Abstract
Background
Reduced DNA repair capacities due to inherited polymorphisms may increase the susceptibility to cancers including gastric cancer. Previous studies investigating the association between Xeroderma Pigmentosum group C (XPC) gene polymorphisms and gastric cancer risk reported inconsistent results. We performed a meta-analysis to summarize the possible association.
Methods
All studies published up to January 2014 on the association between XPC polymorphisms and gastric cancer risk were identified by searching electronic databases PubMed, EMBASE, Cochrane library, and Chinese Biomedical Literature database (CBM). The association between XPC polymorphisms and gastric cancer risk was assessed by odds ratios (ORs) together with their 95% confidence intervals (CIs).
Results
Six studies with 1,355 gastric cancer cases and 2,573 controls were finally included in the meta-analysis. With respect to Lys939Gln polymorphism, we did not observe a significant association when all studies were pooled into the meta-analysis. When stratified by ethnicity, source of control, and study quality, statistical significant association was not detected in all subgroups. With respect to Ala499Val and PAT−/+polymorphisms, we also did not observe any significant association with gastric cancer risk in the pooled analysis.
Conclusions
This meta-analysis based on current evidences suggested that the XPC polymorphisms (Lys939Gln, Val499Arg, and PAT−/+) did not contribute to gastric cancer risk. Considering the limited sample size and ethnicity included in the meta-analysis, further larger scaled and well-designed studies are needed to confirm our results.
Virtual Slides
The virtual slide(s) for this article can be found here: http://www.diagnosticpathology.diagnomx.eu/vs/1485880312555069
Keywords: XPC, Polymorphism, Gastric cancer, Meta-analysis
Background
Gastric cancer is one of the most common cancers and cancer related deaths are highly prevalent worldwide [1]. The development of gastric cancer is a multifactorial and multistep process. Previous epidemiological investigations have identified that high consumption of salty food, low consumption of fresh fruits and vegetables, smoking, drinking, and Helicobacter pylori infection were the major contributors to the development and progression of gastric cancer [2-4]. However, most individuals exposed to these environmental risk factors never develop gastric cancer while many gastric cancer cases develop among individuals without those environmental factors, suggesting that other factors such as genetic factors also play important roles in gastric carcinogenesis.
The DNA repairing system, which was composed of many DNA repair genes, plays an important role in removing damaged genes, maintaining the genomic integrity and preventing carcinogenesis. The xeroderma pigmentosum complementation group C (XPC) is one of the eight core genes (i.e., ERCC1, XPA, XPB, XPC, XPD, XPE, XPF, and XPG) in the nuclear excision repair (NER) pathway of the DNA repairing system. XPC binds to HR23B and forms the XPC-HR23B complex, which is involved in the DNA damage recognition and DNA repair initiation in the NER pathway [5-7], and the binding of XPC to damaged DNA is the rate-limiting step for NER [8]. The XPC gene, which encodes a 940-amino acid protein, spans 33 kb on chromosome 3p25 and contains 16 exons and 15 introns [9]. There are at least 687 reported single nucleotide polymorphisms (SNPs) in the XPC gene region (http://www.ncbi.nlm.nih.gov/projects/SNP). Among all the identified SNPs, three common polymorphisms have been extensively studied: (a) a substitution of alanine for valine in codon 499 (Ala499Val, rs2228000), in the interaction domain of XPC with hHRAD23; (b) an A to C transversion in exon 15 resulting in a lysine-to-glutamine transition at position 939 (Lys939Gln, rs2228001), located in the interaction domain with TFIIH; and (c) a poly AT region on intron 9 (PAT−/+). It was reported that the variant alleles of the three polymorphisms in the XPC genes correlated with relatively high DNA adduct levels in lymphocyte DNA, indicating that these polymorphisms were associated with decreased DNA repair capacity [10,11]. Therefore, it was biologically reasonable to hypothesize a potential relationship between the XPC gene polymorphisms and cancer susceptibility.
To date, the XPC gene polymorphisms (Lys939Gln, Val499Arg, and PAT−/+) to gastric cancer risk have been a research focus in scientific community and have drawn increasing attention. Several original studies have reported the role of XPC polymorphisms in gastric cancer risk [12-17], but the results are inconclusive. For genetic association studies that investigate candidate polymorphisms, sample size is an important influencing factor for study accuracy. Small sample size might have inadequate power to explore a true association of modest effect [18], especially for complex multifactorial disease such as gastric cancer. While combining data from all eligible studies by meta-analysis has the advantage of increasing statistical power and reducing random error and obtaining precise estimates for some potential genetic associations. Therefore, in this study, we conducted a quantitative meta-analysis including all eligible studies.
