Abstract
Doxorubicin (DOX), one of the most effective anticancer drugs, is known to generate progressive cardiac damage, which is due, in part, to DOX-induced reactive oxygen species (ROS). The elevated ROS often induce oxidative protein modifications that result in alteration of protein functions. This study demonstrates that the level of proteins adducted by 4-hydroxy-2-nonenal (HNE), a lipid peroxidation product, is significantly increased in mouse heart mitochondria following DOX treatment. A redox proteomics method involving 2D electrophoresis followed by mass spectrometry and investigation of protein data bases identified several HNE-modified mitochondria proteins, which were verified by HNE-specific immunoprecipitation in cardiac mitochondria from the DOX-treated mice. The majority of the identified proteins are related to mitochondrial energy metabolism. These include proteins in the citric acid cycle (TCA) and electron transport chain (ETC). The enzymatic activities of the HNE-adducted proteins were significantly reduced in DOX-treated mice. Consistent with the decline in the function of the HNE adducted proteins, the respiratory function of cardiac mitochondria as determined by oxygen consumption rate (OCR) was also significantly reduced after DOX treatment. The treatment with Mn(III) meso-tetrakis(N-n-butoxyethylpyridinium-2-yl)porphyrin, MnP, an SOD mimic, averted the doxorubicin-induced mitochondrial dysfunctions as well as the HNE protein adductions. Together, the results demonstrate that free radical-mediated alteration of energy metabolism is an important mechanism mediating DOX-induced cardiac injury suggesting that metabolic intervention may represent a novel approach to preventing cardiac injury after chemotherapy.
Keywords: Doxorubicin; Cardiac injury; ATP synthase, dihydrolipoyl dehydrogenase; Succinate dehydrogenase [ubiquinone] flavoprotein; NADH dehydrogenase [ubiquinone] iron-sulfur protein 2; Oxidative stress; Redox proteomics; Free radicals; metabolism
Introduction
Doxorubicin (DOX), one of the most effective anticancer drugs, has been a therapeutic for a broad spectrum of human cancers for almost half a century, and remains the first choice for many aggressive tumors, such as acute myeloid leukemia [1]. Its clinical application is highly limited due to the dose-related, progressive and irreversible cardiac damage it causes. The mortality of patients who develop congestive heart failure after DOX treatments can be as high as 50% [2], increasing significantly when cumulative doses are more than 500 mg/m2 [3].
Numerous studies have focused on the mechanisms behind DOX-induced cardiotoxic effects and demonstrate multifactorial causes. However, there is consensus that oxidative stress is a primary mechanism of DOX-induced cardiotoxicity and that the stress is attributed to the formation of reactive oxygen species (ROS). DOX generates ROS by various means, mainly through redox cycling. The quinone moiety of DOX can be converted one-electronically to semiquinone by several cellular oxidoreductases [4]. One-electron oxidation of the DOX-semiquinone radical to the DOX-quinone form leads to the generation of a highly reactive superoxide which can be further involved in production of a variety of ROS, including OH·, ROO·, ONOO−, ROOH and H2O2 [5].
ROS are highly reactive with biomolecules, including lipids, proteins, carbohydrates, DNA and RNA, and lead to cellular dysfunction [6]. A major source of ROS–mediated injury is lipid peroxidation, the reaction of ROS with the polyunsaturated fatty acids of lipid membranes. Lipid peroxidation generates a number of cytotoxic and highly reactive byproducts such as aldehydes, alkenals, and hydroxyalkenals. Among lipid peroxidation products, 4-hydroxy-2-nonenal (HNE) is the most abundant [7]. HNE readily reacts with proteins and, at higher concentrations, with DNA [8, 9]. HNE has a high affinity for and covalently attaches to Cys, His, and Lys residues by the Michael addition [10] which leads to dysfunction in the target proteins linked to intracellular signal transduction, aging and many human diseases.
Cardiomyocytes have more mitochondria, both in number and volume, than other cells, and they are the most active cells with regards to the oxidative phosphorylation that is required for their energy needs [11]. The complex that DOX forms with cardiolipin, which resides in the mitochondrial inner membrane [12], places DOX in close proximity to the mitochondrial electron-transport chain. The redox cycling of DOX is mediated through its interaction with NADH dehydrogenase (complex I) of the mitochondrial electron transport chain (ETC) [13]. Thus, active mitochondria are both important sources and primary targets of DOX-induced ROS. Multiple studies indicate that mitochondrial dysfunction is a key factor in the process of DOX-induced pathogenicity [14] [15] [16]. Studies utilizing tissue samples from patients receiving DOX reveal histopathological evidence that suggests disruption of mitochondrial structure and membranes [17]; similar observations were made in animal models and in cells in vitro [8, 18].
HNE protein adductions play important roles in regulating the function of proteins that may lead to cellular dysfunction. In the current study, we used 2D electrophoresis and redox immunochemistry analysis followed by mass spectrometry and interrogation of protein data bases to identify eight HNE adducted proteins in mouse heart mitochondria after DOX treatment. We then verified the consequences of HNE adduction on the functions of those proteins via enzymatic activity assays as well as on mitochondrial function.
Materials and methods
Materials
Doxorubicin was obtained from Bedford Laboratories, Inc. (Bedford, OH). SDHA, DLD and FH antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). MnTnBuOE-2-PyP5+(MnP) was produced by Dr. Ines Batinic-Haberle of the Department of Radiation Oncology, Duke University. N-Acetylcysteine (NAC) and ATP synthase antibody was purchased from Sigma (St. Louis, MO). NUDFS2 and HNE antibodies and complex I and ATP synthase activity assay kits were purchased from Abcam (Cambridge, MA); H9C2, neonatal rat heart cell line, from ATCC (Manassas, VA).
Animal treatment and mitochondrial isolation
All animals were housed in the University of Kentucky Animal Facility and experiments were conducted using procedures approved by Institutional Animal Care in accordance with the NIH Guide for the Care and Use of Laboratory Animals. In-house bred, 9-weeks-old, male, C57BL/6 mice were treated with a single dose of 20 mg/kg of DOX or saline via intraperitoneal injection (IP). 72 h after injection, mice were euthanized.
Heart mitochondria were isolated as described previously [19]. Briefly, freshly isolated hearts were washed in ice-cold isolation buffer (0.225 M mannitol, 0.075 M sucrose, 1 mM EGTA, pH 7.4), and homogenized at 500 rpm with a chilled Teflon pestle for 10 strokes in isolation buffer. The homogenate was centrifuged at 480 g at 4°C for 5 min in a Sorval SS 34 rotor. The resulting supernatant was filtered through double-layered cheesecloth and centrifuged at 7700 g at 4°C for 10 min. The pellet was washed twice by gentle resuspension in 3 mL isolation buffer and centrifuged at 7700 g at 4°C for 10 min. The resulting mitochondria were either used or frozen in liquid nitrogen for further analysis. The purity of mitochondria was examined using Lamin A (nuclear protein) and IκB-α (cytoskeletal protein) on Western blot.
Total protein bound HNE detection
The levels of total protein bound HNE were determined in the Free Radical Biology in Cancer Shared Resource Facility (FRBC SRF) at the University of Kentucky. The mitochondrial samples (pellets) were thawed and resuspended in a small volume (75–150 uL) of ice-cold homogenization buffer (0.32 M Sucrose, 10 mM HEPES pH 7.4, 2 mM EDTA, protease inhibitors). Five microliters of the homogenized sample were mixed and diluted with an equal volume of 12% SDS. Samples were further denatured with 10 µl of modified Laemmli buffer (0.125 M Trizma base, 4% SDS and 20% Glycerol) for 20 min at room temperature. Next, 250 ng of the derivatized protein were loaded in each slot (48-well slot-format Bio-Dot SF Apparatus with nitrocellulose membranes, pore size 0.2 µm, Bio-Rad, Hercules, CA). The antibody reaction was developed by using BCIP (5-bromo-4-chloro-3-indolyl-phosphate) in conjunction with NBT (nitro blue tetrazolium). The immunodetection was performed by using 1:5000 Anti-4 hydroxynonenal antiserum (Alpha Diagnostic International Inc., San Antonio, TX) and goat 1:7500 anti-rabbit IgG (Sigma-Aldrich, St Louis, MO) antibody for the secondary detection. The nitrocellulose membranes were scanned by Photo Scanner (Epson Perfection V600, Long Beach, CA), and slot blot line densities were quantified by the ImageQuant TL software package (GE Healthcare Bio-Sciences, Piscataway, NJ).
Two-dimensional gel electrophoresis and protein mass spectrometry studies
Two-dimensional gel electrophoresis was performed in the core facility of FRBC SRF in a protocol similar to that previously described [20]. Briefly, a duplicate amount (150 µg protein) of each isolated mitochondrial sample underwent trichloroacetic acid (TCA) precipitation and rehydration in 2D gel rehydration media (8 M urea, 2 M thiourea, 2% Chaps, 0.2% biolytes, 50 mM DTT, bromophenol blue dissolved in deionized water made fresh before use). Samples were separated according to their isoelectric point (IP) using 11 cm, pH 3–10, immobilized pH gradient (IPG) strips, using the Isoelectric focusing (IEF) Cell system (Bio-Rad) for the first-dimension separations. For each sample we produced two identical IPG strips with an equal amount of initial protein. After the proteins were separated in the first dimension, every pair of IPG strips was separated according to its molecular migration rate in one gel running box (each box ran two gels simultaneously) with precast Criterion XT 8–16% Bis-Tris Gel w/MOPS for the second-dimension separations. All gels ran under constant voltage (200V) for 65 min. The “twin” gels from each sample were handled as follows: One “twin” gel was stained with Sypro Ruby Stain (Bio-Rad) for total protein and the second “twin” gel was transferred to nitrocellulose and probed with the same antibodies/developer system as for the slot-blots. Images of the “twin” spots from each couple (2D gel – HNE 2D blot) were processed with PDQuest™ 2-D Analysis Software (Bio-Rad) for each group to create master images. Optical densities of the master images for the individual spot signals were analyzed by calculating the average differences in the intensities between the gel spot and corresponding HNE spot signal from the “twin” blot. Next, the ratiometric values were converted to a log scale and two-tailed t-test, assuming unequal variances to identify the spots of the highest statistical significance. After all spots of interest were carefully mapped, gel plugs containing the selected proteins were cut. Each gel plug was trypsinized and peptides were cleaned with ZipTip (Millipore Corp., Bedford, MA) procedures prior to the liquid chromatography–mass spectrometry analysis.
All mass spectra data reported in this study were acquired from the University of Louisville Mass Spectrometry Facility (ULMSF) using nanospray ionization (ESI), with tandem mass spectrometry (ESI-MS/MS) as described previously [21, 22]. MS/MS spectra were searched against the International Protein Index (IPI) database using SEQUEST. IPI accession numbers were cross-correlated with Swiss Prot accession numbers for final protein identification.
