Abstract
We previously demonstrated that APP epigenetically regulates Egr1 expression both in cultured neurons and in vivo. Since Egr1 is an immediate early gene involved in memory formation, we wondered whether other early genes involved in memory were regulated by APP and we studied molecular mechanisms involved. By comparing prefrontal (PF) cortex from wild type (APP+/+) and APP knockout mice (APP−/−), we observed that APP down regulates expression of four immediate early genes, Egr1, c-Fos, Bdnf and Arc. Down regulation of Egr1, c-Fos and Bdnf transcription resulted from a decreased enrichment of acetylated histone H4 on the corresponding gene promoter. Further characterization of H4 acetylation at Egr1 and c-Fos promoters revealed increased acetylation of H4K5 and H4K12 residues in APP−/− mice. Whereas APP affected Egr1 promoter activity by reducing access of the CREB transcription factor, its effect on c-Fos appeared to depend on increased recruitment of HDAC2 histone deacetylase to the gene promoter. The physiological relevance of the epigenetic regulation of Egr1 and c-Fos gene transcription by APP was further analyzed following exposure of mice to novelty. Although transcription of Egr1 and c-Fos was increased following exposure of APP+/+ mice to novelty, such an induction was not possible in APP−/− mice with a high basal level of expression of these immediate early genes. Altogether, these results demonstrate that APP-mediated regulation of c-Fos and Egr1 by different epigenetic mechanisms is needed for their induction during exposure to novelty.
Introduction
One of the first steps involved in memory formation is the rapid induction of immediate early genes (IEGs) transcription in the brain. Egr1 is an IEGs member of early growth response family of zinc fingers transcription factors, widely studied for its role in reconsolidation of memory and its ability to establish long term spatial localization memories [1], [2]. In mouse, Egr1 is needed for late-phase LTP, is involved in long-term memory formation and controls neuronal function in the hippocampus [3], [4].
Expression of c-Fos protein in neurons is induced by a wide range of sensory stimuli [5], [6] and several studies have demonstrated the role of c-Fos in the establishment of neuronal plasticity by regulating downstream gene expression [7], [8]. c-Fos deficient animals show spatial and associative learning deficits correlated with decrease in synaptic plasticity [9]. Although expressed at low levels in the brain, transcription of c-Fos gene sharply increases after exposure to novelty. Both Egr1 and c-Fos mRNA levels are significantly increased 30–45 minutes after exposure to novelty, in particular in the CA1 and CA3 regions of the hippocampus as well as in the PF cortex [10].
Contrary to Egr1 and c-Fos, Arc protein is not a transcription factor but an effector synaptic protein involved in multiple neuronal pathways [11]. Arc induction occurs in the hippocampus and the cortex following exploration of a novel environment [12]. LTP and synaptic activation also induce Arc expression both at the mRNA and protein levels [13].
Bdnf is a member of neurotrophin family involved in neuronal growth and survival [14], in the development of dendritic network modulating synaptic transmission [15] and in the regulation of synaptic plasticity and memory formation [16]. In rodents, at least 22 different Bdnf mRNA are produced by alternative splicing of a primary transcript, and 9 alternative promoters control Bdnf gene transcription, but exon IV promoter is preferentially induced by neuronal activity [17], [18]. Chromatin remodeling also controls Bdnf gene transcription in neurons [19].
We previously demonstrated that the amyloid precursor protein (APP) of Alzheimer disease regulates, at the epigenetic level, the transcription of the Egr1 gene [20]. Induction of IEGs expression is closely related to a final change in chromatin remodeling that allows gene expression [21]. Recruitment of CREB on the Egr1 and c-Fos gene promoters induces an increase in histones acetylation mediated by the CBP/P300 acetyltransferase, and the concomitant depletion of HDACs [19].
Here we show for the first time that APP fosters a low level of Egr1 and c-Fos expression in mouse PF cortex, by inhibiting CREB recruitment and improving HDAC2 recruitment on the corresponding gene promoters. A low constitutive level of Egr1 and c-Fos expression mediated by APP is needed for their induction during exposure of mice to novelty.
Materials and Methods
Animals
Five months old mice C57Bl/6J APP+/+ and APP−/− were used in this study. Mice were obtained from The Jackson laboratory and backcrossed for more than five generations in the CD1 genetic background. All animals had access to food ad libidum and were housed under controlled temperature (22°C) and with a 12 h light cycle (starting at 7 am). One week before experiments, mice were moved to the experimental platform. As previously described, the behavioral test of exposure to novelty was performed in a square openfield (60×60×40 cm) with gray plastic walls. Mice were able to explore this environment during 15 minutes and sacrificed by CO2 inhalation after a resting period of 30 minutes. Another group of mice were directly sacrificed without exposure to the open field. After brain dissection, PF cortex and hippocampus were directly placed in liquid nitrogen, frozen at −80°C or directly crushed and used in further experiments. All manipulations on mice have been approved by the local ethics committee of the catholic University of Louvain and follow the European legislation.