Methods
Search strategy
We conducted a comprehensive literature search in PubMed, EMBASE, Cochrane library, and Chinese Biomedical Literature (CBM) databases up to January 01, 2014 using the following search strategy: (“gastric cancer”) and (“xeroderma pigmentosum complementation group C”, or “XPC”). There was no restriction on time period, sample size, population, language, or type of report. All eligible studies were retrieved and their references were checked for other relevant studies. The literature retrieval was performed in duplication by two authors independently (Qiliu Peng and Yu Lu). When multiple publications reported on the same or overlapping data, we chose the most recent or largest population.
Inclusion and exclusion criteria
Studies were considered if they met the following inclusion criteria: (1) case–control or cohort studies which evaluated the association between XPC polymorphisms and gastric cancer; (2) had an odds ratio (OR) with 95% confidence interval (CI) or other available data for estimating OR (95% CI); and (3) the control population did not contain malignant tumor patients. Studies were excluded if one of the following existed: (1) no control population; (2) duplicate of previous publication; and (3) insufficient information for data extraction.
Data extraction
Information was extracted by two authors independently according to the inclusion criteria listed above. Data extracted from eligible studies included the first author, publication year, ethnicity, country, genotyping methods, matching criteria, source of control, gastric cancer ascertainment, total numbers of cases and controls and genotype frequencies of cases and controls. Ethnic backgrounds were categorized as Caucasian, Asian. To ensure the accuracy of the information extracted, the two authors checked the data extraction results and reached consensus on all of the items. If different results generated, they would check the data again and have a discussion to come to an agreement. If these two authors could not reach a consensus, another author (Xue Qin) was consulted to resolve the dispute.
Quality score assessment
The quality of eligible studies was assessed independently by two authors (Qiliu Peng and Xianjun Lao) according to a set of predefined criteria (Table 1) modified from our previous meta-analysis of molecular association study [19]. The revised criteria cover the representativeness of cases, source of controls, ascertainment of gastric cancer, total sample size, quality control of genotyping methods, and Hardy-Weinberg equilibrium (HWE) in the control population. Disagreements were resolved by consensus. Total scores ranged from 0 (lowest) to 10 (highest). Articles with scores equal to or higher than 7 were considered “high-quality” studies, whereas those with scores less than 7 were considered “low-quality” studies.
Table 1.
Scale for quality assessment
| Criteria | Score |
|---|---|
| Representativeness of cases |
|
| Selected from cancer registry or multiple cancer center sites |
2 |
| Selected from oncology department or cancer institute |
1 |
| Selected without clearly defined sampling frame or with extensive inclusion/exclusion criteria |
0 |
| Source of controls |
|
| Population or community based |
2 |
| Both population-based and hospital-based/healthy volunteers/blood donors |
1.5 |
| Hospital-based controls without gastric cancer |
1 |
| Cancer-free controls without total description |
0.5 |
| Not described |
0 |
| Ascertainment of gastric cancer |
|
| Histologically or pathologically confirmed |
2 |
| Diagnosis of gastric cancer by patient medical record |
1 |
| Not described |
0 |
| Sample size |
|
| >1000 |
2 |
| 200-1000 |
1 |
| <200 |
0 |
| Quality control of genotyping methods |
|
| Clearly described a different genotyping assay to confirm the data |
1 |
| Not described |
0 |
| Hardy-Weinberg equilibrium |
|
| Hardy-Weinberg equilibrium in controls |
1 |
| Hardy-Weinberg disequilibrium in controls |
0.5 |
| No checking for Hardy-Weinberg disequilibrium | 0 |
Statistical analysis
Crude odds ratios (ORs) with their 95% confidence intervals (CIs) were used to assess the strength of association between the XPC polymorphisms and gastric cancer risk. The pooled ORs were performed for co-dominant genetic models, dominant genetic model, and recessive genetic model, respectively. Heterogeneity among studies was checked by the chi-square-based Q-test and I2 statistics [20,21]. If the result of the heterogeneity test was PQ < 0.1 or I2 ≥ 50%, indicating the presence of heterogeneity, a random-effects model (the DerSimonian and Laird method) was used to estimate the summary ORs [22]; otherwise, when the result of the heterogeneity test was PQ ≥ 0.1 and I2 < 50%, indicating the absence of heterogeneity, the fixed-effects model (the Mantel–Haenszel method) was used [23]. Subgroup analyses were performed according to ethnicity, source of control, and study quality. Sensitivity analysis was conducted by sequential omission of individual study to assess the robustness of the results. Publication bias was assessed using a Begg’s funnel plot and Egger’s regression asymmetry test [24]. If publication bias existed, the Duval and Tweedie non-parametric “trim and fill” method was used to adjust for it [25]. The distribution of the genotypes in the control population was tested for HWE using a goodness-of-fit Chi-square test. All the statistical tests were performed using Stata software, version 12.0 (Stata Corp., College Station, TX).