Immunoprecipitation and immunoblotting
Animal heart tissues were homogenized with a chilled Teflon pestle in RIPA buffer (50 mM Tris–HCl pH 7.9, 1% NP-40, 0.1% SDS, 150 mM NaCl, 1 mM EDTA, protease inhibitor cocktail (Calbiochem, Billerica, MA), incubated for 30 min on ice and then centrifuged for 10 min at 16000 xg. The supernatant containing 250 µg of protein in 0.5 mL RIPA buffer was precleaned with protein G-conjugated agarose beads (GE Healthcare, Cincinnati, OH). 2 µg of HNE, or mouse IgG antibody as control, were added. After overnight incubation at 4°C, 40 L of protein G-conjugated agarose beads were added and incubated for 1 h at 4°C. The beads were washed four times with RIPA buffer and resuspended in 60 µL Laemmli buffer. The immunoprecipitated samples were run on SDS-PAGE gels and blotted with specific antibodies at 1:1000 dilution. Western blots were prepared using the same protocol as described previously [23]. H9C2 cells were pretreated with 20 µM of MnP for 1 h, and then with 0.1 µM DOX and the same concentration of MnP for 3 days. The cell lysate was used for immunoprecipitation as described above.
Enzyme activity assays
ATP synthase and complex I activity were analyzed using the ATP synthase microplate kit and complex I activity kit (Abcam, Cambridge, MA), respectively, following the manufacturer’s protocols. Briefly, mitochondrial membrane proteins were isolated and immunocaptured in the wells of the microplate. The ATP synthase activity was measured by monitoring the conversion of ATP to ADP by ATP synthase coupled to the oxidation reaction of NADH to NAD+ with a reduction in absorbance at 340nm. The complex I activity was measured by following the oxidation of NADH to NAD+ and the simultaneous reduction of a dye which leads to increased absorbance at 450nm.
The DLD protein activity was measured by DLD catalyzed NAD+-dependent oxidation of dihydrolipoamide as described by Yan et al. [24]. Briefly, 2 µg of mitochondrial extract or 100 µg of cell lysate were mixed in 200 ul of total reaction volume with 100 mM potassium phosphate, pH 8.0, 1 mM EDTA, 0.6 mg/ml BSA, and 3 mM NAD+. The reaction was initiated by the addition of 3 mM dihydrolipoamide and measured at wavelength 340nm for 5 min.
SDHA activity was measured using a procedure [25] [26] modified by our lab. Homogenated 5 µg of mitochondrial protein or 50 µg of H9C2 cellular protein in 200 L H2O were mixed with 200 µL of reaction buffer (0.1 M potassium phosphate pH 7.4, 0.1 M sodium succinate, 0.05 M sucrose) containing freshly added 1mg/ml of p-iodonitrotetrazolium violet (Sigma). The blank was prepared without sodium succinate. The reaction was incubated at 37°C for 20 min, stopped with 400 L of 5% TCA and extracted with 0.8 mL N-butanol. After centrifugation at 1000 rpm for 5 min, 200 L of upper layer were transferred to a 96-well plate and measured at OD 490nm.
Mitochondrial function measurements
Oxygen consumption in cells and mitochondria was determined using the Seahorse Extracellular Flux (XF-96) analyzer (Seahorse Bioscience, Chicopee, MA). To allow comparison between experiments, data are presented as oxygen consumption rate (OCR) in pmoles/min/mg protein and extracellular acidification rate (ECAR) in mpH/min/mg protein. H9C2 cells were treated with different concentrations of DOX or saline for 48 h, trypsinized and reseeded at 45,000 cells per well in triplicate with the same treatment condition used in the Seahorse Bioscience XF microplates with glucose. After 24 h, basal OCR was measured four times and plotted as a function of cells under the basal condition followed by the sequential addition of oligomycin (1 µg/ml), FCCP (1 µM), antimycin (2 µM) and rotonone (1 µM), as indicated. Freshly isolated mitochondrial samples were plated at 3ug per well in triplicate and immediately measured using the Seahorse analyzer. The progress curve is annotated to show the relative contribution of basal, ATP-linked and maximal oxygen consumption after the addition of FCCP, and the reserve capacity of the samples.
For the ECAR measurements, cells were washed and resuspended in assay media lacking glucose. Basal ECAR was measured four times and plotted as a function of cells in basal condition followed by the sequential addition of glucose (25 mM), oligomycin (1 µg/ml) and 2-DG (25 mM), as indicated. The progress curve is annotated to show the relative contribution of glycolysis, glycolytic capacity and glycolytic reserve of the cells.
WST and Dichlorofluorescein (DCF) assays
Cells were plated at 6000/well or 2500/well for DCF or WST respectively in a 96-wells plate, pretreated with either 20 µM MnP or 1 mM NAC for 1 h, followed by treatment with either 0.1 µM or 0.25 µM DOX and the same concentration of MnP or NAC for 3 days (WST assay) or 1 h ( DCF assay). WST assays were performed by following the manufacturer’s instructions (Biovision, Mountain View, CA). DCF assays were performed as previously described [27]. Briefly, after treatment cells were incubated with 10 µg/ml of carboxy-H2DCFDA (sensitive to oxidation, Invitrogen) and 1 µg/ml of oxidized carboxy-DCFDA (insensitive to oxidation, Invitrogen) in PBS/2%FBS at 37°C for 20 min. The fluorescence in cells preloaded with carboxy-H2DCFDA was normalized to that in cells preloaded with carboxy-DCFDA (ratio of H2DCFDA/DCFDA) to control for the cell number, dye uptake, and ester cleavage differences between different treatment groups.
Statistical analysis
The samples used for 2D gel analysis, total HNE adducted protein detections and protein activity assays are n≥6. The experiments were conducted at least three times to verify the reproducibility of the findings. Statistical analyses were carried out with Statistical Analysis System software (SAS Institute, Cary, NC) and P values were calculated using the Student’s t test.
Results
Quantification of total HNE-bound protein and detection of specific proteins adducted by HNE in mitochondria
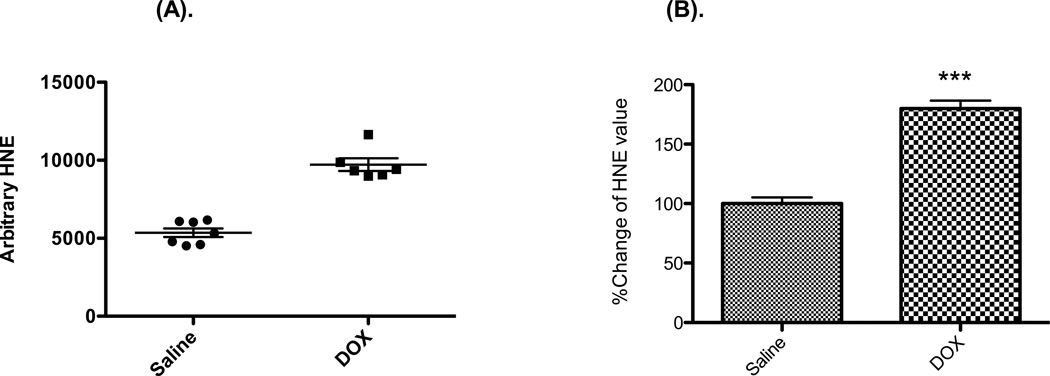
We isolated mitochondria from 9-weeks-old male C57B/L6 mice at 72 h after a single injection of DOX at 20 mg/kg (n≥6). First, we checked the expression of the total HNE adducted protein in mitochondria by using slot-blot apparatus and blotting with HNE antibody. The results show that the total HNE-bound protein level was significantly increased, by 70.4%, in the mitochondria from mice treated with DOX vs. saline (Figure 1).
Figure 1. Total HNE adducted protein is significantly increased in mouse mitochondrial homogenate 3 days after DOX injection.
(A) Absolute values of HNE-bound protein levels by slot blot gel analysis. Mice were treated with DOX at 20 mg/kg for 3 days (n ≥ 6). (B) The normalized % increase of HNE adducted protein in DOX treated vs. saline treated. P < 0.0001 when compared with samples from saline treated mice.
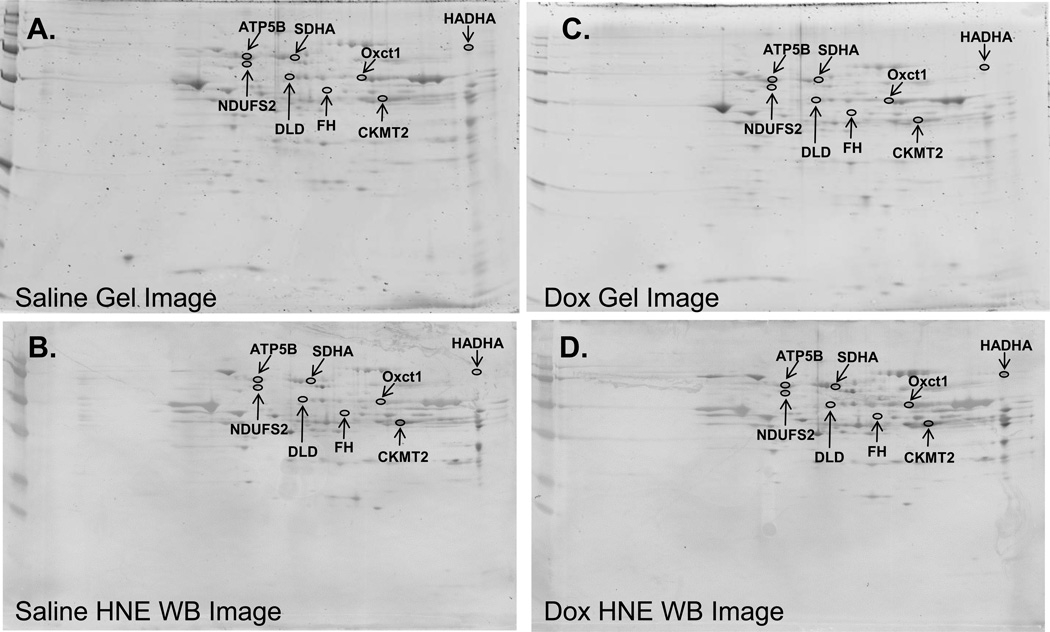
The same samples were used for detection of individual HNE adducted proteins by 2D gel analysis (saline or DOX treated, n=6). Each sample was separated using two 2D gels. One set of the gels was stained for visualization and quantification of proteins and the other one was transferred onto nitrocellulose and was immune detected with HNE antibody (Figure 2). The specific HNE level for each protein was calculated as the ratio of the HNE level on the membrane to the protein level on the gel. Eight spots with the biggest average difference between saline and DOX treatments were identified, excised and analyzed by Mass Spectrometry. As illustrated in Table 1, the Mass Spectrometry-identified proteins were ATP synthase subunit beta (ATP5B), dihydrolipoyl dehydrogenase (DLD), succinate dehydrogenase [ubiquinone] flavoprotein subunit (SDHA), NADH dehydrogenase [ubiquinone] iron-sulfur protein 2 (NDUFS2), fumarate hydratase (FH), trifunctional enzyme subunit alpha (HADHA), creatine kinase S-type (CKMT2), and 3-oxoacid CoA transferase 1 (Oxct1).
Figure 2. HNE adducted proteins identified by redox proteomics.
Representative 2D gels (A and C) and corresponding 2D HNE Western blot images (B and D) from saline treated (A and B) or DOX treated (C and D) (N=6). The identified protein spots are circled and labeled.
Table 1.
