Abstract
Genes with altered DNA methylation can be used as biomarkers for cancer detection and assessment of prognosis. Here we analyzed the methylation status of a colorectal cancer biomarker panel (CNRIP1, FBN1, INA, MAL, SNCA, and SPG20) in 97 cancer cell lines, derived from 17 different cancer types. Interestingly, the genes were frequently methylated also in hematological cancer types and were therefore subjected to analyses in primary tumor samples from the major types of non-Hodgkin lymphomas (NHL) and in healthy controls. In total, the genes CNRIP1, FBN1, INA, MAL, SNCA, and SPG20 were methylated in 53%, 23%, 52%, 69%, 97%, and 92% of the tumor samples, respectively, and were unmethylated in all healthy controls. With the exception of a single tumor sample, a correct prediction of lymphoma or normal sample was made in a blinded analysis of the validation series using a combination of SNCA and SPG20. The combined ROC-curve analysis of these genes resulted in an area under the curve of 0.999 (P = 4.2 × 10−18), and a sensitivity and specificity of 98% and 100%, respectively, across the test and validation series. Interestingly, the promoter methylation of CNRIP1 was associated with decreased overall survival in diffuse large B-cell lymphoma (DLBCL) (P = 0.03).
In conclusion, our results demonstrate that SNCA and SPG20 methylation might be suitable for early detection and monitoring of NHL. Furthermore, CNRIP1 could potentially be used as a prognostic factor in DLBCL.
Keywords: biomarker, CNRIP1, diagnosis, epigenetic, lymphoma, MAL, methylation, prognosis, SNCA, SPG20
Introduction
Non-Hodgkin lymphoma (NHL) is the sixth most common cancer type in the United States with 69 740 new cases per year (2013).1 Over 80% of these cases are diagnosed as B-cell lymphomas. B-cell lymphomas are divided into different types, based on morphology, immunophenotype, and genetic and clinical criteria.2 Lymphomagenesis is a multistep process and the consequence of the accumulation of genetic and epigenetic changes,3 including transcriptional silencing of tumor-suppressor genes by CpG-island promoter hypermethylation. In lymphoma numerous cellular processes, e.g., signal transduction (DLC1, EFNA5, NR0B2), DNA repair (MGMT, MLH1, RASSF1), cell-cycle regulation (p15, p16, RBP1), differentiation (MYOD1), invasion (CDH13), and apoptosis (DAPK) have been shown to be altered by aberrant DNA methylation.4,5 In addition to cancer-type specific aberrations, e.g,. GSTP1-methylation in prostate cancer,6 many tumor suppressors are methylated across several cancer types. RASSF1A is one example, with methylation frequencies ranging from 10% to 90% in solid tumors, including lung, kidney, colorectal, and esophageal cancer, as well as in hematological malignancies such as leukemia and lymphoma.7,8
In the present study we have analyzed 97 cell lines from 17 different cancer types for a DNA-methylation-based biomarker panel consisting of CNRIP1, FBN1, INA, MAL, SNCA, and SPG20.9 In addition to the gastrointestinal tumors,9 the biomarker panel showed a high methylation frequency in hematological cancer cell lines. Using quantitative real-time methylation assays, the biomarker panel was subsequently analyzed in two series of tumor biopsies from patients with various B-cell lymphoma types as well as in samples from healthy control subsets.
Results
Promoter methylation status in cancer cell lines
The methylation frequencies of CNRIP1, FBN1, INA, MAL, SNCA, and SPG20 were analyzed by quantitative methylation-specific PCR (qMSP) in 97 cancer cell lines derived from 17 different cancer tissues. In addition to the cell lines derived from gastro-intestinal cancer,9 the hematological cancer cell lines showed surprisingly high methylation frequencies across all tested genes, indicating that these biomarkers could also perform well in hematological malignancies. In contrast, promoter methylation frequencies were low for testis, ovar-, MPNST, lung, bladder, and kidney cancer cell lines (Fig. 1).
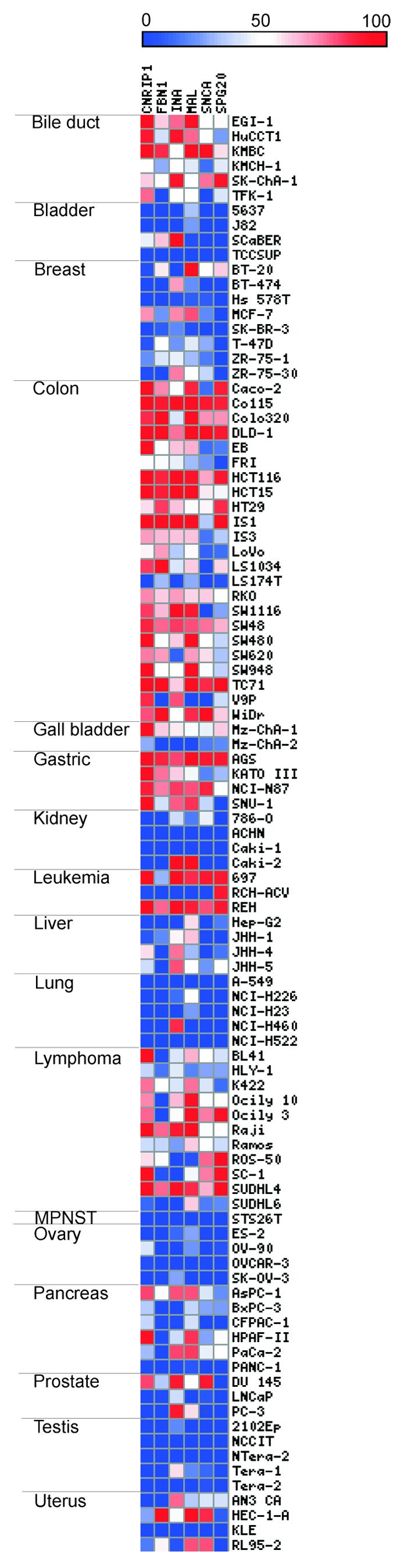
Figure 1. Methylation status of CNRIP1, FBN1, INA, MAL, SNCA, and SPG20 across 97 cancer cell lines. The six biomarkers have been analyzed by quantitative methylation-specific PCR (qMSP) in 97 different cancer cell lines originating from 17 tissues. Each cell line is represented by a square and the color scale indicates the percent of methylation reference (PMR) value. Abbreviations: MPNST, malignant peripheral nerve sheath tumor.