Results
Eligible studies
Based on the search criteria, seven potential relevant studies were identified. One of these articles was excluded because it contained overlapping data [26]. Manual search of references cited in the published studies did not reveal any additional articles. As a result, a total of six studies met the inclusion criteria were included in the meta-analysis [12-17]. The main characteristics of the eligible studies were presented in Table 2. Among them, five studies [12,14-17] including 1,049 cases and 2,026 controls were available for Lys939Gln polymorphism, three studies [14,15,17] with 817 cases and 1438 controls for Val499Arg polymorphism, and two studies [13,15] containing 559 cases and 1159 controls for PAT−/+polymorphism. The sample size of these studies varied considerably, ranging from 222 to 977 individuals. Of all the eligible studies, two were conducted in Caucasians [12,16] and three were in Asians [14,15,17] for Lys939Gln polymorphism; all the three eligible studies for Val499Arg polymorphism were conducted in Asians [14,15,17]; one was conducted in Caucasians [13] and one [15] was in Asians for PAT−/+polymorphism. Three studies were population–based and three were hospital–based studies. One study in the present meta-analysis did not provide definite criteria for the CRC ascertainment [12]. Two genotyping methods were used, including PCR-RFLP and TaqMan assay. The genotype distributions of the controls were consistent with HWE in all of the included studies.
Table 2.
Characteristics of studies included in the meta-analysis
|
First author (Year) |
Country |
Ethnicity |
Sample size (case/control) |
Genotyping methods |
Matching criteria |
Source of control |
GC ascertainment |
SNPs |
HWE(
P
value) |
Quality scores |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lys939Gln | Ala499Val | PAT+/− | ||||||||||
| Palli 2010 |
Italy |
Caucasian |
306/547 |
TaqMan |
Gender |
PB |
Histo- |
PAT+/− |
— |
— |
0.085 |
7 |
| Dong 2008 |
China |
Asian |
253/612 |
PCR-RFLP |
Age and gender |
PB |
Histopatho- |
Lys939Gln, Ala499Val, PAT+/− |
0.699 |
0.217 |
0.786 |
8.5 |
| Engin 2011 |
Turkey |
Caucasian |
106/116 |
PCR-RFLP |
Age and BMI |
HB |
NR |
Lys939Gln |
0.642 |
— |
— |
4.5 |
| Long 2010 |
China |
Asian |
361/616 |
TaqMan |
Age, gender, ethnicity, smoking, and drinking |
HB |
Histopatho- |
Lys939Gln, Ala499Val |
0.446 |
0.673 |
— |
6 |
| Ye 2006 |
Sweden |
Caucasian |
126/472 |
PCR-RFLP |
NR |
PB |
Histo- |
Lys939Gln |
0.540 |
— |
— |
7 |
| Li 2010 | China | Asian | 203/210 | PCR-RFLP | Age, gender, smoking, and drinking | HB | Patho- | Lys939Gln, Ala499Val | 0.173 | 0.462 | — | 7.5 |
GC, Gastric cancer; Histopatho-, Histopathologically confirmed; Histo-, Histologically confirmed; Patho-, Pathologically confirmed; NR, Not reported; PB, Population–based; HB, Hospital–based; HWE, Hardy–Weinberg equilibrium in control population; PCR–RFLP, Polymerase chain reaction-restriction fragment length polymorphism; BMI, Body mass index.
Quantitative analysis
XPC Lys939Gln
The main results of meta-analysis of XPC Lys939Gln polymorphism and gastric cancer risk were present in Table 3. There was no evidence of significant association between XPC Lys939Gln polymorphism and gastric cancer risk when all the eligible studies were pooled into the meta-analysis (Gln/Gln vs. Lys/lys: OR = 1.123, 95% CI = 0.881–1.431, P = 0.349; Gln/Lys vs. Lys/lys: OR = 1.083, 95% CI = 0.917–1.277, P = 0.347; Gln/Gln + Gln/Lys vs. Lys/lys: OR = 1.092, 95% CI = 0.933–1.277, P = 0.273, Figure 1; Gln/Gln vs. Gln/Lys + Lys/lys: OR = 1.046, 95% CI = 0.838-1.307, P = 0.691). In subgroup analyses stratified by ethnicity, source of control, and study quality, statistically significant association was also not observed in all subgroups. Meanwhile, no significant heterogeneity was found in the pooled analysis and subgroup analyses (PQ Values <0.1 and I2 > 50%).
Table 3.
