Abstract
The ZAC1 gene, mapped to the 6q24 region, is part of a network of co-regulated imprinted genes involved in the control of embryonic growth. Loss of methylation at the ZAC1 differentially methylated region (DMR) is associated with transient neonatal diabetes mellitus, a developmental disorder involving growth retardation and diabetes in the first weeks of post-natal life. We assessed whether the degree of methylation of the ZAC1 DMR in leukocytes DNA extracted from cord blood is associated with fetal, birth and post-natal anthropometric measures or with C-peptide concentrations in cord serum. We also searched for an influence of dietary intake and maternal parameters on ZAC1 DMR methylation. We found positive correlations between the ZAC1 DMR methylation index (MI) and estimated fetal weight (EFW) at 32 weeks of gestation, weight at birth and weight at one year of age (respectively, r = 0.15, 0.09, 0.14; P values = 0.01, 0.15, 0.03). However, there were no significant correlations between the ZAC1 DMR MI and cord blood C-peptide levels. Maternal intakes of alcohol and of vitamins B2 were positively correlated with ZAC1 DMR methylation (respectively, r = 0.2 and 0.14; P = 0.004 and 0.04). The influence of ZAC1 seems to start in the second half of pregnancy and continue at least until the first year of life. The maternal environment also appears to contribute to the regulation of DNA methylation.
Keywords: ZAC1/HYMAI imprinted locus, imprinting disorders, transient neonatal diabetes mellitus, C-peptide, metabolism disorders, insulin secretion in vivo, nutrition and epigenetic regulation, pathophysiology and metabolism, fetal development
Introduction
Genomic imprinting is an important epigenetic mechanism and contributes to regulating a subset of mammalian genes. Imprinted genes are themselves key regulators of fetal and post-natal development, metabolism, and behavior. Therefore, faithful epigenetic establishment and maintenance is critical for the future health of the individual.1 The intake of various nutrients, including vitamin B2, 3, 6, 9, and 12, is needed for one-carbon metabolism and is essential for DNA methylation.2,3 In addition, alcohol consumption is associated with alteration of DNA methylation.4 Loss of imprinting (LOI) through loss (LOM) or gain (GOM) of methylation is involved in many human disorders and cancers.5-7 Furthermore, it has been suggested that imprinted genes form a network of co-regulated genes (IGN) involved in the control of embryonic development. Disruption of one of these genes may subsequently result in the deregulation of many other imprinted genes of the IGN.8,9
The imprinted HYMAI/ZAC1 locus, mapped to the 6q24 region, has crucial roles in controlling fetal growth and metabolism. HYMAI encodes an untranslated mRNA, while ZAC1, located 70 kb downstream to the HYMAI gene, encodes a zinc-finger transcription factor (ZAC1) and is part of the IGN.9-13 The HYMAI/ZAC1 locus harbors a differentially methylated region (DMR) that is methylated on the maternal allele. This region plays the role of an imprinting control region (ICR) and restricts the expression of both HYMAI and ZAC1 genes to the paternal allele.14 However, aberrant methylation at HYMAI/ZAC1 locus through genetic [paternal uniparental isodisomy of chromosome 6 (pUPD6) or paternal inheritance of chromosome 6q duplications] or LOM at the ZAC1 DMR (20% of cases) is involved in transient neonatal diabetes mellitus syndrome (TNDM).15 These abnormalities induce biallelic expression of HYMAI and ZAC1 genes. ZAC1 activity is necessary for normal pancreatic islets development.16 TNDM is a developmental disorder associated with growth retardation, failure to thrive, reduced subcutaneous fat, and diabetes in the first months of post-natal life due to lack of normal insulin secretion.13-17 It resolves at the age of 3 mo, on average, but type II diabetes is frequent later in adulthood.18
Birth cohort studies can be used to explore the correlation between genomic imprinting, growth and metabolism in the general population. It has been shown that individuals exposed prenatally to famine during the Dutch Hunger Winter show hypomethylation at the imprinted IGF2 gene.19 By contrast, few data have been published to date about whether pre- and post-natal growth and insulin/C-peptide levels at birth in healthy children are associated with the methylation status at imprinted genes involved in the control of both metabolism and growth.20 In addition, there are scarce data about whether maternal diet, either before or during pregnancy, is associated with variability in the ZAC1 DMR methylation index (MI).21 We report an exploratory analysis of the association between the MI for the ZAC1 DMR in cord blood and both C-peptide levels in cord blood and intra- and extra-uterine growth in healthy children of the French EDEN cohort study (Study of pre- and post-natal child health and development determinants).22 We also tested for associations between the ZAC1 DMR MI and maternal diet, smoking and alcohol consumption both before and during pregnancy.2-4
Results
Population sampling
There was sufficient genomic DNA available for methylation analysis for 254 of the 378 children selected for this study. Compared with the 1896 newborns included in the EDEN study, the 254 babies included in this analysis had slightly higher birth weight (P = 0.03), birth length (P = 0.02), and gestational age (P = 0.003), and more were enrolled in Poitiers (P = 0.01). Table 1 shows the clinical characteristics of the mother-(fetus) child pairs and the dietary (energy and vitamin B intake), smoking, and alcohol-consumption habits of the mothers.