Quantitative promoter DNA methylation analysis in a test series of B-cell lymphoma biopsies and healthy samples
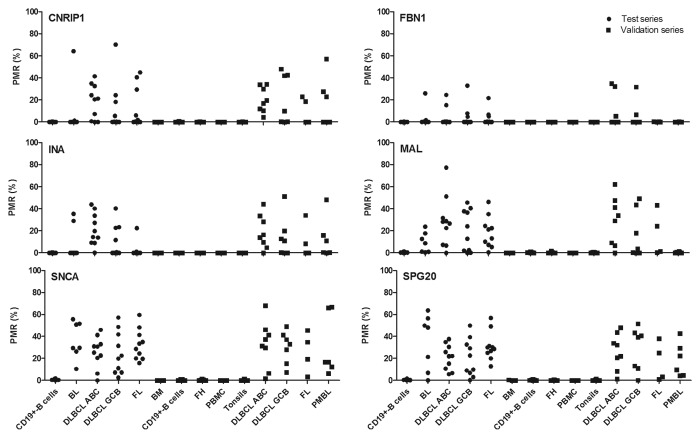
The overall promoter methylation of CNRIP1, FBN1, INA, MAL, SNCA, and SPG20 across all analyzed NHL types was 43%, 24%, 43%, 81%, 97%, and 95%, respectively (Table 1; Fig. 2). All genes were found to be unmethylated in control CD19+-B cells isolated from healthy individuals (with PMR values ranging from 0 to 1.80%). The promoter region of SNCA and SPG20 showed the highest methylation frequencies across all analyzed lymphoma types and 100% of the NHL samples showed methylation of at least one of these two genes, with a specificity of 100%.
Figure 2. Percent promoter methylation of the analyzed genes in the sample series. The biomarker panel has been analyzed in control and tumor samples by quantitative methylation specific PCR (qMSP) in a test and validation series. Each dot represents one sample (lymphoma or control). Abbreviations: BL, Burkitt’s lymphoma; BM, Bone marrow; DLBCL ABC, activated B-cell like diffuse large B-cell lymphoma; DLBCL GCB, germinal center B-cell like diffuse large B-cell lymphoma; FH, follicular hyperplasia; FL, follicular lymphoma; PBMC, peripheral blood mononuclear cells; PMBL, primary mediastinal B-cell lymphoma; PMR, percent methylated reference.
Verification of qMSP results in a validation series - blinded analysis
Using the threshold for scoring methylation-positive samples established from the test series [percent of methylated reference (PMR) ≥ 2 for all genes], we performed a blinded analysis of a validation series, that included NHL samples and additional controls such as bone marrow, follicular hyperplasia (FH), tonsils, CD19+-B cells and peripheral blood mononuclear cells (PBMCs) (Table 1; Fig. 2). With the exception of a single lymphoma sample, the methylation status of SNCA and SPG20 was sufficient to distinguish all patients from healthy donors.
Table 1. Methylation frequencies assessed by qMSP in the test and validation series.
| Test series | BL | DLBCL ABC | DLBCL GCB | FL | NHL |
|---|---|---|---|---|---|
| CNRIP1 | 1/7 (14%) | 7/10 (70%) | 4/10 (40%) | 4/10 (40%) | 16/37(43%) |
| FBN1 | 1/7 (14%) | 2/10 (20%) | 3/10 (30%) | 3/10 (30%) | 9/37 (24%) |
| INA | 2/7 (29%) | 9/10 (90%) | 4/10 (40%) | 1/10 (10%) | 16/37(43%) |
| MAL | 4/7 (57%) | 9/10 (90%) | 8/10 (80%) | 9/10 (90%) | 30/37 (81%) |
| SNCA | 7/7 (100%) | 9/10 (90%) | 10/10 (100%) | 10/10 (100%) | 36/37 (97%) |
| SPG20 | 6/7 (86%) | 10/10 (100%) | 9/10 (90%) | 10/10 (100%) | 35/37 (95%) |
| Biomarker panel | 100% | 100% | 100% | 100% | 100% |
| Validation series | DLBCL ABC | DLBCL GCB | FL | PMBL | NHL |
| CNRIP1 | 8/8 (100%) | 4/7 (57%) | 2/4 (50%) | 3/6 (50%) | 17/25 (68%) |
| FBN1 | 3/8 (38%) | 2/7 (29%) | 0/4 (0%) | 0/6 (0%) | 5/25 (20%) |
| INA | 7/8 (88%) | 4/7 (57%) | 2/4 (50%) | 3/6 (50%) | 16/25 (64%) |
| MAL | 7/8 (88%) | 4/7 (57%) | 2/4 (50%) | 0/6 (0%) | 13/25 (52%) |
| SNCA | 7/8 (88%) | 7/7 (100%) | 4/4 (100%) | 6/6 (100%) | 24/25 (96%) |
| SPG20 | 7/8 (88%) | 6/7 (86%) | 3/4 (75%) | 6/6 (100%) | 22/25 (88%) |
| Biomarker panel | 100% | 100% | 100% | 100% | 100% |
The various healthy controls were unmethylated for all analyzed markers. The biomarker panel was considered to be positive when a minimum of one out of the six analyzed genes was methylated. Abbreviations: Burkitt’s lymphoma (BL), diffuse large B-cell lymphoma (DLBCL) activated B-cell-like type (ABC), germinal center B-cell-like type (GCB), follicular lymphoma (FL), primary mediastinal B-cell lymphoma (PMBL) and non-Hodgkin lymphoma (NHL).