Meta-analysis of the XPC polymorphisms and gastric cancer risk
|
Comparison |
Population |
No. of studies |
Test of association |
Mode |
Test of heterogeneity |
||||
|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P Value | χ 2 | P Q Value | I 2 | ||||
| Lys939Gln |
|
|
|
|
|
|
|
|
|
| Gln/Gln vs. Lys/lys |
Overall |
5 |
1.123 |
0.881-1.431 |
0.349 |
F |
2.69 |
0.612 |
0.0 |
| |
Caucasian |
2 |
1.016 |
0.490-2.109 |
0.965 |
F |
2.31 |
0.128 |
46.8 |
| |
Asian |
3 |
1.140 |
0.857-1.516 |
0.368 |
F |
0.34 |
0.843 |
0.0 |
| |
PB |
2 |
1.260 |
0.877-1.810 |
0.212 |
F |
0.24 |
0.623 |
0.0 |
| |
HB |
3 |
1.026 |
0.741-1.420 |
0.879 |
F |
1.76 |
0.414 |
0.0 |
| |
High quality |
3 |
1.179 |
0.860-1.616 |
0.308 |
F |
0.75 |
0.686 |
0.0 |
| |
Low quality |
2 |
1.048 |
0.718-1.530 |
0.807 |
F |
1.71 |
0.191 |
41.6 |
| Gln/Lys vs. Lys/lys |
Overall |
5 |
1.083 |
0.917-1.277 |
0.347 |
F |
1.21 |
0.876 |
0.0 |
| |
Caucasian |
2 |
0.993 |
0.689-1.432 |
0.972 |
F |
0.87 |
0.352 |
0.0 |
| |
Asian |
3 |
1.107 |
0.919-1.332 |
0.284 |
F |
0.08 |
0.962 |
0.0 |
| |
PB |
2 |
1.026 |
0.795-1.324 |
0.846 |
F |
0.64 |
0.424 |
0.0 |
| |
HB |
3 |
1.126 |
0.906-1.400 |
0.285 |
F |
0.27 |
0.872 |
0.0 |
| |
High quality |
3 |
1.062 |
0.855-1.319 |
0.587 |
F |
0.90 |
0.639 |
0.0 |
| |
Low quality |
2 |
1.112 |
0.861-1.437 |
0.417 |
F |
0.24 |
0.628 |
0.0 |
| Gln/Gln + Gln/Lys vs. Lys/lys |
Overall |
5 |
1.092 |
0.933-1.277 |
0.273 |
F |
0.24 |
0.994 |
0.0 |
| |
Caucasian |
2 |
1.015 |
0.720-1.430 |
0.933 |
F |
0.01 |
0.906 |
0.0 |
| |
Asian |
3 |
1.113 |
0.933-1.327 |
0.235 |
F |
0.00 |
0.998 |
0.0 |
| |
PB |
2 |
1.076 |
0.846-1.369 |
0.551 |
F |
0.18 |
0.667 |
0.0 |
| |
HB |
3 |
1.103 |
0.897-1.356 |
0.351 |
F |
0.03 |
0.986 |
0.0 |
| |
High quality |
3 |
1.087 |
0.886-1.333 |
0.424 |
F |
0.21 |
0.900 |
0.0 |
| |
Low quality |
2 |
1.098 |
0.860-1.401 |
0.453 |
F |
0.02 |
0.881 |
0.0 |
| Gln/Gln vs. Gln/Lys + Lys/lys |
Overall |
5 |
1.046 |
0.838-1.307 |
0.691 |
F |
6.57 |
0.160 |
39.2 |
| |
Caucasian |
2 |
0.921 |
0.344-2.468 |
0.870 |
F |
5.98 |
0.114 |
43.3 |
| |
Asian |
3 |
1.083 |
0.829-1.417 |
0.558 |
F |
0.47 |
0.791 |
0.0 |
| |
PB |
2 |
1.253 |
0.896-1.751 |
0.187 |
F |
0.77 |
0.381 |
0.0 |
| |
HB |
3 |
0.914 |
0.680-1.229 |
0.552 |
F |
3.90 |
0.142 |
48.8 |
| |
High quality |
3 |
1.157 |
0.862-1.553 |
0.331 |
F |
1.66 |
0.435 |
0.0 |
| |
Low quality |
2 |
0.828 |
0.400-1.712 |
0.610 |
F |
3.90 |
0.148 |
44.3 |
| Ala499Val |
|
|
|
|
|
|
|
|
|
| Val/Val vs. Ala/Ala |
Asian |
3 |
0.861 |
0.632-1.172 |
0.341 |
F |
0.57 |
0.754 |
0.0 |
| Val/Ala vs. Ala/Ala |
Asian |
3 |
0.815 |
0.611-1.089 |
0.167 |
F |
4.72 |
0.194 |
37.6 |
| Val/Val + Val/Ala vs. Ala/Ala |
Asian |
3 |
0.823 |
0.630-1.076 |
0.155 |
F |
4.49 |
0.106 |
35.5 |
| Val/Val vs. Val/Ala + Ala/Ala |
Asian |
3 |
0.947 |
0.703-1.274 |
0.717 |
F |
0.04 |
0.981 |
0.0 |
| PAT−/+ |
|
|
|
|
|
|
|
|
|
| +/+vs. −/− |
Overall |
2 |
1.035 |
0.763-1.404 |
0.825 |
F |
0.12 |
0.726 |
0.0 |
| +/− vs. −/− |
Overall |
2 |
0.855 |
0.681-1.073 |
0.176 |
F |
0.00 |
0.975 |
0.0 |
| (+/+) + (+/−) vs. (−/−) |
Overall |
2 |
0.896 |
0.724-1.111 |
0.317 |
F |
0.05 |
0.830 |
0.0 |
| (+/+) vs. (+/−) + (−/−) | Overall | 2 | 1.141 | 0.871-1.495 | 0.338 | F | 0.20 | 0.652 | 0.0 |
OR, odds ratio; CI, confidence intervals; R, random effects model; F, fixed effects model; PB, Population–based; HB, Hospital–based.