Table 1. Clinical characteristics of the 254 mother-child pairs.
| n | Mean (sd) or % | |
|---|---|---|
| Maternal age at delivery | 254 | 29.8 (4.4) |
| Maternal height (cm) | 251 | 163 (6,1) |
| BMI before pregnancy | 251 | 23.2 (4.4) |
| % Obese (BMI > 30 kg/m2) | 251 | 7.2% |
| Maternal weight gain during pregnancy (kg) | 248 | 14.1 (4.94) |
| Maternal smoking in the 2nd and /or 3rd trimester | 254 | 15.0% |
| Maternal dietary intake before pregnancy | ||
| Energy intake (exclusive of alcohol) (Kcal/d) | 226 | 2261 (771) |
| Vitamin B2 (mg/day) | 226 | 2.1 (0.9) |
| Vitamin B3 (mg/day) | 226 | 18.0 (6.6) |
| Vitamin B6 (mg/day) | 226 | 1.9 (0.8) |
| Vitamin B9 (μg/day) | 226 | 365 (180) |
| Vitamin B12 (μg /day) | 226 | 6.6 (2.9) |
| % non-alcohol drinker | 226 | 10.2% |
| Alcohol intake among drinkers (g/d) † | 203 | 3.61 [1.36 – 8.84] |
| Maternal dietary intake in the last three months of pregnancy | ||
| Energy intake (exclusive of alcohol) (Kcal/d) | 213 | 2414 (800) |
| Vitamin B2 (mg/day) | 213 | 2.4 (0.9) |
| Vitamin B3 (mg/day) | 213 | 16.6 (6.1) |
| Vitamin B6 (mg/day) | 213 | 1.9 (0.7) |
| Vitamin B9 (μg/day) | 213 | 357 (160) |
| Vitamin B12 (μg /day) | 213 | 7.2 (3.9) |
| % non alcohol drinker | 213 | 32.4% |
| Alcohol intake among drinkers (g/d)† | 144 | 0.13 [0.02 – 1.45] |
| Fetal characteristics at 20–24 WA | ||
| Femur length (mm) | 251 | 38.7 (3.0) |
| Estimated Fetal Weight (g) | 247 | 522 (99) |
| Fetal characteristics at 30–34 WA | ||
| Femur length (mm) | 243 | 61.8 (3.0) |
| Estimated Fetal Weight (g) | 241 | 1993 (309) |
| Birth characteristics | ||
| Gestational age (WA) | 254 | 39.5 (1.5) |
| Birth weight (g) | 254 | 3346 (469) |
| Birth weight z-score** | 0.06 (0.98) | |
| Birth length (cm) | 250 | 49.9 (2.4) |
| Birth length z-score** | 0.16 (1.23) | |
| Head circumference (cm) | 248 | 34.6 (1.3) |
| Head circumference z-score** | 0.36 (1.02) | |
| BMI (kg/m2) | 250 | 13.5 (1.3) |
| BMI z-score** | 250 | 0.01 (1.01) |
| C peptide in cord blood (nmol/L) | 252 | 0.85 (0.47) |
| ZAC1 methylation index in cord blood | 254 | 51.5 (4.1) |
| Child characteristics at one year | ||
| Weight (kg) | 251 | 9.85 (1.12) |
| Weight z-score** | 0.37 (0.87) | |
| Length (cm) | 251 | 74.8 (2.7) |
| Length z-score** | −0.16 (0.99) | |
| Head circumference (cm) | 228 | 46 (1.44) |
| Head circumference z-score** | 0.63 (0.97) | |
| BMI (kg/m2) | 251 | 17.6 (1.6) |
| BMI z-score** | 0.64 (1.00) |
The values reported are means (sd) ** Z score according to WHO standards (http://www.who.int/growthref/en/). †For alcohol consumption, descriptive statistics represent median [q1 – q3] given that data are not normally distributed.
Correlation between ZAC1 DMR methylation and anthropometric markers
The methylation quantitation at the ZAC1 DMR has been performed with a quantitative method ASMM RTQ-PCR23 and the methylation index represents the ratio between the methylated and unmethylated alleles as detailed in material and methods section. The mean cord blood ZAC1 DMR MI in the newborns was 52 ± 4.08% (normal range = mean ± 2SD = 44–60%). Three subjects were considered as outliers according to the normal range calculated from the entire population: one had partial hypomethylation (30%) and two had partial hypermethylation (63% and 65%) (Fig. 1).
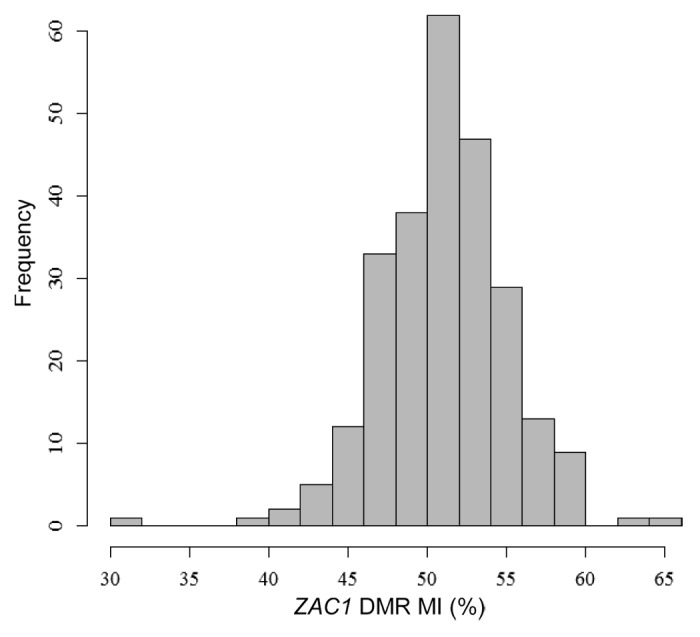
Figure 1. Distribution of the ZAC1 DMR methylation index, scored as a percentage (n = 254).