RNA Extraction and Quantitative Real Time PCR
Total RNA was purified using Trizol method (Tripure, Roche). Reverse transcription (RT) and quantitative (q) real time PCR (q-RT PCR) were performed with the iScript cDNA synthesis Kit and the iQ SYBR Green supermix using a iCycler MyIQ2 multicolor Real-Time PCR detection system (Biorad). The relative amplification of cDNA fragments was calculated by the 2-ΔΔCt method. q-RT PCR primer sequences used were as follows: Egr1 Forward: TCCTCTCCATCACATGCCTG, Egr1 Reverse: CACTCTGACACATGCTCCAG, c-Fos Forward: GATGTTCTCGGGTTTCAACG, c-Fos Reverse: GGAGAAGGAGTCGGCTGG. GAPDH Forward: ACCCAGAAGACTGTGGATGG, GAPDH Reverse: ACACATTGGGGGTAGGAACA, Arc Forward: GCTGAGCTCTGCTCTTCTTCA, Arc Reverse: GGTGAGCTGAAGCCACAAAT Bdnf Forward: GCGGACCCATGGGACTCT, Bdnf Reverse: CTGCTGCTGTAGTGACCGA.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed using the EZ-ChIP Assay kit (Millipore) according to manufacturer instructions and as described previously [20]. Chromatin was isolated and pooled from the two PF cortices of a single mouse. ChIP experiments were performed using a minimum of 3 mice per group. Chromatin was sheared in an ice bath by a 25 cycles of 30 sec on/off sonication using the “Bioruptor UCD-20” sonicator (Diagenode). Samples were kept on ice during 30 s between two pulses. An aliquot of precleared chromatin was collected as the input. The samples were incubated overnight at 4°C with the antibodies of interest: 5 µg anti-H4Ac (Millipore), 5 µg anti-H3Ac (Millipore), 5 µl anti-H2BAc (Abcam); 10 µl anti-Tip60 (SantaCruz), 5 µl anti-HDAC2 (Abcam), 10 µl anti-CREB total (Millipore), 3 µl of acetylated H4K5, K12, K16 (Active Motif). The immunoprecipitated chromatin was analyzed by quantitative PCR with primers designed to amplify short regions of the promoters of genes of interest.
qPCR primers were as follows: Egr1 Forward: GTGCCCACCACTCTTGGAT, Egr1 Reverse: CGAATCGGCCTCTATTTCAA, c-Fos Forward: GAAAGCCTGGGGCGTAGAGT, c-Fos Reverse: CCTCAGCTGGCGCCTTTAT, Arc Forward: CAGCATAAATAGCCGCTGGT, Arc Reverse: AGTGTGGCAGGCTCGTC, Bdnf exIV Forward: GCGCGGAATTCTGATTCTGGTAAT, Bdnf exIV Reverse: GAGAGGGCTCCACGCTGCCTTGACG. GAPDH Forward: AGAGAGGGAGGAGGGGAAATG, GAPDH Reverse: AACAGGGAGGAGCAGAGAGCAC. The quantification method used is based on the ratio between immunoprecipitated chromatin and input chromatin.
Protein Analysis
Nuclear extracts of mouse PF cortex were prepared in 0.25 M sucrose buffer (sucrose 0.25 M, Tris 50 mM, EDTA 1 mM, imidazole 3 mM, pH 7.0 + proteases inhibitor cocktail). Samples were centrifuged 10 min at 250 g and nuclear fraction was resuspended in Laemmli buffer. All samples were sonicated before protein assay (BCA Pierce, Thermoscientific) and Western blotting was performed on 20 µg of protein lysates. Membranes were incubated overnight at 4°C with the primary antibodies; anti-HDAC2 1∶1500 (Abcam), anti-H3 1∶10000 (Millipore), anti-tubulin 1∶4000 (Sigma), anti-CREB and phospho-CREB 1∶1000 (Millipore). Washes with PBS-Tween (0.005%) were followed by incubation with secondary antibody (1∶10 000 anti-mouse or anti-rabbit IgG) (GE Healthcare) coupled to horseradish peroxidase and revealed by ECL. For quantification, the membranes were stripped and reincubated with an anti-tubulin or an anti-H3 antibody. Immunoreactive bands were quantified with an electrophoresis Gel Doc 2000 imaging system coupled to a Quantity one software (Bio-Rad).
Statistical Analysis
All results were expressed as mean ± standard deviation (SD) values. Statistical significance was determined by student’s t-test for two-group comparisons or one-way analysis of variance (ANOVA) followed by Bonferroni’s multiple comparisons test for multi-group comparison.
Results
1. APP Decreases the Transcription of 4 Different IEGs in the Mouse PF Cortex
RNA was prepared from the hippocampus and PF cortex of APP+/+ and APP−/− mice, and qRT-PCR were performed to quantify IEGs mRNAs. Results presented in figure 1A indicate a 2.5 increase in Egr1 mRNA levels in PF cortex of APP−/− mice, confirming our previous results [20]. We wondered whether transcription of other IEGs involved in memory formation could also be regulated by APP. Results presented figure 1B indicate a significant increase in c-Fos, Arc and Bdnf mRNA levels in APP−/− mice. Levels of IEGs mRNA were normalized to GAPDH mRNA levels, which were not regulated by APP, as 3 other housekeeping genes (Actin, peptidylprolyl isomerase A, and β-glucuronidase) (Figure S1). In addition, expression of 2 other IEGs (c-Jun and Homer-1a) was not affected by APP (Figure S1).
Figure 1. APP regulates IEGs expression in mouse PF cortex.
A) Egr1 mRNA levels were quantified by q-RT PCR in APP+/+ and APP−/− PF cortex (n = 9), and in the hippocampus (n = 6). B) q-RT PCR method was used to quantify mRNA levels of c-Fos, Arc and Bdnf in APP+/+ and APP−/− PF cortex (n = 6). Values were normalized to the GAPDH mRNA, and expressed as percentage of APP+/+, mean ± SD. Student’s t-test: ***p<0.001,**p<0.01, *p<0.05.
These results therefore suggest that APP is able to regulate transcription of several IEGs involved in memory formation.