Figure 1.
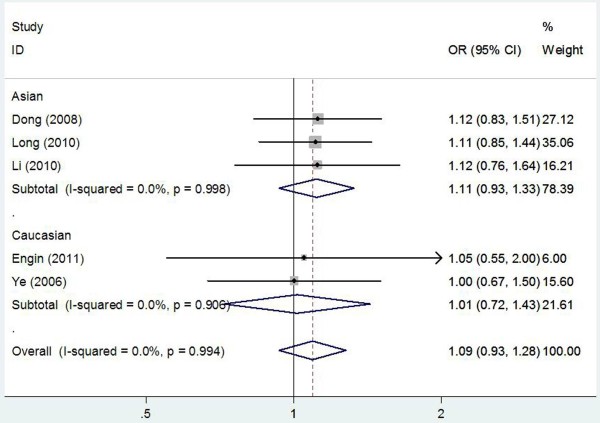
Forest plot of subgroup analysis by ethnicity on the association between XPC Lys939Gln polymorphism and gastric cancer risk using a fixed-effect model (dominant model Gln/Gln + Gln/Lys vs. Lys/lys).
XPC Val499Arg
The main results of meta-analysis of XPC Val499Arg polymorphism and gastric cancer risk were summarized in Table 3. We did not found any significant association between XPC Val499Arg polymorphism and gastric cancer risk when all the eligible studies were pooled into the meta-analysis (Val/Val vs. Ala/Ala: OR = 0.861, 95% CI = 0.632–1.172, P = 0.341; Val/Ala vs. Ala/Ala: OR = 0.815, 95% CI = 0.611–1.089, P = 0.167; Val/Val + Val/Ala vs. Ala/Ala: OR = 0.823, 95% CI = 0.630–1.076, P = 0.155; Val/Val vs. Val/Ala + Ala/Ala: OR = 0.947, 95% CI = 0.703-1.274, P = 0.717). In addition, no significant heterogeneity was found in the pooled analysis (PQ Values <0.1 and I2 > 50%). We did not perform subgroup analysis because of the limited number of studies available for XPC Val499Arg polymorphism.
XPC PAT−/+
The results of meta-analysis of XPC PAT−/+polymorphism and gastric cancer risk were summarized in Table 3. Overall, there were no significant associations of XPC PAT−/+polymorphism and gastric cancer risk when all the eligible studies were pooled into the meta-analysis (+/+vs. −/−: OR = 1.035, 95% CI = 0.763–1.404, P = 0.825; +/− vs. −/−: OR = 0.855, 95% CI = 0.681–1.073, P = 0.176; (+/+) + (+/−) vs. (−/−): OR = 0.896, 95% CI = 0.724–1.111, P = 0.317; (+/+) vs. (+/−) + (−/−): OR = 1.141, 95% CI = 0.871-1.495, P = 0.338). Moreover, statistical significant heterogeneity was also not found in the pooled analysis. We also did not perform subgroup analysis because only two studies were available for XPC PAT−/+polymorphism.
Sensitivity analysis
A single study involved in the meta-analysis was deleted each time to reflect the influence of the individual data-set to the pooled OR, and the corresponding pooled OR was not materially altered (Figure 2), indicating that our results were statistically robust.
Figure 2.