The promoter-methylation frequency of CNRIP1, FBN1, INA, MAL, SNCA, and SPG20 across all NHL types in the validation series was 68%, 20%, 64%, 52%, 96%, and 88%, respectively. All genes were unmethylated in the healthy samples (with PMR values ranging from 0 to 1.43%) as well as in the follicular hyperplasia samples (Fig. 2). No statistical difference was seen between the test and validation series. The association between promoter methylation status and gene expression has previously been reported for all genes in a series of cancer cell lines.9-11
Receiver operating characteristics (ROC) curves
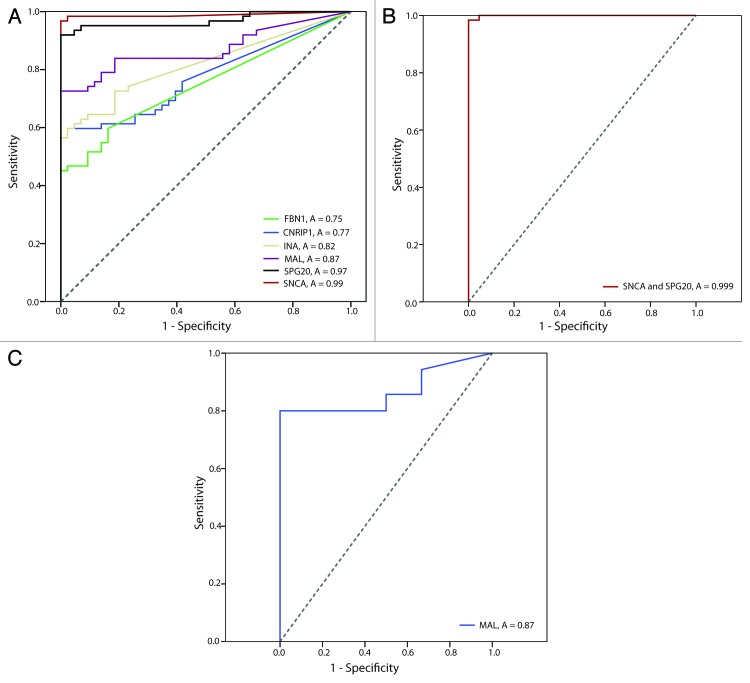
Receiver operating characteristics (ROC) curves were generated based on the PMR values from the qMSP analyses in the combined test and validation series. The area under the curve for SNCA, SPG20, MAL, INA, CNRIP1, and FBN1 was 0.99, 0.97, 0.87, 0.82, 0.77, and 0.75, respectively (Fig. 3 A). Combining SNCA and SPG20 (based on the sum of the PMR values) resulted in an area under the curve of 0.999 (Fig. 3B).
Figure 3. Receiver Operating Characteristics (ROC) curves for individual and combined markers in lymphoma patients vs. healthy donors. The area under the ROC curve (AUC; A) represents how accurate the individual and combined biomarkers can discriminate between lymphomas and normal samples. (A) Lymphoma patients vs. healthy donors for individual genes. (B) Lymphoma patients vs. healthy donors for the genes SNCA and SPG20. (C) DLBCL ABC (activated B-cell like diffuse large B-cell lymphoma) and GCB (germinal center B-cell like diffuse large B-cell lymphoma) vs. PMBL (primary mediastinal B-cell lymphoma) for the MAL gene.
Because the expression of the MAL protein has been reported to be restricted to primary mediastinal B-cell lymphoma (PMBL),12 we expected MAL to be unmethylated among these samples. Indeed, all PMBLs analyzed in the present study had unmethylated MAL promoters, in contrast to the other diffuse large B-cell lymphoma (DLBCL) activated B-cell like (ABC); 88% and DLBCL germinal center B-cell like (GCB); 71%). We further used ROC-curve analysis to estimate how suitable the methylation status of MAL was for separating PMBLs from DLBCL ABC and GCB, which resulted in an area under the curve of 0.87 (Fig. 3C).
Methylation status as a predictor of poor outcome in B-cell lymphoma patients
The methylation status of each gene was analyzed for predicting clinical outcome in DLBCL patients. We observed that promoter methylation of CNRIP1 was significantly associated with a worse overall survival (P = 0.03) in DLBCL (Log-rank test; Fig. 4). The combined promoter methylation of CNRIP1 and MAL achieved a P value of 0.01. For MAL alone as well as INA alone, a trend toward poor overall survival was seen for methylated gene promoters, although this was not significant (P = 0.075 and P = 0.055, respectively).
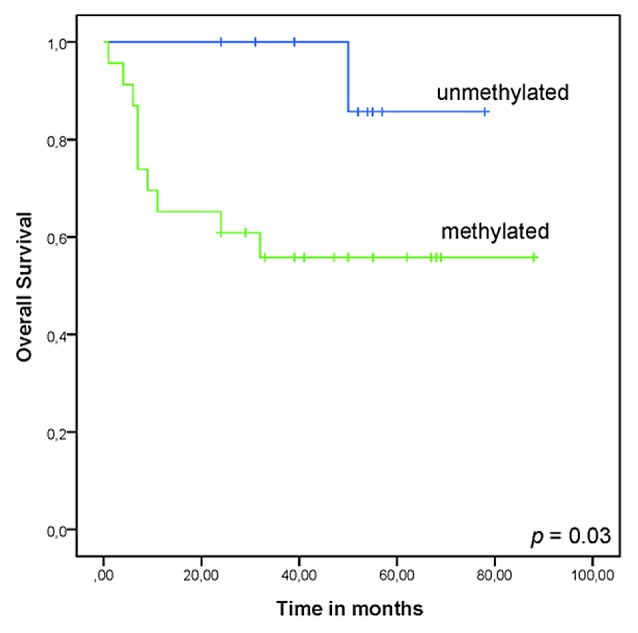
Figure 4.CNRIP1 methylation status has prognostic value in DLBCL ABC (activated B-cell like diffuse large B-cell lymphoma) and GCB (germinal center B-cell like diffuse large B-cell lymphoma). The overall survival was analyzed using the Kaplan-Meier-method and the log-rank test. Survival was calculated in months from date of diagnosis to last follow up.
Discussion
In the present study, we demonstrated that the promoters of CNRIP1, FBN1, INA, MAL, SNCA, and SPG20 were frequently methylated in lymphoma biopsies from patients with various types of B-cell lymphomas (BL, DLBCL ABC, DLBCL GCB, PMBL, and FL). At the same time these genes showed no methylation across normal cell types from healthy donors, as well as follicular hyperplasia samples; indicating that the methylation was cancer specific. Interestingly, the methylation status of SNCA and SPG20 was sufficient to separate lymphoma biopsies from healthy donors in a blinded analysis with an accuracy of 98%.