2. APP Affects Enrichment of Acetylated Histone H4 on Genes Promoters
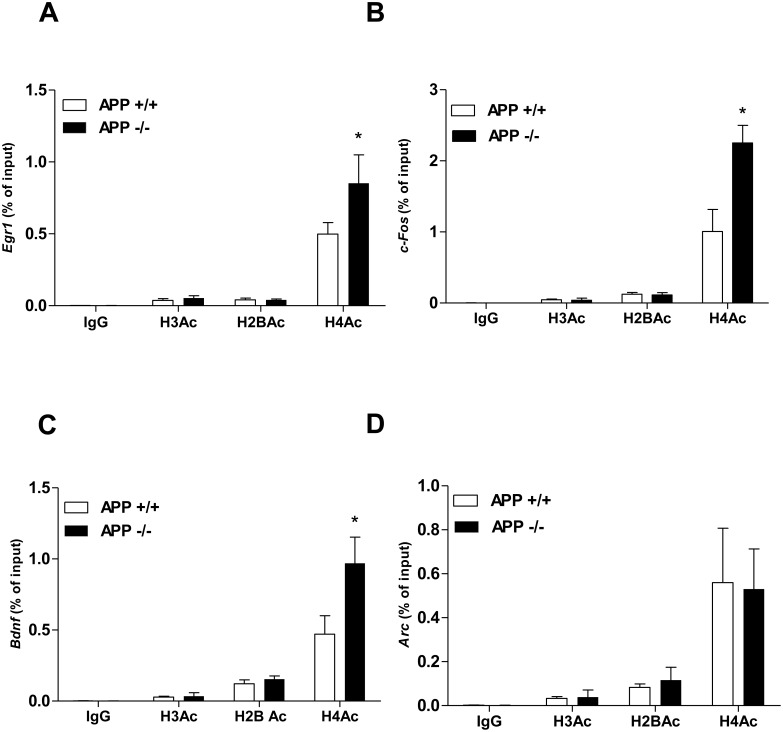
We previously observed that the control of Egr1 gene transcription by APP is independent of the APP intracellular domain (AICD) but occurs at the epigenetic level [20]. Therefore, we wondered whether APP could control transcription of other IEGs by the same mechanism. To that aim, ChIP experiments were performed using anti-acetylated histones antibodies. Results presented in figure 2A clearly demonstrated a specific enrichment of acetylated H4, but not H3 and H2B, on the Egr1 promoter in APP−/− mice, confirming our previous results [20]. Interestingly, the same specific enrichment of acetylated H4 was measured on both the c-Fos (Figure 2B) and Bdnf (Figure 2C) gene promoters in APP−/− mice. No modification in H3Ac, H2BAc and H4Ac enrichment on the GAPDH gene promoter was observed in APP+/+ and APP−/− experimental conditions (Figure S2). Enrichment of acetylated H4 was not observed on the Arc gene promoter in APP−/− mice (Figure 2D), indicating that a different molecular mechanism is involved in the APP-mediated regulation of Arc gene expression. This conclusion, together with a much weaker up regulation of Bdnf expression in APP−/− mice (Figure 1) led us to focus on the molecular mechanisms involved in the regulation of Egr1 and c-Fos expression by APP.
Figure 2. Analysis of histone acetylation by ChIP assays on IEGs promoters.
ChIP experiments were performed with chromatin obtained from APP+/+ and APP−/− PF cortex. Immunoprecipitation was completed with antibody recognizing normal mouse IgG as negative control, or anti- H3Ac, H2BAc and H4Ac antibodies. The quantification of immunoprecipitated chromatin and the normalization versus total chromatin (input) was assessed by real-time qPCR with primers designed on A) Egr1, B) c-Fos, C) Arc and D) Bdnf exon IV promoters; *p<0.05. All results were obtained from at least 3 mice per group and per antibody, and are expressed as mean ± SD.
3. APP Specifically Regulates Acetylation of Histone H4 at Lysines 5 and 12
Acetylation of histone H4 can occur at different positions including lysines 5, 12 and 16. To identify the positions that are acetylated on H4 enriched on the Egr1 and c-Fos gene promoters in APP−/− mice, ChIP experiments were performed using anti-H4K5Ac, -H4K12Ac and -H4K16Ac specific antibodies. Results presented in figure 3A indicate that histones H4 enriched on the Egr1 gene promoter in APP−/− mice were acetylated on positions K5 and K12, but not on K16. Interestingly, the same profile of acetylation was found for histone H4 enriched on the c-Fos gene promoter in APP−/− mice (Figure 3B). These results suggest that APP regulates Egr1 and c-Fos gene transcription by similar molecular mechanisms.
Figure 3. H4K5 and H4K12 are enriched at Egr1 and c-Fos gene promoter in APP−/− mice.
ChIP method was used to evaluate the enrichment of H4K12Ac, H4K5Ac, and H4K16Ac. A) Egr1 promoter. B) c-Fos promoter. Data represent the level of enrichment normalized as percentage of APP+/+. ***p<0.001. All results derive from at least 3 mice per group and per antibody, and are expressed as mean ± SD.
4. Regulation of Egr1 Gene Transcription by APP is CREB Dependent
A schematic representation of the Egr1 gene promoter is given in figure 4A [22]. The histone acetyltransferase Tip60 is able to acetylate histone H4 at positions H4K5 and H4K12 [23] and interacts with AICD [24]. Therefore, an interaction between Tip60 and APP could inhibit H4K5 and H4K12 acetylation in APP+/+ mice. To test this hypothesis, ChIP experiments were performed using anti-Tip60 antibodies. Tip60 is able to bind to the promoter of KAI1 gene and regulates its transcription [25], [26]. In Chip experiments, we indeed demonstrated an enrichment of Tip60 on the promoter of the KAI1 gene (Figure S3). However, results presented in figure 4B do not show any enrichment of Tip60 on the Egr1 gene promoter in APP−/− mice, allowing us to rule out the implication of Tip60 in this regulation. On the other hand, acetylation of H4K5 and H4K12 is mediated by CBP/P300, which associates with phosphorylated CREB DNA binding protein [27]. In addition, the Egr1 promoter contains two CREB responsive elements (CRE) (Figure 4A). Therefore we attempted to immunoprecipitate CREB on Egr1 promoter and we confirmed that this transcription factor was significantly more enriched on the Egr1 gene promoter in APP−/− mice (figure 4C). To test whether APP was able to down regulate CREB expression or to inhibit its phosphorylation, nuclear extracts from APP+/+ and APP−/− mice were analyzed in Western blotting using anti-CREB and anti-S133PhosphoCREB specific antibodies. Results presented in figure 4D indicate the same CREB/PhosphoCREB ratio in APP+/+ and APP−/− mice, ruling out modification of CREB expression and phosphorylation by APP. The recruitment of CREB on a gene promoter is often followed by local depletion of several HDACs [28], [29]. As HDAC2 epigenetically regulates transcription of several IEGs including Egr1 [30], we also performed ChIP experiments using HDAC2 antibodies. Results presented in figure 4E do not show any significant enrichment of HDAC2 on the Egr1 gene promoter in APP−/− mice. Altogether, these results indicate that APP fosters low level of Egr1 expression in mouse brain by inhibiting recruitment of CREB without affecting enrichment of HDAC2 on the Egr1 gene promoter.
