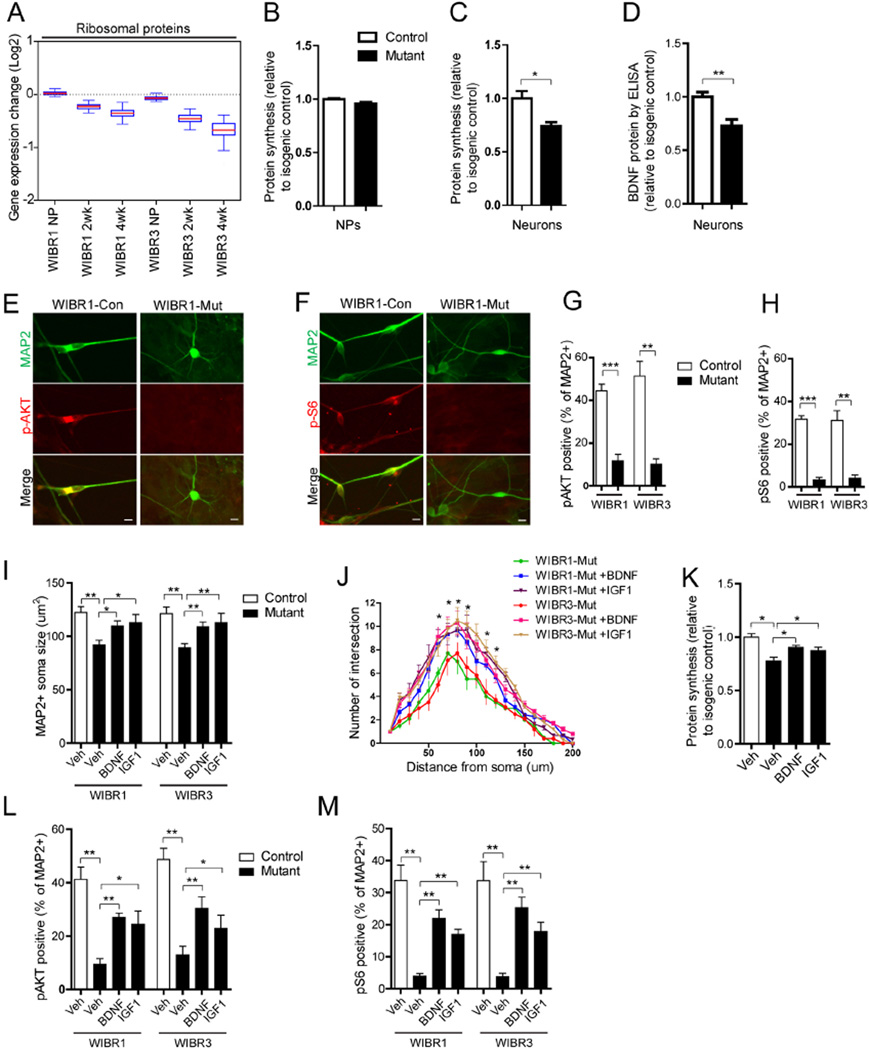
A prominent group of genes affected in MECP2 mutant neurons were ribosomal proteins (Figure 5A) which were collectively down-regulated at 4-weeks of differentiation (Figure S4A–B). Combining with the reduced level of rRNAs (Figure 3D), these findings predicted impaired capacity for translation. To evaluate nascent protein synthesis, we measured 35S-Cys/Met incorporation during a 30-minute period. Figure 5B shows that, while mutant NPs had similar levels of incorporation as their isogenic controls, 3-week-differentiated mutant neurons displayed significantly reduced incorporation (Figure 5C) suggesting impaired protein synthesis.
Figure 5. Reduced protein synthesis and AKT/mTOR activity in MECP2 mutant neurons were rescued by BDNF and IGF1 treatment.
A) Boxplots of transcriptional changes of ribosomal proteins between mutant and control cells.
B and C) MECP2 mutant neurons (B), but not NPs (C), have reduced protein synthesis, as measured by 35S-Cys/Met incorporation.
D) MECP2 mutant neurons have reduced levels of secreted BDNF protein, as measured by BDNF ELISA.
E to H) MAP2-positive MECP2 mutant neurons have reduced levels of phospho-AKT (E and G) and phospho-S6 (F and H), compared to their isogenic controls. Scale bar, 10um.
