Abstract
To rigorously assess the tools and protocols that can be used to understand and predict macromolecular recognition, and to gain more structural insight into three newly-discovered allosteric binding sites on a critical drug target involved in the treatment of HIV infections, the Olson and Levy labs collaborated on the SAMPL4 challenge. This computational blind challenge involved predicting protein-ligand binding against the three allosteric sites of HIV integrase (IN), a viral enzyme for which two drugs (that target the active site) have been approved by the FDA. Positive control cross-docking experiments were utilized to select 13 receptor models out of an initial ensemble of 41 different crystal structures of HIV IN. These 13 models of the targets were selected using our new “Rank Difference Ratio” metric. The first stage of SAMPL4 involved using Virtual Screens to identify 62 active, allosteric IN inhibitors out of a set of 321 compounds. The second stage involved predicting the binding site(s) and crystallographic binding mode(s) for 57 of these inhibitors. Our team submitted four entries for the first stage that utilized: 1) AutoDock Vina plus visual inspection; 2) a new Common Pharmacophore Engine; 3) BEDAM replica exchange re-scoring simulations, and a Consensus approach that combined the predictions of all three strategies. Even with the SAMPL4’s very challenging compound library that displayed a significantly lower amount of structural diversity than most of the libraries that are conventionally employed in prospective Virtual Screens, these approaches produced hit rates of 24%, 25%, 34%, and 27%, respectively, on a set with 19% declared binders. Our only entry for the second stage challenge was based on the results of AD Vina plus visual inspection, and it ranked third place overall according to several different metrics provided by the SAMPL4 organizers. The successful results displayed by these approaches highlight the utility of the computational structure-based drug discovery tools and strategies that are being developed to advance the goals of the newly-created, multi-institution, NIH-funded center called the “HIVE” (for HIV Interaction and Viral Evolution Center).
Introduction
For an introduction to HIV infection, AIDS epidemics and integrase/LEDGF inhibition, we refer to the general introduction paper in this issue[1].
Our laboratory has been involved in HIV-related research for more than 20 years, with computational and drug design efforts targeting HIV Protease (PR) and more recently HIV integrase (IN)[2-7] This work has led to the identification of several potential new allosteric sites on HIV protease. Our main computational effort uses the FightAids@Home project,(http://fightaidsathome.scripps.edu/) in collaboration with IBM World Community Grid, where volunteer’s computer power is used to perform high-throughput virtual screenings of millions of commercially-available compounds versus HIV-related targets.
In 2012, we formed the HIV Interaction and Viral Evolution (HIVE) Center (http://hive.scripps.edu/) whose goal is to characterize at the atomic level the structural and dynamic relationships between interacting macromolecules in the HIV life cycle to understand the mechanistic evolution of drug resistance. The Center involves 13 individual research groups from six different institutions. One recent collaborative effort between the two HIVE Center computational groups, the Olson lab and the Levy lab, has been established to reduce the number of false positive results from large virtual screens. These virtual screens are used to recommend acquisition or synthesis of promising compounds for subsequent wet-lab analysis. Thus, the higher the rate of true positives from computational experiments, the less time, effort and money is wasted on testing compounds that are not true binders. To accomplish this we are developing a procedure that takes the top hits selected from virtual screens using either AutoDock or AutoDock Vina and evaluates their binding free energy using the replica exchange molecular dynamics computation, BEDAM, from the Levy Lab (described in detail in a companion paper in this issue[8]). Initial retrospective analysis of this procedure on HIV protease allosteric site-binders has shown to be very promising (paper in preparation). When the SAMPL4 Challenge was announced it provided a useful blind data set with unpublished results upon which we could further test our methodology.
Moreover, the SAMPL4 Challenge organizers had chosen to use data from studies on three allosteric sites of the HIV Integrase[1]. While we had not previously worked on these HIV target sites, two other member labs in our HIVE Center, the Engelman and Kvaratskhelia groups, focus on allosteric inhibition of Integrase, and the SAMPL4 Challenge presented an opportunity to initiate a computational effort in that area, and promote further HIVE Center collaborations. Thus we decided to participate in the SAMPL4 Challenge.
Before the Challenge began, the participants were informed that most of the SAMPL4 compounds were known to bind to (at least) the LEDGF site of IN, but some of the compounds were known to bind to at least one of the two additional allosteric sites of IN, which were referred to as the “FBP” site (for Fragment Binding Pocket) and the “Y3” site (see Figure 1). Like the LEDGF site, the FBP site is also located at the dimer interface of the catalytic core domain (CCD) of IN. There are two LEDGF sites per IN CCD dimer, two FBP sites per IN CCD dimer, and also two Y3 sites per IN CCD dimer. However, the Y3 site is entirely contained within each monomer of the core domain and is located underneath the very flexible 140s loop (i.e., Gly140-Gly149). The top of the 140s loop flanks the active site region, and the composition, conformation, and flexibility of the 140s loop is known to be critical to IN activity [9-11] Since most of the inhibitors in the SAMPL4 library (i.e., the “true positives”) are LEDGF binders, and since the previously-published inhibitors of the LEDGF site (e.g., the ALLINIs) are more advanced and more well-characterized[12-15], most of our effort focused on the LEDGF site. Most of this paper will focus on the results versus the LEDGF site, as well.
Figure 1.
Integrase functional structure and architecture. The three domain structure of a monomer of DNA target structure complex of IN is displayed. The SAMPL4 reference structure of the HIV IN Catalytic Core Domain dimer (CCD, PDB ID: 3NF8) was superimposed onto PFV IN crystal structure (PDB ID: 3OS1) to show the relative arrangement of the domain components. The CCD domain is represented as ribbon model with each monomer colored green and in cyan, respectively; the other domains are represented as semi-transparent surfaces: C-Terminal Domain (CTD, yellow); N-Terminal Domain (NTD, light blue); host DNA (salmon). The CDQ allosteric ligand from 3NF8 is displayed as sticks (white) to highlight the three allosteric sites of HIV IN involved in the SAMPL4 challenge: LEDGF, Y3, and FBP. The IN inhibitor Raltegravir (RLT) bound in the PFV IN active site is shown with black outline. The active catalytic state of HIV IN is a tetramer formed by a dimer of dimers (not shown).
Methods
Positive control cross-docking studies
The Challenge organizers gave as a reference structure an integrase catalytic domain dimer with ligands bound to all three allosteric sites (PDB 3NF8). With myriad HIV IN structures available and 3 sites to consider we decided to inspect the interactions and rankings of known binders of the HIV IN allosteric sites to select an optimally informative subset of structures as targets for the virtual screening. Thus, co-crystallized ligands for each site were cross-docked from the collection of IN structures.