Although the list of methylated genes in NHL is constantly increasing, only a few have been shown to be methylated across the various types.13-20 This might, in part, be explained by the fact that most methylation studies are focusing on a single NHL type21-23 or, alternatively, the differences between two NHL types.24,25 Genes frequently methylated across the majority of NHL biopsies, and not restricted to specific NHL types, represent novel biomarkers for NHL in general and may become valuable diagnostic and/or disease monitoring tools. Promoter methylation of the DLC1 gene was recently proposed as a non-invasive epigenetic biomarker for lymphoma.26 This gene is methylated across several tumor types, e.g., gastric and breast cancer,27,28 in addition to different Hodgkin’s and non-Hodgkin lymphomas with methylation frequencies ranging from 60% to 90%. Ying and co-workers validated the tumor specificity of DLC1 promoter methylation in various lymphomas and controls.26 Using an MSP-assay, they were able to detect 44% of primary HL biopsies. Another study reporting on the methylation of p57KIP2 suggested that this is a sensitive biomarker to detect minimal residual disease in DLBCL. They were able to discriminate 80% of the DLBCL patients from healthy controls.29 In the present study we showed that the genes SNCA and SPG20 were methylated in close to all (98%) NHL samples comprising five different types. Furthermore, the genes showed 100% specificity with no methylation in various sources of healthy donors. Receivers operating characteristic (ROC) curves are frequently used to estimate how suitable potential biomarkers are in separating malignant from non-malignant samples. In the present study, combining the methylation status of SNCA and SPG20 resulted in close to the maximum possible area under the ROC curve (0.999). We have altogether analyzed 62 NHL patients including the major types of NHL (FL and DLBCL) as well as BL and PMBL. Although this covers about 60% of all NHL cases we cannot exclude that the methylation frequency of the genes in question might differ in the NHL types not included here.
In contrast with all other NHL types, the MAL gene promoter of PMBLs was unmethylated. Although only six PMBLs were included in the study, this methylation pattern is in accordance with previously reported data, showing that the MAL protein is expressed in PMBLs,12,30 but not in other NHL types. Of interest, the MAL protein is expressed by most normal B lymphocytes. Loss of expression in most NHL is most likely due to gene promoter methylation as shown in this study. Why the MAL promoter is not methylated leaving MAL expression intact in PMBL is not known. Even though it has been shown that the MAL gene is a known tumor suppressor in colorectal cancer,10 and hypermethylated also in breast cancer,31 the functional role of MAL expression in PMBL remains unclear.
Epigenetically silenced genes frequently represent tumor-suppressor genes, e.g. CDKN2, RB, and DAPK.32,33 Several of the epigenetically altered genes identified in lymphoma have additionally been shown to have a functional role in lymphomagenesis, including KLF4, BMP6, and TP73.17,34-36 Although it is tempting to speculate that the analyzed genes in this study might have a role in tumor development, these genes have so far not been directly linked to lymphomagenesis. Nevertheless, reports show that FBN1 and SPG20 are involved in the TGF-β and BMP signaling pathways in different cell types,37 and we recently showed that the Spartin protein, encoded by SPG20, is an important player in cytokinesis.11 Furthermore, FBN1, SPG20, and SNCA have been shown to be methylated in prostate,38 renal,39 and breast cancer,40 respectively. SNCA has also been shown to be hypomethylated in the brain and blood of patients with Parkinson disease.41-43 The protein encoded by INA has previously been shown to regulate cell-cell interactions, which is a fundamental component of tumor growth and invasion.3,44 Hence, a role in lymphomagenesis for these genes cannot be excluded.
In addition to serving as diagnostic markers, gene-specific DNA methylation has been shown to be useful for prognostication of several cancer types.45 For instance, the methylation of CDKN2A and CDKN2B is characteristic for different hematological malignancies; leukemias are frequently associated with CDKN2A methylation whereas lymphomas often harbor CDKN2B methylation.5 The prognostic value of both genes has been shown in the various hematological malingnancies.21,46-48 In a recent study, De et al. used a methylation heterogeneity score to demonstrate that the extent of aberrant DNA methylation is associated with worse overall survival and progression-free survival in DLBCL.49 Furthermore, in DLBCL patients, promoter methylation of MGMT was associated with an improved overall as well as progression-free survival.50-52 In the present study, methylation of CNRIP1 showed a significant prognostic value in DLBCL of GCB and ABC gene expression phenotype where promoter methylation led to a worse overall survival in the analyzed patient cohort. To confirm these data, further studies including larger patient cohorts are warranted. Due to new treatment regimens, especially the introduction of rituximab in therapy protocols, several of the established markers have lost their prognostic potential.53,54 Therefore, identification of new prognostic markers that can guide the treatment strategy for NHL patients in the rituximab era is a major goal. Additionally, such biomarkers can be used to monitor cancer therapy with demethylating drugs, that have already shown promising results for acute myeloid leukemia (AML) and, in particular, myelodysplastic syndromes (MDS).55-57
In summary, the promoter methylation of SPG20 and SNCA showed high sensitivity and an excellent ability to discriminate lymphoma from healthy control samples. Furthermore, the promoter methylation of CNRIP1 was significantly associated with poor overall survival.
Materials and Methods
Cancer cell lines and patient material
DNA from 97 cell lines derived from 17 different tumor tissues (gastrointestinal cancer [bile duct, colon, gall bladder, gastric, liver, pancreas], breast cancer, germ-line [testicular] cancer, gynecological cancer [ovary, uterus], hematological malignancies [leukemia, lymphoma], lung cancer, malignant peripheral nerve sheath tumor, urological cancer [bladder, kidney, prostate]) was included in the present study. All cell lines were recently authenticated using the AmpFLSTR Identifiler PCR Amplification Kit (Applied Biosystems) and verified by comparing the STR loci results to the cell line databases (ATCC, DSMZ). The non-commercially available cell lines have also been tested and the STR loci results can be given on request.
The patient materials consisted of a test and a validation series and the material was obtained from the Department of Pathology, Oslo University Hospital, Norway (45 cases) and from the Department of Pathology, University Hospital of Leuven, Belgium (17 cases). The test series comprised 37 patients diagnosed with B-cell lymphoma (DLBCL ABC, n = 10; DLBCL GCB, n = 10; FL, n = 10; and BL, n = 7) and CD19+-B cells isolated from buffy coats of 10 healthy donors.
The validation series included material from 25 patients (DLBCL ABC, n = 8; DLBCL GCB, n = 7, FL, n = 4, and PMBL, n = 6) as well as several control samples representing different B-cell developmental stages (bone marrow, n = 3; tonsils, n = 10; peripheral blood mononuclear cells (PBMC), n = 10, CD19+-B cells isolated from buffy coat, n = 10) and follicular hyperplasia samples (FH), (n = 9). Due to the rareness of BL we were unable to include patient material from this group in the validation series.