I) BDNF or IGF1 treatment partially rescues the soma size defect in MECP2 mutant MAP2-positive neurons.
J) BDNF or IGF1 treatment increases neurite complexity in MECP2 mutant Synapsin-GFP-positive neurons.
K) BDNF or IGF1 treatment enhances protein synthesis in MECP2 mutant neurons as measured by 35S-Cys/Met incorporation.
L and M) Quantitative analyses of MAP2-positive neurons co-stained with antibodies against phospho-AKT or phospho-S6 demonstrate that BDNF or IGF1 treatment partially ameliorated the AKT and S6 signaling defects in MECP2 mutant neurons.
Results are mean +/− SEM. *p<0.05, **p<0.01, ***p<0.005. See also Figure S4.
We have previously reported that MECP2 mutant mice could be partially rescued either by over-expression of BDNF, or by systemic delivery of IGF1 (Chang et al., 2006; Tropea et al., 2009), consistent with a reduced BDNF mRNA and protein level in brains of Mecp2 mutant mice (Chang et al., 2006). Indeed, MECP2 mutant neurons had both lower basal as well as KCl-induced levels of BDNF mRNA (Figure 4C–4D). Also, MECP2 mutant neurons secreted reduced levels of BDNF protein as measured by ELISA (Figure 5D). TrkB and IGF1R, the high affinity receptors for BDNF and IGF1, were abundant in human neurons and were expressed at similar levels in control and mutant neurons as evaluated by quantitative RT-PCR (Figure S4C). We further examined the activities of key intra-cellular signaling pathways downstream of these receptors focusing on the AKT/mTOR pathway because of its known central function in regulating translation (Zoncu et al., 2011). Mutant neurons displayed severely diminished levels of phospho-AKT and phospho-S6 as seen by immuno-staining (Figure 5E–5H).
We next investigated whether treatment with BDNF or IGF1 could activate the AKT and S6 pathway and affect the in vitro mutant phenotype. Three-week treatment with BDNF or IGF1 significantly rescued the soma size and dendritic complexity deficits seen in MECP2 mutant neurons (Figure 5I–5J). Although BDNF and IGF1 treatment did not significantly affect the total RNA content per cell (not shown), nascent protein synthesis as evaluated by 35S-Cys/Met incorporation levels was elevated in treated mutant neurons (Figure 5K). Furthermore, BDNF and IGF1 treatment significantly increased the percentage of MAP2+ mutant neurons that co-stained for phospho-AKT and phospho-S6 (Figure 5L–5M). Thus, these results strongly support the notion that reduced levels of new protein synthesis in MECP2 mutant neurons was accompanied with impaired activities of the AKT/mTOR pathways and that treatment with exogenous growth factors promoted protein synthesis via enhancing AKT/mTOR signaling activities.