Mouse mitochondrial cardiac proteins showing increased HNE levels at 3 days after DOX injection.
| Protein Identified | Accession # |
Coverage | Number of peptidesa |
Score | MW (kDa) |
pI | t-testc | Foldb change |
|---|---|---|---|---|---|---|---|---|
| ATP synthase subunit beta | P56480 | 17.6 | 6 | 31.0 | 56.3 | 5.34 | 0.036 | 10.9 |
| Dihydrolipoyl dehydrogenase | O08749 | 22.2 | 9 | 57.8 | 54.2 | 7.90 | 0.054 | 8.94 |
| Succinate dehydrogenase [ubiquinone] flavoprotein subunit | Q8K2B3 | 15.7 | 6 | 42.3 | 72.5 | 7.37 | 0.057 | 7.20 |
| Trifunctional enzyme subunit alpha | Q8BMS1 | 24.0 | 16 | 137 | 82.6 | 9.14 | 0.038 | 4.97 |
| Creatine kinase S-type | Q6P8J7 | 41.0 | 13 | 118 | 47.4 | 8.40 | 0.051 | 4.40 |
| Isoform Cytoplasmic of Fumarate hydratase | P97807-2 | 12.4 | 4 | 27.1 | 50.0 | 7.94 | 0.029 | 3.66 |
| Succinyl-CoA:3-ketoacid-coenzyme A transferase 1 | Q3UJQ9 | 15.8 | 4 | 12.5 | 52.2 | 8.94 | 0.057 | 3.46 |
| NADH dehydrogenase [ubiquinone] iron-sulfur protein 2 | Q91WD5 | 48.0 | 16 | 145 | 52.6 | 6.99 | 0.058 | 1.98 |
The number of peptide sequences identified by nanospray ESI-MS/MS of tryptic peptides.
The fold-change in spot density from DOX treated compared with Saline treated.
The t value associated with fold-change calculated using a Student’s t test.
Under basal aerobic conditions, heart muscle gets 60% of its energy from fat, 35% from carbohydrates, and 5% from amino acids and ketone bodies [28]. Among the identified proteins, HADHA is a subunit of the mitochondrial trifunctional protein, which catalyzes the last three steps of mitochondrial beta-oxidation of long chain fatty acids. Octx1 plays a central role in ketone body catabolism by catalyzing the reversible transfer of coenzyme A from succinyl-CoA to acetoacetate. CKMT2 is responsible for the transfer of high-energy phosphate from mitochondria to the cytosolic carrier creatine. The HADHA protein has been reported as being modified by HNE in the ventilator muscles of rats after LPS induction [29] and its function was reduced after DOX treatment in rat and mouse [30, 31]. Interestingly, the other five proteins directly participate in the TCA cycle, a system critical for the maintenance of cardiomyocyte function [32, 33]. Therefore, in the current study we focused on the five TCA cycle component proteins.
Verification of HNE adduction by immunoprecipitation and examination of expression levels of the HNE adducted proteins by Western blot analysis
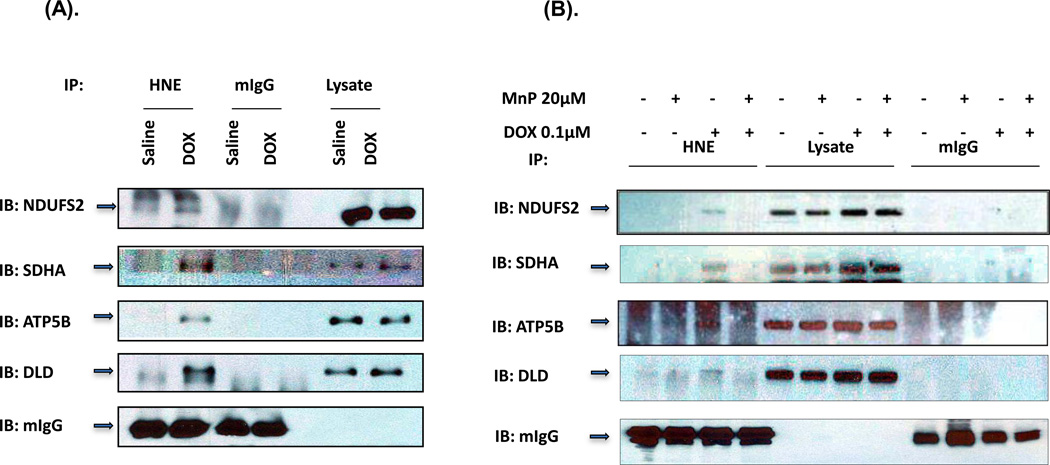
To verify HNE adduction to the proteins, we performed immunoprecipitation with HNE antibody followed by Western blot analysis with specific antibodies to each of the five proteins. In these experiments, we used whole-heart homogenates from mice treated with saline or DOX for 3 days. The data demonstrate that the ATP5B, DLD, SDHA and NDUFS2 were immunoprecipitated by HNE antibody but not by the control IgG antibody (Figure 3A). However, we were unable to immunoprecipitate FH protein by HNE antibody (data not shown). Our results confirm that the ATP5B, DLD, SDHA and NDUFS2 were modified by HNE.
Figure 3. Immunoprecipitation of HNE adducted proteins.
(A) Murine heart homogenates from mice treated with either saline or DOX for 3 days were immunoprecipitated with mouse HNE antibody (lanes 1 & 2) or mouse IgG antibody (lanes 3 & 4) as control. 20 µg of total heart homogenates from IP samples were loaded in lane 5 & 6. The immunoprecipitates were immunoblotted (IB) with antibodies specific for the indicated proteins. The experiment was repeated 3 times to verify results. (B) H9C2 cells pretreated with 20 µM MnP followed with 0.1µM DOX and MnP for 3 days. 500 µg cell lysate was used for each immunoprecipitation. Lane 1–4 were immunoprecipitions with mouse HNE antibody, lane 5–8 were 20 µg of each cell lysate, and lane 9–12 were immunoprecipitations with mouse IgG antibody.
To further verify whether HNE adduction of the proteins is due to DOX-induced ROS, we performed immunoprecipitation on H9C2 cells treated with DOX in the presence or absence of 20 µM MnTnBuOE-2-PyP 5+(MnP). MnP is an optimized SOD mimetic which mimics well the thermodynamics and kinetics of the catalysis of superoxide dismutation by Superoxide Dismutase enzyme (SOD). It further is a lipophilic analog which has enhanced mitochondrial accumulation, while reduced cellular toxicity [34–37]. As shown in Figure 3B, the MnP treatment protected the proteins from HNE adduction.
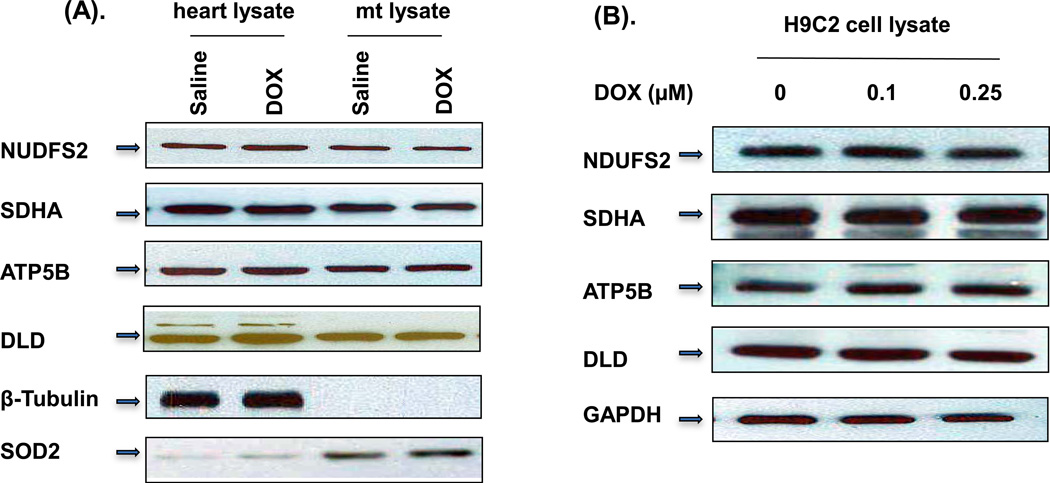
To investigate whether DOX treatment would alter the expression level of the four identified proteins, Western blot analysis was performed on mouse whole-heart homogenates as well as mitochondrial homogenates. As demonstrated in Figure 4A, the protein expression levels were not affected by DOX treatment in either whole heart or mitochondrial homogenates. To further verify the results, we performed Western blot analysis on H9C2 cell extracts (Figure 4B). The cells were treated with 0.1 or 0.25 µM of DOX for 72 h. Our H9C2 data support the finding that DOX treatment did not change the expression levels of the proteins in mice.
Figure 4. Western blot analysis of the identified HNE adducted proteins.
(A) 30 µg of total mouse heart homogenate and 10 µg of mouse mitochondrial homogenate from 3-days DOX-treated mice were loaded in gels and blotted with different antibodies as indicated. β-Tubulin and SOD2 were used as total heart homogenate and mitochondrial fraction loading controls, respectively. (B) Total H9C2 cell extracts treated with indicated concentration of DOX for 3 days were loaded and blotted with indicated antibodies. The experiments were repeated 4 times.
Examination of enzyme activity
HNE modification of proteins often leads to altered protein activity [38, 39]. Therefore, we next checked the activity of the individual proteins.
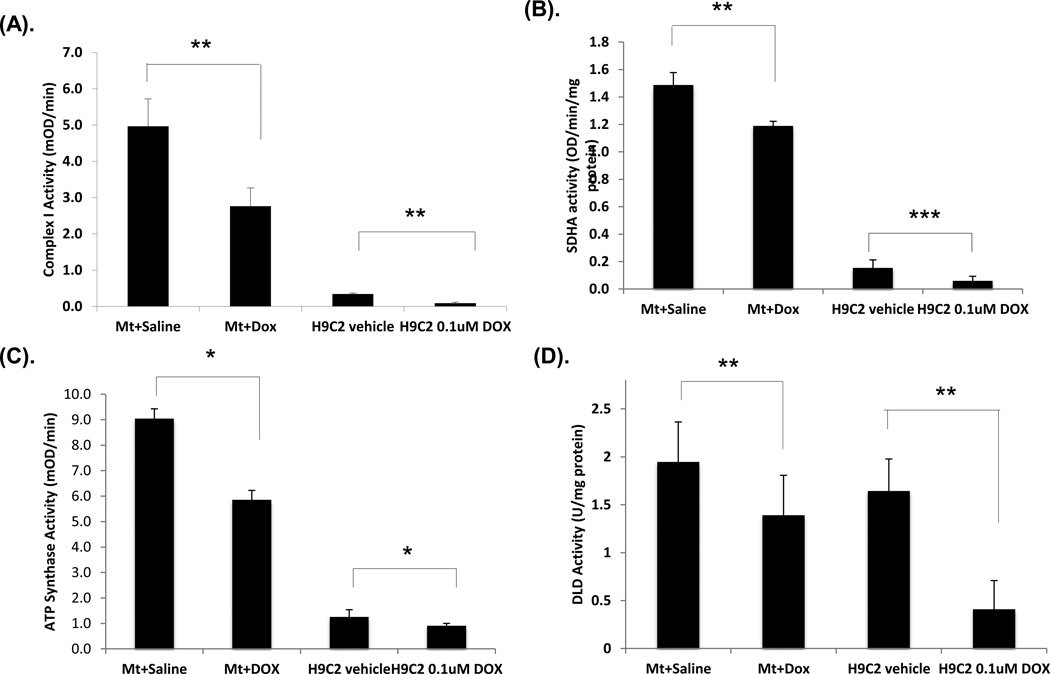
Mammalian mitochondrial complex I is composed of at least 43 different subunits. The NDUFS2 subunit is the third largest subunit, located in the extra-membranous part of complex I, near the membrane domain [40]. Disruption of this subunit results in the complete absence of the peripheral arm of complex I and in mitochondrial complex I deficiency [40]. Since the NDUFS2 subunit is the component of complex I, we measured the complex I activity. The level of complex I activity was reduced by 36% in mouse mitochondrial homogenates after DOX treatment and 77% in H9C2 cell extracts with 0.1 µM DOX treatment. The reduced activity in H9C2 cells treated with DOX was dose dependent (data not shown).