To prepare for the positive control cross-docking studies, the Protein Data Bank[16,17] was searched for the available crystal structures of IN bound to an allosteric inhibitor. When a particular crystal structure displayed both “A” and “B” form coordinates for residues that were within one of the three allosteric sites of IN (or within the shell of residues that surround one of these sites), then that complex was split into two separate target files (i.e., PDBID_A and PDBID_B). If no “_A” or “_B” is listed, then that crystal structure had no alternate conformations in the regions surrounding the allosteric sites. The LEDGF site is represented by 64 different receptor models, the FBP is involved in 32 different receptor models, and the Y3 site is described by 10 different receptor models. These 106 receptor models of 41 crystallographic complexes were superimposed onto the coordinate reference frame provided by SAMPL4 (alignment by alpha carbons performed with PyMOL) and then organized according to which of the three allosteric sites had a ligand bound to it (using visual inspection). All hydrogen atoms were added to the proteins using the MolProbity server, which adjusts the pKa’s of the titratable residues, optimizes the hydrogen bond network, and allows His, Gln, and Asn residues to flip if doing so lowers the energy of the system.[18,19] All hydrogen atoms were added to the ligands using Avogadro.[20] Gasteiger-Marsili charges were added to the models of both the ligands and the targets,[21] and then the non-polar hydrogens were merged onto their respective heavy atoms using AutoDockTools and Raccoon.[22,23] The docking studies were all performed using AutoDock Vina 1.1.2.[24] See Figure 2 for a summary of the workflow employed in the positive control cross-docking experiments.
Figure 2.
Summary of the workflow used in the positive control cross-docking experiments with AutoDock Vina. This protocol was used to select the targets that were involved in the subsequent Virtual Screens with the SAMPL4 compounds.
Only the small molecule allosteric inhibitors of IN were selected for the positive control cross-docking studies, the cyclic peptide inhibitor complexes being removed from the set. These positive control small molecules were utilized in the cross-docking experiments, with two ligand models for each ligand. One model started with a pose close to the crystallographic conformation and position, while the second model began with a randomized position, orientation, and conformation, generated by using the “randomize_only” function in AutoDock Vina).[24] 42 ligand models were known (observed crystallographically) to be LEDGF binders, 33 ligand models were known to bind the FBP site, and 5 ligand models were known to bind the Y3 site. These 80 ligand models and their 2 forms of randomized and non-randomized structures yielded 160 total ligand models for use in the positive control dockings. All 160 ligand models were given filenames that began with the site from which they were extracted, to make the subsequent workflow easier to organize and analyze. When a particular ligand crystallized in more than one type of allosteric site, it was included in the set of ligands for each of those sites. The aligned, crystallographic conformations of the positive control ligands from each site were used as inputs to train the “Common Pharmacophore Engine” approach (see Table 1) (discussed below).
Table I.
Common Pharmacophore Engine’s training sets for each allosteric site.
| FBP | |
| Receptor (PDB ID) | Ligand (HET ID) |
| s4AH9 | 0MB |
| s3VQ4 | 0NX |
| s3AO4 | 833 |
| s4AHS | AKH |
| s3AO2 | AVX |
| s3AO5 | BBY |
| s3AO3 | BMC |
| s3AO3 | BMC |
| s3VQQ | BTE |
| s3AO1 (top) | BZX |
| s3AO1 (bottom) | BZX |
| s3NF8 | CDQ |
| s3VQP | DBJ |
| s3VQB | FBG |
| s3VQE | FMQ |
| s4AHR | I2E |
| s4AHU | ICO |
| s3VQ5 | MMJ |
| s3VQD | MOK |
| s3VQC | MPK |
| s3OVN | MPV |
| s4AHT | Q6T |
| s4AHV | Z5P |
| s3ZT2 | ZT2 |
| s3ZT4 | ZT2 |
| LEDGF | |
| Receptors (PDB ID) | Ligands (HET ID) |
| 3ZCM | PX3 |
| 3ZSY | OM3 |
| 3ZSZ | OM2 |
| 3ZT1 | OM1 |
| 3ZSQ | O4N |
| 3ZSR | O3N |
| 3ZSO | O2N |
| 3ZSX | N44 |
| 3NF6 | IMV |
| 3NF8 | CDQ |
| 3NFA | CBJ |
| 3VQ8 | BCU |
| 3VQ4 | 0NX |
| 3ZT3 | ZT4 |
| 3ZT4 | ZT2 |
| 3ZT2 | ZT2 |
| 3ZT0 | ZT0 |
| 3ZSW | ZSW |
| 3ZSV | ZSV |
| 3VQ7 | SNU |
| Y3 | |
| Receptor (PDB ID) | Ligand (HET ID) |
| 3NF6 | IMV |
| 3NF7 | CIW |
| 3NF8 | CDQ |
| 3NF9 | CD9 |
| 3NFA | CBJ |
The following crystal structures had ligands that crystallized in both the LEDGF site and the FBP site: 3ZT2 (“A” and “B” forms), 3ZT4 (“A” and “B” forms), 3VQ4, and 3VQ7. Similarly, the following crystal structures had ligands that crystallized in both the LEDGF site and the Y3 site: 3NF6 (“A” and “B” forms) and 3NFA (“A” and “B” forms). For 3NF8 (which also had “A” and “B” forms), the fragment “CDQ” crystallized in all three allosteric sites. (See Figure 3 for images that display the crystallographic binding mode of CDQ with each site and for a depiction of the grid boxes that were used in the AutoDock Vina calculations against each site). For both the positive controls and the Virtual Screens of the SAMPL4 compounds, 4 CPU’s were used per docking calculation (on the TSRI Linux cluster), the grid box used for each site had a size of 30×30×30 Å3, and the grids were centered on an atom in CDQ from the 3NF8.pdb reference (in the relative coordinate frame provided by SAMPL4). For the FBP site the nitrogen atom of CDQ was used for the center (x,y,z = 3.426, −21.239, −1.559), for the LEDGF site the C11 atom was used for the center (x,y,z = 11.095, −46.191, 0.368), and for the Y3 site the N atom of CDQ was used for the center (x,y,z = 9.137, −25.675, −22.414). Since large grid boxes were used, the “exhaustiveness” setting in AD Vina was increased to 20.