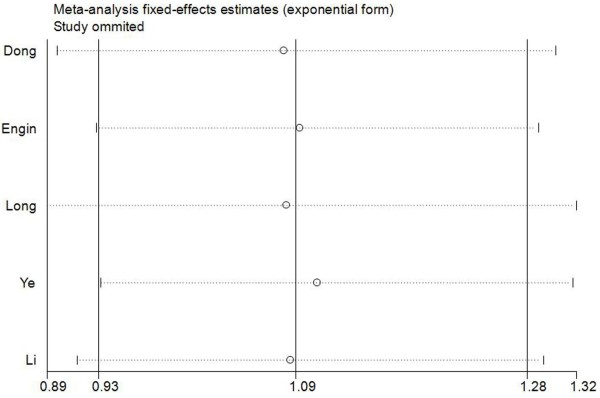
Sensitivity analysis for XPC Lys939Gln polymorphism and gastric cancer risk (dominant model Gln/Gln + Gln/Lys vs. Lys/lys). This figure shows the influence of individual studies on the summary OR. The middle vertical axis indicates the overall OR and the two vertical axes indicate its 95% CI. Every hollow round indicates the pooled OR when the left study is omitted in this meta-analysis. The two ends of every broken line represent the 95% CI.
Publication bias
Begger’s funnel plot and Egger’s test were performed to assess the publication bias of the included studies for XPC Lys939Gln and XPC Val499Arg polymorphisms. The shape of the funnel plot did not reveal any evidence of obvious asymmetry. Then, the Egger’s test was used to provide statistical evidence of funnel plot symmetry. The results still did not suggest any evidence of publication bias for XPC Lys939Gln (P = 0.152 for Gln/Gln vs. Lys/lys; P = 0.778 for Gln/Lys vs. Lys/lys; P = 0.339 for Gln/Gln + Gln/Lys vs. Lys/lys; P = 0.282 for Gln/Gln vs. Gln/Lys + Lys/lys) and XPC Val499Arg (P = 0.948 for Val/Val vs. Ala/Ala; P = 0.959 for Val/Ala vs. Ala/Ala; P = 0.937 for Val/Val + Val/Ala vs. Ala/Ala; P = 0.852 for Val/Val vs. Val/Ala + Ala/Ala). We did not carry out Begger’s funnel plot and Egger’s test for XPC PAT−/+polymorphism because only two studies were available for this polymorphism.
Discussion
Previous studies evaluating the association between XPC polymorphisms (Lys939Gln, Val499Arg, and PAT−/+) and gastric cancer risk have provided inconsistent results, and most of these studies involved no more than a few hundred gastric cancer cases, which is too few to assess any genetic effects reliably. Meta-analysis has been recognized as an important way to detect the effect of selected genetic polymorphisms on disease risk precisely and to identify potential important sources of between-study heterogeneity. Hence, we performed this meta-analysis including all published studies to investigate the association between the XPC polymorphisms and gastric cancer risk. To the best of our knowledge, this is the first comprehensive meta-analysis of genetics studies on the association between XPC polymorphisms and gastric cancer risk. Our results suggested that the XPC polymorphisms (Lys939Gln, Val499Arg, and PAT−/+) were not associated with gastric cancer risk when all studies were pooled together. In subgroup analyses stratified by ethnicity, source of control, and study quality, statistical significant association was also not observed in all subgroups.
DNA repair mechanisms are important pathways in the removal of DNA adducts from damaged genomic sites and play crucial roles in the carcinogenesis of cancers [27]. XPC is a key member of the NER pathway. It binds to HR23B and forms the stable XPC-HR23B complex and is involved in the recognition and initiation of the genome repair of the NER pathway [5-7]. Mutations of the XPC genes may increase gastric cancer susceptibility by causing a severe depression of NER and consequently altering DNA repair activity [28]. However, in our study, no significant association between variant genotypes of XPC polymorphisms and gastric cancer risk was observed in the pooled analysis and subgroup analyses, which was inconsistent with the hypothesis above. Several potential concerns should be discussed for the non-significant associations between XPC polymorphisms and gastric cancer susceptibility. First, gastric cancer is a multi-factorial disease resulting from complex interactions between environmental and genetic factors [29,30]. It is possible that the variants at this locus have some modest effects on gastric cancer. Environmental factors, such as living habits and exposure to carcinogens, however, may also play a role in gastric cancer development. Thus, no regard of these factors may confer the non-significance for the independent role of XPC polymorphisms in gastric cancer development. Second, there are eight core genes (i.e., ERCC1, XPA, XPB, XPC, XPD, XPE, XPF, and XPG) in the NER pathway of the DNA repairing system, variants in these genes may interfere with each other in associated functions. This could cover the true associations of XPC gene polymorphisms with gastric cancer. Therefore, other variants as gastric cancer risk factors should be induced as co-variants to determine their true effects. The lack of considering above confounding factors might affect the significance of their results. Moreover, the null result may be due to the limited number of studies included in the meta-analysis, which had insufficient statistical power to detect a slight effect or may have generated a fluctuated risk estimate. Therefore, the negative results of the association between XPC polymorphisms and gastric cancer risk should be interpreted with caution.