The median observation time was 36 months, eight out of 62 patients (13%) died. Clinical information about the included patients can be found in Table 2. Patients with DLBCL were treated with CHOP-like therapy plus rituximab, patients with BL according to an intensified chemotherapy regimen with rituximab (GMALL 2002) and FL patients, if in need of therapy, with rituximab monotherapy, CVP plus rituximab or CHOP plus rituximab. All samples analyzed in the present study have been collected at diagnosis, prior to patient treatment. Furthermore, all patients have signed an informed consent and the project has been approved by the Regional Ethics Committee (S-05145).
Table 2. Patient characteristics.
| Test series | Validation series | |
|---|---|---|
| Number of patients | 37 | 25 |
| Number of BL | 7 | 0 |
| IPI low (0–2) | 4 | |
| IPI high (3–5) | 3 | |
| Stage 1–2 | 4 | |
| Stage 3–4 | 3 | |
| Number of DLBCL ABC | 10 | 8 |
| IPI low (0–2) | 4 | 2 |
| IPI high (3–5) | 3 | 2 |
| Stage 1–2 | 3 | 2 |
| Stage 3–4 | 7 | 6 |
| Number of DLBCL GCB | 10 | 7 |
| IPI low (0–2) | 7 | 3 |
| IPI high (3–5) | 2 | 1 |
| Stage 1–2 | 3 | 3 |
| Stage 3–4 | 6 | 4 |
| Number of FL | 10 | 4 |
| FLIPI low (0–2) | 7 | 1 |
| FLIPI high (3–5) | 3 | 1 |
| Stage 1–2 | 3 | 1 |
| Stage 3–4 | 7 | 3 |
| Number of PMBL | 0 | 6 |
| IPI low (0–2) | 2 | |
| IPI high (3–5) | 0 | |
| Stage 1–2 | 4 | |
| Stage 3–4 | 2 | |
| Male/female quotient | 3,1 | 0,9 |
| Median age, year (range) | 61 (34–73) | 55 (29–81) |
The international prognostic index (IPI) and follicular lymphoma IPI (FLIPI) or stage status could not be obtained from every patient. Abbreviations: Burkitt’s lymphoma (BL), diffuse large B-cell lymphoma (DLBCL) activated B-cell type (ABC), germinal center B-cell type (GCB), follicular lymphoma (FL), and primary mediastinal B-cell lymphoma (PMBL).
Nucleic acid isolation
DNA from lymphoma biopsies was isolated using the AllPrep DNA/RNA Kit from Qiagen. DNA from cell lines was isolated either with Phenol-Chloroform, the AllPrep DNA/RNA/Protein Kit from Qiagen or the Maxwell® 16 Tissue DNA Purification Kit from Promega. Resulting DNA concentration and quality was measured using the ND-1000 Nanodrop.
Bisulfite treatment
The EpiTect bisulfite kit from Qiagen was used to treat 1.3 µg DNA from each sample, following the manufacturer’s protocol. After standard clean-up, the bisulfite-treated DNA was eluted in 40 µl elution buffer. The treatment results in the conversion of unmethylated cytosines to uracils, which are amplified as thymines during PCR. Methylated cytosines are not converted during this protocol, and are therefore left as cytosines.
Quantitative methylation-specific PCR (qMSP)
For the qMSP reaction we used 30 ng of bisulfite-treated DNA for each individual 384-well plate reaction. In addition, TaqMan Universal PCR Master Mix No AmpErase UNG (Applied Biosystems), 0.9 µM of forward and reverse primer and 0.2 µM probe was used in the qMSP reaction with a total well-volume of 10 µl (patient samples) or 20 µl (cancer cell lines). Sequences for primers and probes are provided in Table S1. The PCR reaction was performed with the following conditions: incubation step at 95 °C for 10 min, followed by 45 cycles of 95 °C for 15 s and 60 °C for 1 min. The samples were run in triplicates on a 7900HT Fast Real-Time PCR System from Applied Biosystems, and the median value was used for further calculations. Samples with amplification after cycle 35 were censored according to the manufacturer’s protocol. Serial dilutions (32.5 – 0.052 ng) of bisulfite-converted completely methylated DNA (IVD; CpGenome Universal Methylated DNA, Millipore) were used to construct a standard curve to determine the quantity of fully methylated DNA in each reaction. Each reaction was normalized for DNA input using ALU-C4 as a reference.58 For each gene in each sample, the amount of DNA methylation (PMR, percent of methylated reference) was calculated relative to the fully methylated reference (IVD) according to the following: PMR = [(gene/ALU)sample/(gene/ALU)IVD]* 100. The samples were finally dichotomized into a methylated or unmethylated group based on a fixed threshold (PMR ≥ 2), selected based on the highest PMR value across the healthy controls from the test series. This threshold ensured a high specificity; all samples with PMR of two or higher were considered to be methylated. Finally, in addition to several water blanks, the following controls were included in each plate: bisulfite modified unmethylated DNA (normal blood) which served as a methylation negative control, as well as unmodified DNA, which served as a quality control for primer and probes (to ensure that unmodified DNA is not amplified and misinterpreted as methylated DNA).
Prediction of survival by analysis of the promoter-methylation status
The effect of promoter-methylation status on survival was estimated for the 35 DLCBL patients of subtypes ABC and GCB from the combined test and validation data set (for other subtypes, the number of patients/events were too low for analyses). The analysis was performed for the CNRIP1, FBN1, INA, and MAL genes; for SNCA and SPG20 the numbers of non-methylated cases were too low.
Overall survival was calculated from the date of diagnosis until the last follow up or death from any cause. Survival curves were calculated using the Kaplan-Meier-method and were compared using the log-rank test.
Supplementary Material
Disclosure of Potential Conflicts of Interest
A US provisional patent application has been filed (INVEN-32454/US-1/PRO) covering CNRIP1, FBN1, INA, MAL, SNCA, and SPG20 as biomarkers for hematological cancers.
Acknowledgments
Financial support: This work was supported by grants from the South-Eastern Norway Regional Health Authority (ES: no. 39232, funding NB as PhD. GEL: no. 39535), and The Norwegian Cancer Society (ES: no. 33260 and GEL: PR-2008-0163).