SDHA encodes a major catalytic subunit of succinate-ubiquinone oxidoreductase in complex II located in the inner mitochondrial membrane. It also participates in the TCA cycle, catalyzing succinate to fumarate conversion. Our results show that the SDHA activity in mouse heart mitochondrial homogenates and H9C2 cell extracts dropped 17.4% and 51.8%, respectively, after DOX treatment (Figure 5B).
Figure 5. Enzymatic activity of mitochondrial complex I, SDHA, ATP synthase and DLD.
Mitochondrial homogenates or H9C2 cell extracts treated with saline or DOX (mice n = 6, cells repeated at least 3 times) were used. The amount of protein from mitochondrial homogenate or H9C2 extracts used for each reaction in different assays: complex I (20 µg or 100 µg), SDHA (5 µg or 50 µg), DLD (2 µg or 50 µg) or ATP synthase (0.5 µg or 100µg), respectively. (A, B, C, D) are the activities for complex I, SDHA, ATP synthase or DLD, respectively. Mt represents mitochondrial homogenate. Data are expressed as mean ± SD. P-value (Student’s t test) <0.05 is considered significant.
ATP synthase subunit beta is an enzyme encoded by the ATP5B gene, one of the five subunits in the catalytic portion of complex V. The enzyme synthesizes ATP from ADP in the presence of a proton gradient across the mitochondrial inner membrane generated by the electron transport chain. In this experiment, we used the Elisa-based ATP synthase activity kit which first captures the ATP synthase complex in the reaction wells and then measures the activity by oxidation of NADH to NAD+. Our measurements show that ATP synthase activity was reduced by 27.4% in H9C2 cells treated with DOX and 44% in mitochondrial homogenates (Figure 5C).
DLD is a flavoprotein encoded by the DLD gene. The enzyme is a component in the alpha-ketoglutarate dehydrogenase complex (KGDC), which catalyzes the decarboxylation of alpha-ketoglutarate into succinyl-CoA in the TCA cycle, and the pyruvate dehydrogenase complex, which catalyzes pyruvate to Acetyl-CoA before the TCA cycle. The DLD activity was determined by measuring NAD+-dependent oxidation of dihydrolipoamide [24]. We used heart mitochondrial homogenates and H9C2 cell extracts treated with saline or DOX. The activity of DLD was reduced by 28.5% and 75.1%, respectively (Figure 5D).
Examination of mitochondrial function
Our results show that DOX treatment reduced the activities of several enzymes in the TCA cycle. In order to determine whether HNE adduction of the proteins in mitochondrial complexes is related to mitochondrial dysfunction, we examined mitochondrial function by using a Seahorse extracellular Flux (XF-96) analyzer. We assessed OCR on isolated crude mitochondria from mice, as well as OCR and ECAR from H9C2 cells treated with DOX.
OCR measures oxygen-dependent mitochondrial respiration. Oligomycin injections inhibit ATP synthesis by blocking ETC complex V, which distinguishes the oxygen consumption devoted to ATP synthesis from the oxygen consumption required to overcome the natural proton leak across the inner mitochondrial membrane. The injection of FCCP decouples ATP synthesis and hydrogen ion transport, leading to rapid consumption of energy and oxygen (mitochondrial maximal capacity) without the generation of ATP. FCCP treatment is also used to calculate the reserve oxygen consumption capacity of mitochondria under stress conditions [32, 41]. The reserve capacity was calculated with the difference between maximal OCR and basal OCR. Rotenone and antimycin were used to inhibit the function of complex I and III, respectively. The application of the two drugs completely inhibite the function of ETC.
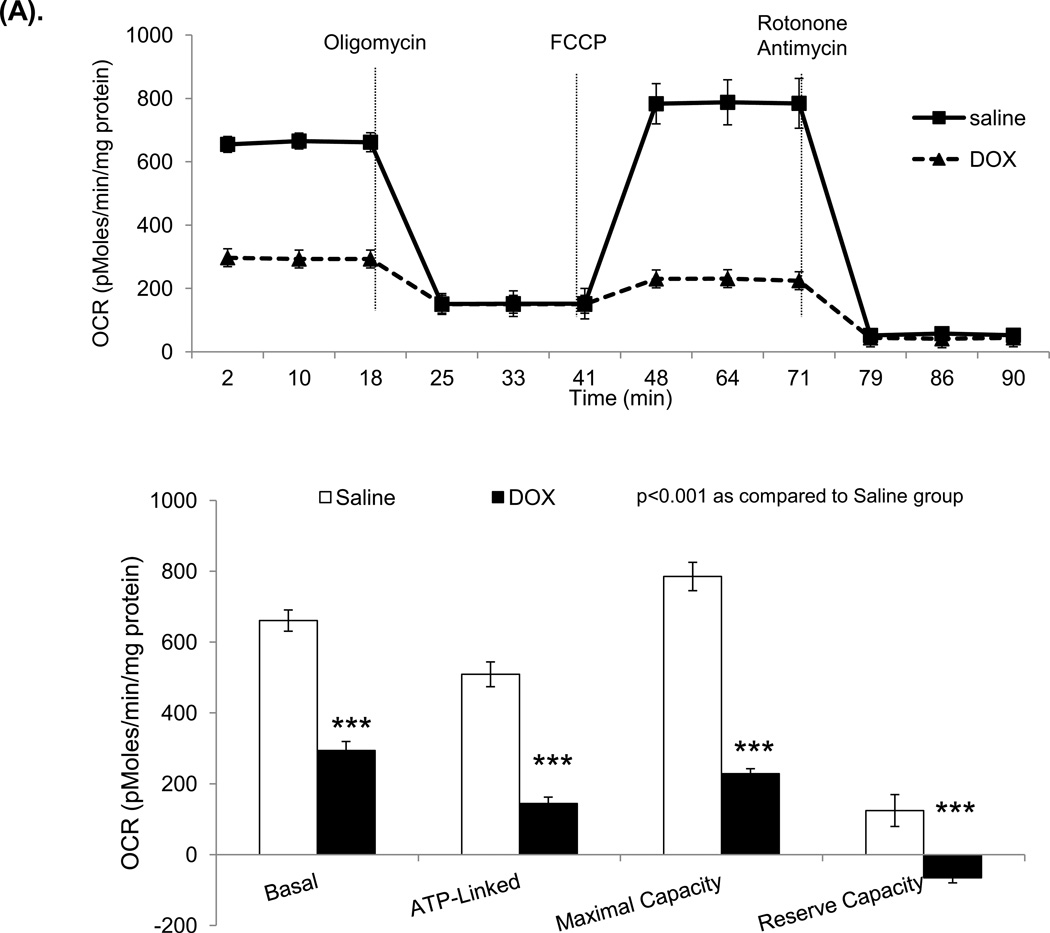
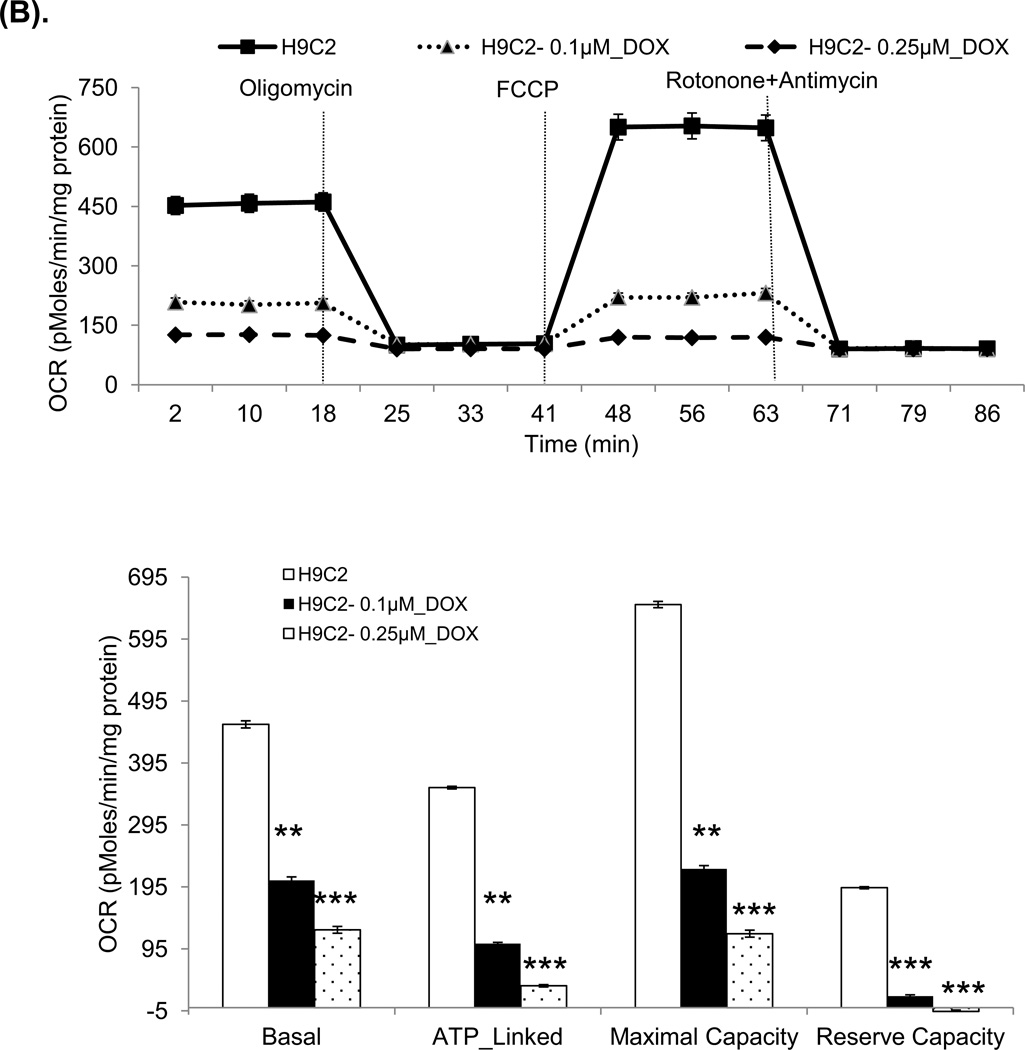