Figure 4. CREB is better recruited on the Egr1 gene promoter in APP−/− mice.
A) Schematic representation of the structure of Egr1 gene promoter containing several binding sites for transcription factors and localization of primers utilized for q-PCR (CREB: cAMP response element, CRE: cAMP response elements SRF: Serum Response factor, SRE: Serum Response Element, SP1: specificity protein). B) Tip60 binding to Egr1 gene promoter in APP+/+ and APP−/− PF cortex was assessed by ChIP using anti-Tip60 antibody. C) CREB binding to Egr1 promoter in APP+/ and APP−/− mice. IgG was used as negative control. Equal amounts of ChIP and input DNA were used for qRT-PCR analysis on the Egr1 gene promoter. Results show a significantly lower enrichment of CREB in APP+/+ mice, *p<0.05. Enrichment values were normalized to input values and represented the average of three or more experiments per group. Results are expressed as mean ± SD. D) Ratio between total and phosphorylated CREB was detected by western blot analysis of nuclear extracts from PF cortex of APP+/+ and APP−/− mice, 5 months of age (n = 5). Typical blot is shown, CREB/P-CREB ratio were quantified, and expressed as percentage of the APP+/+ mice. Data are expressed as mean ± SD. E) ChIP assay on Egr1 promoter using HDAC2 antibody.
5. Regulation of c-Fos Gene Transcription by APP is HDAC2 Dependent
The c-Fos gene promoter also contains a single CRE site (Figure 5A). The same profile of acetylation of histone H4 enriched on c-Fos and Egr1 gene promoters in APP−/− mice (Figure 3B) suggested that APP might regulate Egr1 and c-Fos gene transcription by similar molecular mechanisms. Therefore, we first measured recruitment of CREB on the c-Fos gene promoter in APP+/+ and APP−/− mice. Results of ChIP experiments (Figure 5B) show an enrichment of CREB on c-Fos promoter, but no difference between APP+/+ and APP−/− mice, suggesting that basal regulation of c-Fos gene transcription by APP is not CREB dependent although c-Fos is regulated by CREB [31]. Since HDAC2 was previously demonstrated to inhibit c-Fos gene transcription [30], we performed ChIP experiments using anti-HDAC2 antibodies. Results presented in figure 5C clearly indicate a significant enrichment of HDAC2 on the c-Fos gene promoter in APP+/+ mice. Western blot analysis of nuclear extracts prepared from the PF cortex of APP+/+ and APP−/− mice with anti-HDAC2 antibodies did not show any modification of HDAC2 nuclear or cytoplasmic content, indicating that APP does not modify neither HDAC2 expression nor its nuclear localization (Figure 5D). Altogether, these results indicate that APP fosters a low level of c-Fos expression in mouse brain by increasing enrichment of HDAC2 on the c-Fos gene promoter.
Figure 5. HDAC2 is enriched on the c-Fos gene promoter in APP−/− mice.
A) Schematic representation of the structure of c-Fos promoter containing only one CREB binding site and the localization of the primers utilized for q-PCR. (CREB: cAMP response element, CRE: cAMP response elements SRF: Serum Response factor, SRE: Serum Response Element, AP1: Activator protein 1, SIE: sis-inducible element, STAT: Signal Transducer and Activator of Transcription). B) Study of CREB binding to c-Fos gene promoter in APP+/and APP−/− mice. C) ChIP assay on c-Fos gene promoter using anti-HDAC2 antibody. Results show a significant lower enrichment of HDAC 2 in PF cortex of APP−/− mouse, Student’s t-test: **p<0.01. IgG was used as negative control. Equal amounts of ChIP and input chromatin were used for qRT-PCR analysis on the c-Fos gene promoter. Enrichment values were normalized to input values and represented the average of three or more mice per experiment. Results are expressed as mean ± SD. D) HDAC2 protein expression was detected by western blot analysis of nuclear (N) or cytoplasmic (C) extracts obtained from PF cortex of APP+/+ and APP−/− mice, 5 months of age (n = 5). Typical blot is shown with anti-histone H3 and anti-tubulin used as a loading control. Data are normalized against histone H3 to assess the level of nuclear HDAC2 and expressed as mean ± SD.
6. Exposure of Mice to Novelty Induces IEGs Transcription in APP+/+ but not in APP−/− Mice
Egr1 and c-Fos are inducible transcription factors that are needed for synaptic plasticity and establishment of long term memory. We reasoned that induction of IEGs transcription needs a constitutive low level of expression, as measured for Egr1 and c-Fos in APP+/+ mice. APP+/+ and APP−/− mice were exposed to novelty during a spatial exploration of an open field, in an experimental protocol that was previously described to enhance c-Fos and Egr1 mRNA levels in mouse PF cortex [10], [32]. After a short period of exploration of 15 min and a resting period of 30 min, the mRNA levels of IEGs were quantified in APP+/+ and APP−/− mice. Results presented in figure 6 clearly demonstrated that induction of both Egr1 and c-Fos gene transcription was possible in APP+/+ but not in APP−/− mice. These results suggest that the low level of IEGs transcripts measured in APP+/+ allow their induction during exposure of mice to novelty, while such an induction is not possible in APP−/− mice showing a high basal level of IEGs expression.