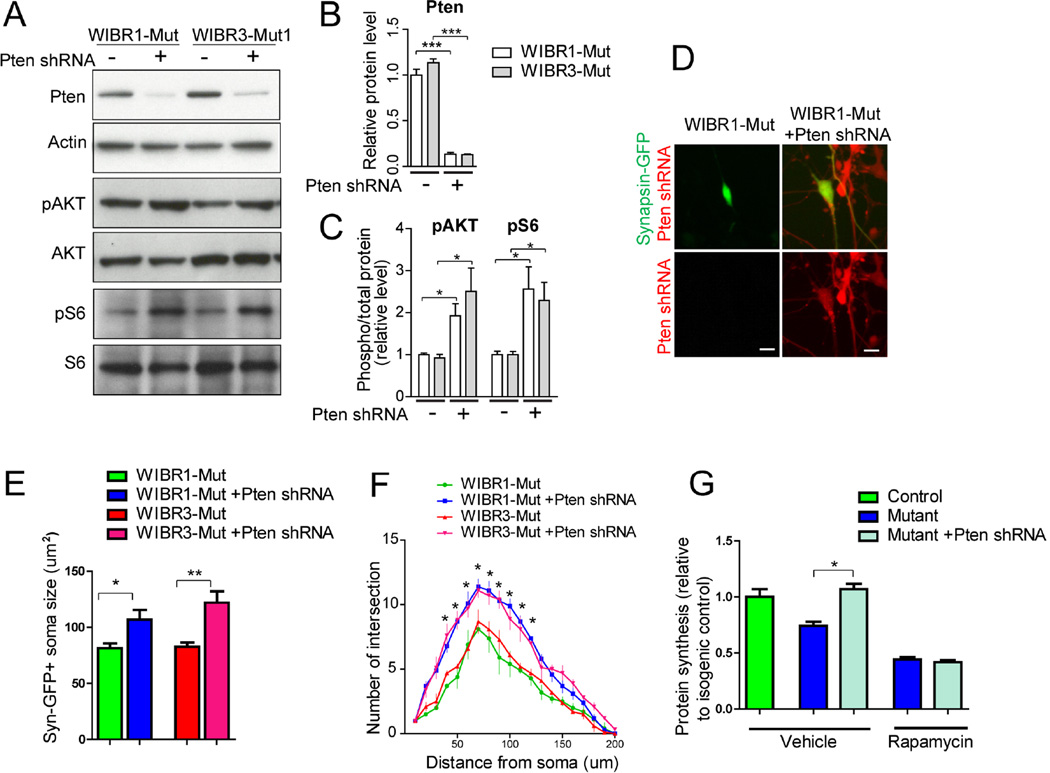
To further investigate the effects of promoting protein synthesis on the mutant phenotypes, we used a genetic approach to activate the AKT/mTOR pathway. PTEN is a known negative regulator of PI3K, an upstream component of the AKT/mTOR pathway (Sansal and Sellers, 2004). Ablation of PTEN has been shown to lead to constitutive activation of the AKT/mTOR pathway in a variety of tissues, including the brain (Kwon et al., 2006). We transduced control and mutant NPs with lentiviruses encoding a PTEN shRNA that has been proven to knock down PTEN efficiently (Luikart et al., 2011). Immuno-blotting confirmed significant reductions in PTEN protein, and corresponding increases in the levels of phospho-AKT and phospho-S6 proteins upon transduction (Figure 6A–6C). This lentivirus construct contains an mCherry reporter, which allows the identification of individual PTEN knockdown cells. Figures 6D to 6F show that PTEN knockdown significantly increased soma size and neurite arborization as compared to vector treated cells. This was accompanied by an increase in nascent protein synthesis as measured by 35S-Cys/Met incorporation (Figure 6G). These results demonstrate that genetic activation of the AKT/mTOR pathway in mutant neurons promoted protein synthesis and was sufficient to ameliorate the disease-related cellular impairments.
Figure 6. PTEN knockdown rescues protein synthesis and morphological defects in MECP2 mutant neurons.
A to C) Immuno-blot analysis for PTEN (A and B), phospho-AKT and phospho-S6 (A and C) demonstrate effective knockdown of PTEN and activation of AKT/S6 pathway using PTEN shRNA.
D) Representative images of Synapsin-GFP-positive MECP2 mutant neurons treated with vector (left) or PTEN shRNA-mCherry (right). Scale bar, 10um.
E and F) PTEN-shRNA increased soma size (E) and neurite complexity (F) in MECP2 mutant neurons.
G) PTEN-shRNA enhanced protein synthesis in MECP2 mutant neurons, as measured by 35S-Cys/Met incorporation. Inhibition of mTOR signaling by rapamycin suppressed PTEN knockdown-induced increase in protein synthesis.
Results are mean +/− SEM. *p<0.05, **p<0.01, ***p<0.005.