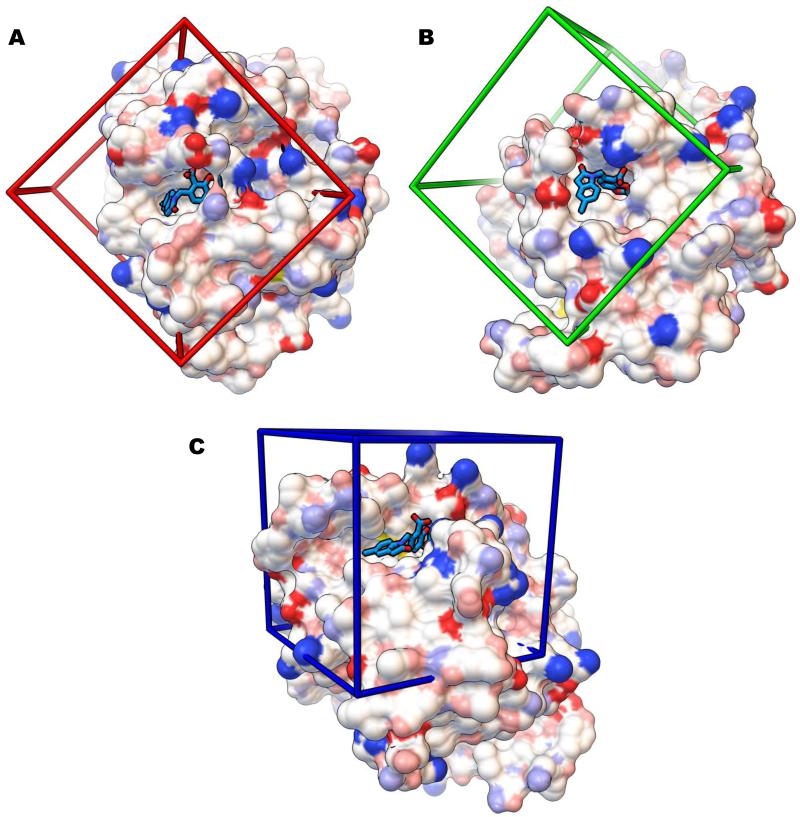
Figure 3.
Grid boxes (30 × 30 × 30 Å3) utilized in the positive control cross-docking studies and in the Virtual Screens of the SAMPL4 compounds. Each image contains the solvent-excluded molecular surface of the HIV integrase catalytic core domain (colored with the “David Goodsell” convention) from the crystal structure PDB ID 3NF8, in which the fragment “CDQ” (shown as sticks with turquoise carbon atoms) is bound. Shown are the LEDGF (A), “FBP” (B), and “Y3” (C) sites.
Selecting the models for each of the targets
The combination of the three sets of positive control compounds were cross-docked against all of the models for each of the 3 allosteric sites (see Figure 2 for a summary of the workflow). Interaction-based filters, based on key hydrogen bonds displayed by many of the ligands in the crystal structures for each particular site, were applied to the AD Vina results against each allosteric site. The filtering was done using in-house python scripts that were created during the development of Raccon2 and Fox.[23] The filters were only applied to the top-ranked AD Vina mode per model of each compound (for both the positive control experiments and the subsequent Virtual Screens of the SAMPL4 compounds). For example, about a dozen different sets of interaction-based filters were investigated for the LEDGF results, and two particular filters were selected, since they harvested a reasonable number of docked modes per target for visual inspection. For the LEDGF site, the filter requirements consisted of a minimum of 2 predicted hydrogen bonds to IN, and either: (Fa) a hydrogen bond with Glu170; or (Fb) a hydrogen bond to the backbone amino group of His171 (similar to the ALLINIs).
The method used for choosing receptors for virtual screening of the IN models for the LEDGF site described here is the same for the FBP site. For each LEDGF target, the top-ranked docked mode of all of the positive control ligands (from all 3 sites) that passed a particular filter were sorted according to the estimated Free Energy of Binding as calculated by AD Vina. This sorting process determined the absolute rank for each compound whether they be known LEDGF-site binders or decoys (observed to bind at the FBP or Y3 sites). The ligands that crystallized in the LEDGF site were then extracted from that sorted list, and their order in that LEDGF-site specific list determined their relative rank. Receptor models were chosen from statistics and visual analysis of the rankings of the site-specific binders (Supplemental Information, Figure 1). Subsequently, this ad hoc visualization process formalized into our “Rank Difference Ratio” (RDR) procedure where the relative ranking of the appropriate positive control ligands was used with the corresponding absolute ranking as the Rank Difference Ratio metric for a receptor i, as follows:
where Rabs,j and Rrel,j are the absolute and relative rankings, respectively, for ligand j. For example, a LEDGF target model for which all of the LEDGF ligands ranked better than the ligands from the other two sites, the absolute and relative rankings for all ligands would be the same and the receptor would thus have an RDR value of 0. In Figure 4 we plot RDR vs. total number of site-specific hits. The receptor models having a low RDR value and a high number of LEDGF-site hits demonstrates that this formalized RDR metric matches the receptors chosen by the original ad hoc procedure. For targets that displayed similar RDR values using a particular filter, receptor models were selected to maximize structural diversity. Further, if two forms of a structure had a difference of total number of LEDGF-site hits less than 2, the receptor model with the lower RDR value was chosen. A similar strategy was used to select the FBP targets. For the small set of Y3 receptor models, the median statistic of rankings was sufficient to choose the best receptor model, 3NF8_B.
Figure 4.
Hydrogen bond interactions of AVX17561 (sticks with green carbon atoms) docked with the LEDGF site (white ribbon) of HIV integrase. Three residues (sticks with pink carbon atoms) are shown with hydrogen bonds (magenta dotted lines) to the ligand model. Two hydrogen bonds (Glu170 and His171) were required by the interaction filters.
The set of 106 receptor models representing the FBP, LEDGF, and Y3 sites enabled the selection of the following number of targets per site: 6 crystal structures of the LEDGF site, 6 structures of the FBP site, and 1 crystal structure of the Y3 site. The LEDGF targets selected were: 3ZSO_B, 3ZT4_B, 3ZT1_A, 3ZT3_A, 3NF8_A, and 3ZCM_A.[25,26] The FBP targets selected were: 3AO1, 3AO2, 3VQD, 3VQE_A, 3VQ4, and 3VQ7.[27,28] The Y3 target selected was 3NF8_B.[26]
Virtual Screen of the SAMPL4 compounds using AutoDock Vina
AutoDock Vina (AD Vina) was used to screen the SAMPL4 compound library against these 13 receptor models of IN.[24] See Figure 5 for a summary of the workflow used for the LEDGF targets. A similar strategy was utilized for the FBP and Y3 sites. The same grid box size, location, and settings for AD Vina from the positive controls were also used in these Virtual Screens (see Figure 3). The 321 compounds provided by SAMPL4 were used as inputs by the Levy Lab for LigPrep and Epik [29,30] at a pH of 7 ±2, which generated additional tautomers and protonation states of some compounds to produce a final set of 451 models of the SAMPL4 compounds. All 451 ligand models of the compounds were docked against the 6 LEDGF targets, 6 FBP targets, and 1 Y3 target, using the TSRI Linux cluster. The same filters used in the positive control dockings were applied to the results of the SAMPL4 dockings.
Figure 5.