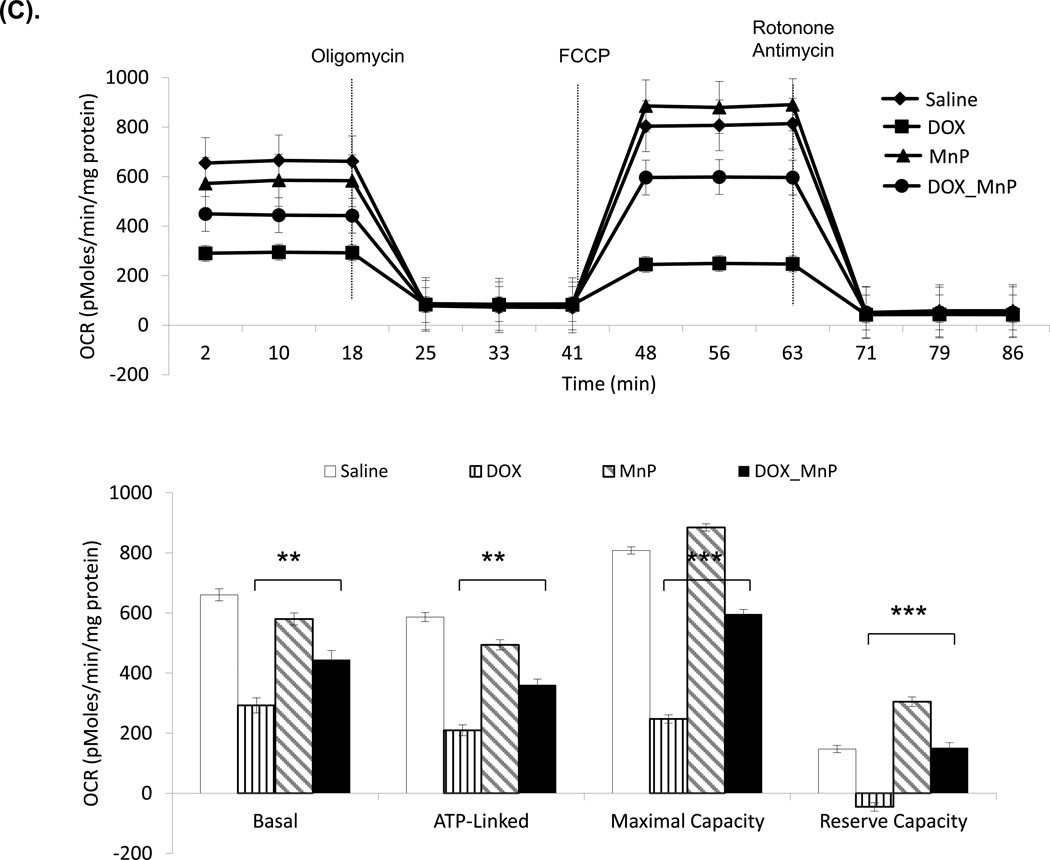
These experiments used freshly isolated mouse mitochondria after three-day treatment of DOX and H9C2 cells treated with DOX for 3 days. The results show reduced basal oxygen consumption that is consistent with our discovery of damaged complex I. As expected, the significantly reduced OCR in ATP-linked maximal capacity as well as reserve capacity after DOX treatment supports our findings of deficiency in complex II and V activities (Figure 6A and 6B). The DOX effect was dose-dependent in H9C2 cells. As shown in Figure 6C, treating mice with MnP prevented DOX induced damage in mitochondrial respiration.
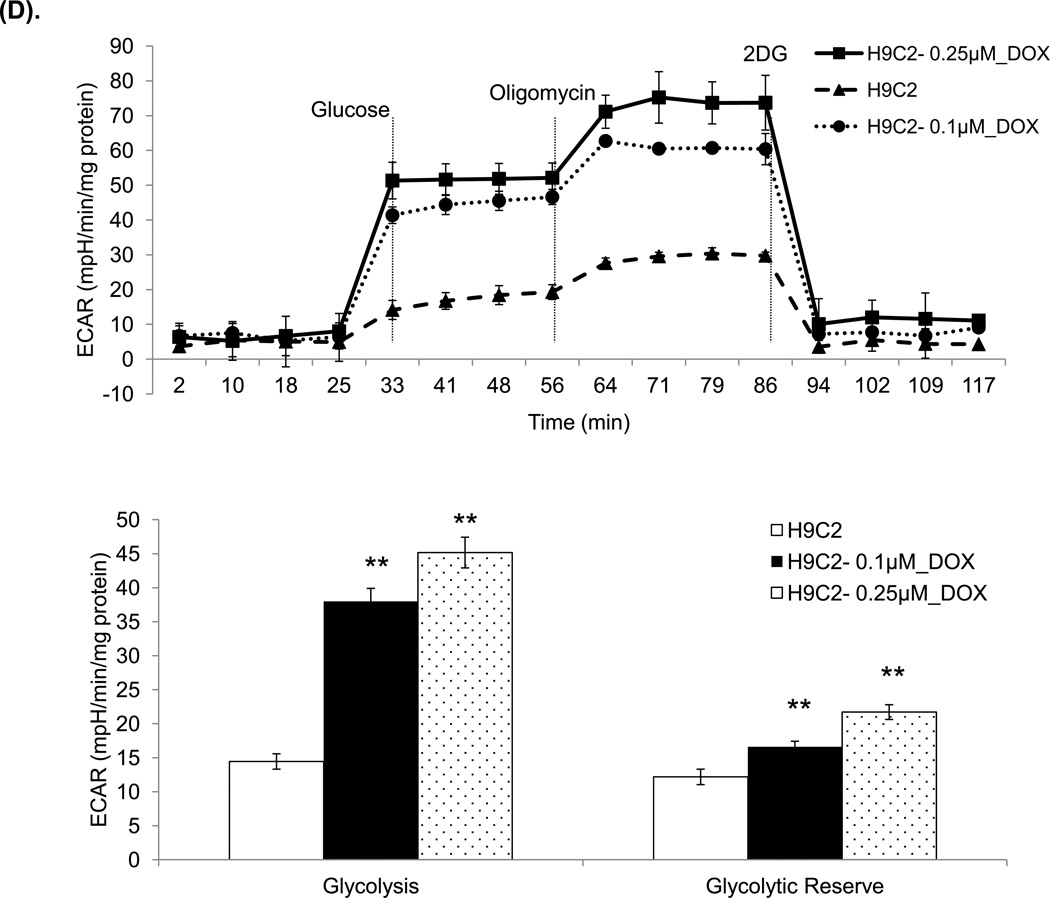
Figure 6. Oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in mouse mitochondria and H9C2 cells.
The Seahorse XF96 analyzer in the presence of the mitochondrial inhibitors (oligomycin, 1 µg/ml, FCCP 1 µM, or rotonone 1 µM and antimycin 2 µM) was used in the assays for OCR and ECAR. (A) The OCR of mitochondria freshly isolated from mouse heart at 3 days after saline or DOX injection. The units (pMoles/min/mg protein) for basal OCR, ATP-linked oxygen consumption, the maximum OCR after the addition of FCCP, and the reserve capacity of the cells were calculated and plotted. (B) The OCR of H9C2 cells after 3 days saline or DOX treatment and corresponding quantitation. (C) The OCR of H9C2 cells pretreated with MnP for 1 h and 3 days treatment of DOX together with MnP. (D) ECAR of H9C2 cells with the same treatment as (B). Cells were cultured for 2 h in the absence of glucose. Three sequential injections of D-glucose (2 g/L), oligomycin (1 µM), and 2-Deoxyglucose (100 mM) provided ECAR associated with glucose consumption. Glycolysis was defined as ECAR following the addition of D-glucose. The glycolytic capacity, defined as ECAR following the addition of oligomycin, and glycolytic reserve capacity, defined as the difference in ECAR following the addition of oligomycin following the addition of 2DG, were calculated and plotted.
ECAR measures extracellular acidification rate generated from glycolysis independent of oxygen. We also assessed the differences in glycolysis-induced ECAR among the samples. The measurement after injection of oligomycin represents glycolytic reserve. The injection of 2-DG, a glycolysis inhibitor, shut down the entire glycolytic process. The results of extracellular proton flux reveal that DOX-treated H9C2 cells showed a significant increase in extracellular acidification rates in glycolysis and glycolytic reserve (Figure 6D), suggesting that these cells exhibited enhanced glycolysis.
Assessment of ROS generation by treatment of DOX with or without two different classes of redox-active compounds
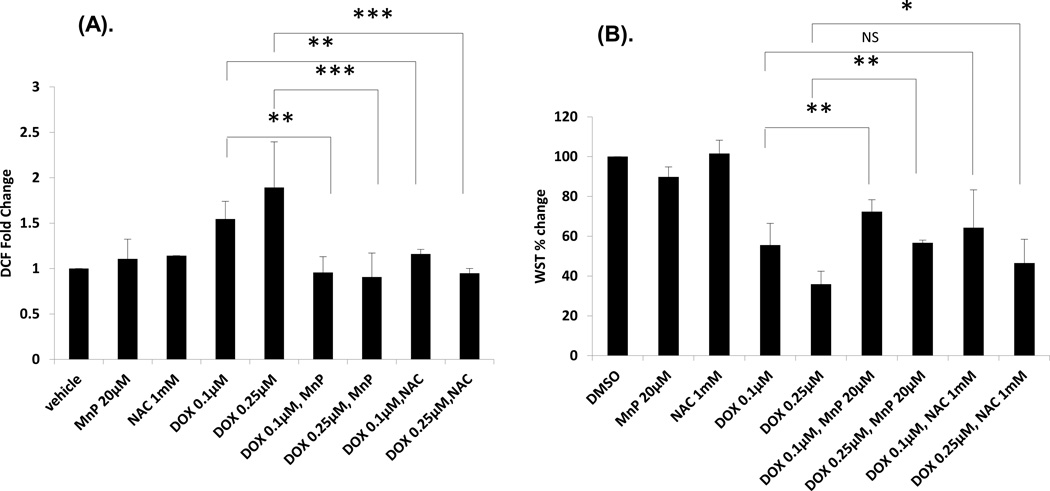
Elevated ROS in cells is considered as one of the major mechanisms of DOX-induced toxicity. HNE is one of the most active lipid peroxidation products generated via ROS-mediated injury. We next did a DCF assay to check the ROS levels in DOX-treated H9C2 cells in the presence or absence of antioxidants. As shown in Figure 7A, 0.1 µM or 0.25 µM of DOX treatment rapidly (after one hour treatment) induced normalized carboxy-H2DCFDA fluorescence, a general indicator of cellular ROS level. Moreover, MnP or NAC treatment significantly reduced cellular ROS level upon DOX treatment. Our results also suggest that DOX-induced elevation of cellular ROS is a relatively early event.
Figure 7. DCF and WST assays on H9C2 cells.
(A) The fold change of DCF on H9C2 cells pretreated with MnP or NAC for 1 h and then treated with DOX / MnP or NAC for 1 h. (B) The % change of WST on H9C2 cells pretreated with MnP or NAC for 1 h and then treated with DOX / MnP or NAC for 3 days.