MECP2 mutant neurons display impaired mitochondrial function
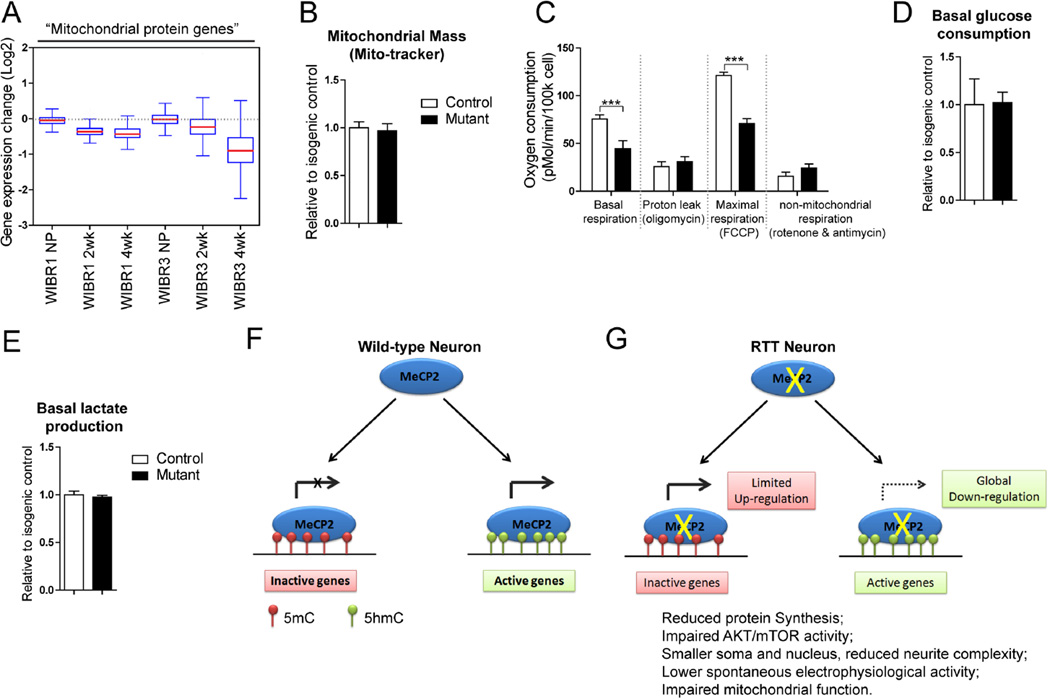
The expression of all genes predicted to encode mitochondrial proteins (Pagliarini et al., 2008) was significantly down-regulated in mutant neurons (Figure 7A). This is consistent with mitochondrial protein genes as a group being highly expressed (Figure S5A). A survey of all genes encoding mitochondrial ribosomal proteins revealed that similar to cytoplasmic ribosomal protein genes (Figure S4), the majority of these genes were also down-regulated (Figure S5B–C). Consistent with this observation, expression of the UQCRC1 gene, which was previously suggested to be up-regulated in MECP2 mutant mice (Kriaucionis et al., 2006), was in fact down-regulated in MECP2 mutant neurons (Figure S5D). To investigate the physiological consequences of these transcriptional changes, we measured cellular oxygen consumption. Despite having normal mitochondrial mass per cell as measure by mitochondria-selective dye uptake, mutant neurons displayed a 40% reduction in their basal oxygen consumption, and a similar reduction in the maximal respiration rate observed upon FCCP addition (Figure 7B–C). These results strongly suggest that the capacity of the mitochondrial electron transport chain is reduced in mutant neurons. Given that mitochondrial mass was similar between control and mutant neurons, this capacity could be limited by the availability of substrate due to an overall decrease in glucose metabolism. To explore this possibility further we measured rates of glucose consumption and lactate production in these cells (Figure 7D–E). No differences in glucose consumption or lactate production were observed in these cells. These data are consistent with the decrease in respiration being explained by down-regulation of mitochondrial gene expression resulting in fewer electron transport chain units.
Figure 7. MECP2 mutant human neurons show impaired mitochondrial function.
A) Boxplots of transcriptional changes of mitochondrial protein genes between mutant and control cells.
B) Quantification analyses of mitochondrial mass as measured by mito-tracker fluorescence, normalized to Hoechst intensity.
C) Measurement of oxygen consumption reveals reduced respiration at the basal and FCCP uncoupled states in MECP2 mutant neurons.
D and E) Quantification of the basal consumption of glucose and production of lactate in control and MECP2 mutant neurons.
(F and G) A model of MECP2 regulated gene expression in human neurons. MECP2 is present on both inactive genes (marked by 5mC) and active genes (marked by 5hmC). In RTT neurons, deletion of MECP2 induces up-regulation of a small group of lowly expressed genes, indicating a repressor function for MECP2 for these genes. The main effect of MECP2 deletion, however, is a genome-wide down-regulation of a majority of genes, with preferential effects on those that are highly expressed. This demonstrates a global activator role for MECP2 (G). This global transcriptional repression in RTT neurons leads to reduced protein synthesis, AKT/mTOR activity, soma and nucleus size, neurite complexity, electrophysiological activity and mitochondrial function.
Results are mean +/− SEM. ***p<0.005. See also Figure S5.