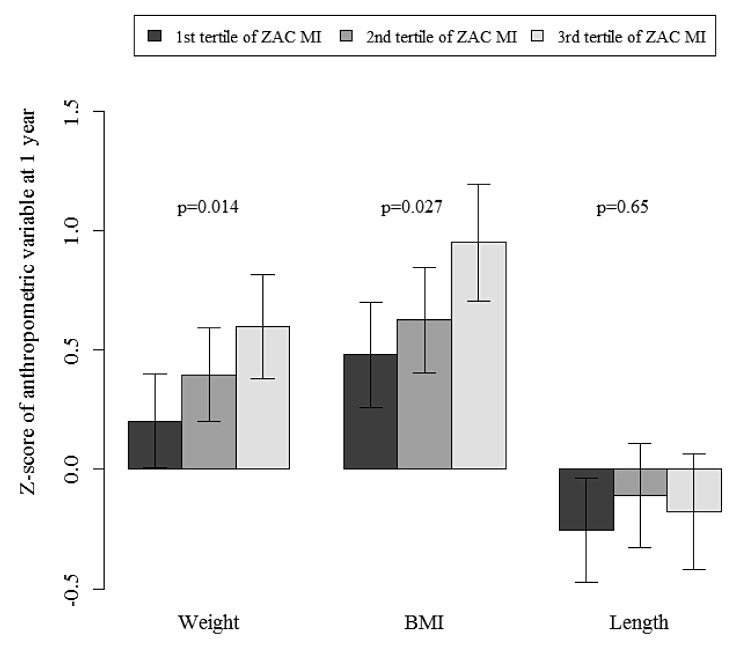
Table 2 shows the correlation coefficients between the cord blood methylation index of the ZAC1 DMR and both C-peptide concentrations in cord blood, and fetal and infant auxological measures. The methylation index of the ZAC1 DMR was not significantly correlated with EFW or femoral length at 22 wk gestation. The ZAC1 MI correlated positively with the EFW at 32 wk gestation but not with femoral length. There was a weaker and non-significant correlation between MI and birth weight. However, at one year of age, the ZAC1 DMR MI correlated positively with weight and BMI Z scores (respectively, r = 0.14 and 0.15; P value = 0.03 and 0.01, after adjustment for gestational age at birth, child gender and exact age at the one-year examination). The distribution of weight at one-year old and BMI Z scores by tertiles of MI of the ZAC1 DMR is depicted in Figure 2. A higher ZAC1 DMR MI was associated with a larger increase in estimated fetal weight SDS from 22 to 32 wk of gestation (r = 0.13, P = 0.04), but not with the change of Z score weight from birth to one year (r = 0.05, P = 0.47). Neither length at birth, length SDS at one year of age, or C-peptide concentration in cord serum were correlated with the MI of the ZAC1 DMR.
Table 2. Correlation coefficients between the cord blood methylation index for the ZAC1 DMR and both cord blood concentration of C peptide and fetal and infant auxological markers.
| Auxological markers | Correlation coefficient | P value | Partial correlation coefficient* | P value |
|---|---|---|---|---|
| Weight | ||||
| EFW 20 wk SD-score | −0.004 | 0.94 | −0.003 | 0.95 |
| EFW 30 wk SD-score | 0.19 | 0.002 | 0.15 | 0.01 |
| Birth weight z-score | 0.09 | 0.15 | 0.08 | 0.23 |
| Birth BMI z-score | 0.11 | 0.09 | 0.10 | 0.13 |
| Weight z-score at 1 y | 0.14 | 0.03 | 0.14 | 0.03 |
| BMI z-score at 1 y | 0.16 | 0.009 | 0.15 | 0.01 |
| Length | ||||
| Femur length 20 wk SD-score | −0.03 | 0.59 | −0.068 | 0.28 |
| Femur length 30 wk SD-score | 0.10 | 0.10 | 0.08 | 0.21 |
| Birth length z-score | 0.03 | 0.58 | 0.04 | 0.51 |
| Length z-score at 1 y | 0.05 | 0.48 | 0.017 | 0.79 |
| C peptide concentration | 0.025 | 0.69 | 0.03 | 0.63 |
Adjusted for center, child’s gender and gestational age (ultrasound and birth data) or child’s age (one-year data); EFW, estimated fetal weight.
Figure 2. Mean anthropometric z-scores by tertiles of the ZAC1 DMR methylation index (n = 251). Mean values adjusted for gender and study center with standard errors are presented, global P values for Anova tests are shown for each variable.
Considering the three outliers, none of them showed significant difference C-peptide concentrations in cord blood, and fetal and infant auxological parameters were comparable to the rest of the population. The omission of these patients from the analysis did not change the statistical results. The subject with ZAC1 DMR hypomethylation did not develop TNDM.
ZAC1 DMR methylation and maternal parameters correlation
We tested whether maternal environment and characteristics influenced the ZAC1 DMR MI: various variables known to affect fetal growth (maternal height, BMI, weight gain, smoking during pregnancy, and gestational age) were investigated. Only maternal BMI was associated with the ZAC1 DMR MI (r = 0.13, P = 0.03). The association between the child’s prenatal and post-natal growth and the ZAC1 DMR MI was thus further adjusted for maternal BMI. This resulted in a slight decrease in the strength of the correlation (EFW 30 wk SD score: r = 0.12, P = 0.06; one-year BMI z-score: r = 0.14, P = 0.03).
Impact of nutrition on ZAC1 DMR methylation
We tested for correlations between the ZAC1 DMR methylation index and maternal dietary variables (Table 3). Vitamin B2 (riboflavin) and alcohol intake were positively correlated with the ZAC1 DMR MI (r = 0.14, P = 0.04 and r = 0.11, P = 0.09 prior pregnancy and r = 0.11, P = 0.09 and r = 0.20, P = 0.004 last 3 mo of pregnancy, respectively). The ZAC1 DMR MI was not significantly associated with dietary vitamin B9 intake or with folic acid supplementation alone and/or the use of a combination of micronutrients either prior to or during pregnancy (data not shown). Further adjustment for alcohol, vitamin B2 intake did not change the association between the MI and fetal or post-natal anthropometric measures.