Authorship Contributions
Bethge N and Honne H performed the experimental analyses. Scoring of data was done by Bethge N, Honne H, and Andresen K supervised by Lind G; Bethge N and Lind G. drafted the manuscript; Trøen G and Eknæs M participated in sample preparation; Liestøl K and Bethge N performed the statistical analyses; Holte H collected clinical samples, provided patient data and took part in scientific discussions; Delabie J made diagnoses and reviewed those and took part in scientific discussions; Lothe RA, Smeland EB, and Lind G conceived and designed the study, contributed in data interpretation and in writing the manuscript. All authors have read and approved the final manuscript.
Glossary
Abbreviations:
- AML
acute myeloid leukemia
- BL
Burkitt’s lymphoma
- BM
bone marrow
- DLBCL ABC
activated B-cell like (ABC) diffuse large B-cell lymphoma
- DLBCL GCB
germinal center B-cell like (GCB)
- FH
follicular hyperplasia
- FL
follicular lymphoma
- MDS
myelodysplasic syndromes
- MSP
methylation specific PCR
- NHL
Non-Hodgkin lymphoma
- PBMC
peripheral blood mononuclear cell
- PMBL
primary mediastinal B-cell lymphoma
- PMR
percent of methylated reference
References
- 1.American Cancer Society. Cancer Facts & Figures 2013 American Cancer Society. 2013 [Google Scholar]
- 2.Seto M. Genetic and epigenetic factors involved in B-cell lymphomagenesis. Cancer Sci. 2004;95:704–10. doi: 10.1111/j.1349-7006.2004.tb03249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Shi H, Guo J, Duff DJ, Rahmatpanah F, Chitima-Matsiga R, Al-Kuhlani M, Taylor KH, Sjahputera O, Andreski M, Wooldridge JE, et al. Discovery of novel epigenetic markers in non-Hodgkin’s lymphoma. Carcinogenesis. 2007;28:60–70. doi: 10.1093/carcin/bgl092. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M. Profiling aberrant DNA methylation in hematologic neoplasms: a view from the tip of the iceberg. Clin Immunol. 2003;109:80–8. doi: 10.1016/S1521-6616(03)00208-0. [DOI] [PubMed] [Google Scholar]
- 6.Harden SV, Sanderson H, Goodman SN, Partin AAW, Walsh PC, Epstein JI, Sidransky D. Quantitative GSTP1 methylation and the detection of prostate adenocarcinoma in sextant biopsies. J Natl Cancer Inst. 2003;95:1634–7. doi: 10.1093/jnci/djg082. [DOI] [PubMed] [Google Scholar]
- 7.Agathanggelou A, Cooper WN, Latif F. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005;65:3497–508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- 8.Hesson LB, Cooper WN, Latif F. The role of RASSF1A methylation in cancer. Dis Markers. 2007;23:73–87. doi: 10.1155/2007/291538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lind GE, Danielsen SA, Ahlquist T, Merok MA, Andresen K, Skotheim RI, Hektoen M, Rognum TO, Meling GI, Hoff G, et al. Identification of an epigenetic biomarker panel with high sensitivity and specificity for colorectal cancer and adenomas. Mol Cancer. 2011;10:85. doi: 10.1186/1476-4598-10-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lind GE, Ahlquist T, Kolberg M, Berg M, Eknaes M, Alonso MA, Kallioniemi A, Meling GI, Skotheim RI, Rognum TO, et al. Hypermethylated MAL gene - a silent marker of early colon tumorigenesis. J Transl Med. 2008;6:13. doi: 10.1186/1479-5876-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lind GE, Raiborg C, Danielsen SA, Rognum TO, Thiis-Evensen E, Hoff G, Nesbakken A, Stenmark H, Lothe RA. SPG20, a novel biomarker for early detection of colorectal cancer, encodes a regulator of cytokinesis. Oncogene. 2011;30:3967–78. doi: 10.1038/onc.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Copie-Bergman C, Gaulard P, Maouche-Chrétien L, Brière J, Haioun C, Alonso MA, Roméo PH, Leroy K. The MAL gene is expressed in primary mediastinal large B-cell lymphoma. Blood. 1999;94:3567–75. [PubMed] [Google Scholar]
- 13.Pike BL, Greiner TC, Wang X, Weisenburger DD, Hsu YH, Renaud G, Wolfsberg TG, Kim M, Weisenberger DJ, Siegmund KD, et al. DNA methylation profiles in diffuse large B-cell lymphoma and their relationship to gene expression status. Leukemia. 2008;22:1035–43. doi: 10.1038/leu.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Riain C, O’Shea DM, Yang Y, Le Dieu R, Gribben JG, Summers K, Yeboah-Afari J, Bhaw-Rosun L, Fleischmann C, Mein CA, et al. Array-based DNA methylation profiling in follicular lymphoma. Leukemia. 2009;23:1858–66. doi: 10.1038/leu.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang XM, Greiner TC, Bibikova M, Pike BL, Siegmund KD, Sinha UK, Müschen M, Jaeger EB, Weisenburger DD, Chan WC, et al. Identification and functional relevance of de novo DNA methylation in cancerous B-cell populations. J Cell Biochem. 2010;109:818–27. doi: 10.1002/jcb.22461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor KH, Kramer RS, Davis JW, Guo J, Duff DJ, Xu D, Caldwell CW, Shi H. Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer Res. 2007;67:8511–8. doi: 10.1158/0008-5472.CAN-07-1016. [DOI] [PubMed] [Google Scholar]
- 17.Daibata M, Nemoto Y, Bandobashi K, Kotani N, Kuroda M, Tsuchiya M, Okuda H, Takakuwa T, Imai S, Shuin T, et al. Promoter hypermethylation of the bone morphogenetic protein-6 gene in malignant lymphoma. Clin Cancer Res. 2007;13:3528–35. doi: 10.1158/1078-0432.CCR-06-2766. [DOI] [PubMed] [Google Scholar]
- 18.Eberth S, Schneider B, Rosenwald A, Hartmann EM, Romani J, Zaborski M, Siebert R, Drexler HG, Quentmeier H. Epigenetic regulation of CD44 in Hodgkin and non-Hodgkin lymphoma. BMC Cancer. 2010;10:517. doi: 10.1186/1471-2407-10-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grønbaek K, Ralfkiaer U, Dahl C, Hother C, Burns JS, Kassem M, Worm J, Ralfkiaer EM, Knudsen LM, Hokland P, et al. Frequent hypermethylation of DBC1 in malignant lymphoproliferative neoplasms. Mod Pathol. 2008;21:632–8. doi: 10.1038/modpathol.2008.27. [DOI] [PubMed] [Google Scholar]
