Abstract
Metalloglycomics - the effects of defined coordination compounds on oligosaccharides and their structure and function – opens new areas for bioinorganic chemistry and expands its systematic study to the third major class of biomolecules after DNA/RNA and proteins.
In this communication we demonstrate proof-of-concept that the strong binding of polynuclear platinum complexes (PPCs) to oligosaccharides provides a new approach to glycan-based targeting by protection against enzymatic cleavage. Proteoglycans are composed of structurally diverse glycosaminoglycans (GAGs) such as heparin and heparan sulphate covalently attached to proteins. They are present on both the cell surface as well as in the extracellular matrix and the basement membrane, bind to a wide variety of proteins and exercise important normal physiological functions such as cell-cell and cell-extracellular matrix adhesion and are receptors for adhesion molecules and growth factors.1 Heparan Sulphate proteoglycans (HSPGs) are degraded by mammalian and bacterial enzymes. In the case of the mammalian endoglycosidase heparanase, degradation releases angiogenic and growth factors leading to tumor cell migration, growth and angiogenesis. The bacterial lyase heparinase is important as a carbon source and degradation of heparin and heparan sulfate leads to biologically active oligosaccharides with significant clinical and pharmaceutical implications. Proteoglycans and their associated enzymes are significant emerging drug targets of high biological relevance.2–4 Design of mimetics for competitive enzyme inhibition involves the complex synthesis of small (tetra/penta) oligosaccharides. Relevant examples are the paradigmatic pentasaccharide Fondaparinux, the fully synthetic methyl glycoside of the antithrombin III (ATIII)-activating pentasaccharide sequence of heparin,5 and PI-88, a yeast-derived mixture of highly sulfated, monophosphorylated mannose oligosaccharides.6
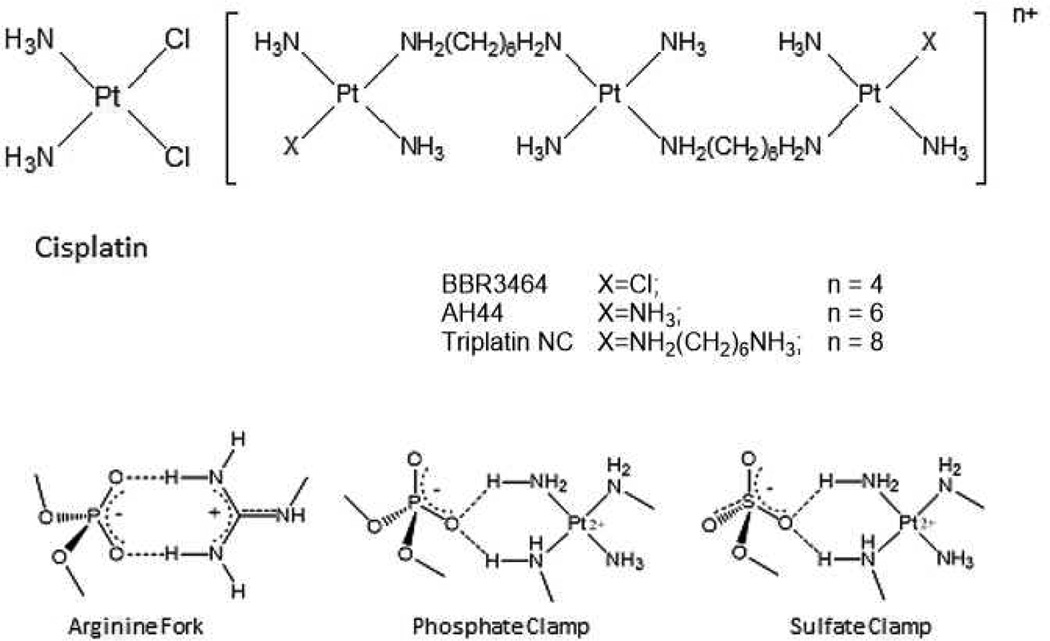
HSPGs are the receptors for cellular internalization of polycationic, arginine-rich peptides (protein transduction domains, PTDs) through molecular recognition of the sulphate backbone of the oligosaccharide.7,8,9 Nona-L-arginine (R9) is the most efficacious known PTD.7 PPC-HSPG interactions also mediate the cellular internalization of the polynuclear platinum drugs, a unique mechanism not shared with cisplatin or oxaliplatin.10,11 PPCS are competitive inhibitors of HSPG-polyarginine binding, confirmed using the fluorescent nonaarginine derivative TAMRA-R9.10 Given the measured affinity of TAMRA-R9 to heparin is Kd = 109 nM9, similar to typical receptor-ligand interactions, PPCs must have similar high affinity.10 The interactions between the amine groups of the triplatinum compounds and the phosphate groups of the DNA backbone are very similar to those of the guanidine groups on arginine (Figure 1). Conceptualizing polynuclear platinum complexes as “polyarginine mimics’ has been very useful in drawing analogies between the DNA recognition modes of the arginine fork and the phosphate clamp, a third mode of ligand-DNA binding discrete from the classical intercalator and minor groove binders.12,13,14 These considerations further suggested extension of the analogy to isostructural sulphate and membrane biomolecule interactions.