Selection of the 6 crystal structures that were used as targets for the LEDGF site during the Virtual Screen of the SAMPL4 compounds. The X-axis indicates the number of docked models of LEDGF ligands that passed through a particular filter during the positive control cross-docking experiments, while the Y-axis plots the “Rank Difference Ratio” metric that quantifies how well the LEDGF ligands ranked, in relation to the FBP and Y3 ligands. 84 models of small molecule LEDGF ligands were docked to 66 LEDGF-site receptor models and then 2 interaction filters (Fa: a hydrogen bond with Glu170; Fb: a hydrogen bond to the backbone amino group of His171) were applied. After normalization of data between the two sets of filtered data, only the best for is receptor source is visualized here.
The sets of docked modes harvested by either of these two filters per target were combined and visually inspected. The hydrogen bond python filter used only the donor-acceptor distance as a criterion (the donor-hydrogen-acceptor angles were manually measured during the visual inspection process). Positive factors considered during visual inspection included the hydrogen bonds exceeding the number specified in the filters, potentially favorable interactions of nearby side-chains if they flexed, and agreement of position between a set of stereoisomers. Negative factors included solvent-exposure of non-polar atoms or large portions of the ligand. Though somewhat subjective in nature, these criteria were used to make final choices of hits and later determine a confidence level for each ligand as required by SAMPL. The range of confidence was 1-5 with 5 being the highest confidence. The docked modes that passed the visual inspection process against any of the LEDGF targets were combined, to generate a set of 69 unique compounds that were predicted to be LEDGF binders.
For FBP, two filters were also combined to harvest ligands for visual inspection. In one filter, the top-ranked docked mode needed to have a minimum of 2 hydrogen bonds and a ligand efficiency value less than −0.30 kcal/mol/heavy atom. In the other filter, the compound had to display a minimum of 3 hydrogen bonds and a ligand efficiency value better than −0.25 kcal/mol/heavy atom. The interaction-based filters from the positive control dockings that used key hydrogen bonds to the FBP site were too stringent; very few or no docked compounds passed that filter. Twenty-five compounds passed the visual inspection process and were predicted to bind the FBP site.
For the Y3 receptor model, two different filters were combined to choose docking modes for visual inspection. One filter required the docked compounds to have 1 hydrogen bond with Lys and 1 additional unspecified hydrogen bond. The second filter only required the docked compounds to possess 4 hydrogen bonds. A set of 7 compounds passed the visual inspection process and were predicted to bind to the Y3 site of IN.
The predicted binding modes of all of our candidate inhibitors that passed the visual inspection process against the 13 target models were sent to the Levy lab, to be used as inputs for their BEDAM replica exchange simulations[31]. (See the companion paper on the BEDAM method for results and a discussion of the Consensus approach[8].) In addition, the filtered binding modes predicted by AD Vina were also re-ranked by the new Common Pharmacophore Engine method (below), which identified an alternate set of candidate inhibitors.
Common Pharmacophore Engine
We have developed a 3D pharmacophore model[32,33] based on the AutoDock forcefield/atom type set. The pharmacophore is based on the conversion of explicit chemical groups into basic chemical features: hydrogen bond acceptors and donor, aromatic rings, aliphatic carbons, and halogens. Each feature is represented by a combination of the three-dimensional location of a given feature with respect to the ligand structure and a series of properties specific to each feature (i.e., hydrogen bond direction vector, aromatic ring plane). Each feature can also include a tolerance setting for its properties and location. This simplifies the comparison of chemical structures and provides a rapid method for quantitatively scoring structural similarity. By using pharmacophore representations it is possible to generate a common pharmacophore model that recapitulates the most representative set of chemical features of a series of ligands bound to the same site. The choice of features to represent the pharmacophore is performed after geometrical clustering. A common pharmacophore engine generates similar feature clusters that are processed to produce an average representation. Isolated features are discarded. In addition to the aforementioned chemical features, the engine also generates special sets (i.e., an “honorable mention” set) representing recurrent atom type clusters present in the ligand set, like halogens or sulfur.
The common pharmacophore can then be used to re-score ligands docked to the binding site, providing a new quantitative score based on the similarity of binding patterns between docked results and known ligands. This pharmacophore model has been successfully applied to identify high micromolar allosteric inhibitors of HIV-1 protease, including several crystallographic hits (unpublished data). Further details about the method and the pharmacophore engine will be described in a future publication.
Generation of common pharmacophores
In order to generate the common pharmacophores for the three different sites (LEDGF, FBP, and Y3), the three different training sets of ligand-receptor complexes were aligned (by alpha carbons in PyMOL) to the 3NF8 structure provided as an input for the challenge. For each site, ligand structures were extracted and their overlapped conformations were processed with the common pharmacophore engine. Features present in at least two ligands were considered, directionality was disabled for the hydrogen bond and aromatic ring features, no maximum features count was used, and all features had the same weight and radius tolerance (1.5 Å). These settings were used to increase the tolerance of the common pharmacophores and to provide a generic filter to prioritize docked poses of ligands that displayed an interaction pattern similar to known ligands. A summary of the different training sets used for the generation of each common pharmacophore is shown in Table I. The number and nature of the features generated for each pharmacophore is strictly correlated with the number of ligands available for each site and their structural diversity.
LEDGF common pharmacophore
The LEDGF common pharmacophore was calculated from19 different ligands (see Table I) and contains the following features: 5 aromatic rings, 2 hydrogen bond donors, 6 hydrogen bond acceptors, 5 aliphatic carbons, and 1 halogen (see Figure 7A).
Figure 7.
“Common Pharmacophore Engine” representation for each of the three allosteric sites of HIV integrase. The SAMPL4 reference structure 3NF8 is represented as white cartoons with the crystallographic ligand CDQ represented as. Pharmacophore features (see Table I for full listing) are shown as semi-transparent spheres, with solid sphere centroids: aromatic features (orange); aliphatic carbon (gray); hydrogen bond (HB) acceptor (red), HB donor (white); and halogen (green). Shown are the pharmacophore models of the LEDGF (A), FBP (B), and Y3 (C) sites.
FBP common pharmacophore
The FBP common pharmacophore was calculated from 22 different ligands (see Table I) and contains the following features: 3 aromatic rings, 3 hydrogen bond donors, 9 hydrogen bond acceptors, 3 aliphatic carbons, and 2 halogens (see Figure 7B).
Y3 common pharmacophore
The Y3 common pharmacophore was calculated from 5 different ligands (see Table I) and contains the following features: 3 aromatic rings, 4 hydrogen bond acceptors, 2 aliphatic carbons, and 1 halogen (see Figure 7C).
Results and Discussion
Since this was a blind challenge, the official classification of the success of these methods was determined by the SAMPL4 organizers, using metrics that they selected and applied. (See [1] for details used to calculate these metrics). The results presented here were plotted by the SAMPL4 organizers, and the graphs in Figures 8-14 were generated by them. For these graphs in Figures 8-14, our different predictions had the following submission ID numbers: (A) AutoDock Vina plus visual inspection, entry 133; (B) Common Pharmacophore Engine (without visual inspection), entry 134; (C) BEDAM replica exchange binding free energy simulations (of the AD Vina docked modes that passed the visual inspection process in A), entry 135; and (D) Consensus approach that combined A-C, entry 136. See the companion paper on the BEDAM entry by the Levy Lab[8] to learn the details regarding the methods and results for C and D.