- 20.Hagiwara K, Nagai H, Li Y, Ohashi H, Hotta T, Saito H. Frequent DNA methylation but not mutation of the ID4 gene in malignant lymphoma. J Clin Exp Hematop. 2007;47:15–8. doi: 10.3960/jslrt.47.15. [DOI] [PubMed] [Google Scholar]
- 21.Amara K, Trimeche M, Ziadi S, Laatiri A, Hachana M, Korbi S. Prognostic significance of aberrant promoter hypermethylation of CpG islands in patients with diffuse large B-cell lymphomas. Ann Oncol. 2008;19:1774–86. doi: 10.1093/annonc/mdn374. [DOI] [PubMed] [Google Scholar]
- 22.Bennett LB, Schnabel JL, Kelchen JM, Taylor KH, Guo J, Arthur GL, Papageorgio CN, Shi H, Caldwell CW. DNA hypermethylation accompanied by transcriptional repression in follicular lymphoma. Genes Chromosomes Cancer. 2009;48:828–41. doi: 10.1002/gcc.20687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi JH, Li Y, Guo J, Pei L, Rauch TA, Kramer RS, Macmil SL, Wiley GB, Bennett LB, Schnabel JL, et al. Genome-wide DNA methylation maps in follicular lymphoma cells determined by methylation-enriched bisulfite sequencing. PLoS One. 2010;5:e13020. doi: 10.1371/journal.pone.0013020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaknovich R, Geng H, Johnson NA, Tsikitas L, Cerchietti L, Greally JM, Gascoyne RD, Elemento O, Melnick A. DNA methylation signatures define molecular subtypes of diffuse large B-cell lymphoma. Blood. 2010;116:e81–9. doi: 10.1182/blood-2010-05-285320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SM, Lee EJ, Ko Y-H, Lee SH, Maeng L, Kim K-M. Prognostic significance of O6-methylguanine DNA methyltransferase and p57 methylation in patients with diffuse large B-cell lymphomas. APMIS. 2009;117:87–94. doi: 10.1111/j.1600-0463.2008.00017.x. [DOI] [PubMed] [Google Scholar]
- 26.Ying J, Li H, Murray P, Gao Z, Chen YW, Wang Y, Lee KY, Chan AT, Ambinder RF, Srivastava G, et al. Tumor-specific methylation of the 8p22 tumor suppressor gene DLC1 is an epigenetic biomarker for Hodgkin, nasal NK/T-cell and other types of lymphomas. Epigenetics. 2007;2:15–21. doi: 10.4161/epi.2.1.3883. [DOI] [PubMed] [Google Scholar]
- 27.Seng TJ, Low JSW, Li H, Cui Y, Goh HK, Wong MLY, Srivastava G, Sidransky D, Califano J, Steenbergen RD, et al. The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene. 2007;26:934–44. doi: 10.1038/sj.onc.1209839. [DOI] [PubMed] [Google Scholar]
- 28.Low JSW, Tao Q, Ng KM, Goh HK, Shu XS, Woo WL, Ambinder RF, Srivastava G, Shamay M, Chan AT, et al. A novel isoform of the 8p22 tumor suppressor gene DLC1 suppresses tumor growth and is frequently silenced in multiple common tumors. Oncogene. 2011;30:1923–35. doi: 10.1038/onc.2010.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Nagai H, Ohno T, Yuge M, Hatano S, Ito E, Mori N, Saito H, Kinoshita T. Aberrant DNA methylation of p57(KIP2) gene in the promoter region in lymphoid malignancies of B-cell phenotype. Blood. 2002;100:2572–7. doi: 10.1182/blood-2001-11-0026. [DOI] [PubMed] [Google Scholar]
- 30.Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, Chan WC, Zhao T, Haioun C, Greiner TC, et al. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med. 2003;198:851–62. doi: 10.1084/jem.20031074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horne HN, Lee PS, Murphy SK, Alonso MA, Olson JA, Jr., Marks JR. Inactivation of the MAL gene in breast cancer is a common event that predicts benefit from adjuvant chemotherapy. Mol Cancer Res. 2009;7:199–209. doi: 10.1158/1541-7786.MCR-08-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayslip J, Montero A. Tumor suppressor gene methylation in follicular lymphoma: a comprehensive review. Mol Cancer. 2006;5:44. doi: 10.1186/1476-4598-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai HC, Baylin SB. Cancer epigenetics: linking basic biology to clinical medicine. Cell Res. 2011;21:502–17. doi: 10.1038/cr.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guan H, Xie L, Leithäuser F, Flossbach L, Möller P, Wirth T, Ushmorov A. KLF4 is a tumor suppressor in B-cell non-Hodgkin lymphoma and in classic Hodgkin lymphoma. Blood. 2010;116:1469–78. doi: 10.1182/blood-2009-12-256446. [DOI] [PubMed] [Google Scholar]
- 35.Yusuf I, Kharas MG, Chen J, Peralta RQ, Maruniak A, Sareen P, Yang VW, Kaestner KH, Fruman DA. KLF4 is a FOXO target gene that suppresses B cell proliferation. Int Immunol. 2008;20:671–81. doi: 10.1093/intimm/dxn024. [DOI] [PubMed] [Google Scholar]
- 36.Kawano S, Miller CW, Gombart AF, Bartram CR, Matsuo Y, Asou H, Sakashita A, Said J, Tatsumi E, Koeffler HP. Loss of p73 gene expression in leukemias/lymphomas due to hypermethylation. Blood. 1999;94:1113–20. [PubMed] [Google Scholar]
- 37.Nistala H, Lee-Arteaga S, Smaldone S, Siciliano G, Carta L, Ono RN, Sengle G, Arteaga-Solis E, Levasseur R, Ducy P, et al. Fibrillin-1 and -2 differentially modulate endogenous TGF-β and BMP bioavailability during bone formation. J Cell Biol. 2010;190:1107–21. doi: 10.1083/jcb.201003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Yu Q, Cho AH, Rondeau G, Welsh J, Adamson E, Mercola D, McClelland M. Survey of differentially methylated promoters in prostate cancer cell lines. Neoplasia. 2005;7:748–60. doi: 10.1593/neo.05289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slater AA, Alokail M, Gentle D, Yao M, Kovacs G, Maher ER, Latif F. DNA methylation profiling distinguishes histological subtypes of renal cell carcinoma. Epigenetics. 2013;8:252–67. doi: 10.4161/epi.23817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan PS, Venkataramu C, Ibrahim A, Liu JC, Shen RZ, Diaz NM, Centeno B, Weber F, Leu YW, Shapiro CL, et al. Mapping geographic zones of cancer risk with epigenetic biomarkers in normal breast tissue. Clin Cancer Res. 2006;12:6626–36. doi: 10.1158/1078-0432.CCR-06-0467. [DOI] [PubMed] [Google Scholar]