Figure 1.
Structures of glycan-interacting polynuclear platinum complexes, and structural analogies between phosphate and sulphate clamps mediated by the complexes and/or arginine.
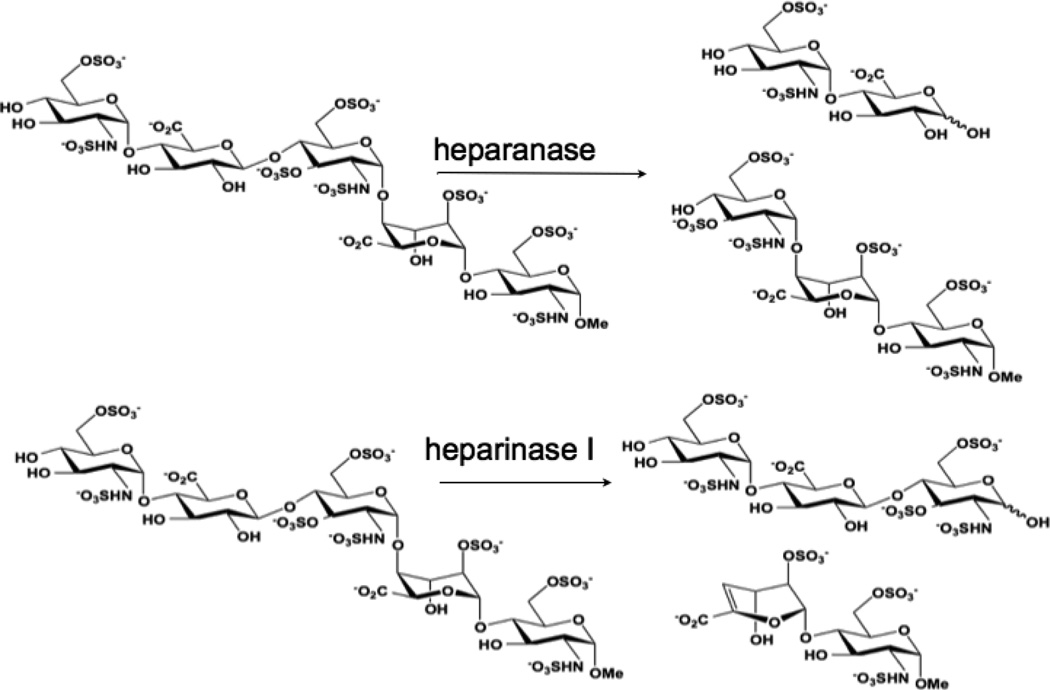
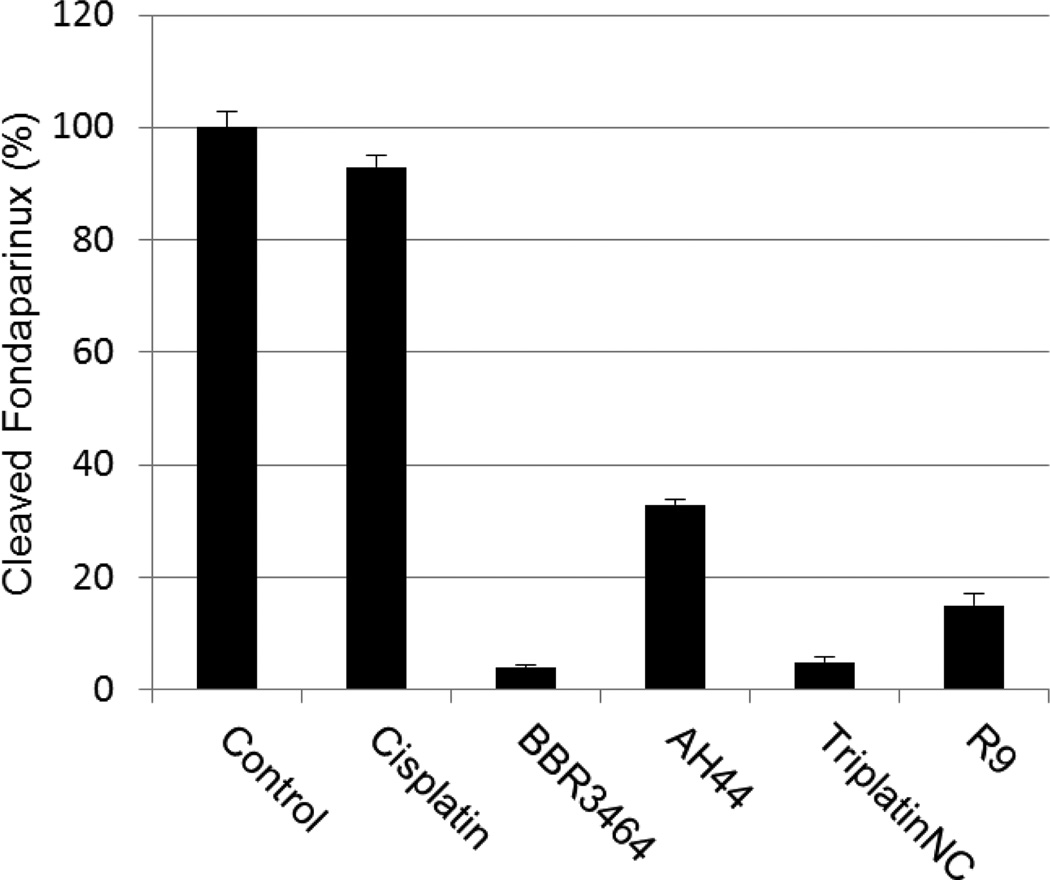
We therefore asked the question - “What are the functional consequences of strong Pt-GAG binding?” The cleavage patterns for mammalian heparanase and bacterial heparinase I (often used as a model for the mammalian enzyme) are shown in Figure 2. Colorimetric assays for enzymatic activity and suitable for kinetic analysis and inhibitor screening have been developed.15 We therefore adapted the assay to examine the inhibitory effect of platinum complexes on the enzymatic (heparinase) degradation of Fondaparinux. The pentasaccharide substrate was incubated with platinum complex prior to enzyme exposure and cleavage measured versus control in absence of added complex. Inhibition of heparinase cleavage is effective in a charge and concentration-dependent manner for the non-covalent compounds (Figure 3). The 8+ compound TriplatinNC is more effective than the 6+ compound AH44. These results are consistent with the greater efficacy of TriplatinNC compared to AH44 to compete with TAMRA-R9 for HSPG binding.10
Figure 2.
Cleavage patterns of Fondaparinux by mammalian (heparanase) and bacterial (heparinase) enzymes.
Figure 3.
Inhibition of heparinase I Fondiparinux cleavage (3h incubation) by polynuclear platinum complexes and the arginine-rich R9 protein (1:3 stoichiometry).