Cardiomyocytes have a high number of mitochondria and their function depends heavily on mitochondrial respiration. We next did a WST assay to determine the survival of H9C2 cells treated with DOX with or without antioxidants. The results show (Figure 7B) that 20 µM of MnP or 1 mM NAC partially but significantly protected the survival of H9C2 cells treated with DOX at 0.25 M concentration. However, MnP, but not NAC, significantly reduced cardiomocyte death at lower DOX concentrations. These results suggest that redox active compounds, able to reduce levels of reactive species via their antioxidative actions, play an important role in the attenuation of DOX-induced toxicity in vitro.
Discussion
This study is the first global approach to identify specific HNE-adducted proteins associated with the TCA cycle and ETC chain after DOX treatment in mice. We chose to use a single high dose (20 mg/kg) of DOX, which is equivalent to a high-dose single injection in cancer patients, such as those with small-cell lung cancer [42], and has been used in a variety of animal models [23, 43, 44]. In our previous studies, we observed mitochondrial ultrastructural damage five days after this treatment [45]. Rosenoff et al. also reported cardiomyopathy in mice four days after this dosage, which is similar to the delayed DOX-induced cardiomyopathy noted in humans [43].
At least 20% of cardiomyocyte volume is composed of mitochondria in human and 32% in mouse [46] [47], enabling efficient energy production via oxidative phosphorylation. At basal metabolic rates, more than 95% of energy that sustains cardiac function and viability is derived from aerobic metabolism [28]. Damaged oxidative phosphorylation in mitochondria can lead to reliance on anaerobic glycolysis which may provide a short-term solution, but prolonged dependence most likely results in a severe energy deficiency that ultimately creates a bioenergetics crisis and leads to cardiac failure [14].
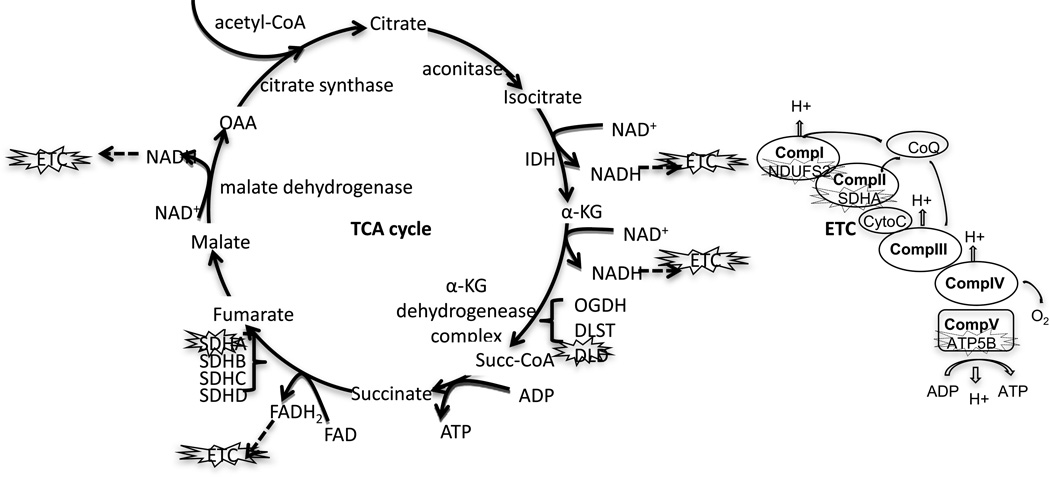
Our data are consistent with those observed for the HNE adduction of proteins in diabetes and cardiovascular diseases, such as ischemic heart and congestive heart failure [48–50]. It has been reported that treatment of isolated crude animal mitochondria by HNE induces mitochondrial respiration deficiency [41, 49–51]. HNE modification of SDHA [52], DLD [29], ATP5B [53] as well as NDUFS2 [54] has been identified in vitro or in vivo under oxidative conditions by different reseachers. However, the exact proteins modified by HNE in mitochondrial respiration after DOX or HNE treatment have not been reported. Using global analysis, we found that several major HNE-adducted proteins were involved in the TCA cycle and ETC (Figure 8). In addition, we show that the activities of complex I, II, and V were decreased by DOX. Furthermore, our bioenergetics results demonstrate that mitochondrial oxidative respiration was severely damaged and glycolysis was increased after DOX treatment, suggesting a shift of energy production in the damaged hearts.
Figure 8. A schematic demonstration of the identified HNE adducted proteins in the TCA cycle and ETC.
The main steps in the TCA cycle and ETC are presented. The HNE-adducted and functionally affected proteins are marked.
Mitochondrial complex I is a multi-subunit protein complex and an entry point for electrons into the respiratory chain in mitochondria. Mutations of the NDUFS2 gene are associated with mitochondrial complex I deficiency and cardiomyopathy [55]. Complex I is also a major source of cellular ROS and one of the three major DOX metabolism pathways [56]. The semiquinone to quinone cycling of DOX is carried out in complex I, which leads to the formation of superoxide and other ROS products [14] [57]. This laboratory and others have reported decreased complex I activity after DOX treatment [4, 44, 58, 59]. Our current results extend these findings and demonstrate, for the first time, that the NUDSF2 subunit is adducted by HNE after DOX treatment suggesting that this adduction may be a cause of DOX-induced complex I deficiency. There are seven Fe-sulfur protein subunits in mitochondrial complex I. The differential susceptibility of these proteins to oxidative modification upon DOX treatment is not clear. NUDSF2 and NUDFS7 are the membrane proximal subunites of the seven and the last proteins in the electron transport sequence in complex I. It is possible that the relative position of these Fe–sulfur proteins to membrane may play a role in the modification by a lipid proxidation like HNE.
SDHA is one of the four subunits in ETC complex II, and participates in the TCA cycle to catalyze succinate to fumarate conversion. Lashin et al. have reported that complex II activity was reduced in diabetic rat hearts and identified one of the subunits in complex II as having been modified by HNE [60]. Defects in SDHA cause cardiomyopathy dilated type 1GG (CMD1GG) [61]. CMD1GG is a disorder characterized by ventricular dilation and impaired systolic function, resulting in congestive heart failure, which shares a pathology similar to DOX-induced cardiac damage. Our previous study in DOX-treated mouse heart tissue also demonstrates a decline in complex II activity after DOX treatment, but the protein responsible for the defective complex II activity was unknown. In the current study, we identify SDHA as a HNE-modified protein in complex II suggesting that HNE adduction of SDHA may contribute to the reduced function of complex II in both mouse mitochondria and H9C2 cells treated with DOX.
ATP synthase/complex V synthesizes ATP from ADP by phosphorylation in the presence of oxygen and a proton gradient across the membrane. Complex V is composed of two structural domains, F(1) which contains the extra-membranous catalytic core and F(0) which contains the membrane proton channel. ATP5B is in one of the five subunits in F(1). DOX has been shown to inhibit the F0F1 proton pump of mitochondria [62, 63], but the exact target for the effect of DOX remains unknown. In this study, we demonstrate that ATP5B is adducted by HNE and that ATP synthase complex activity was reduced after DOX treatment suggesting ATP5B be the target for DOX-mediated inactivation of F0F1 function.
DLD is a component of the pyruvate dehydrogenase complex (catalyzing pyruvate to acetyl-CoA), the alpha-ketoglutarate dehydrogenase complex (catalyzing α-Ketoglutarate to succinate) in the TCA cycle, and the branched-chain alpha-keto acid dehydrogenase complex (involved in the breakdown of amino acids—leucine, isoleucine, and valine). The enzyme is redox sensitive [24] and has two redox-reactive cysteine residues at its active center that are indispensable for its catalytic function [64]. It has been reported that DLD was modified by HNE in the ventilatory muscles of rats after LPS treatment [29]. Our finding that HNE adduction of DLD is associated with the impaired activity of this protein is consistent with the decline in oxidative metabolism of cardiac tissue after DOX treatment.
In our mitochondrial function study, the oxygen consumption rate was significantly reduced by DOX. It is worth mentioning that our data demonstrate that DOX reduces basal oxygen consumption, which differs from other reports showing that stress overload as well as ischemia in isolated cardiomyocytes results in increased basal oxygen consumption [65]. However this reduction is consistent with the DOX induced damage to complex I and defective ETC activity. Thus, stress overload and DOX-induced effect might have different outcomes.
DOX, though an older chemotherapeutic drug, remains in active use for cancer treatments, despite its known cardiac toxicity, because of its therapeutic efficacy [1]. While attempts have been made to develop newer analogues that are less cardiotoxic, the resulting products are not as efficient as DOX [1]. Thus, identification of selective targets in DOX-induced cardiotoxicity may lead to potential therapeutic interventions. In addition to DOX, nearly 50% of the current FDA approved anticancer drugs are known to generate ROS [66]. Thus, the cardiovascular side effect of DOX may share common mechanisms with other anticancer drugs known to cause cardiac injury. We and others have previously shown that overexpression of antioxidant enzymes, such as manganese superoxide dismutase [45] and catalase [67], can protect the heart from DOX cardiotoxicity in transgenic mice. It should also be noted that while DOX causes oxidative stress via redox cycling of its quinone moiety and the consequent accumulation of ROS is well-documented, it has been shown recently that DOX induced DNA double strand breaks by poisoning of topoisomerase 2beta and transcriptome changes are responsible for defective mitochondrial biogenesis and ROS formation regardless of anthracycline redox cycling [68]. Thus, it is possible that the lipid peroxidation and protein adduction caused by HNE might be both the determinants and the consequnces of mitochondrial dysfunction and cardiotoxicity. However, our DCF results show that DOX induced ROS increase is rather an early event. Although classical antioxidants and ROS scavengers (such as NAC and vitamin E), usually very effective in acute DOX cardiotoxicity experiments, have failed in clinical trials, there were discrepancies in the concentrations/doses of antioxidants used in vitro and in vivo as well as their local concentrations in mitochondria [69]. Thus, the redox-active compounds, which, like MnP efficiently accumulate in mitochondria are powerful scavengers of reactive species [70] should be considered for future clinical studies. The data obtained from the current study provide specific markers for identifing and testing the cardioprotective effect of various antioxidants, mimetics or agents that selectively protect against DOX-induced cardiac injury and would benefit a large number of patients.
Supplementary Material
Highlights.
Doxorubicin increased total HNE adducted proteins in cardiac mitochondria.
Redox proteomics identified several HNE targetted cardiac mitochondria proteins important for energy metabolism.
Doxorubicin reduced the activities of complex I, SDHA, ATP synthase and DLD in ETC.
MnP averted Doxorubicin induced mitochondrial dysfunction and HNE adductions.
Acknowledgements
This work is supported by NIH Grants CA 049797, CA 139843, Cancer Center Support Grant (CCSG) P30CA177558 from NCI and the Edward P. Evans Foundation. The authors thank the University of Kentucky Free Radical Biology in Cancer Shared Resource Facility for its support. We also thank Dr. Liang-Jun Yan of the University of North Texas Health Science Center for the kind gift of synthesized dihydrolipoamide, as well as for his excellent technical help.