Figure 8.
Area under the curve (AUC) performance of the Phase 1 entries in the SAMPL4 challenge provided by the SAMPL organizers. Red boxes highlight the entries for the Common Pharmacophore Engine (134) and AutoDock Vina plus visual inspection (133).
Figure 14.
Fraction of ligand poses that were successfully predicted by AutoDock Vina. RMSD data (in Å) provided by the SAMPL organizers.
Virtual Screens using AD Vina with visual inspection (Phase 1 of Challenge): identifying IN binders
Phase 1 involved predicting which of the SAMPL4 compounds actually bind to any of the three allosteric sites of HIV integrase. The Common Pharmacophore Engine’s predictions (without human intervention or visual inspection) are listed as entry number 134 and ranked 4th place, while the predictions from AutoDock Vina with visual inspection are listed as entry number 133 and ranked 7th place according to the AUC metric. Although the Common Pharmacophore Engine’s predictions ranked better than the results from AutoDock Vina with visual inspection, their respective AUC values were within the standard deviations of each other. However, using the Recognition Factor metric, the performance of the Common Pharmacophore Engine (which ranked 3rd place) was clearly superior to the predictions from AD Vina with visual inspection (which ranked 7th place) when considering the standard deviations.
For the general metric used to judge most prospective Virtual Screens in the published literature, the Common Pharmacophore Engine had a hit rate of 24.85% (42/169), and AD Vina with visual inspection had a similar hit rate of 23.76% (24/101). Given the intrinsic approximations of empirical scoring functions, docking methods are usually less sensitive to congeneric compounds, while they are more effective in identifying potential hits in chemically diverse libraries.[34] Therefore, in the context of the low diversity library provided in this challenge, the overall success rate of these docking-based methods is high.
Pharmacophore Results (Phase 1 of Challenge): identifying IN binders
Every set of docking results generated with AutoDock Vina for each binding site in the 13 targets were re-scored with the corresponding common pharmacophore. The pharmacophore score (ps) ranged from 100.0 (all features matched) to 0.0 (no feature matched). For the LEDGF and FBP sites, where six different receptor models were targeted for each site, the pharmacophore score, ps, was calculated separately for each set. The 6 sets (per site) were then combined by weighting the score of each ligand versus a given receptor pose by its positional ranking in the set (pstot = Σpsi / ranki). The pharmacophore score for each binding site was then used to rank the ligands and identify the binders. Depending on their ps ranking, the ligands were subdivided into three classes, with high (top 19), medium (20-50) and low (>50) confidence, respectively. Accordingly to the challenge guidelines, ligands in the high confidence class were considered binders with a confidence value of 5, ligands in the medium confidence class were considered as binders with a confidence value of 3, and ligands in the low confidence class were considered as non-binders with a confidence value of 1. The overall success rate in hit identification for binders versus non-binders using the pharmacophore score was 25%. Results for the Common Pharmacophore Engine approach (i.e., entry 134), using different metrics calculated by the SAMPL4 organizers, are presented: Area Under the Curve (see Figure 8), Recognition Factor (see Figure 9), enrichment factor (see Figure 10), and BEDROC (see Figure 11).
Figure 9.
“Recognition Factor” performance of the Phase 1 entries in the SAMPL4 challenge provided by the SAMPL organizers. Red boxes highlight the entries for the Common Pharmacophore Engine (134) and AutoDock Vina plus visual inspection (133).
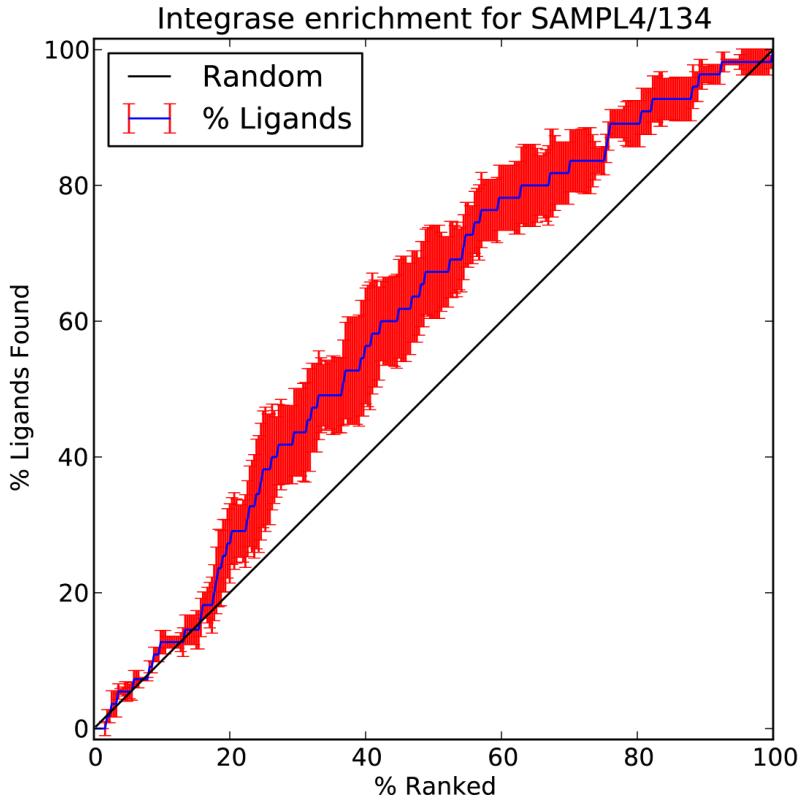
Figure 10.
Enrichment factor of SAMPL4 entry 134 corresponding to the Common Pharmacophore Engine’s score.
Figure 11.
BEDROC score of the Phase 1 entries in the SAMPL4 challenge provided by the SAMPL organizers. Red boxes highlight the entries for the Common Pharmacophore Engine (134) and AutoDock Vina plus visual inspection (133).
Virtual Screens with AD Vina plus visual inspection (Phase 2 of Challenge): identifying the binding site and binding pose within the three allosteric sites of IN
Phase 2 of the SAMPL4 challenge involved predicting the binding site (or sites) and binding mode (or modes) of the 57 SAMPL4 compounds that crystallized in an allosteric site of HIV IN. Since this was a blind contest, our team submitted all of our predictions for Phase 1 before we obtained the list of compounds that were involved in Phase 2.