- 41.Matsumoto L, Takuma H, Tamaoka A, Kurisaki H, Date H, Tsuji S, Iwata A. CpG demethylation enhances alpha-synuclein expression and affects the pathogenesis of Parkinson’s disease. PLoS One. 2010;5:e15522. doi: 10.1371/journal.pone.0015522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jowaed A, Schmitt I, Kaut O, Wüllner U. Methylation regulates alpha-synuclein expression and is decreased in Parkinson’s disease patients’ brains. J Neurosci. 2010;30:6355–9. doi: 10.1523/JNEUROSCI.6119-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sx A. Xu Q, Hu Yc, Song Cy, Guo Jf, Shen L et al. Hypomethylation of SNCA in blood of patients with sporadic Parkinson's disease. Journal of the Neurological Sciences 2013, ISSN 0022-510X. [DOI] [PubMed]
- 44.Schimmack S, Lawrence B, Svejda B, Alaimo D, Schmitz-Winnenthal H, Fischer L, Büchler MW, Kidd M, Modlin I. The clinical implications and biologic relevance of neurofilament expression in gastroenteropancreatic neuroendocrine neoplasms. Cancer. 2012;118:2763–75. doi: 10.1002/cncr.26592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duffy MJ, Napieralski R, Martens JWM, Span PN, Spyratos F, Sweep FCGJ, Brunner N, Foekens JA, Schmitt M, EORTC PathoBiology Group Methylated genes as new cancer biomarkers. Eur J Cancer. 2009;45:335–46. doi: 10.1016/j.ejca.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 46.Chim CS, Liang R, Tam CYY, Kwong YL. Methylation of p15 and p16 genes in acute promyelocytic leukemia: potential diagnostic and prognostic significance. J Clin Oncol. 2001;19:2033–40. doi: 10.1200/JCO.2001.19.7.2033. [DOI] [PubMed] [Google Scholar]
- 47.Shiozawa E, Takimoto M, Makino R, Adachi D, Saito B, Yamochi-Onizuka T, Yamochi T, Shimozuma J, Maeda T, Kohno Y, et al. Hypermethylation of CpG islands in p16 as a prognostic factor for diffuse large B-cell lymphoma in a high-risk group. Leuk Res. 2006;30:859–67. doi: 10.1016/j.leukres.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 48.Villuendas R, Sánchez-Beato M, Martínez JC, Saez AI, Martinez-Delgado B, García JF, Mateo MS, Sanchez-Verde L, Benítez J, Martínez P, et al. Loss of p16/INK4A protein expression in non-Hodgkin’s lymphomas is a frequent finding associated with tumor progression. Am J Pathol. 1998;153:887–97. doi: 10.1016/S0002-9440(10)65630-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De S, Shaknovich R, Riester M, Elemento O, Geng H, Kormaksson M, Jiang Y, Woolcock B, Johnson N, Polo JM, et al. Aberration in DNA methylation in B-cell lymphomas has a complex origin and increases with disease severity. PLoS Genet. 2013;9:e1003137. doi: 10.1371/journal.pgen.1003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee GW, Kang JH, Kim IS, Kim HG, Ko GH, Lee JH, Kim DC, Song DH, Yang JW, Lee JS. Is inactivation of O6-methylguanine DNA methyltransferase still a favorable prognostic factor of patients with diffuse large B-cell lymphoma in the era of R-CHOP chemotherapy? Leuk Lymphoma. 2009;50:1992–8. doi: 10.3109/10428190903312462. [DOI] [PubMed] [Google Scholar]
- 51.Esteller M, Gaidano G, Goodman SN, Zagonel V, Capello D, Botto B, Rossi D, Gloghini A, Vitolo U, Carbone A, et al. Hypermethylation of the DNA repair gene O(6)-methylguanine DNA methyltransferase and survival of patients with diffuse large B-cell lymphoma. J Natl Cancer Inst. 2002;94:26–32. doi: 10.1093/jnci/94.1.26. [DOI] [PubMed] [Google Scholar]
- 52.Uccella S, Cerutti R, Placidi C, Marchet S, Carnevali I, Bernasconi B, Proserpio I, Pinotti G, Tibiletti MG, Furlan D, et al. MGMT methylation in diffuse large B-cell lymphoma: validation of quantitative methylation-specific PCR and comparison with MGMT protein expression. J Clin Pathol. 2009;62:715–23. doi: 10.1136/jcp.2009.064741. [DOI] [PubMed] [Google Scholar]
- 53.Sehn LH. Optimal use of prognostic factors in non-Hodgkin lymphoma. Hematology Am Soc Hematol Educ Program. 2006:295–302. doi: 10.1182/asheducation-2006.1.295. [DOI] [PubMed] [Google Scholar]
- 54.Kwong YL. Predicting the outcome in non-Hodgkin lymphoma with molecular markers. Br J Haematol. 2007;137:273–87. doi: 10.1111/j.1365-2141.2007.06571.x. [DOI] [PubMed] [Google Scholar]
- 55.Lübbert M, Rüter BH, Claus R, Schmoor C, Schmid M, Germing U, Kuendgen A, Rethwisch V, Ganser A, Platzbecker U, et al. A multicenter phase II trial of decitabine as first-line treatment for older patients with acute myeloid leukemia judged unfit for induction chemotherapy. Haematologica. 2012;97:393–401. doi: 10.3324/haematol.2011.048231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fenaux P, Mufti GJ, Hellstrom-Lindberg E, Santini V, Finelli C, Giagounidis A, Schoch R, Gattermann N, Sanz G, List A, et al. International Vidaza High-Risk MDS Survival Study Group Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 2009;10:223–32. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schoofs T, Müller-Tidow C. DNA methylation as a pathogenic event and as a therapeutic target in AML. Cancer Treat Rev. 2011;37(Suppl 1):S13–8. doi: 10.1016/j.ctrv.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 58.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, Ehrlich M, Laird PW. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–36. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.