Discussion
Our study shows that compared to their isogenic controls, human neurons that lack MECP2 have smaller nuclei and soma, as well as less complex dendritic arborization. This decrease in cell size was coupled with a previously un-appreciated reduction in total RNA on a per cell level. In agreement with a recent report of decreased RNA synthesis in MECP2 mutant mouse neurons (Yazdani et al., 2012), our findings demonstrate that MECP2 mutant human neurons, but not NPs, have reduced total RNA and rRNA. More importantly, unbiased global transcription analyses revealed a striking genome-wide down-regulation in mutant human neurons. These findings highlight a drastic decline in the integrity of the neuronal transcriptional program in the absence of MECP2. Furthermore, although the downward shift in gene expression is widespread, the transcriptional activity of genes directly correlated with their probability to be down-regulated upon MECP2 deletion. In addition, genes known to have higher ratios of 5hmC compared to 5mC were more likely down-regulated in an MECP2-dependent manner in neurons but not NPs. This preferential down-regulation of active genes strongly supports a crucial role for MECP2 in facilitating active transcription in human neurons.
One of the key conclusions from our work is that MECP2 can act as a global activator of gene expression in neurons, a conclusion that contrasts with the widely accepted notion of MECP2 acting as a transcriptional repressor (Ebert et al., 2013; Lyst et al., 2013). Our results are based on gene expression analyses using the addition of synthetic RNA spike-in standards, which allowed unbiased gene expression analysis in equal numbers of cells. In contrast, traditional gene expression measurements are based on the assumption that control and experimental cells have a similar amount of total RNA and thus are normalized to input RNA. This assumption leads to erroneous conclusions if, as in RTT neurons, the amount of total RNA differs between experimental and control cells. The present and previous results (Lin et al., 2012; Loven et al., 2012) emphasize the necessity of a “per-cell” perspective for interpreting genome-wide gene expression data, particularly when the experimental manipulation could result in perturbations of global transcription.
It was surprising that control and mutant cells derived from the female WIBR3 ESCs displayed higher levels of gene expression changes than the male WIBR1-derived cells. While we have no mechanistic explanation for this finding, global differences in DNA methylation between male and female mouse ESCs have been reported previously (Zvetkova et al., 2005), and female human ESCs and iPSCs also display variable gene expression (Anguera et al., 2012; Mekhoubad et al., 2012). The lack of MECP2 mRNA and protein expression in female WIBR3-mut cells demonstrates that the MECP2 locus on the inactive X-chromosome remained silent and the global transcriptional down-regulation and the preferential vulnerability of active genes were independently identified in mutant male and female cells. Thus, our findings ascertain an unambiguous role for MECP2 in facilitating active transcription on a genome-wide scale.
Furthermore, we demonstrated that mutant neurons display several novel disease-related physiological changes as compared to their isogenic controls. Using multi-electrode arrays, we followed the course of neuronal maturation and identified a significant reduction in spontaneous network electric activity in mutant neurons. Prompted by the finding that many mitochondrial protein genes were down-regulated in the absence of MECP2, we investigated respiration in whole neuronal population, and identified significantly impaired metabolic function in mutant neurons. This echoes earlier observations that RTT shares hallmarks of mitochondrial encephalopathies, and was thought to be a mitochondrial disorder characterized by dysregulation of lactate metabolism (Cornford et al., 1994). These findings provide novel insights into the manifestation of disease symptoms for RTT and potential future avenues of therapeutic intervention.
Exogenous growth factors such as BDNF and IGF1 are effective in mitigating disease-related symptoms in MECP2 mutant mice (Chang et al., 2006; Kline et al., 2010; Tropea et al., 2009) and in patient iPSC-derived neurons (Marchetto et al., 2010). The underlying mechanism for these reversals remains to be identified. Using isogenic human cells we demonstrated that MECP2 mutant neurons have significantly lower protein synthesis activity. Furthermore, our study identified a severe defect in the activity of the AKT/mTOR signaling cascade, a crucial pathway known to directly modulate protein synthesis and is induced by BDNF and IGF1 (Ricciardi et al., 2011). Genetic activation of the AKT/mTOR pathway by PTEN ablation ameliorated the protein synthesis and the neural morphological defects in MECP2 mutant neurons, offering further evidence that protein synthesis impairment is central to RTT pathology. Thus, our results provide mechanistic insights into impairments seen in MECP2 mutant neurons and suggest the unifying hypothesis that a defect in the global control of transcription and translation is a fundamental cause of RTT-related symptoms.