Some limitations of this meta-analysis should be acknowledged. First, only published studies were included in the meta-analysis. It is possible that some related unpublished studies that might meet the inclusion criteria were missed; therefore, publication bias may have been present, even though statistical analysis indicated this not to be the case. Second, our results were based on unadjusted estimates and a more precise analysis could be conducted if more individual data were available; this would allow for adjustment by other covariates such as the quantity of salty food consumption, drinking, smoking and Helicobacter pylori infection; Third, in the pooled analyses for XPC Val499Arg and XPC PAT−/+polymorphisms, the number of studies included was relatively small, not having enough statistical power to investigate a real association of the polymorphisms with gastric cancer susceptibility. However, our meta-analysis also had some advantages. First, a substantial number of cases and controls were pooled from different studies, which significantly increased the statistical power compared with the individual studies. Second, no heterogeneity and publication bias was detected in our meta-analysis, indicating that the pooled results were precise and reliable.
Conclusions
In conclusion, the result of this meta-analysis based on current evidences suggests that the XPC polymorphisms (Lys939Gln, Val499Arg, and PAT−/+) may not contribute to gastric cancer risk. However, it is necessary to conduct large sample studies using standardized unbiased genotyping methods, homogeneous gastric cancer patients, and well-matched controls. Moreover, gene–gene and gene–environment interactions should also be considered in the analysis. Such studies taking these factors into account may eventually lead to our better, comprehensive understanding of the association between the XPC polymorphisms and gastric cancer risk.
Abbreviations
HWE: Hardy–Weinberg equilibrium; XPC: Xeroderma Pigmentosum group C; NER: Nuclear excision repair; SNP: Single nucleotide polymorphism; OR: Odds ratio; CI: Confidence interval.
Competing interest
The authors declare that they have no competing interest.
Authors’ contributions
QP, XQ performed the literature search, data extraction, statistical analysis and drafted the manuscript. QP, YL, XL, CM, and RL participated in data extraction. SL, XQ, ZC supervised the literature search, data extraction, statistical analysis and drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
Qiliu Peng, Email: Pengql45@163.com.
Zhiping Chen, Email: cczzpp919@163.com.
Yu Lu, Email: 253869943@qq.com.
Xianjun Lao, Email: 153816478@qq.com.
Cuiju Mo, Email: 523731863@qq.com.
Ruolin Li, Email: 41918940@qq.com.
Xue Qin, Email: qinxue919@126.com.
Shan Li, Email: lis8858@126.com.
Funding
This research was supported by National Natural Science Foundation of China (No. 81260302).
References
- Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354–362. doi: 10.3748/wjg.v12.i3.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathbone M, Rathbone B. Helicobacter pylori and gastric cancer. Recent Results Cancer Res. 2011;185:83–97. doi: 10.1007/978-3-642-03503-6_5. [DOI] [PubMed] [Google Scholar]
- Moy KA, Fan Y, Wang R, Gao YT, Yu MC, Yuan JM. Alcohol and tobacco use in relation to gastric cancer: a prospective study of men in Shanghai, China. Cancer Epidemiol Biomarkers Prev. 2010;19:2287–2297. doi: 10.1158/1055-9965.EPI-10-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZF, Kurtz RC, Yu GP, Sun M, Gargon N, Karpeh M Jr, Fein JS, Harlap S. Adenocarcinomas of the esophagus and gastric cardia: the role of diet. Nutr Cancer. 1997;27:298–309. doi: 10.1080/01635589709514541. [DOI] [PubMed] [Google Scholar]
- Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. 1997;272:23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- Thoma BS, Vasquez KM. Critical DNA damage recognition functions of XPC-hHR23B and XPA-RPA in nucleotide excision repair. Mol Carcinog. 2003;38:1–13. doi: 10.1002/mc.10143. [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2:223–232. doi: 10.1016/S1097-2765(00)80132-X. [DOI] [PubMed] [Google Scholar]