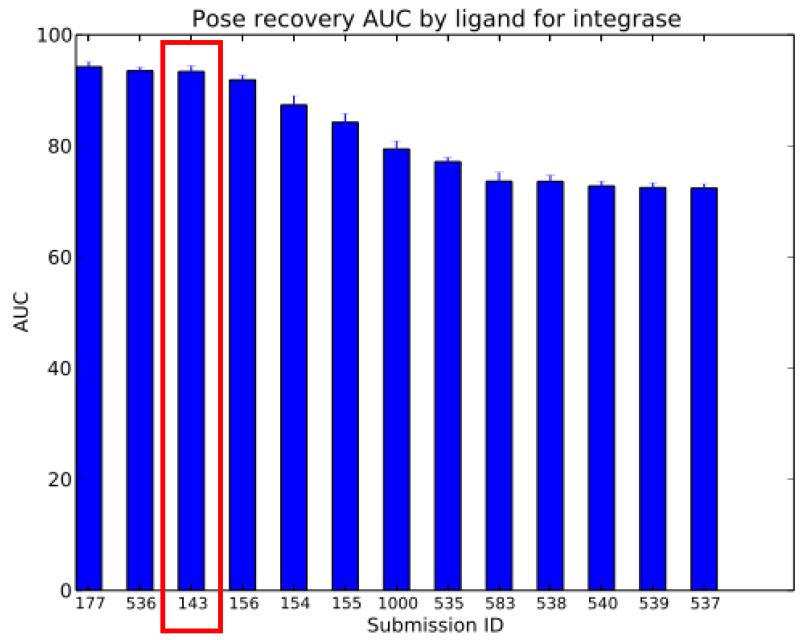
AutoDock Vina with visual inspection was the only entry that we submitted for Phase 2, and it is listed as entry number 143. Generally, AutoDock Vina placed between 2nd and 5th places over 6 different metrics. (See Figure 4 in the Overview article.) According to the “Pose recovery Area Under the Curve by ligand” metric (see Figure 12), AD Vina ranked 3rd place. But considering the standard deviations, it was nearly identical to the 2nd place entry and similar to the 1st place entry. Using the “RMSD by ligand” box plot data (see Figure 13), AD Vina again ranked 3rd place. However, when comparing the medians, AD Vina was actually 1st place (lowest median). Similarly, if only the data distributions within the 1st and 3rd quartiles are compared and outliers disregarded, AD Vina ranked 2nd place (with 1st and 3rd quartile values lower than those of 2nd place Entry 536). Thus, the binding modes predicted by AD Vina with visual inspection against these three allosteric sites of IN were relatively accurate, which contributed to the success of the BEDAM replica exchange re-scoring entry for Phase 1[8].
Figure 12.
“Pose Recovery Area Under the Curve” performance of the Phase 2 entries in the SAMPL4 challenge provided by the SAMPL organizers. The AutoDock Vina plus visual inspection process (143, red box) was our only entry for Phase 2.
Figure 13.
“RMSD per pose performance of the Phase 2 entries in the SAMPL4 challenge provided by the SAMPL organizers. The AutoDock Vina plus visual inspection process (143, red box) was our only entry for Phase 2.
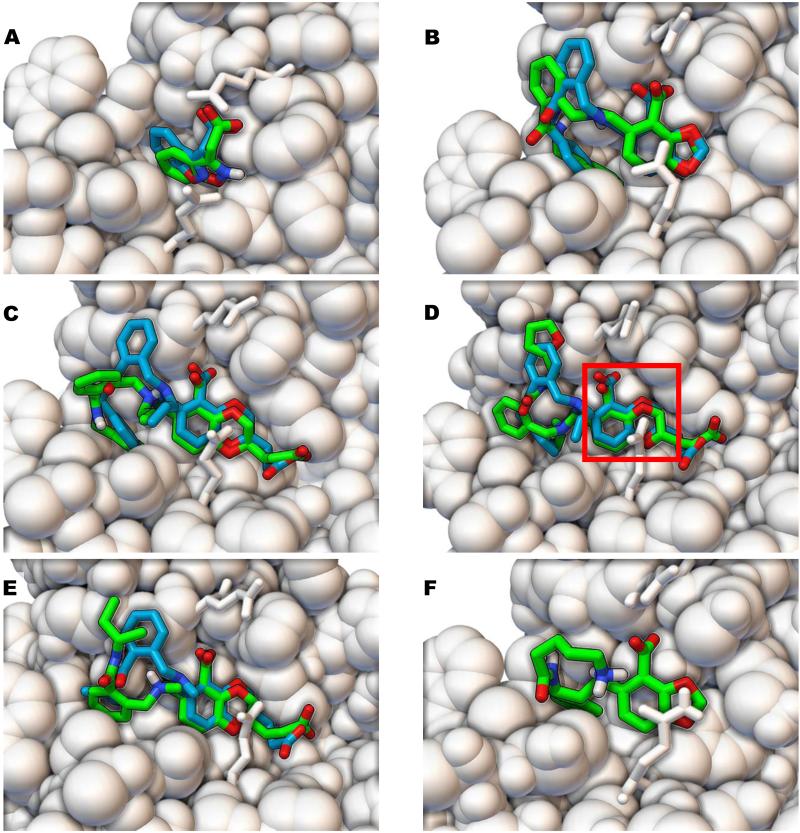
Using the “Success fraction by RMSD, by ligand” plot (see Figure 14), approximately 33% of the ligands were docked within an RMSD of 3 Å of the crystallographic binding mode(s). For a visual comparison between the docked modes of true positives identified in Phase 1 against the LEDGF site and their crystallographic binding modes (provided after we submitted our entry for Phase 2), see Figure 15. In these molecular images (created with PMV 1.5.6 release candidate 3)[22,35], some of the best predictions are displayed as well as some representative results. In all of these molecular images in Figure 15, the fragment hit or the scaffold region (within the larger derivatives based on extending the fragment hits) docked accurately with AD Vina and displayed some of the same interactions that the published ALLINIs display. The fragment AVX17679 docked within 1.3 Å of its crystallographic pose, while the larger compounds AVX17684m and AVX38753 docked within 1.7 and 2.1 Å of their crystallographic binding modes, respectively. Even when AVX38783 and AVX38784 docked with larger RMSD values of 4.5 Å and 3.4 Å, respectively, (see Figure 15, D-E), their scaffolds superimposed well (RMSD of 1.1 Å and 1.4 Å) with the crystallographic pose. AVX17561 is an interesting case where the docked scaffold matches crystal data (RMSD is 1.2 Å without the lysine like group, co-crystallized ligand structure not shown), but the hydrophobic tail is solvated. The binding modes predicted by AD Vina earned 3rd place in the pose prediction with a relative performance that was very close to the 1st and 2nd place entries, using the metrics provided by SAMPL4.
Figure 15.