The critical dependence on MECP2 in maintaining gene expression appears to be a phenomenon unique to differentiated neurons and was not seen in NPs. The critical role of MECP2 in neurons is supported by previous studies in mice showing that ablation of MECP2 specifically in post-mitotic neurons was sufficient to produce RTT symptoms (Chao et al., 2010; Chen et al., 2001). Thus, the mechanism by which re-expression of MECP2 reverses RTT-phenotypes may reside in its ability to reinstate proper gene expression regulation thus reversing the downstream transcriptional and translational impairments. Consistent with the notion that the RTT phenotype is reversible, BDNF, IGF1 addition and PTEN knockdown rescued the disease-related phenotype in mutant neurons suggesting that boosting protein synthesis by activation of the AKT/mTOR pathway may represent a potential therapeutic strategy for RTT.
Experimental Procedures
TALEN construct and genome editing in human ESCs
TALENs-mediated gene targeting was performed as previously described (Hockemeyer et al., 2011). The DNA binding sites of the TALENs used for targeting the exon 3 of MECP2 are gcagccatcagcccaccact and ctctgctttgcctgcct.
Cell culture and neural differentiation
HESC lines WIBR1, WIBR3 were previously described and cultured as previously described (Lengner et al., 2010). To induce neural differentiation, increasing amounts of N2 media (25, 50, 75, 100%) were added to mTeSR medium every other day whilst maintaining 2.5uM dorsomorphin. NPs were passaged at day 20–24 using accutase and cultured in N2 media with 20ng/ml bFGF. NPs were further differentiated in differentiation medium without bFGF. For morphological and electrophysiological analyses, cells were lifted using accutase and replated onto coverglass or culture dish.
RNA preparation, microarray analysis and quantitative RT-PCR
Total RNA extraction and RNA electrophoresis was performed as previously described (Lin et al., 2012). RNA spike-in controls was added to total RNA extracted from equal numbers of cells, as previously described and following manufacturer’s recommendations (Loven et al., 2012). RNA was reverse transcribed using Superscript III reverse transcriptase (Invitrogen) with random hexamer primers. Transcript abundance was determined by quantitative PCR using SYBR Green PCR mix (Applied Biosystems). Raw Ct values were normalized to GAPDH or ERCC spike-in standards.
Protein assays
Total protein extraction and immuno-blotting was performed as previously described (Li et al., 2008). Metabolic labeling of newly synthesis protein (Thoreen et al., 2012) was performed by labeling with 165uCi of EasyTag EXPRESS 35S protein labeling mix (Life Technologies) for 30min. The concentration of secreted BDNF protein in culture medium was measured using a BDNF ELISA Kit (Promega) (Li et al., 2012).
Supplementary Material
Acknowledgements
The authors thank Ping Xu, Raaji Alagappan, Tenzin Lungjangwa, Dongdong Fu, Daniel B. Dadon, Kibibi Ganz and Ruth Flannery for technical support; Drs. Bryan Luikart and David Sabatini for PTEN shRNA lentivirus constructs and access to Seahorse XF24; Dr. Carson Thoreen for advice on the protein synthesis assay; Drs. Peter Rahl, Bingbing Yuan and Jeong-Ah Kwon for assistance with microarray; and members of the Jaenisch lab for helpful discussions. Y.L. was a Simons Postdoctoral Fellow. Y.L. was supported in part by a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation, and the IRSF. A.W.C. was supported by a Croucher scholarship. J.L. was supported by a Swedish Research Council Postdoctoral Fellowship (VR-B0086301). D.O. and R.Y. were supported by NIH grant R01-HG002668. M.V.H and C.A.L. acknowledge support from P30-CA14051, the Koch Institute and Curt Marble Cancer Research Fund. R.J. was supported by NIH grants HD 045022, R37-CA084198, SFARI and the ELA foundation.
Footnotes
Accession numbers
The microarray data are available in the Gene Expression Omnibus (GEO) database under the accession number GSE50584.
Supplemental Information
Supplemental Information includes Supplemental Experimental Procedures and five figures.