Table 3. Correlation coefficients between the cord blood methylation index for the ZAC1 DMR and maternal dietary variables prior to, and during the last three months of, pregnancy.
| Dietary variables | Prior to pregnancy | P value | Last 3 mo of pregnancy | P value |
|---|---|---|---|---|
| Energy intake* | 0.10 | 0.14 | 0.09 | 0.15 |
| Vitamin B2 | 0.14 | 0.04 | 0.11 | 0.09 |
| Vitamin B3 | 0.04 | 0.60 | 0.08 | 0.22 |
| Vitamin B6 | 0.04 | 0.49 | 0.04 | 0.53 |
| Vitamin B9 | 0.02 | 0.74 | 0.04 | 0.56 |
| Vitamin B12 | 0.11 | 0.08 | 0.02 | 0.79 |
| Alcohol intake | 0.11 | 0.09 | 0.20 | 0.004 |
Exclusive of alcohol.
Discussion
We report the first study describing the relationship in healthy newborns between the methylation index at the imprinted HYMAI/ZAC1 locus on chromosome 6q24 and each of anthropometric measures, the C-peptide concentration in cord blood and the maternal environment (dietary intake and maternal parameters). Methylation of the ZAC1 DMR was positively associated with fetal and post-natal weight and BMI but not with length. Furthermore, methylation also increased with maternal BMI, and maternal intake of alcohol and of a diet rich in vitamins B2, involved in one-carbon metabolism.
The study sample was a subgroup of mothers and infants from the general population enrolled in the EDEN cohort. Women who agreed to participate in the cohort had a higher level of education and were more often employed than the general population of women who gave birth at the same time.24 The subsample included in the present analysis had heavier babies on average, because cord blood was more frequently available for research following uncomplicated deliveries of healthy babies. If ZAC1 methylation is associated with fetal growth the restriction of methylation analyses to this sub-sample may have limited the ability to detect overall associations between ZAC1 methylation and the phenotypic measurements. Our results can however be considered relevant to the healthy population of mothers and their infants.
ZAC1 is an imprinted gene expressed from the paternal allele (the maternal allele is methylated and consequently not expressed) and encodes a zinc finger transcription factor. It is widely expressed in many human tissues during both fetal and post-natal periods.25 ZAC1 can either activate or repress its target genes, depending on the configuration of its binding sites. The ZAC1 protein plays an antiproliferative role by promoting cell cycle arrest and apoptosis in various cell lineages during development.10-12 Knockdown of the endogenous Zac1 in murine cells results in cell proliferation in dose-dependent manner.26 In humans, complete loss of methylation at the ZAC1 DMR is associated with TNDM syndrome.14,27 These observations highlight the crucial role of ZAC1 in controlling both growth and metabolism.
The loss of methylation at the ZAC1 DMR identified in TNDM patients most probably induces biallelic overexpression of the ZAC1 gene. Mackay and coworkers demonstrated that ZAC1 is expressed from both alleles in fibroblasts from TNDM patients with ZAC1 DMR abnormality, demonstrating its involvement in the etiology of the disease.28 Hoffmann’s group reported that doubling Zac1 expression reduced the expression of Rasgrf1 and consequently reduced insulin secretion29; also, this effect on Rasgrf1 expression and insulin secretion was dose dependent. By contrast, ZAC1 expression is reduced in human ovarian cancer with hypermethylation at the ZAC1 DMR, and treatment of the hypermethylated cell lines with a demethylating agent reactivates ZAC1 expression.30 These various findings suggest that ZAC1 expression is proportionally associated with the methylation status of its DMR and that the effect of ZAC1 protein on cell proliferation and metabolism is dose dependent. Furthermore, the ZAC1 gene is part of a network of co-regulated imprinted genes involved in controlling fetal and post-natal development9; thus, variation of ZAC1 expression may induce variations in the expression of its co-regulated genes, which may in turn affect growth homeostasis.
Our data are in part consistent with these observations: we describe a positive association between the degree of methylation of the ZAC1 DMR and each of fetal weight, and infant weight and BMI. In addition, we also show that the effects of ZAC1 methylation variation started in the second period of pregnancy and persisted until at least one year of age. However, the ZAC1 DMR MI was not correlated with pre- or post-natal length. In TNDM patients, both birth weight and birth length were affected.31 It should be noted that in TNDM patients, the LOM found at the ZAC1 DMR is total15 whereas in our study population, we observed smaller variations of the MI (52 ± 4%). This suggests that only severe abnormalities of ZAC1 methylation have a significant impact on pre- and post-natal length.
The C-peptide is considered to be a good marker of β-cell function. Indeed, proinsulin produced by β-cells is cleaved to give C-peptide and insulin, which are then secreted in equimolar amounts. We chose to measure C-peptide rather than insulin because the C-peptide concentration is not altered by hemolysis, whereas insulin degradation is increased by even small amounts of hemolysis.32 TNDM patients with 6q24 abnormalities present with hyperglycemia during the few weeks of post-natal life due to a failure of insulin production.15,33,34 In our study population, the ZAC1 DMR MI was not significantly associated with the cord blood concentration of C-peptide. This may have been because many other genetic, epigenetic, and environmental factors have positive or negative effects on pancreatic development and consequently insulin production. Indeed, Regnault et al. demonstrated a significant effect of the maternal intrauterine environment in the cohort we studied: the cord blood concentration of C-peptide at birth correlated positively with maternal BMI pre-pregnancy and maternal glycemia.22 Furthermore, it seems that ZAC1 may be involved in the differentiation rather than the proliferation of β-cell islets: its overexpression may affect the normal development of the β-cells, which in turn may impair their ability to produce insulin.16 Interestingly, a transgenic mouse line that weakly expressing genes of the TNDM locus, blood glucose was normal in the neonates reflecting a gene dose-dependent phenotype.16 Our observations are consistent with this finding by Ma and colleagues. In our population, the methylation index variations at the ZAC1 DMR may be too small to cause substantial effects on ZAC1 expression, which could in turn impair insulin secretion. Alternatively, the effect of ZAC1 may be specifically important during the early post-natal period when pancreatic β-cells undergo a period of intense proliferation and apoptosis, before the equilibrium between insulin production and weight is reached.35 This is consistent with the transient neonatal hyperglycemia of TNDM patients and could also explain the lack of association with C-peptide concentration at birth.