Figure 6. APP+/+ but not APP−/− mice induce Egr1 and c-Fos expression after exposure to novelty.
A) Effect of exposure to novelty on the levels of Egr1 and c-Fos transcripts. qRT PCR were performed on mRNA extracted from APP+/+ and APP−/− mice exposed (Novelty) or not (Home cage) to the open field (n = 6 or more per group). All values were normalized to the GAPDH mRNA, and expressed as percentage of the APP+/+. Results are expressed as mean ± SD. Student’s t-test. (***p<0.001).
Discussion
The main finding of this study is that APP fosters low expression of c-Fos and Egr1 in mouse PF cortex, in which induction of several IEGs is involved in memory formation [33]–[35]. APP-mediated down regulation of c-Fos and Egr1 by two distinct epigenetic mechanisms is needed to induce properly transcription of these IEGs upon exposure of mice to novelty.
We extended our observation of the epigenetic regulation of Egr1 gene transcription by APP to c-Fos, Bdnf and Arc. All of those genes belong to the IEGs family of transcription or neurotrophic factors needed for memory formation [2], [7], [36], [37]. The ability of APP to increase gene transcription by an AICD dependent mechanism has been previously demonstrated, although still debated [38]–[43]. Here, we show for the first time that APP represses transcription of a group of genes, all related to synaptic plasticity. The moderate effect of APP on IEGs expression that we measured could explain why these IEGs have never been reported as APP target genes in microarray experiments in which the cut off of differences in gene expression is usually higher.
Induction of IEGs through acetylation of histones H3, H4 and H2B is well documented [44], and acetylation of histone H3K14, H4K12 or phosphorylation of H3S10 via ERK activation are associated with chromatin relaxation leading to IEGs transcription and memory formation [45]–[47]. We have previously demonstrated that trichostatin A, a specific HDAC inhibitor, was able to induce Egr1 gene transcription in both APP+/+ and APP−/− neurons, although induction was significantly higher in APP−/− neurons, in agreement with higher H4 acetylation at the Egr1 gene promoter in these cells [20].
Although induction of IEGs has been widely studied, little is known about basal regulation of IEGs transcription in the hippocampus and the cortex. Basal expression of IEGs in the brain is referred as an expression induced by physiological synaptic input [48]. We demonstrate here that APP fosters a low level of expression of the four IEGs studied. APP represses transcription of Egr1, c-Fos, and Bdnf by decreasing enrichment of acetylated H4, but not H3 nor H2B histones on the corresponding gene promoters. APP-mediated repression of Arc gene transcription appears to be controlled by other molecular mechanisms. SAHA, a nonspecific HDAC inhibitor, was demonstrated to up regulate c-Fos and Bdnf expression, while Arc induction was much more moderate [49], indicating that Arc expression should be regulated by other mechanisms than histone modification.
We observed a very similar acetylation profile of H4K5 and H4K12, but not H4K16, on Egr1 and c-Fos gene promoters, which suggested that APP might regulate both genes by similar mechanisms. H4K5 and H4K12 are known to be acetylated by Tip60 and CBP/P300 histone acetyltransferase activities [23]. As Tip60 interacts with AICD and the adaptor protein Fe65 [24], APP could inhibit H4K5 and H4K12 acetylation in APP+/+ mice. We analyzed a possible interaction of Tip60 with Egr1 promoter but failed to detect any significant interaction or any enrichment in APP−/− mice. Therefore, the contribution of CBP/P300 in the acetylation of H4K5 and H4K12 present on the Egr1 and c-Fos gene promoters was further investigated.
CBP and p300 are two highly related histone acetyltransferases that share many biological functions and interact with phosphorylated cAMP response element binding protein (CREB) [50]. CRE sites are present on Egr1 and c-Fos gene promoters, and regulation of Egr1 and c-Fos gene transcription by CREB is well established [19], [21], [31], [51]. We demonstrate that regulation of Egr1 gene transcription by APP is CREB dependent. In APP−/− mice, CREB is better recruited on the Egr1 gene promoter, arguing for a CBP/P300-mediated increase in H4K5 and H4K12 acetylation of H4 present on the Egr1 promoter. The presence of CREB on a gene promoter has been associated with the removal of HDACs [28]. HDAC2 deacetylates H4K5 and H4K12, and is described as a negative actor in memory formation and synaptic plasticity [30]. Even if we were able to detect enrichment of HDAC2 on Egr1 gene promoter, the level of HDAC2 detected was the same in APP+/+ and APP−/− mice.
Surprisingly, regulation of c-Fos gene transcription by APP occurs by a different mechanism, since the same recruitment of CREB on the c-Fos gene promoter was measured in both APP+/+ and APP−/− mice but less HDAC2 was found on the c-Fos gene promoter in APP−/− mice. APP decreased CREB recruitment on Egr1 gene promoter and increased HDAC2 recruitment on c-Fos gene promoter, without any modification of either CREB and HDAC2 expression or CREB phosphorylation. The presence of two CRE sites in the Egr1 gene promoter could explain why APP-mediated regulation of this gene is more dependent on CREB recruitment.
Our results indicate that APP fosters a low expression of a group of IEGs involved in memory formation. Consequently, overexpression of APP could have important consequences on memory formation. Interestingly, it was previously demonstrated that in APP transgenic mice, overexpression of mutated APP decreases the basal levels of IEGs mRNA and impairs the proper induction of IEGs transcription upon exposure to novelty [32]. Similarities in the cognitive declines observed in both APP−/− and APP transgenic mice [52]–[54] could be related to their impairment in inducing properly IEGs transcription. Moreover, impairment of LTP observed in APP transgenic mice have been associated with dysregulation of histone H4 acetylation [55], [56].
Basal expression of IEGs is important for normal synaptic activity, but their rapid induction is needed to activate transcription of many genes playing a key role in establishment of long term memory [7]. An important function of APP is to epigenetically foster low level of expression of IEGs, allowing rapid induction of their transcription.