Reference
- Akbarian S. Diseases of the mind and brain: Rett's syndrome. Am J Psychiatry. 2002;159:1103. doi: 10.1176/appi.ajp.159.7.1103. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Ananiev G, Williams EC, Li H, Chang Q. Isogenic pairs of wild type and mutant induced pluripotent stem cell (iPSC) lines from Rett syndrome patients as in vitro disease model. PLoS One. 2011;6:e25255. doi: 10.1371/journal.pone.0025255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguera MC, Sadreyev R, Zhang Z, Szanto A, Payer B, Sheridan SD, Kwok S, Haggarty SJ, Sur M, Alvarez J, et al. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell. 2012;11:75–90. doi: 10.1016/j.stem.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateup HS, Johnson CA, Denefrio CL, Saulnier JL, Kornacker K, Sabatini BL. Excitatory/inhibitory synaptic imbalance leads to hippocampal hyperexcitability in mouse models of tuberous sclerosis. Neuron. 2013;78:510–522. doi: 10.1016/j.neuron.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shachar S, Chahrour M, Thaller C, Shaw CA, Zoghbi HY. Mouse models of MeCP2 disorders share gene expression changes in the cerebellum and hypothalamus. Hum Mol Genet. 2009;18:2431–2442. doi: 10.1093/hmg/ddp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Q, Khare G, Dani V, Nelson S, Jaenisch R. The disease progression of Mecp2 mutant mice is affected by the level of BDNF expression. Neuron. 2006;49:341–348. doi: 10.1016/j.neuron.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, Jaenisch R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- Cheung AY, Horvath LM, Grafodatskaya D, Pasceri P, Weksberg R, Hotta A, Carrel L, Ellis J. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum Mol Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornford ME, Philippart M, Jacobs B, Scheibel AB, Vinters HV. Neuropathology of Rett syndrome: case report with neuronal and mitochondrial abnormalities in the brain. J Child Neurol. 1994;9:424–431. doi: 10.1177/088307389400900419. [DOI] [PubMed] [Google Scholar]
- Ebert DH, Gabel HW, Robinson ND, Kastan NR, Hu LS, Cohen S, Navarro AJ, Lyst MJ, Ekiert R, Bird AP, et al. Activity-dependent phosphorylation of MeCP2 threonine 308 regulates interaction with NCoR. Nature. 2013;499:341–345. doi: 10.1038/nature12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacometti E, Luikenhuis S, Beard C, Jaenisch R. Partial rescue of MeCP2 deficiency by postnatal activation of MeCP2. Proc Natl Acad Sci U S A. 2007;104:1931–1936. doi: 10.1073/pnas.0610593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan C, Li HH, Kwan HC, Francke U. Cerebellar gene expression profiles of mouse models for Rett syndrome reveal novel MeCP2 targets. BMC Med Genet. 2007;8:36. doi: 10.1186/1471-2350-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline DD, Ogier M, Kunze DL, Katz DM. Exogenous brain-derived neurotrophic factor rescues synaptic dysfunction in Mecp2-null mice. J Neurosci. 2010;30:5303–5310. doi: 10.1523/JNEUROSCI.5503-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S, Paterson A, Curtis J, Guy J, Macleod N, Bird A. Gene expression analysis exposes mitochondrial abnormalities in a mouse model of Rett syndrome. Mol Cell Biol. 2006;26:5033–5042. doi: 10.1128/MCB.01665-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kron M, Howell CJ, Adams IT, Ransbottom M, Christian D, Ogier M, Katz DM. Brain activity mapping in Mecp2 mutant mice reveals functional deficits in forebrain circuits, including key nodes in the default mode network, that are reversed with ketamine treatment. J Neurosci. 2012;32:13860–13872. doi: 10.1523/JNEUROSCI.2159-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler S, Kilic E, Bahr M. Human synapsin 1 gene promoter confers highly neuron-specific long-term transgene expression from an adenoviral vector in the adult rat brain depending on the transduced area. Gene Ther. 2003;10:337–347. doi: 10.1038/sj.gt.3301905. [DOI] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, Li Y, Baker SJ, Parada LF. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, Muffat J, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–883. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Li Y, Luikart BW, Birnbaum S, Chen J, Kwon CH, Kernie SG, Bassel-Duby R, Parada LF. TrkB regulates hippocampal neurogenesis and governs sensitivity to antidepressive treatment. Neuron. 2008;59:399–412. doi: 10.1016/j.neuron.2008.