Although methylation of imprinted genes is thought to occur mainly during gametogenesis and further maintained during the lifelong of the individual,36,37 there are indications that maternal nutrition during critical windows of fetal development may affect the methylation of imprinted genes, thereby altering their regulation with subsequent consequences on feto-placental development. Work concerning the Dutch famine cohort shows that IGF2 DMR0 is hypomethylated in subjects born to mothers whose nutrition was compromised around the time of conception or in the first trimester of pregnancy.19 A recent study of a protein-restricted mouse model of nutritional programming concluded, however, that imprinted genes may be resistant to developmental programming by maternal nutrition.38,39 By contrast, a randomized controlled trial of periconceptional supplementation in Gambia found sex-specific hypomethylation at IGF2R (in girls) and GTL2 (in boys) in cord blood but not for ZAC1 DMR.21 This discrepancy could be due to the difference in the size of the two populations studied: n = 58 in the former study and n = 254 in our study. We tested whether maternal nutrition before and during pregnancy had an impact on the ZAC1 DMR MI. Interestingly, we found a positive association between ZAC1 DMR methylation status and maternal intake of alcohol and vitamins B2. These compounds are indirectly involved in DNA methylation.2-4 Furthermore, we also found a positive association between the ZAC1 DMR MI and maternal BMI. These observations indicate that the maternal environment may influence methylation at imprinted loci in humans.
In conclusion, our exploratory study in a cohort of healthy subject suggests that ZAC1 DMR methylation contributes to the control of fetal and post-natal growth. The effect was seen from the second semester of pregnancy and involved fetal, birth, and infant weight and BMI, but not length. It may therefore be involved in adipose tissue growth. Although the association with the cord blood concentration of C-peptide was not significant, this does not exclude the possibility that ZAC1 is involved in the regulation of insulin during post-natal life. The contribution of maternal nutrition during embryogenesis and fetal development to the methylation of imprinted genes is currently the subject of debate. Here, we observed a positive association between ZAC1 DMR methylation and maternal intake of alcohol, and vitamins B2 and 12.
Methods
The data reported are from the EDEN population-based prospective cohort study of children, the general aim of which is to study the pre- and early post-natal determinants of child development and health. The study has been approved by the Medical Ethics Committee of the Kremlin Bicêtre Hospital. Written consent was obtained from the mother both for herself at enrolment and for her newborn child after delivery.
Study design
Pregnant women attending a prenatal visit at the departments of Obstetrics and Gynaecology of the University Hospitals of Nancy and Poitiers (France) before 24 wk of gestation were invited to participate. Enrolment started in 2003, in February in Poitiers and in September in Nancy; enrolment lasted 27 mo in each center. Exclusion criteria were twin pregnancies, known diabetes before pregnancy, not being able to speak and read French, and intention to move away from the region. Among eligible women, 55% agreed to participate. Thus, 2002 women were enrolled in the study (969 in Poitiers, 1033 in Nancy).
Data at birth were available for 1896 of the 2002 women included in the study after accounting for those who decided to withdraw from the study, those lost to follow-up and miscarriages or fetal deaths. A subsample of the cohort was selected for bone mass investigations, and research on growth factors and methylation of growth-related genes. This subsample comprised children of all mothers enrolled after April 2005 who had a follow-up of at least one year after birth. This subsample included 378 children [158 (42%) Poitiers, 220 (58%) Nancy].
Clinical measurements
Standard ultrasound fetal measurements were recorded from routine examinations performed at 22 wk (20 to 24) and at 32 wk (30 to 34) of gestation. Measurements included biparietal diameter, head, and abdominal circumferences and femur length. All ultrasound examinations were performed by a small number of specialists who standardized procedures before the study. At a visit between 24 and 28 wk of gestation by midwife research assistants, maternal height was measured with a wall Seca 206 stadiometer to the nearest 0.2 cm and maternal weight was measured using electronic Terraillon SL 351 scales (Hanson Ltd) to the nearest 0.1 kg. Weight before pregnancy, educational level and smoking habits during pregnancy were obtained by interview. Pre-pregnancy BMI was computed as reported weight (kg)/ measured height squared (m2). According to the reference values of the International Obesity Task Force, maternal obesity was defined as a BMI of 30 kg/m2 and above.
Gestational age at delivery was determined from the date of the last menstrual period or early ultrasound assessment. In the two obstetric departments, electronic Seca scales (Seca 737 in Nancy and Seca 335 in Poitiers) were used to measure the infant’s weight and a wooden somatometer (Testut) to measure length. Birth weight and length were extracted from clinical records. When the child was 1 y old, research midwives measured the weight of the mothers alone, then holding their infant, wearing light clothes using the same Terraillon SL-351 scales used for weighing the mother after birth. Infant weight was obtained by subtraction. A somatometer was used to measure infant length with a precision of 5 mm (NM Medical). Questionnaires at 4, 8, and 12 mo were used to assess breastfeeding.
Dietary data
Mothers completed two food frequency questionnaires (FFQs) similar to the questionnaire developed for the French population for a previous study. This food frequency questionnaire has been validated against a series of 24 h recalls.40 It inquires about the intake of 137 different foods or food groups with a 7-item scale ranking from never to more than once a day.
A first FFQ (completed at recruitment, at 15 WG on average) addressed the usual diet during the year prior to pregnancy; the second FFQ (completed in the first few days following delivery) was related to food intake during the last three months of pregnancy. To compute energy and nutrient intakes, we multiplied, for each food, the intake frequency by the nutrient composition for a portion as described previously.41
The first FFQ also included questions about the use of dietary supplements within three months prior to pregnancy and the second FFQ about the use of dietary supplements during gestation. Questions inquired about the duration of use during pregnancy and whether or not the supplements were taken as a mixture. If not, a list of vitamins and minerals was provided for the type of supplements taken to be declared.