Abbreviations
- DOX
doxorubicin
- ATP5B
ATP synthase subunit beta
- DLD
dihydrolipoyl dehydrogenase
- SDHA
succinate dehydrogenase [ubiquinone] flavoprotein subunit
- NDUFS2
NADH dehydrogenase [ubiquinone] iron-sulfur protein 2
- ROS
Reactive oxygen species
- HNE
4-hydroxy-2-nonenal
- TCA
citric acid cycle
- ETC
electron transport chain
- OCR
oxygen consumption rate
- ECAR
extracellular acidification rate
- ATP5B
ATP synthase subunit beta
- DLD
dihydrolipoyl dehydrogenase
- SDHA
succinate dehydrogenase [ubiquinone] flavoprotein subunit
- NDUFS2
NADH dehydrogenase [ubiquinone] iron-sulfur protein 2
- FH
fumarate hydratase
- HADHA
trifunctional enzyme subunit alpha
- CKMT2
creatine kinase S-type
- Oxct1
3-oxoacid CoA transferase 1
- BMX-001
Mn(III) meso-tetrakis(N-n-butxyethylpyridinum-2-yl) porphyrin
- MnP
MnTnBuOE-2-PyP5+
- NAC
N-Acetylcysteine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pang B, Qiao X, Janssen L, Velds A, Groothuis T, Kerkhoven R, Nieuwland M, Ovaa H, Rottenberg S, van Tellingen O, Janssen J, Huijgens P, Zwart W, Neefjes J. Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat Commun. 2013;4:1908. doi: 10.1038/ncomms2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chatterjee K, Zhang J, Honbo N, Karliner JS. Doxorubicin cardiomyopathy. Cardiology. 2010;115:155–162. doi: 10.1159/000265166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singal PK, Iliskovic N. Doxorubicin-induced cardiomyopathy. N Engl J Med. 1998;339:900–905. doi: 10.1056/NEJM199809243391307. [DOI] [PubMed] [Google Scholar]
- 4.Marcillat O, Zhang Y, Davies KJ. Oxidative and non-oxidative mechanisms in the inactivation of cardiac mitochondrial electron transport chain components by doxorubicin. Biochem J. 1989;259:181–189. doi: 10.1042/bj2590181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minotti G, Recalcati S, Mordente A, Liberi G, Calafiore AM, Mancuso C, Preziosi P, Cairo G. The secondary alcohol metabolite of doxorubicin irreversibly inactivates aconitase/iron regulatory protein-1 in cytosolic fractions from human myocardium. FASEB J. 1998;12:541–552. doi: 10.1096/fasebj.12.7.541. [DOI] [PubMed] [Google Scholar]
- 6.Stadtman ER, Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Chem Res Toxicol. 1997;10:485–494. doi: 10.1021/tx960133r. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y, Sharma R, Sharma A, Awasthi S, Awasthi YC. Lipid peroxidation and cell cycle signaling: 4-hydroxynonenal, a key molecule in stress mediated signaling. Acta Biochim Pol. 2003;50:319–336. [PubMed] [Google Scholar]
- 8.Porta EA, Joun NS, Matsumura L, Nakasone B, Sablan H. Acute adriamycin cardiotoxicity in rats. Res Commun Chem Pathol Pharmacol. 1983;41:125–137. [PubMed] [Google Scholar]
- 9.al-Shabanah OA, Badary OA, Nagi MN, al-Gharably NM, al-Rikabi AC, al-Bekairi AM. Thymoquinone protects against doxorubicin-induced cardiotoxicity without compromising its antitumor activity. J Exp Clin Cancer Res. 1998;17:193–198. [PubMed] [Google Scholar]
- 10.Perluigi M, Coccia R, Butterfield DA. 4-Hydroxy-2-nonenal, a reactive product of lipid peroxidation, and neurodegenerative diseases: a toxic combination illuminated by redox proteomics studies. Antioxidants & redox signaling. 2012;17:1590–1609. doi: 10.1089/ars.2011.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou L, O'Rourke B. Cardiac mitochondrial network excitability: insights from computational analysis. American journal of physiology. Heart and circulatory physiology. 2012;302:H2178–H2189. doi: 10.1152/ajpheart.01073.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goormaghtigh E, Huart P, Praet M, Brasseur R, Ruysschaert JM. Structure of the adriamycin-cardiolipin complex. Role in mitochondrial toxicity. Biophys Chem. 1990;35:247–257. doi: 10.1016/0301-4622(90)80012-v. [DOI] [PubMed] [Google Scholar]
- 13.Davies KJ, Doroshow JH. Redox cycling of anthracyclines by cardiac mitochondria. I. Anthracycline radical formation by NADH dehydrogenase. The Journal of biological chemistry. 1986;261:3060–3067. [PubMed] [Google Scholar]
- 14.Berthiaume JM, Wallace KB. Adriamycin-induced oxidative mitochondrial cardiotoxicity. Cell Biol Toxicol. 2007;23:15–25. doi: 10.1007/s10565-006-0140-y. [DOI] [PubMed] [Google Scholar]
- 15.Grimsrud PA, Xie H, Griffin TJ, Bernlohr DA. Oxidative stress and covalent modification of protein with bioactive aldehydes. The Journal of biological chemistry. 2008;283:21837–21841. doi: 10.1074/jbc.R700019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho FS, Burgeiro A, Garcia R, Moreno AJ, Carvalho RA, Oliveira PJ. Doxorubicin-Induced Cardiotoxicity: From Bioenergetic Failure and Cell Death to Cardiomyopathy. Med Res Rev. 2013 doi: 10.1002/med.21280. [DOI] [PubMed] [Google Scholar]
- 17.Lefrak EA, Pitha J, Rosenheim S, Gottlieb JA. A clinicopathologic analysis of adriamycin cardiotoxicity. Cancer. 1973;32:302–314. doi: 10.1002/1097-0142(197308)32:2<302::aid-cncr2820320205>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 18.Green PS, Leeuwenburgh C. Mitochondrial dysfunction is an early indicator of doxorubicin-induced apoptosis. Biochim Biophys Acta. 2002;1588:94–101. doi: 10.1016/s0925-4439(02)00144-8. [DOI] [PubMed] [Google Scholar]
- 19.Velez JM, Miriyala S, Nithipongvanitch R, Noel T, Plabplueng CD, Oberley T, Jungsuwadee P, Van Remmen H, Vore M, St Clair DK. p53 Regulates oxidative stress-mediated retrograde signaling: a novel mechanism for chemotherapy-induced cardiac injury. PLoS One. 2011;6:e18005. doi: 10.1371/journal.pone.0018005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reed T, Perluigi M, Sultana R, Pierce WM, Klein JB, Turner DM, Coccia R, Markesbery WR, Butterfield DA. Redox proteomic identification of 4-hydroxy-2-nonenal-modified brain proteins in amnestic mild cognitive impairment: insight into the role of lipid peroxidation in the progression and pathogenesis of Alzheimer's disease. Neurobiol Dis. 2008;30:107–120. doi: 10.1016/j.nbd.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Jungsuwadee P, Cole MP, Sultana R, Joshi G, Tangpong J, Butterfield DA, St Clair DK, Vore M. Increase in Mrp1 expression and 4-hydroxy-2-nonenal adduction in heart tissue of Adriamycin-treated C57BL/6 mice. Mol Cancer Ther. 2006;5:2851–2860. doi: 10.1158/1535-7163.MCT-06-0297. [DOI] [PubMed] [Google Scholar]
- 22.Fiorini A, Sultana R, Forster S, Perluigi M, Cenini G, Cini C, Cai J, Klein JB, Farr SA, Niehoff ML, Morley JE, Kumar VB, Allan Butterfield D. Antisense directed against PS-1 gene decreases brain oxidative markers in aged senescence accelerated mice (SAMP8) and reverses learning and memory impairment: A proteomics study. Free radical biology & medicine. 2013;65C:1–14. doi: 10.1016/j.freeradbiomed.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, Daosukho C, Opii WO, Turner DM, Pierce WM, Klein JB, Vore M, Butterfield DA, St Clair DK. Redox proteomic identification of oxidized cardiac proteins in adriamycin-treated mice. Free radical biology & medicine. 2006;41:1470–1477. doi: 10.1016/j.freeradbiomed.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Yan LJ, Sumien N, Thangthaeng N, Forster MJ. Reversible inactivation of dihydrolipoamide dehydrogenase by mitochondrial hydrogen peroxide. Free Radic Res. 2013;47:123–133. doi: 10.3109/10715762.2012.752078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pennington RJ. Biochemistry of dystrophic muscle. Mitochondrial succinate-tetrazolium reductase and adenosine triphosphatase. Biochem J. 1961;80:649–654. doi: 10.1042/bj0800649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takiyyuddin MA, Cervenka JH, Hsiao RJ, Barbosa JA, Parmer RJ, O'Connor DT, Chromogranin A. Storage and release in hypertension. Hypertension. 1990;15:237–246. doi: 10.1161/01.hyp.15.3.237. [DOI] [PubMed] [Google Scholar]
- 27.Sun Y, St Clair DK, Xu Y, Crooks PA, St Clair WH. A NADPH oxidase-dependent redox signaling pathway mediates the selective radiosensitization effect of parthenolide in prostate cancer cells. Cancer Res. 2010;70:2880–2890. doi: 10.1158/0008-5472.CAN-09-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barret KE, Barman SM, Boitano S, Brooks HL. Ganong's Review of Medical Physiology, 22th Edition (LANGE Basic Science) 2005. [Google Scholar]
- 29.Hussain SN, Matar G, Barreiro E, Florian M, Divangahi M, Vassilakopoulos T. Modifications of proteins by 4-hydroxy-2-nonenal in the ventilatory muscles of rats. Am J Physiol Lung Cell Mol Physiol. 2006;290:L996–L1003. doi: 10.1152/ajplung.00337.2005. [DOI] [PubMed] [Google Scholar]
- 30.Robison TW, Giri SN, Wilson DW. Effects of chronic administration of doxorubicin on myocardial creatine phosphokinase and antioxidant defenses and levels of lipid peroxidation in tissues and plasma of rats. Journal of biochemical toxicology. 1989;4:87–94. doi: 10.1002/jbt.2570040204. [DOI] [PubMed] [Google Scholar]
- 31.Mihm MJ, Coyle CM, Schanbacher BL, Weinstein DM, Bauer JA. Peroxynitrite induced nitration and inactivation of myofibrillar creatine kinase in experimental heart failure. Cardiovasc Res. 2001;49:798–807. doi: 10.1016/s0008-6363(00)00307-2. [DOI] [PubMed] [Google Scholar]
- 32.Sansbury BE, Jones SP, Riggs DW, Darley-Usmar VM, Hill BG. Bioenergetic function in cardiovascular cells: the importance of the reserve capacity and its biological regulation. Chem Biol Interact. 2011;191:288–295. doi: 10.1016/j.cbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bugger H, Guzman C, Zechner C, Palmeri M, Russell KS, Russell RR., 3rd Uncoupling protein downregulation in doxorubicin-induced heart failure improves mitochondrial coupling but increases reactive oxygen species generation. Cancer Chemother Pharmacol. 2011;67:1381–1388. doi: 10.1007/s00280-010-1441-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajic Z, Tovmasyan A, Spasojevic I, Sheng H, Lu M, Li AM, Gralla EB, Warner DS, Benov L, Batinic-Haberle I. A new SOD mimic, Mn(III) ortho N-butoxyethylpyridylporphyrin, combines superb potency and lipophilicity with low toxicity. Free radical biology & medicine. 2012;52:1828–1834. doi: 10.1016/j.freeradbiomed.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adachi K, Fujiura Y, Mayumi F, Nozuhara A, Sugiu Y, Sakanashi T, Hidaka T, Toshima H. A deletion of mitochondrial DNA in murine doxorubicin-induced cardiotoxicity. Biochem Biophys Res Commun. 1993;195:945–951. doi: 10.1006/bbrc.1993.2135. [DOI] [PubMed] [Google Scholar]