Comparison of the AutoDock Vina docking poses to the crystallographic binding modes of ligands that bound to the LEDGF site of HIV integrase. The crystallographic binding modes of the ligands are displayed as sticks with turquoise carbon atoms, while the binding modes predicted by AutoDock Vina are rendered with green carbon atoms. HIV integrase is shown as white CPK spheres, with residues Glu170 and Gln95 displayed as white sticks for clarity. AVX17679 (A), AVX17684m (B), AVX38753 (C), AVX38783 (D), and AVX38784 (E) is displayed with RMSD values compared to the crystallographic poses of 1.3 Å, 1.7 Å, 2.1 Å, 4.5 Å, and 3.4 Å, respectively. RMSD values for only the scaffold (red box in D) were 1.4 Å for AVX38784 and 1.1 Å for AVX38784. The docking pose of AVX17561 (F) is also shown (without the crystallographic structure) having an RMSD value of 2.3 Å; however, the RMSD value is 0.8 Å if the lysine-like tail and amide group are disregarded.
Conclusion
The positive control cross-docking experiments performed with AutoDock Vina indicated that at least 13 different crystal structures of HIV integrase (6 LEDGF models, 6 FBP models, and 1 Y3 model) displayed reasonable predictive power for identifying the appropriate ligands that are known to bind each of the three sites. Our new Rank Difference Ratio metric appears helpful when selecting targets out of a large ensemble of different receptor models and should be further investigated as an alternative and/or complementary way of selecting snapshots of targets for Relaxed Complex Scheme experiments[10,36-39] and Virtual Screens.
The binding modes calculated by AD Vina (when docking the 451 models of the 321 SAMPL4 compounds against these 13 targets) were accurate enough to enable the following: (1) achieving a hit rate of 24% using visual inspection of the poses predicted by AD Vina; (2) obtaining a hit rate of 24% by re-ranking the docked poses using our new Common Pharmacophore Engine (without any visual inspection); and (3) by using the docked poses that passed the visual inspection process as inputs for BEDAM replica exchange re-scoring calculations, the hit rate was further improved to 34%.
These results have helped to validate the efficacy of the virtual screening pipeline that we are developing. In particular we have established the following conclusions: 1) The Common Pharmacophore Engine approach was more efficient than the visual inspection process in terms of the amount of human time involved and definitely merits further exploration and development; 2) The binding modes predicted by AD Vina were of sufficient accuracy to serve effectively as input to the free energy calculation of BEDAM. 3)The BEDAM post-processing of virtual screening results provided a significant improvement in false positive reduction
Considering that SAMPL4 involved (A) three different allosteric sites of a flexible enzyme and (B) a very challenging library of compounds that displayed a low amount of structural diversity and that contained ligands that could bind to more than one type of allosteric site, the hit rates that our collaborative, multi-institution HIVE team achieved were impressive.
We have applied our new knowledge of structural detail about these three allosteric sites of HIV integrase motivating us to test and hone our tools and protocols. These aspects will help advance the goals we are pursuing as part of the new HIVE center, which is devoted to understanding and defeating the multidrug-resistant strains of HIV that are constantly evolving and spreading. In addition, the 13 models of these allosteric sites of HIV integrase that we identified in our positive control cross-docking experiments are currently being used as targets for the FightAIDS@Home project.
Supplementary Material
Supplementary Figure 1. Raw data used to choose LEDGF receptor models. Visual analysis was used for receptor evaluation since AUC values from ROC curves were too similar. After Phase 1 and 2 submissions to SAMPL, the visual analysis was formalized in creating the Rank Difference Ratio metric (see Fig. 4). For A and B the relative ranks for LEDGF ligands are listed in column 1, and the absolute ranks versus each target are listed in the subsequent columns underneath the PDB ID for that receptor model. The receptor models that were selected as targets are highlighted in magenta in row 1. For each block of 10 rows; the minimum (in green), average (in white), and maximum (in red) values of the absolute ranks were calculated and colored. More predictive targets have more green and white in each block and less red, they have more blocks (i.e., more LEDGF ligands were ranked higher than decoys), and the numbers in each cell will be closer to the relative rankings (“line numbers” in column 1). The compounds that were harvested in (A) had to pass the following filter: a minimum of 2 hydrogen bonds to IN and a hydrogen bond to the backbone amino group of Glu170. The compounds that were harvested in (B) had to pass the following filter: a minimum of 2 hydrogen bonds to IN and a hydrogen bond to the backbone amino group of His171.
Figure 6.
Summary of the workflow used in the in the SAMPL4 challenge for the LEDGF site.
Acknowledgments
We thank the I.T. staff at The Scripps Research Institute (especially Jean-Christophe Ducom, and Lisa Dong) for maintaining a great Linux cluster and for giving the Levy lab access to it for their BEDAM calculations. This research was funded by the HIVE center grant (P50 GM103368) and by the AutoDock grant (R01 GM069832).
We thank Dr. Ron Levy and Dr. Emilio Gallicchio for their collaboration on the SAMPL4 Challenge using their BEDAM software.
References
- 1.Mobley DL, Liu S, Lim NM, Wymer KL, Perryman AL, Forli S, Deng N, Su J, Branson K, Olson AJ. J Comput Aided Mol Des. doi: 10.1007/s10822-014-9723-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tiefenbrunn T, Forli S, Happer M, Gonzalez A, Tsai Y, Soltis M, Elder JH, Olson AJ, Stout CD. Chem Biol Drug Des. 2013 doi: 10.1111/cbdd.12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tiefenbrunn T, Forli S, Baksh MM, Chang MW, Happer M, Lin YC, Perryman AL, Rhee JK, Torbett BE, Olson AJ, Elder JH, Finn MG, Stout CD. ACS Chem Biol. 2013 doi: 10.1021/cb300611p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin YC, Perryman AL, Olson AJ, Torbett BE, Elder JH, Stout CD. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 6):540. doi: 10.1107/S0907444911011681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perryman AL, Zhang Q, Soutter HH, Rosenfeld R, McRee DE, Olson AJ, Elder JE, Stout CD. Chem Biol Drug Des. 2010;75(3):257. doi: 10.1111/j.1747-0285.2009.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vajragupta O, Boonchoong P, Morris GM, Olson AJ. Bioorg Med Chem Lett. 2005;15(14):3364. doi: 10.1016/j.bmcl.2005.05.032. [DOI] [PubMed] [Google Scholar]