Laboratory measurements
At birth, cord blood samples were collected by midwives. DNA was extracted from leukocytes by standard procedures with the QIAamp DNA Blood Mini Kit (QIAGEN). After centrifugation, serum was aliquoted and stored at –80 °C until thawing for analyses. The median time between cord blood collection and DNA freezing was 22 h (interquatile range: 5–33 h).
DNA methylation analysis
Methylation of the ZAC1 DMR was analyzed by allele-specific methylated multiplex real-time quantitative PCR (ASMM RTQ-PCR) as previously described.23,42,43 The methylation index (MI) was calculated as the ratio between the methylated and unmethylated alleles as follows: [(amount of methylated allele × 100)/(amount of methylated allele + amount of unmethylated allele)]. The intra- and inter-assay coefficients of variation (calculated from five independent runs) were 1.7% and 2.2%, respectively.
C-peptide measurement
C-peptide (nanomoles per liter, nmol/L) was determined by immunochemiluminescent immunoassays performed on the LIAISON platform manufactured by DiaSorin (Sallugia, Italy). The detection limit was 0.01 nmol/L. Intra- and inter-assay coefficients of variation were 4.0 and 6.8%, respectively
Statistical analysis
Calculated variables: Gestational age and sex specific standard deviations were constructed for all fetal growth measurements based on data from the whole EDEN study population (n = 1799 to 1921 depending on the measurements). All ultrasound measures are expressed as internal SD-scores in the analysis. Estimated Fetal Weight (EFW) was calculated using the formula of Hadlock et al.44 The birth and one year anthropometric measurements were expressed as Z-scores according to the WHO growth standards (http://www.who.int/growthref/en/).45 The breastfeeding information in the various questionnaires were used to create a variable “any breastfeeding since birth.” Nutrient density was calculated as nutrient intake (in g/day) per 100 Kcal of energy intake. Mothers were classified into three smoking habit groups: mothers who did not smoke during pregnancy, mothers who stopped smoking in the first trimester and mothers who smoked in the second and/or third trimester.
Results are expressed as means (SD), unless otherwise indicated. To compare characteristics of the subsample with those of the remaining EDEN population, or methylation status by supplement use, we used a t test or the Mann-Whitney U-test for continuous variables, and the chi-square test for categorical variables. Associations between the ZAC1 DMR MI in cord blood and fetal and infant anthropometric, dietary or biological variables were assessed with Spearman’s rank partial correlations. Correlations were adjusted for center, child’s gender and gestational age or child’s age. All statistical tests were two-sided, and a P value of less than 0.05 was considered significant. All analyses were performed with SAS version 9.2 software.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the heads of the maternity units in Poitiers and Nancy, the local investigators, the EDEN mother-child study group for their contribution to the EDEN study (www.eden.vjf.inserm.fr) and most of all the participating families. We acknowledge all funding sources for the EDEN study: Fondation pour la Recherche Médicale (FRM), the French Ministry of Research, the Federative Research Institutes and Cohort Program, INSERM Human Nutrition National Research Program, and Diabetes National Research Program (through a collaboration with the French Association of Diabetic Patients (AFD)), the French Ministry of Health, the French Agency for Environment Security (AFSSET), the French National Institute for Population Health Surveillance (InVS), Paris–Sud University, the French National Institute for Health Education (INPES), Nestlé, Mutuelle Générale de l’Education Nationale (MGEN), the French-speaking association for the study of diabetes and metabolism (ALFEDIAM), the National Agency for Research (ANR non thematic program), and the National Institute for Research in Public health (IRESP: TGIR cohorte santé 2008 program), recurrent Institut National de la Santé et de la Recherche Médicale (INSERM) and Université Pierre et Marie Curie (UPMC) funding; and Agence Nationale de la Recherche (ANR) (EPIFEGRO 2010). Azzi S was supported by ANR (EPIFEGRO 2010) and Sas T by an ESPE sabbatical grant and a Growth Hormone Research society grant.
References
- 1.Gicquel C, El-Osta A, Le Bouc Y. Epigenetic regulation and fetal programming. Best Pract Res Clin Endocrinol Metab. 2008;22:1–16. doi: 10.1016/j.beem.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 2.Kirkbride JB, Susser E, Kundakovic M, Kresovich JK, Davey Smith G, Relton CL. Prenatal nutrition, epigenetics and schizophrenia risk: can we test causal effects? Epigenomics. 2012;4:303–15. doi: 10.2217/epi.12.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim U, Song MA. Dietary and lifestyle factors of DNA methylation. Methods Mol Biol. 2012;863:359–76. doi: 10.1007/978-1-61779-612-8_23. [DOI] [PubMed] [Google Scholar]
- 4.Wilhelm-Benartzi CS, Houseman EA, Maccani MA, Poage GM, Koestler DC, Langevin SM, Gagne LA, Banister CE, Padbury JF, Marsit CJ. In utero exposures, infant growth, and DNA methylation of repetitive elements and developmentally related genes in human placenta. Environ Health Perspect. 2012;120:296–302. doi: 10.1289/ehp.1103927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler MG. Genomic imprinting disorders in humans: a mini-review. J Assist Reprod Genet. 2009;26:477–86. doi: 10.1007/s10815-009-9353-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirasawa R, Feil R. Genomic imprinting and human disease. Essays Biochem. 2010;48:187–200. doi: 10.1042/bse0480187. [DOI] [PubMed] [Google Scholar]