Supporting Information
Housekeeping genes and other IEGs expressions are not regulated by APP. GAPDH mRNA levels were quantified by q-RT PCR in APP+/+ and APP−/− PF cortex (n = 6) and normalized versus Actin B) q-RT PCR method was used to assess mRNA levels of the housekeeping genes Actin, peptidylprolyl isomerase A (Ppia) and β-glucuronidase (Gusb) in APP+/+ and APP−/− PF cortex (n = 6). C) mRNA levels of the IEGs c-Jun and Homer-1a were quantified by q-RT PCR in in APP+/+ and APP−/− PF cortex (n = 6). Values were normalized to the GAPDH mRNA, and expressed as percentage of APP+/+, mean ± SD. Primers sequences used were as follows: Ppia FOR: CAGACGCCACTGTCGCTTT; Ppia REV: TGTCTTTGGAACTTTGTCTGCAA, Gusb FOR: ACTGACACCTCCATGTATCCCAAG, Gusb REV: CAGTAGGTCACCAGCCCGATG, c-Jun FOR: TGAAAGCTGTGTCCCCTGTC; c-Jun REV: ATCACAGCACATGCCACTTC, Homer 1a FOR: GAAGTCGCAGGAGAAGATG, Homer1a REV: TGATTGCTGAATTGAATGTGTACC.
(TIF)
Analysis of histone acetylation by ChIP assays on GAPDH promoters. ChIP experiments were performed on chromatin obtained from APP+/+ and APP−/− PF cortex. Immunoprecipitation was completed with antibody recognizing normal mouse IgG as negative control, anti H3Ac, H2BAc and H4Ac. The quantification of immunoprecipitated DNA and the normalization versus total DNA (input) was assessed by real-time qPCR with primers designed on GAPDH. All results were obtained from at least 3 or more mice per group and per antibody, and are expressed as mean ± SD.
(TIF)
Tip60 is enriched in KAI1 promoter. Tip60 binding to KAI1 gene promoter in APP+/+ and APP−/− PF cortex was assessed by ChIP using anti-Tip60 antibody, with primers designed in KAI1 promoter region. Primers sequences used were as follows: KAI1 FOR: ACCGTTAGGCAGCGCCGTGAG; KAI1 Rev: CTTGGGAAGGCGGTGCGCTC. IgG was used as negative control. Results show a significant enrichment of Tip60 in APP+/+ mice, **p<0.01. Enrichment values were normalized to input values and are the average of three or more experiments per group. Results are expressed as mean ± SD.
(TIF)
Funding Statement
This work was supported by FRIA funding of the Belgian Fonds pour la Recherche Scientifique (FNRS-FRS), by Interuniversity Attraction Poles Programme-Belgian State-Belgian Science Policy, The Belgian Fonds de la Recherche Scientifique Médicale, the Fondation pour la Recherche sur la Maladie d’Alzheimer (SAOFRA) and the Programme d’excellence « Marshall » Diane convention. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jones MW, Errington ML, French PJ, Fine A, Bliss TVP, et al. (2001) A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nature Neuroscience 4: 289–296. [DOI] [PubMed] [Google Scholar]
- 2. Bozon B, Davis S, Laroche S (2002) Regulated transcription of the immediate-early gene Zif268: mechanisms and gene dosage-dependent function in synaptic plasticity and memory formation. Hippocampus 12: 570–577. [DOI] [PubMed] [Google Scholar]
- 3. Veyrac A, Gros A, Bruel-Jungerman E, Rochefort C, Kleine Borgmann FB, et al. (2013) Zif268/egr1 gene controls the selection, maturation and functional integration of adult hippocampal newborn neurons by learning. Proc Natl Acad Sci U S A 110: 7062–7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Renaudineau S, Poucet B, Laroche S, Davis S, Save E (2009) Impaired long-term stability of CA1 place cell representation in mice lacking the transcription factor zif268/egr1. Proc Natl Acad Sci USA 106: 11771–11775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hunt SP, Pini A, Evan G (1987) Induction of c-fos-like protein in spinal cord neurons following sensory stimulation. Nature 328: 632–634. [DOI] [PubMed] [Google Scholar]
- 6. Rusak B, Robertson HA, Wisden W, Hunt SP (1990) Light pulses that shift rhythms induce gene expression in the suprachiasmatic nucleus. Science 248: 1237–1240. [DOI] [PubMed] [Google Scholar]
- 7. Guzowski JF (2002) Insights into immediate-early gene function in hippocampal memory consolidation using antisense oligonucleotide and fluorescent imaging approaches. Hippocampus 12: 86–104. [DOI] [PubMed] [Google Scholar]
- 8. Kovács KJ (2008) Measurement of immediate-early gene activation- c-fos and beyond. J Neuroendocrinol 20: 665–672. [DOI] [PubMed] [Google Scholar]
- 9. Fleischmann A, Hvalby O, Jensen V, Strekalova T, Zacher C, et al. (2003) Impaired long-term memory and NR2A-type NMDA receptor-dependent synaptic plasticity in mice lacking c-Fos in the CNS. J Neurosci 23: 9116–9122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rinaldi A, Romeo S, Agustín-Pavón C, Oliverio A, Mele A (2010) Distinct patterns of Fos immunoreactivity in striatum and hippocampus induced by different kinds of novelty in mice. Neurobiol Learn Mem 94: 373–381. [DOI] [PubMed] [Google Scholar]
- 11. Shepherd JD, Bear MF (2011) New views of Arc, a master regulator of synaptic plasticity. Nat Neurosci 14: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vazdarjanova A, Ramirez-Amaya V, Insel N, Plummer TK, Rosi S, et al. (2006) Spatial exploration induces Arc, a plasticity-related immediate-early gene, only in calcium/calmodulin-dependent protein kinase II-positive principal excitatory and inhibitory neurons of the rat forebrain. J Comp Neurol 498: 317–329. [DOI] [PubMed] [Google Scholar]
- 13. Steward O, Wallace CS, Lyford GL, Worley PF (1998) Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 21: 741–751. [DOI] [PubMed] [Google Scholar]