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yui D, Luikart BW, McKay RM, Rubenstein JL, Parada LF. Conditional ablation of brain-derived neurotrophic factor-TrkB signaling impairs striatal neuron development. Proc Natl Acad Sci U S A. 2012;109:15491–15496. doi: 10.1073/pnas.1212899109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Orlando DA, Sigova AA, Lin CY, Rahl PB, Burge CB, Levens DL, Lee TI, Young RA. Revisiting global gene expression analysis. Cell. 2012;151:476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart BW, Schnell E, Washburn EK, Bensen AL, Tovar KR, Westbrook GL. Pten knockdown in vivo increases excitatory drive onto dentate granule cells. J Neurosci. 2011;31:4345–4354. doi: 10.1523/JNEUROSCI.0061-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyst MJ, Ekiert R, Ebert DH, Merusi C, Nowak J, Selfridge J, Guy J, Kastan NR, Robinson ND, de Lima Alves F, et al. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat Neurosci. 2013;16:898–902. doi: 10.1038/nn.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A, Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Nuber UA, Kriaucionis S, Roloff TC, Guy J, Selfridge J, Steinhoff C, Schulz R, Lipkowitz B, Ropers HH, Holmes MC, et al. Up-regulation of glucocorticoid-regulated genes in a mouse model of Rett syndrome. Hum Mol Genet. 2005;14:2247–2256. doi: 10.1093/hmg/ddi229. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirooznia M, Wang T, Avramopoulos D, Valle D, Thomas G, Huganir RL, Goes FS, Potash JB, Zandi PP. SynaptomeDB: an ontology-based knowledgebase for synaptic genes. Bioinformatics. 2012;28:897–899. doi: 10.1093/bioinformatics/bts040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciardi S, Boggio EM, Grosso S, Lonetti G, Forlani G, Stefanelli G, Calcagno E, Morello N, Landsberger N, Biffo S, et al. Reduced AKT/mTOR signaling and protein synthesis dysregulation in a Rett syndrome animal model. Hum Mol Genet. 2011;20:1182–1196. doi: 10.1093/hmg/ddq563. [DOI] [PubMed] [Google Scholar]
- Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol. 2004;22:2954–2963. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- Scala E, Longo I, Ottimo F, Speciale C, Sampieri K, Katzaki E, Artuso R, Mencarelli MA, D'Ambrogio T, Vonella G, et al. MECP2 deletions and genotype-phenotype correlation in Rett syndrome. Am J Med Genet A. 2007;143A:2775–2784. doi: 10.1002/ajmg.a.32002. [DOI] [PubMed] [Google Scholar]
- Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova M, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Jaenisch R. Medicine. iPSC disease modeling. Science. 2012;338:1155–1156. doi: 10.1126/science.1227682. [DOI] [PubMed] [Google Scholar]
- Su D, Cha YM, West AE. Mutation of MeCP2 alters transcriptional regulation of select immediate-early genes. Epigenetics. 2012;7:146–154. doi: 10.4161/epi.7.2.18907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropea D, Giacometti E, Wilson NR, Beard C, McCurry C, Fu DD, Flannery R, Jaenisch R, Sur M. Partial reversal of Rett Syndrome-like symptoms in MeCP2 mutant mice. Proc Natl Acad Sci U S A. 2009;106:2029–2034. doi: 10.1073/pnas.0812394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor M, Akbarian S, Chen RZ, Jaenisch R. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci U S A. 2002;99:15536–15541. doi: 10.1073/pnas.242566899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullai JW, Schaffer ME, Mullenbrock S, Sholder G, Kasif S, Cooper GM. Immediate-early and delayed primary response genes are distinct in function and genomic architecture. J Biol Chem. 2007;282:23981–23995. doi: 10.1074/jbc.M702044200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urdinguio RG, Lopez-Serra L, Lopez-Nieva P, Alaminos M, Diaz-Uriarte R, Fernandez AF, Esteller M. Mecp2-null mice provide new neuronal targets for Rett syndrome. PLoS One. 2008;3:e3669. doi: 10.1371/journal.pone.0003669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, Thatcher KN, Farnham PJ, Lasalle JM. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci U S A. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdani M, Deogracias R, Guy J, Poot RA, Bird A, Barde YA. Disease modeling using embryonic stem cells: MeCP2 regulates nuclear size and RNA synthesis in neurons. Stem Cells. 2012;30:2128–2139. doi: 10.1002/stem.1180. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhou J, Su P, Li D, Tsang S, Duan E, Wang F. High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor beta superfamily receptors. Stem Cells. 2010;28:1741–1750. doi: 10.1002/stem.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvetkova I, Apedaile A, Ramsahoye B, Mermoud JE, Crompton LA, John R, Feil R, Brockdorff N. Global hypomethylation of the genome in XX embryonic stem cells. Nat Genet. 2005;37:1274–1279. doi: 10.1038/ng1663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.