- 7.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 8.Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forné T, Jammes H, Ainscough JF, Surani MA, Journot L, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–21. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 9.Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11:711–22. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Valente T, Auladell C. Expression pattern of Zac1 mouse gene, a new zinc-finger protein that regulates apoptosis and cellular cycle arrest, in both adult brain and along development. Mech Dev. 2001;108:207–11. doi: 10.1016/S0925-4773(01)00492-0. [DOI] [PubMed] [Google Scholar]
- 11.Valente T, Junyent F, Auladell C. Zac1 is expressed in progenitor/stem cells of the neuroectoderm and mesoderm during embryogenesis: differential phenotype of the Zac1-expressing cells during development. Dev Dyn. 2005;233:667–79. doi: 10.1002/dvdy.20373. [DOI] [PubMed] [Google Scholar]
- 12.Varrault A, Ciani E, Apiou F, Bilanges B, Hoffmann A, Pantaloni C, Bockaert J, Spengler D, Journot L. hZAC encodes a zinc finger protein with antiproliferative properties and maps to a chromosomal region frequently lost in cancer. Proc Natl Acad Sci U S A. 1998;95:8835–40. doi: 10.1073/pnas.95.15.8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arima T, Drewell RA, Oshimura M, Wake N, Surani MA. A novel imprinted gene, HYMAI, is located within an imprinted domain on human chromosome 6 containing ZAC. Genomics. 2000;67:248–55. doi: 10.1006/geno.2000.6266. [DOI] [PubMed] [Google Scholar]
- 14.Arima T, Drewell RA, Arney KL, Inoue J, Makita Y, Hata A, Oshimura M, Wake N, Surani MA. A conserved imprinting control region at the HYMAI/ZAC domain is implicated in transient neonatal diabetes mellitus. Hum Mol Genet. 2001;10:1475–83. doi: 10.1093/hmg/10.14.1475. [DOI] [PubMed] [Google Scholar]
- 15.Temple IK, Shield JP. Transient neonatal diabetes, a disorder of imprinting. J Med Genet. 2002;39:872–5. doi: 10.1136/jmg.39.12.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma D, Shield JP, Dean W, Leclerc I, Knauf C, Burcelin R Ré, Rutter GA, Kelsey G. Impaired glucose homeostasis in transgenic mice expressing the human transient neonatal diabetes mellitus locus, TNDM. J Clin Invest. 2004;114:339–48. doi: 10.1172/JCI19876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diatloff-Zito C, Nicole A, Marcelin G, Labit H, Marquis E, Bellanné-Chantelot C, Robert JJ. Genetic and epigenetic defects at the 6q24 imprinted locus in a cohort of 13 patients with transient neonatal diabetes: new hypothesis raised by the finding of a unique case with hemizygotic deletion in the critical region. J Med Genet. 2007;44:31–7. doi: 10.1136/jmg.2006.044404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arthur EI, Zlotogora J, Lerer I, Dagan J, Marks K, Abeliovich D. Transient neonatal diabetes mellitus in a child with invdup(6)(q22q23) of paternal origin. Eur J Hum Genet. 1997;5:417–9. [PubMed] [Google Scholar]
- 19.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–9. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim AL, Ng S, Leow SC, Choo R, Ito M, Chan YH, Goh SK, Tng E, Kwek K, Chong YS, et al. Epigenetic state and expression of imprinted genes in umbilical cord correlates with growth parameters in human pregnancy. J Med Genet. 2012;49:689–97. doi: 10.1136/jmedgenet-2012-100858. [DOI] [PubMed] [Google Scholar]
- 21.Cooper WN, Khulan B, Owens S, Elks CE, Seidel V, Prentice AM, Belteki G, Ong KK, Affara NA, Constância M, et al. DNA methylation profiling at imprinted loci after periconceptional micronutrient supplementation in humans: results of a pilot randomized controlled trial. FASEB J. 2012;26:1782–90. doi: 10.1096/fj.11-192708. [DOI] [PubMed] [Google Scholar]
- 22.Regnault N, Botton J, Heude B, Forhan A, Hankard R, Foliguet B, Hillier TA, Souberbielle JC, Dargent-Molina P, Charles MA, EDEN Mother-Child Cohort Study Group Higher cord C-peptide concentrations are associated with slower growth rate in the 1st year of life in girls but not in boys. Diabetes. 2011;60:2152–9. doi: 10.2337/db10-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azzi S, Steunou V, Rousseau A, Rossignol S, Thibaud N, Danton F, Le Jule M, Gicquel C, Le Bouc Y, Netchine I. Allele-specific methylated multiplex real-time quantitative PCR (ASMM RTQ-PCR), a powerful method for diagnosing loss of imprinting of the 11p15 region in Russell Silver and Beckwith Wiedemann syndromes. Hum Mutat. 2011;32:249–58. doi: 10.1002/humu.21403. [DOI] [PubMed] [Google Scholar]
- 24.Albouy-Llaty M, Thiebaugeorges O, Goua V, Magnin G, Schweitzer M, Forhan A, Lelong N, Slama R, Charles MA, Kaminski M, EDEN Mother–Child Cohort Study Group Influence of fetal and parental factors on intrauterine growth measurements: results of the EDEN mother-child cohort. Ultrasound Obstet Gynecol. 2011;38:673–80. doi: 10.1002/uog.9006. [DOI] [PubMed] [Google Scholar]
- 25.Valleley EM, Cordery SF, Bonthron DT. Tissue-specific imprinting of the ZAC/PLAGL1 tumour suppressor gene results from variable utilization of monoallelic and biallelic promoters. Hum Mol Genet. 2007;16:972–81. doi: 10.1093/hmg/ddm041. [DOI] [PubMed] [Google Scholar]