- 36.Miriyala S, Spasojevic I, Tovmasyan A, Salvemini D, Vujaskovic Z, St Clair D, Batinic-Haberle I. Manganese superoxide dismutase, MnSOD and its mimics. Biochim Biophys Acta. 2012;1822:794–814. doi: 10.1016/j.bbadis.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Batinic-Haberle I, Tovmasyan A, Roberts ER, Vujaskovic Z, Leong KW, Spasojevic I. SOD Therapeutics: Latest Insights into Their Structure-Activity Relationships and Impact on the Cellular Redox-Based Signaling Pathways. Antioxidants & redox signaling. 2013 doi: 10.1089/ars.2012.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gewirtz DA. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem Pharmacol. 1999;57:727–741. doi: 10.1016/s0006-2952(98)00307-4. [DOI] [PubMed] [Google Scholar]
- 39.Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev. 2004;56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 40.Gianni L, Herman EH, Lipshultz SE, Minotti G, Sarvazyan N, Sawyer DB. Anthracycline cardiotoxicity: from bench to bedside. J Clin Oncol. 2008;26:3777–3784. doi: 10.1200/JCO.2007.14.9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hill BG, Dranka BP, Zou L, Chatham JC, Darley-Usmar VM. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem J. 2009;424:99–107. doi: 10.1042/BJ20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piscitelli SC, Rodvold KA, Rushing DA, Tewksbury DA. Pharmacokinetics and pharmacodynamics of doxorubicin in patients with small cell lung cancer. Clin Pharmacol Ther. 1993;53:555–561. doi: 10.1038/clpt.1993.69. [DOI] [PubMed] [Google Scholar]
- 43.Rosenoff SH, Olson HM, Young DM, Bostick F, Young RC. Adriamycin-induced cardiac damage in the mouse: a small-animal model of cardiotoxicity. J Natl Cancer Inst. 1975;55:191–194. doi: 10.1093/jnci/55.1.191. [DOI] [PubMed] [Google Scholar]
- 44.Chaiswing L, Cole MP, St Clair DK, Ittarat W, Szweda LI, Oberley TD. Oxidative damage precedes nitrative damage in adriamycin-induced cardiac mitochondrial injury. Toxicol Pathol. 2004;32:536–547. doi: 10.1080/01926230490502601. [DOI] [PubMed] [Google Scholar]
- 45.Yen HC, Oberley TD, Vichitbandha S, Ho YS, St Clair DK. The protective role of manganese superoxide dismutase against adriamycin-induced acute cardiac toxicity in transgenic mice. J Clin Invest. 1996;98:1253–1260. doi: 10.1172/JCI118909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.David H, Meyer R, Marx I, Guski H, Wenzelides K. Morphometric characterization of left ventricular myocardial cells of male rats during postnatal development. J Mol Cell Cardiol. 1979;11:631–638. doi: 10.1016/0022-2828(79)90377-8. [DOI] [PubMed] [Google Scholar]
- 47.Schaper J, Meiser E, Stammler G. Ultrastructural morphometric analysis of myocardium from dogs, rats, hamsters, mice, and from human hearts. Circ Res. 1985;56:377–391. doi: 10.1161/01.res.56.3.377. [DOI] [PubMed] [Google Scholar]
- 48.Hill BG, Awe SO, Vladykovskaya E, Ahmed Y, Liu SQ, Bhatnagar A, Srivastava S. Myocardial ischaemia inhibits mitochondrial metabolism of 4-hydroxy-trans-2-nonenal. Biochem J. 2009;417:513–524. doi: 10.1042/BJ20081615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eaton P, Li JM, Hearse DJ, Shattock MJ. Formation of 4-hydroxy-2-nonenal-modified proteins in ischemic rat heart. Am J Physiol. 1999;276:H935–H943. doi: 10.1152/ajpheart.1999.276.3.H935. [DOI] [PubMed] [Google Scholar]
- 50.Benderdour M, Charron G, DeBlois D, Comte B, Des Rosiers C. Cardiac mitochondrial NADP+-isocitrate dehydrogenase is inactivated through 4-hydroxynonenal adduct formation: an event that precedes hypertrophy development. The Journal of biological chemistry. 2003;278:45154–45159. doi: 10.1074/jbc.M306285200. [DOI] [PubMed] [Google Scholar]
- 51.Chen J, Henderson GI, Freeman GL. Role of 4-hydroxynonenal in modification of cytochrome c oxidase in ischemia/reperfused rat heart. J Mol Cell Cardiol. 2001;33:1919–1927. doi: 10.1006/jmcc.2001.1454. [DOI] [PubMed] [Google Scholar]
- 52.Aitken RJ, Whiting S, De Iuliis GN, McClymont S, Mitchell LA, Baker MA. Electrophilic aldehydes generated by sperm metabolism activate mitochondrial reactive oxygen species generation and apoptosis by targeting succinate dehydrogenase. The Journal of biological chemistry. 2012;287:33048–33060. doi: 10.1074/jbc.M112.366690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guo J, Prokai-Tatrai K, Nguyen V, Rauniyar N, Ughy B, Prokai L. Protein targets for carbonylation by 4-hydroxy-2-nonenal in rat liver mitochondria. J Proteomics. 2011;74:2370–2379. doi: 10.1016/j.jprot.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choksi KB, Nuss JE, Deford JH, Papaconstantinou J. Age-related alterations in oxidatively damaged proteins of mouse skeletal muscle mitochondrial electron transport chain complexes. Free radical biology & medicine. 2008;45:826–838. doi: 10.1016/j.freeradbiomed.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loeffen J, Elpeleg O, Smeitink J, Smeets R, Stockler-Ipsiroglu S, Mandel H, Sengers R, Trijbels F, van den Heuvel L. Mutations in the complex I NDUFS2 gene of patients with cardiomyopathy and encephalomyopathy. Ann Neurol. 2001;49:195–201. doi: 10.1002/1531-8249(20010201)49:2<195::aid-ana39>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 56.Thorn CF, Oshiro C, Marsh S, Hernandez-Boussard T, McLeod H, Klein TE, Altman RB. Doxorubicin pathways: pharmacodynamics and adverse effects. Pharmacogenet Genomics. 2011;21:440–446. doi: 10.1097/FPC.0b013e32833ffb56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pawlowska J, Tarasiuk J, Wolf CR, Paine MJ, Borowski E. Differential ability of cytostatics from anthraquinone group to generate free radicals in three enzymatic systems: NADH dehydrogenase, NADPH cytochrome P450 reductase, and xanthine oxidase. Oncol Res. 2003;13:245–252. doi: 10.3727/096504003108748294. [DOI] [PubMed] [Google Scholar]
- 58.Nicolay K, de Kruijff B. Effects of adriamycin on respiratory chain activities in mitochondria from rat liver, rat heart and bovine heart. Evidence for a preferential inhibition of complex III and IV. Biochimica et biophysica acta. 1987;892:320–330. doi: 10.1016/0005-2728(87)90236-2. [DOI] [PubMed] [Google Scholar]
- 59.Lebrecht D, Kokkori A, Ketelsen UP, Setzer B, Walker UA. Tissuespecific mtDNA lesions and radical-associated mitochondrial dysfunction in human hearts exposed to doxorubicin. J Pathol. 2005;207:436–444. doi: 10.1002/path.1863. [DOI] [PubMed] [Google Scholar]
- 60.Lashin OM, Szweda PA, Szweda LI, Romani AM. Decreased complex II respiration and HNE-modified SDH subunit in diabetic heart. Free radical biology & medicine. 2006;40:886–896. doi: 10.1016/j.freeradbiomed.2005.10.040. [DOI] [PubMed] [Google Scholar]
- 61.Levitas A, Muhammad E, Harel G, Saada A, Caspi VC, Manor E, Beck JC, Sheffield V, Parvari R. Familial neonatal isolated cardiomyopathy caused by a mutation in the flavoprotein subunit of succinate dehydrogenase. Eur J Hum Genet. 2010;18:1160–1165. doi: 10.1038/ejhg.2010.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olson RD, Mushlin PS, Brenner DE, Fleischer S, Cusack BJ, Chang BK, Boucek RJ., Jr Doxorubicin cardiotoxicity may be caused by its metabolite, doxorubicinol. Proc Natl Acad Sci U S A. 1988;85:3585–3589. doi: 10.1073/pnas.85.10.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mordente A, Meucci E, Silvestrini A, Martorana GE, Giardina B. New developments in anthracycline-induced cardiotoxicity. Curr Med Chem. 2009;16:1656–1672. doi: 10.2174/092986709788186228. [DOI] [PubMed] [Google Scholar]
- 64.Brautigam CA, Chuang JL, Tomchick DR, Machius M, Chuang DT. Crystal structure of human dihydrolipoamide dehydrogenase: NAD+/NADH binding and the structural basis of disease-causing mutations. J Mol Biol. 2005;350:543–552. doi: 10.1016/j.jmb.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 65.Smith DR, Stone D, Darley-Usmar VM. Stimulation of mitochondrial oxygen consumption in isolated cardiomyocytes after hypoxia-reoxygenation. Free Radic Res. 1996;24:159–166. doi: 10.3109/10715769609088013. [DOI] [PubMed] [Google Scholar]
- 66.Chen Y, Jungsuwadee P, Vore M, Butterfield DA, St Clair DK. Collateral damage in cancer chemotherapy: oxidative stress in nontargeted tissues. Mol Interv. 2007;7:147–156. doi: 10.1124/mi.7.3.6. [DOI] [PubMed] [Google Scholar]
- 67.Kang YJ, Sun X, Chen Y, Zhou Z. Inhibition of doxorubicin chronic toxicity in catalase-overexpressing transgenic mouse hearts. Chem Res Toxicol. 2002;15:1–6. doi: 10.1021/tx015532n. [DOI] [PubMed] [Google Scholar]
- 68.Zhang S, Liu X, Bawa-Khalfe T, Lu LS, Lyu YL, Liu LF, Yeh ET. Identification of the molecular basis of doxorubicin-induced cardiotoxicity. Nature medicine. 2012;18:1639–1642. doi: 10.1038/nm.2919. [DOI] [PubMed] [Google Scholar]
- 69.van Dalen EC, Caron HN, Dickinson HO, Kremer LC. Cardioprotective interventions for cancer patients receiving anthracyclines. Cochrane Database Syst Rev. 2011:CD003917. doi: 10.1002/14651858.CD003917.pub3. [DOI] [PubMed] [Google Scholar]
- 70.Bloodsworth A, O'Donnell VB, Batinic-Haberle I, Chumley PH, Hurt JB, Day BJ, Crow JP, Freeman BA. Manganese-porphyrin reactions with lipids and lipoproteins. Free radical biology & medicine. 2000;28:1017–1029. doi: 10.1016/s0891-5849(00)00194-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.