- 14. Barde YA, Edgar D, Thoenen H (1982) Purification of a new neurotrophic factor from mammalian brain. EMBO J 1: 549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McAllister AK, Lo DC, Katz LC (1995) Neurotrophins regulate dendritic growth in developing visual cortex. Neuron 15: 791–803. [DOI] [PubMed] [Google Scholar]
- 16. Mizuno M, Yamada K, Olariu A, Nawa H, Nabeshima T (2000) Involvement of brain-derived neurotrophic factor in spatial memory formation and maintenance in a radial arm maze test in rats. J Neurosci 20: 7116–7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T (2007) Dissecting the human Bdnf locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics 90: 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T (2007) Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res 85: 525–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cortés-Mendoza J, Díaz de León-Guerrero S, Pedraza-Alva G, Pérez-Martínez L (2013) Shaping synaptic plasticity: the role of activity-mediated epigenetic regulation on gene transcription. Int J Dev Neurosci 31: 359–369. [DOI] [PubMed] [Google Scholar]
- 20. Hendrickx A, Pierrot N, Tasiaux B, Schakman O, Brion JP, et al. (2013) Epigenetic induction of Egr1 expression by the amyloid precursor protein during exposure to novelty. PLoS One 8: e74305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Flavell SW, Greenberg ME (2008) Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci 31: 563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Russell DL, Doyle KM, Gonzales-Robayna I, Pipaon C, Richards JS (2003) Egr-1 induction in rat granulosa cells by follicle-stimulating hormone and luteinizing hormone: combinatorial regulation by transcription factors cyclic adenosine 3′,5′-monophosphate regulatory element binding protein, serum response factor, sp1, and early growth response factor-1. Mol Endocrinol 17: 520–533. [DOI] [PubMed] [Google Scholar]
- 23. Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- 24. Cao X, Sudhof TC (2001) A transcriptively active complex of APP with Fe65 and histone acetyltransferase Tip60 (vol 293, pg 115, 2001). Science 293: 1436–1436. [DOI] [PubMed] [Google Scholar]
- 25. Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, et al. (2002) Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell 110: 55–67. [DOI] [PubMed] [Google Scholar]
- 26. Rowe A, Weiske J, Kramer TS, Huber O, Jackson P (2008) Phorbol ester enhances KAI1 transcription by recruiting Tip60/Pontin complexes. Neoplasia 10: 1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lubin FD, Gupta S, Parrish RR, Grissom NM, Davis RL (2011) Epigenetic mechanisms: critical contributors to long-term memory formation. Neuroscientist 17: 616–632. [DOI] [PubMed] [Google Scholar]
- 28. Riccio A (2010) Dynamic epigenetic regulation in neurons: enzymes, stimuli and signaling pathways. Nat Neurosci 13: 1330–1337. [DOI] [PubMed] [Google Scholar]
- 29. Peixoto L, Abel T (2013) The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology 38: 62–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guan J-S, Haggarty SJ, Giacometti E, Dannenberg J-H, Joseph N, et al. (2009) HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459: 55–U58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sassone-Corsi P, Visvader J, Ferland L, Mellon PL, Verma IM (1988) Induction of proto-oncogene fos transcription through the adenylate cyclase pathway: characterization of a cAMP-responsive element. Genes Dev 2: 1529–1538. [DOI] [PubMed] [Google Scholar]
- 32. Dickey CA, Gordon MN, Mason JE, Wilson NJ, Diamond DM, et al. (2004) Amyloid suppresses induction of genes critical for memory consolidation in APP+PS1 transgenic mice. J Neurochem 88: 434–442. [DOI] [PubMed] [Google Scholar]
- 33. DeVito LM, Lykken C, Kanter BR, Eichenbaum H (2010) Prefrontal cortex: role in acquisition of overlapping associations and transitive inference. Learn Mem 17: 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yochiy A, Britto LR, Hunziker MH (2012) Novelty, but not operant aversive learning, enhances Fos and Egr-1 expression in the medial prefrontal cortex and hippocampal areas of rats. Behav Neurosci 126: 826–834. [DOI] [PubMed] [Google Scholar]
- 35. Dias R, Honey RC (2002) Involvement of the rat medial prefrontal cortex in novelty detection. Behav Neurosci 116: 498–503. [DOI] [PubMed] [Google Scholar]
- 36. Bertaina-Anglade V, Tramu G, Destrade C (2000) Differential learning-stage dependent patterns of c-Fos protein expression in brain regions during the acquisition and memory consolidation of an operant task in mice. Eur J Neurosci 12: 3803–3812. [DOI] [PubMed] [Google Scholar]
- 37. Hall J, Thomas KL, Everitt BJ (2000) Rapid and selective induction of Bdnf expression in the hippocampus during contextual learning. Nat Neurosci 3: 533–535. [DOI] [PubMed] [Google Scholar]
- 38. Hébert SS, Serneels L, Tolia A, Craessaerts K, Derks C, et al. (2006) Regulated intramembrane proteolysis of amyloid precursor protein and regulation of expression of putative target genes. EMBO Rep 7: 739–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pardossi-Piquard R, Petit A, Kawarai T, Sunyach C, Alves da Costa C, et al. (2005) Presenilin-dependent transcriptional control of the Abeta-degrading enzyme neprilysin by intracellular domains of betaAPP and APLP. Neuron 46: 541–554. [DOI] [PubMed] [Google Scholar]