- Janicijevic A, Sugasawa K, Shimizu Y, Hanaoka F, Wijgers N, Djurica M, Hoeijmakers JH, Wyman C. DNA bending by the human damage recognition complex XPC-HR23B. DNA Repair (Amst) 2003;2:325–336. doi: 10.1016/S1568-7864(02)00222-7. [DOI] [PubMed] [Google Scholar]
- Li L, Peterson C, Legerski R. Sequence of the mouse XPC cDNA and genomic structure of the human XPC gene. Nucleic Acids Res. 1996;24:1026–1028. doi: 10.1093/nar/24.6.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–1530. [PubMed] [Google Scholar]
- Perez-Mayoral J, Pacheco-Torres AL, Morales L, Acosta-Rodriguez H, Matta JL, Dutil J. Genetic polymorphisms in RAD23B and XPC modulate DNA repair capacity and breast cancer risk in Puerto Rican women. Mol Carcinog. 2013;52(Suppl 1):E127–138. doi: 10.1002/mc.22056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin AB, Karahalil B, Engin A, Karakaya AE. DNA repair enzyme polymorphisms and oxidative stress in a Turkish population with gastric carcinoma. Mol Biol Rep. 2011;38:5379–5386. doi: 10.1007/s11033-011-0690-9. [DOI] [PubMed] [Google Scholar]
- Palli D, Polidoro S, D'Errico M, Saieva C, Guarrera S, Calcagnile AS, Sera F, Allione A, Gemma S, Zanna I, Filomena A, Testai E, Caini S, Moretti R, Gomez-Miguel MJ, Nesi G, Luzzi I, Ottini L, Masala G, Matullo G, Dogliotti E. Polymorphic DNA repair and metabolic genes: a multigenic study on gastric cancer. Mutagenesis. 2010;25:569–575. doi: 10.1093/mutage/geq042. [DOI] [PubMed] [Google Scholar]
- Long XD, Ma Y, Huang YZ, Yi Y, Liang QX, Ma AM, Zeng LP, Fu GH. Genetic polymorphisms in DNA repair genes XPC, XPD, and XRCC4, and susceptibility to Helicobacter pylori infection-related gastric antrum adenocarcinoma in Guangxi population, China. Mol Carcinog. 2010;49:611–618. doi: 10.1002/mc.20630. [DOI] [PubMed] [Google Scholar]
- Dong Z, Guo W, Zhou R, Wan L, Li Y, Wang N, Kuang G, Wang S. Polymorphisms of the DNA repair gene XPA and XPC and its correlation with gastric cardiac adenocarcinoma in a high incidence population in North China. J Clin Gastroenterol. 2008;42:910–915. doi: 10.1097/MCG.0b013e3180f6262c. [DOI] [PubMed] [Google Scholar]
- Ye W, Kumar R, Bacova G, Lagergren J, Hemminki K, Nyren O. The XPD 751Gln allele is associated with an increased risk for esophageal adenocarcinoma: a population-based case–control study in Sweden. Carcinogenesis. 2006;27:1835–1841. doi: 10.1093/carcin/bgl017. [DOI] [PubMed] [Google Scholar]
- Li D, Zhang C, Li K, Chen Z, Xu C, Ning T. The association of gastric cancer with XPC gene haplotype. Chin J Exp Surg. 2010;27:747–750. [Google Scholar]
- Casas JP, Hingorani AD, Bautista LE, Sharma P. Meta-analysis of genetic studies in ischemic stroke: thirty-two genes involving approximately 18,000 cases and 58,000 controls. Arch Neurol. 2004;61:1652–1661. doi: 10.1001/archneur.61.11.1652. [DOI] [PubMed] [Google Scholar]
- Peng Q, Lao X, Chen Z, Lai H, Deng Y, Wang J, Mo C, Sui J, Wu J, Zhai L, Yang S, Qin X, Li S. TP53 and MDM2 Gene Polymorphisms, Gene-Gene interaction, and Hepatocellular Carcinoma Risk: evidence from an updated meta-analysis. PLoS One. 2013;8:e82773. doi: 10.1371/journal.pone.0082773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56:455–463. doi: 10.1111/j.0006-341X.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- Zhou RM, Li Y, Wang N, Zhang XJ, Dong XJ, Guo W. Correlation of XPC Ala499Val and Lys939Gln polymorphisms to risks of esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma. Ai Zheng. 2006;25:1113–1119. [PubMed] [Google Scholar]
- Agalliu I, Kwon EM, Salinas CA, Koopmeiners JS, Ostrander EA, Stanford JL. Genetic variation in DNA repair genes and prostate cancer risk: results from a population-based study. Cancer Causes Control. 2010;21:289–300. doi: 10.1007/s10552-009-9461-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J, Hoeijmakers JH. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21:453–460. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- Liu X, Xiong H, Li J, He Y, Yuan X. Correlation of hK6 expression with tumor recurrence and prognosis in advanced gastric cancer. Diagn Pathol. 2013;8:62. doi: 10.1186/1746-1596-8-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Zhang J, Song S, Dai D. Promoter methylation and expression of TIMP3 gene in gastric cancer. Diagn Pathol. 2013;8:110. doi: 10.1186/1746-1596-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]