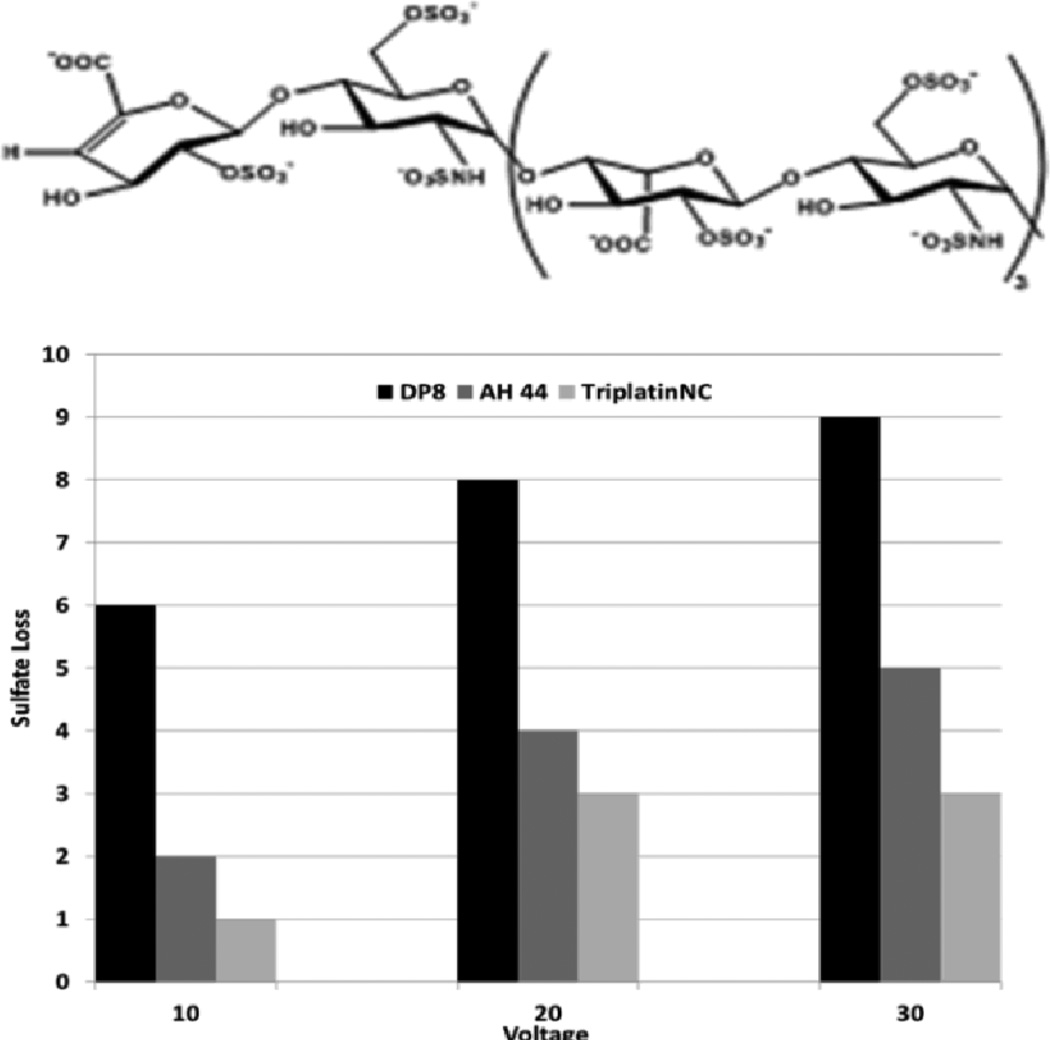
Time course studies show that whereas the non-covalent compounds instantly inhibited activity with little or no variation with time, BBR3464 (4+) inhibition reached a maximum after 3 hours co-incubation with Fondaparinux. BBR3464 also shows increased efficacy of inhibition compared to the 6+ non-covalent AH44. Both the slower response and greater inhibition may be attributed to a contribution from covalent binding by Pt-Cl substitution (only possible for BBR3464). In agreement, we note that the aquation kinetics in 15 mM sulphate of the prototypical dinuclear compound [{trans-PtCl(NH3)2}2(μ-NH2(CH2)6NH2)]2+ (structurally analogous to BBR3464) confirmed covalent binding to SO42−.16 The major difference between binding of phosphate and sulfate is the very high k−L for loss of sulphate, suggesting that when formed the sulphato species will be substitution-labile. Incubation of the platinum compounds with the enzyme prior to addition of Fondaparinux does not affect the assay results, indicating that there is no direct action of the compounds on the enzyme. Cisplatin does not inhibit cleavage, consistent with the fact that it is not a substrate for HSPG-mediated internalization.10 Given the binding of arginine-rich sequences to heparan sulphate,7,8,9 we examined the efficiency of the R9-protein itself in this assay. The polyarginine also inhibits heparinase activity, but not to the same extent as BBR3464 or TriplatinNC. Confirming the potential for high-affinity binding to oligosaccharides, ESI-MS spectra of a model DP8 octasaccharide in the presence of the highly charged 6+ (AH44) and 8+ (TriplatinNC) ions showed clear evidence of 1:1 adducts and stabilization toward sulphate loss (Figure 4). The interaction is by its nature non-covalent and is the first demonstration of a platinum compound interaction with a sulphated polysaccharide. The sulfonated moieties on octasaccharides are quite labile and the spectrum of free octasaccharide shows a series of peaks corresponding to sequential sulphonate loss.17,18 In contrast, under the same conditions, the initial MS of the adducts show little loss compared to free polymer. ESI-MS/MS of the 1:1 adducts at increasing energies also shows a stabilization toward sulphonate loss, (Figures S1–S6). The stabilization is dependent on size and charge of the non-covalent platinum compound with the 8+ compound significantly more effective with a difference of up to 7 sulphate groups protected versus free oligosaccharide.