- 26.Pagotto U, Arzberger T, Ciani E, Lezoualc’h F, Pilon C, Journot L, Spengler D, Stalla GK. Inhibition of Zac1, a new gene differentially expressed in the anterior pituitary, increases cell proliferation. Endocrinology. 1999;140:987–96. doi: 10.1210/en.140.2.987. [DOI] [PubMed] [Google Scholar]
- 27.Gardner RJ, Mackay DJ, Mungall AJ, Polychronakos C, Siebert R, Shield JP, Temple IK, Robinson DO. An imprinted locus associated with transient neonatal diabetes mellitus. Hum Mol Genet. 2000;9:589–96. doi: 10.1093/hmg/9.4.589. [DOI] [PubMed] [Google Scholar]
- 28.Mackay DJ, Coupe AM, Shield JP, Storr JN, Temple IK, Robinson DO. Relaxation of imprinted expression of ZAC and HYMAI in a patient with transient neonatal diabetes mellitus. Hum Genet. 2002;110:139–44. doi: 10.1007/s00439-001-0671-5. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann A, Spengler D. Transient neonatal diabetes mellitus gene Zac1 impairs insulin secretion in mice through Rasgrf1. Mol Cell Biol. 2012;32:2549–60. doi: 10.1128/MCB.06637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamikihara T, Arima T, Kato K, Matsuda T, Kato H, Douchi T, Nagata Y, Nakao M, Wake N. Epigenetic silencing of the imprinted gene ZAC by DNA methylation is an early event in the progression of human ovarian cancer. Int J Cancer. 2005;115:690–700. doi: 10.1002/ijc.20971. [DOI] [PubMed] [Google Scholar]
- 31.Metz C, Cavé H, Bertrand AM, Deffert C, Gueguen-Giroux B, Czernichow P, Polak M, NDM French Study Group. Neonatal diabetes mellitus Neonatal diabetes mellitus: chromosomal analysis in transient and permanent cases. J Pediatr. 2002;141:483–9. doi: 10.1067/mpd.2002.127089. [DOI] [PubMed] [Google Scholar]
- 32.O’Rahilly S, Burnett MA, Smith RF, Darley JH, Turner RC. Haemolysis affects insulin but not C-peptide immunoassay. Diabetologia. 1987;30:394–6. doi: 10.1007/BF00292540. [DOI] [PubMed] [Google Scholar]
- 33.Fösel S. Transient and permanent neonatal diabetes. Eur J Pediatr. 1995;154:944–8. doi: 10.1007/BF01958635. [DOI] [PubMed] [Google Scholar]
- 34.Temple IK, Gardner RJ, Robinson DO, Kibirige MS, Ferguson AW, Baum JD, Barber JC, James RS, Shield JP. Further evidence for an imprinted gene for neonatal diabetes localised to chromosome 6q22-q23. Hum Mol Genet. 1996;5:1117–21. doi: 10.1093/hmg/5.8.1117. [DOI] [PubMed] [Google Scholar]
- 35.Bock T, Kyhnel A, Pakkenberg B, Buschard K. The postnatal growth of the beta-cell mass in pigs. J Endocrinol. 2003;179:245–52. doi: 10.1677/joe.0.1790245. [DOI] [PubMed] [Google Scholar]
- 36.Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev. 2002;117:15–23. doi: 10.1016/S0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 37.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14:R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 38.Ivanova E, Chen JH, Segonds-Pichon A, Ozanne SE, Kelsey G. DNA methylation at differentially methylated regions of imprinted genes is resistant to developmental programming by maternal nutrition. Epigenetics. 2012;7:1200–10. doi: 10.4161/epi.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radford EJ, Isganaitis E, Jimenez-Chillaron J, Schroeder J, Molla M, Andrews S, Didier N, Charalambous M, McEwen K, Marazzi G, et al. An unbiased assessment of the role of imprinted genes in an intergenerational model of developmental programming. PLoS Genet. 2012;8:e1002605. doi: 10.1371/journal.pgen.1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deschamps V, de Lauzon-Guillain B, Lafay L, Borys JM, Charles MA, Romon M. Reproducibility and relative validity of a food-frequency questionnaire among French adults and adolescents. Eur J Clin Nutr. 2009;63:282–91. doi: 10.1038/sj.ejcn.1602914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drouillet P, Forhan A, De Lauzon-Guillain B, Thiébaugeorges O, Goua V, Magnin G, Schweitzer M, Kaminski M, Ducimetière P, Charles MA. Maternal fatty acid intake and fetal growth: evidence for an association in overweight women. The ‘EDEN mother-child’ cohort (study of pre- and early postnatal determinants of the child’s development and health) Br J Nutr. 2009;101:583–91. doi: 10.1017/S0007114508025038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Azzi S, Rossignol S, Steunou V, Sas T, Thibaud N, Danton F, Le Jule M, Heinrichs C, Cabrol S, Gicquel C, et al. Multilocus methylation analysis in a large cohort of 11p15-related foetal growth disorders (Russell Silver and Beckwith Wiedemann syndromes) reveals simultaneous loss of methylation at paternal and maternal imprinted loci. Hum Mol Genet. 2009;18:4724–33. doi: 10.1093/hmg/ddp435. [DOI] [PubMed] [Google Scholar]
- 43.Maupetit-Méhouas S, Azzi S, Steunou V, Sakakini N, Silve C, Reynes C, Perez de Nanclares G, Keren B, Chantot S, Barlier A, et al. Simultaneous hyper- and hypomethylation at imprinted loci in a subset of patients with GNAS epimutations underlies a complex and different mechanism of multilocus methylation defect in pseudohypoparathyroidism type 1b. Hum Mutat. 2013;34:1172–80. doi: 10.1002/humu.22352. [DOI] [PubMed] [Google Scholar]
- 44.Hadlock FP, Harrist RB, Sharman RS, Deter RL, Park SK. Estimation of fetal weight with the use of head, body, and femur measurements--a prospective study. Am J Obstet Gynecol. 1985;151:333–7. doi: 10.1016/0002-9378(85)90298-4. [DOI] [PubMed] [Google Scholar]
- 45.de Onis M, Garza C, Victora CG, Onyango AW, Frongillo EA, Martines J. The WHO Multicentre Growth Reference Study: planning, study design, and methodology. Food Nutr Bull. 2004;25(Suppl):S15–26. doi: 10.1177/15648265040251S103. [DOI] [PubMed] [Google Scholar]