- 40. Belyaev ND, Nalivaeva NN, Makova NZ, Turner AJ (2009) Neprilysin gene expression requires binding of the amyloid precursor protein intracellular domain to its promoter: implications for Alzheimer disease. EMBO Rep 10: 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ryan KA, Pimplikar SW (2005) Activation of GSK-3 and phosphorylation of CRMP2 in transgenic mice expressing APP intracellular domain. J Cell Biol 171: 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim HS, Kim EM, Lee JP, Park CH, Kim S, et al. (2003) C-terminal fragments of amyloid precursor protein exert neurotoxicity by inducing glycogen synthase kinase-3beta expression. FASEB J 17: 1951–1953. [DOI] [PubMed] [Google Scholar]
- 43. Aydin D, Filippov MA, Tschäpe JA, Gretz N, Prinz M, et al. (2011) Comparative transcriptome profiling of amyloid precursor protein family members in the adult cortex. BMC Genomics 12: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bousiges O, Vasconcelos AP, Neidl R, Cosquer B, Herbeaux K, et al. (2010) Spatial memory consolidation is associated with induction of several lysine-acetyltransferase (histone acetyltransferase) expression levels and H2B/H4 acetylation-dependent transcriptional events in the rat hippocampus. Neuropsychopharmacology 35: 2521–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bousiges O, Neidl R, Majchrzak M, Muller MA, Barbelivien A, et al. (2013) Detection of histone acetylation levels in the dorsal hippocampus reveals early tagging on specific residues of H2B and H4 histones in response to learning. PLoS One 8: e57816. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46. Reul JM (2014) Making Memories of Stressful Events: A Journey Along Epigenetic, Gene Transcription, and Signaling Pathways. Front Psychiatry 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, et al. (2004) Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem 279: 40545–40559. [DOI] [PubMed] [Google Scholar]
- 48. Herdegen T, Leah JD (1998) Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Res Brain Res Rev 28: 370–490. [DOI] [PubMed] [Google Scholar]
- 49. Koppel I, Timmusk T (2013) Differential regulation of Bdnf expression in cortical neurons by class-selective histone deacetylase inhibitors. Neuropharmacology 75: 106–115. [DOI] [PubMed] [Google Scholar]
- 50. Kuo MH, Allis CD (1998) Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20: 615–626. [DOI] [PubMed] [Google Scholar]
- 51. Lonze BE, Ginty DD (2002) Function and regulation of CREB family transcription factors in the nervous system. Neuron 35: 605–623. [DOI] [PubMed] [Google Scholar]
- 52. Taylor CJ, Ireland DR, Ballagh I, Bourne K, Marechal NM, et al. (2008) Endogenous secreted amyloid precursor protein-alpha regulates hippocampal NMDA receptor function, long-term potentiation and spatial memory. Neurobiol Dis 31: 250–260. [DOI] [PubMed] [Google Scholar]
- 53. Dawson GR, Seabrook GR, Zheng H, Smith DW, Graham S, et al. (1999) Age-related cognitive deficits, impaired long-term potentiation and reduction in synaptic marker density in mice lacking the beta-amyloid precursor protein. Neuroscience 90: 1–13. [DOI] [PubMed] [Google Scholar]
- 54. Seabrook GR, Smith DW, Bowery BJ, Easter A, Reynolds T, et al. (1999) Mechanisms contributing to the deficits in hippocampal synaptic plasticity in mice lacking amyloid precursor protein. Neuropharmacology 38: 349–359. [DOI] [PubMed] [Google Scholar]
- 55. Francis YI, Fà M, Ashraf H, Zhang H, Staniszewski A, et al. (2009) Dysregulation of histone acetylation in the APP/PS1 mouse model of Alzheimer’s disease. J Alzheimers Dis 18: 131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, et al. (2010) Altered Histone Acetylation Is Associated with Age-Dependent Memory Impairment in Mice. Science 328: 753–756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Housekeeping genes and other IEGs expressions are not regulated by APP. GAPDH mRNA levels were quantified by q-RT PCR in APP+/+ and APP−/− PF cortex (n = 6) and normalized versus Actin B) q-RT PCR method was used to assess mRNA levels of the housekeeping genes Actin, peptidylprolyl isomerase A (Ppia) and β-glucuronidase (Gusb) in APP+/+ and APP−/− PF cortex (n = 6). C) mRNA levels of the IEGs c-Jun and Homer-1a were quantified by q-RT PCR in in APP+/+ and APP−/− PF cortex (n = 6). Values were normalized to the GAPDH mRNA, and expressed as percentage of APP+/+, mean ± SD. Primers sequences used were as follows: Ppia FOR: CAGACGCCACTGTCGCTTT; Ppia REV: TGTCTTTGGAACTTTGTCTGCAA, Gusb FOR: ACTGACACCTCCATGTATCCCAAG, Gusb REV: CAGTAGGTCACCAGCCCGATG, c-Jun FOR: TGAAAGCTGTGTCCCCTGTC; c-Jun REV: ATCACAGCACATGCCACTTC, Homer 1a FOR: GAAGTCGCAGGAGAAGATG, Homer1a REV: TGATTGCTGAATTGAATGTGTACC.
(TIF)
Analysis of histone acetylation by ChIP assays on GAPDH promoters. ChIP experiments were performed on chromatin obtained from APP+/+ and APP−/− PF cortex. Immunoprecipitation was completed with antibody recognizing normal mouse IgG as negative control, anti H3Ac, H2BAc and H4Ac. The quantification of immunoprecipitated DNA and the normalization versus total DNA (input) was assessed by real-time qPCR with primers designed on GAPDH. All results were obtained from at least 3 or more mice per group and per antibody, and are expressed as mean ± SD.
(TIF)
Tip60 is enriched in KAI1 promoter. Tip60 binding to KAI1 gene promoter in APP+/+ and APP−/− PF cortex was assessed by ChIP using anti-Tip60 antibody, with primers designed in KAI1 promoter region. Primers sequences used were as follows: KAI1 FOR: ACCGTTAGGCAGCGCCGTGAG; KAI1 Rev: CTTGGGAAGGCGGTGCGCTC. IgG was used as negative control. Results show a significant enrichment of Tip60 in APP+/+ mice, **p<0.01. Enrichment values were normalized to input values and are the average of three or more experiments per group. Results are expressed as mean ± SD.
(TIF)