Figure 4.
Sulphate loss in the octasaccharide DP8 by binding to polynuclear platinum complexes at varying ESI-MS/MS voltages.
There are several significant aspects to our findings. Firstly, the ability to inhibit oligosaccharide degradation with these PPC “metalloshields” presents an exciting alternative approach to enzyme inhibition, distinct from the complex design and synthetic chemistry of oligosaccharide mimics. Specifically, the proof of concept demonstrated here may be extended to inhibition of the heparanase/growth factor interaction complementary to the design of the short-chain oligosaccharide competitive inhibitors. Heparanase is over-expressed in tumors and there is significant correlation between metastatic potential and heparanase activity.3,19 The definitive ‘end-point” of inhibition of heparanase and growth factor binding to heparan sulphate is the inhibition of angiogenesis. It is relevant in this context that the small cationic highly arginine-rich protamine is a specific inhibitor of angiogenesis and in vitro inhibits the angiogenic activity of FGF-2.20 Note the further extension of the PPC-polyarginine analogy in this case. Secondly, the mass spectral results are entirely consistent with strong-PPC-oligosaccharide binding with increased stability of the sulfate group to dissociation which (a) verifies the complexation with sulphate moieties in preference to elsewhere on the glycosidic backbone and (b) may have biological consequences in its own right by reducing the “effective” sulphate concentration on the membrane surface.2 Thirdly, the oligosaccharide binding is of fundamental interest in describing the molecular nature of the PPC-HSPG interaction and thus the cellular internalization of PPCs. The model further differentiates the biological profile of polynuclear and mononuclear platinums.10,21,22 Glycans are aberrantly expressed in many tumors, regulating many important events in tumor progression including proliferation, invasion, angiogenesis and metastasis.2,23 The model thus suggests the possibility of a systematic approach to tumor selectivity of platinum complexes through glycan targeting of those tumors with known glycan overexpression.23 Our recent demonstration of the utility of the multi-element NanoSIMS technology for cellular imaging opens the possibility for the influence of HSPGs in sub-cellular distribution and trafficking to be examined.24 Finally, BBR3464 remains the only “non-classical” platinum drug to enter human clinical trials and is the prototypical PPC.18,19 TriplatinNC has demonstrated interesting biological activity in its own right. The complex is cytotoxic at micromolar concentrations, similar to cisplatin, but is unaffected by serum degradation25 and displays antitumor activity in a mouse A2008 ovarian cancer model. These are remarkable results for a non-covalent compound and, with the results presented here, is another paradigm shift for the design of biologically active platinum agents.
Overall these results highlight that the study of defined coordination compounds with oligosaccharides – metalloglycomics - has rich and multiple applications in a new area of endeavor in the field of bioinorganic chemistry distinct from protein and DNA/RNA interactions. The inherent ability to alter oxidation state, coordination number and geometry, and substitution lability of ligands allows study of a wide variety of structural types to enhance shielding and enzyme inhibition. Divalent metal ions such as Ca2+ and Zn2+ are implicated in many protein-carbohydrate interactions although the biological activities remain mostly unknown.17
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health RO1CA78754.
Footnotes
Electronic Supplementary Information (ESI) available: Details of enzyme inhibition assay and mass spectral details including fragmentation patterns of platinum complex-adducted oligosaccharide.