- 7.Perryman AL, Forli S, Morris GM, Burt C, Cheng Y, Palmer MJ, Whitby K, McCammon JA, Phillips C, Olson AJ. J Mol Biol. 2010;397(2):600. doi: 10.1016/j.jmb.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallicchio E, Deng N, He P, Perryman AL, Santiago DN, Forli S, Olson AJ, Levy R. J Comput Aided Mol Des. doi: 10.1007/s10822-014-9711-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenwald J, Le V, Butler SL, Bushman FD, Choe S. Biochemistry. 1999;38(28):8892. doi: 10.1021/bi9907173. [DOI] [PubMed] [Google Scholar]
- 10.Perryman AL, Forli S, Morris GM, Burt C, Cheng Y, Palmer MJ, Whitby K, McCammon JA, Phillips C, Olson AJ. Journal of Molecular Biology. 2010;397(2):600. doi: 10.1016/j.jmb.2010.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dewdney TG, Wang Y, Kovari IA, Reiter SJ, Kovari LC. Journal of Structural Biology. (0) doi: 10.1016/j.jsb.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 12.Kessl JJ, Jena N, Koh Y, Taskent-Sezgin H, Slaughter A, Feng L, de Silva S, Wu L, Le Grice SFJ, Engelman A, Fuchs JR, Kvaratskhelia M. Journal of Biological Chemistry. 2012;287(20):16801. doi: 10.1074/jbc.M112.354373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsiang M, Jones GS, Niedziela-Majka A, Kan E, Lansdon EB, Huang W, Hung M, Samuel D, Novikov N, Xu Y, Mitchell M, Guo H, Babaoglu K, Liu X, Geleziunas R, Sakowicz R. Journal of Biological Chemistry. 2012;287(25):21189. doi: 10.1074/jbc.M112.347534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christ F, Shaw S, Demeulemeester J, Desimmie BA, Marchand A, Butler S, Smets W, Chaltin P, Westby M, Debyser Z, Pickford C. Antimicrobial Agents and Chemotherapy. 2012;56(8):4365. doi: 10.1128/AAC.00717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jurado KA, Wang H, Slaughter A, Feng L, Kessl JJ, Koh Y, Wang W, Ballandras-Colas A, Patel PA, Fuchs JR, Kvaratskhelia M, Engelman A. Proceedings of the National Academy of Sciences. 2013;110(21):8690. doi: 10.1073/pnas.1300703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Research. 2000;28(1):235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Feng Z, Gilliland GL, Iype L, Jain S, Fagan P, Marvin J, Padilla D, Ravichandran V, Schneider B, Thanki N, Weissig H, Westbrook JD, Zardecki C. Acta Crystallographica Section D. 2002;58(6 Part 1):899. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 18.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. Acta Crystallographica Section D. 2010;66(1):12. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC. Nucleic Acids Research. 2007;35(suppl 2):W375. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. J Cheminform. 2012;4(1):17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gasteiger J, Marsili M. Tetrahedron. 1980;36(22):3219. [Google Scholar]
- 22.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. Journal of Computational Chemistry. 2009;30(16):2785. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forli S. Raccoon. Molecular Graphics Laboratory, The Scripps Research Institute; La Jolla, CA: [Accessed 2013]. 2010. < http://autodock.scripps.edu/resources/raccoon>. [Google Scholar]
- 24.Trott O, Olson AJ. J Comput Chem. 2010;31(2):455. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peat TS, Rhodes DI, Vandegraaff N, Le G, Smith JA, Clark LJ, Jones ED, Coates JAV, Thienthong N, Newman J, Dolezal O, Mulder R, Ryan JH, Savage GP, Francis CL, Deadman JJ. PLoS ONE. 2012;7(7):e40147. doi: 10.1371/journal.pone.0040147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhodes DI, Peat TS, Vandegraaff N, Jeevarajah D, Le G, Jones ED, Smith JA, Coates JA, Winfield LJ, Thienthong N, Newman J, Lucent D, Ryan JH, Savage GP, Francis CL, Deadman JJ. Antivir Chem Chemother. 2011;21(4):155. doi: 10.3851/IMP1716. [DOI] [PubMed] [Google Scholar]
- 27.Wielens J, Headey SJ, Deadman JJ, Rhodes DI, Le GT, Parker MW, Chalmers DK, Scanlon MJ. ChemMedChem. 2011;6(2):258. doi: 10.1002/cmdc.201000483. [DOI] [PubMed] [Google Scholar]
- 28.Wielens J, Headey SJ, Rhodes DI, Mulder RJ, Dolezal O, Deadman JJ, Newman J, Chalmers DK, Parker MW, Peat TS, Scanlon MJ. J Biomol Screen. 2013;18(2):147. doi: 10.1177/1087057112465979. [DOI] [PubMed] [Google Scholar]
- 29.Greenwood JR, Calkins D, Sullivan AP, Shelley JC. J Comput Aided Mol Des. 2010;24(6-7):591. doi: 10.1007/s10822-010-9349-1. [DOI] [PubMed] [Google Scholar]
- 30.LigPrep. 2.6. Schrödinger LLC; New York, NY: 2013. [Google Scholar]
- 31.Gallicchio E, Lapelosa M, Levy RM. J Chem Theory Comput. 2010;6(9):2961. doi: 10.1021/ct1002913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pharmacophore perception, development, and use in drug design. International University Line; La Jolla, CA: 1999. [Google Scholar]
- 33.Pharmacophores and Pharmacophore Searches. Wiley-VCH; 2006. [Google Scholar]
- 34.Zhu T, Cao S, Su P-C, Patel R, Shah D, Chokshi HB, Szukala R, Johnson ME, Hevener KE. Journal of Medicinal Chemistry. 2013;56(17):6560. doi: 10.1021/jm301916b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanner MF. J Mol Graph Model. 1999;17(1):57. [PubMed] [Google Scholar]
- 36.Lin J-H, Perryman AL, Schames JR, McCammon JA. Journal of the American Chemical Society. 2002;124(20):5632. doi: 10.1021/ja0260162. [DOI] [PubMed] [Google Scholar]
- 37.Amaro RE, Baron R, McCammon JA. J Comput Aided Mol Des. 2008;22(9):693. doi: 10.1007/s10822-007-9159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nichols SE, Baron R, Ivetac A, McCammon JA. Journal of Chemical Information and Modeling. 2011;51(6):1439. doi: 10.1021/ci200117n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin JH, Perryman AL, Schames JR, McCammon JA. Biopolymers. 2003;68(1):47. doi: 10.1002/bip.10218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Raw data used to choose LEDGF receptor models. Visual analysis was used for receptor evaluation since AUC values from ROC curves were too similar. After Phase 1 and 2 submissions to SAMPL, the visual analysis was formalized in creating the Rank Difference Ratio metric (see Fig. 4). For A and B the relative ranks for LEDGF ligands are listed in column 1, and the absolute ranks versus each target are listed in the subsequent columns underneath the PDB ID for that receptor model. The receptor models that were selected as targets are highlighted in magenta in row 1. For each block of 10 rows; the minimum (in green), average (in white), and maximum (in red) values of the absolute ranks were calculated and colored. More predictive targets have more green and white in each block and less red, they have more blocks (i.e., more LEDGF ligands were ranked higher than decoys), and the numbers in each cell will be closer to the relative rankings (“line numbers” in column 1). The compounds that were harvested in (A) had to pass the following filter: a minimum of 2 hydrogen bonds to IN and a hydrogen bond to the backbone amino group of Glu170. The compounds that were harvested in (B) had to pass the following filter: a minimum of 2 hydrogen bonds to IN and a hydrogen bond to the backbone amino group of His171.