Notes and references
- 1.Gandhi AS, Mancera RL. Chem. Biol. Drug Des. 2008;72:455–482. doi: 10.1111/j.1747-0285.2008.00741.x. [DOI] [PubMed] [Google Scholar]
- 2.Fuster MM, Esko JD. Nature Reviews Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 3.Vlodavsky I, Goldschmidt O, Zcharia E, Atzman R, Rangini-Guatta Z, Elkin M, Peretz T, Friedmann Y. Cancer Biology. 2002;12:121–129. doi: 10.1006/scbi.2001.0420. [DOI] [PubMed] [Google Scholar]
- 4.Yip GW, Smollisch M, Götte M. Mol. Cancer Ther. 2006;5:2139–2148. doi: 10.1158/1535-7163.MCT-06-0082. [DOI] [PubMed] [Google Scholar]
- 5.Petitou M, van Boeckel CA. Angew. Chem. Intl. Ed. 2004;43:3118–3133. doi: 10.1002/anie.200300640. [DOI] [PubMed] [Google Scholar]
- 6.Parish CR, Freeman C, Brown KJ, Francis DJ, Cowden WB. Cancer Res. 1999;59:3433–3441. [PubMed] [Google Scholar]
- 7.Fuchs SM, Raines RT. Cell Mol Life Sci. 2006;63:1819–1822. doi: 10.1007/s00018-006-6170-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belting M. Trends Biochem Sci. 2003;28:145–151. doi: 10.1016/S0968-0004(03)00031-8. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs SM, Raines RT. Biochemistry. 2004;43:2438–2444. doi: 10.1021/bi035933x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva H, Frézard F, Peterson EJ, Kabolizadeh P, Ryan JJ, Farrell NP. Mol Pharm. 2012;9:1795–1802. doi: 10.1021/mp300098t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall MD, Okabe M, Shen DW, Liang XJ, Gottesman MM. Annu. Rev. Pharmacol. Toxicol. 2008;48:495–453. doi: 10.1146/annurev.pharmtox.48.080907.180426. [DOI] [PubMed] [Google Scholar]
- 12.Calnan BJ, Tidor B, Biancalana S, Hudson D, Frankel AD. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- 13.Komeda S, Moulaei T, Woods KK, Chikuma M, Farrell NP, Williams LD. J Am Chem Soc. 2006;128:16092–16103. doi: 10.1021/ja062851y. [DOI] [PubMed] [Google Scholar]
- 14.Komeda S, Moulaei T, Chikuma M, Odani A, Kipping R, Farrell NP, Williams LD. Nucleic Acids Res. 2011;39:325–336. doi: 10.1093/nar/gkq723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammond E, Li CP, Ferro V. Anal Biochem. 2010;396:112–116. doi: 10.1016/j.ab.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Ruhayel RA, Corry BA, Braun C, Thomas DS, Berners-Price SJ, Farrell NP. Inorg. Chem. 2010;49:10815–10819. doi: 10.1021/ic100576k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo Y, Schenauer MR, Leary JA. Inter. J. Mass Spectrom. 2011;303:191–198. doi: 10.1016/j.ijms.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chai WG, Luo JL, Lim CK, Lawson AM. Anal. Chem. 1998;70:2060–2066. doi: 10.1021/ac9712761. [DOI] [PubMed] [Google Scholar]
- 19.Levy-Adam F, Ilan N, Vlodavsky I. Semin. Cancer Biol. 2010;20:153–160. doi: 10.1016/j.semcancer.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor S, Folkman J. Nature. 1982;297:307–312. doi: 10.1038/297307a0. [DOI] [PubMed] [Google Scholar]
- 21.Mangrum JB, Farrell NP. J. Chem. Soc. Chem. Comm. 2010;46:6640–6650. doi: 10.1039/c0cc01254h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farrell NP. Drugs of the Future. 2012;37:795–806. [Google Scholar]
- 23.Coombe DR, Kett WC. Handbook Exp. Pharmacol. 2012;207:361–383. doi: 10.1007/978-3-642-23056-1_16. [DOI] [PubMed] [Google Scholar]
- 24.Wedlock LE, Kilburn MR, Liu R, Shaw JA, Berners-Price SJ, Farrell NP. J. Chem. Soc. Chem. Comm. 2013;49:6944–6946. doi: 10.1039/c3cc42098a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benedetti BT, Peterson EJ, Kabolizadeh P, Martinez AR, Kipping R, Farrell NP. Mol. Pharm. 2011;8:940–948. doi: 10.1021/mp2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.