Abstract
Pluripotency of embryonic stem cells (ESCs) is defined by their ability to differentiate into three germ layers and derivative cell types1-3 and is established by an interactive network of proteins including OCT4 (also known as POU5F1; ref. 4), NANOG (refs 5,6), SOX2 (ref. 7) and their binding partners. The forkhead box O (FoxO) transcription factors are evolutionarily conserved regulators of longevity and stress response whose function is inhibited by AKT protein kinase. FoxO proteins are required for the maintenance of somatic and cancer stem cells8-13; however, their function in ESCs is unknown. We show that FOXO1 is essential for the maintenance of human ESC pluripotency, and that an orthologue of FOXO1 (Foxo1) exerts a similar function in mouse ESCs. This function is probably mediated through direct control by FOXO1 of OCT4 and SOX2 gene expression through occupation and activation of their respective promoters. Finally, AKT is not the predominant regulator of FOXO1 in human ESCs. Together these results indicate that FOXO1 is a component of the circuitry of human ESC pluripotency. These findings have critical implications for stem cell biology, development, longevity and reprogramming, with potentially important ramifications for therapy.
ESC pluripotency is maintained by OCT4 (octamer-binding transcription factor 4), NANOG and SOX2 (SRY-box containing protein 2), which form a feedback regulatory circuit positively regulating their own genes and activating genes encoding critical components of pluripotency while repressing genes important for developmental processes14. Identification of key regulators of ESC pluripotency provided a foundation for somatic cell reprogramming15-17 and is likely to have a critical impact on the use of human ESCs (hESCs) in regenerative medicine.
FoxO proteins are mammalian orthologues of DAF-16 (abnormal dauer formation protein 16), an essential protein in the regulation of stress response and ageing in Caenorhabditis elegans18. FoxO proteins are primarily phosphorylated and negatively regulated by AKT serine/threonine protein kinase downstream of the PI(3)K (phosphatidylinositol-3-OH kinase) signalling pathway19,20. A number of kinases other than AKT also phosphorylate and regulate FoxO proteins either positively or negatively21-23. In addition to phosphorylation, FoxO proteins are subject to several post-translational modifications such as acetylation24-26, ubiquitylation27, methylation28 and redox modulation29. The combined output of this stringent multilayer regulation determines the subcellular localization of FoxO proteins and ultimately their transcriptional activity30,31.
FoxO proteins exert key biological functions (reviewed in ref. 30) that seem overlapping but non-redundant, as evidenced by distinct phenotypes of their respective knockout models32,33 as well as studies in primary stem and progenitor cells9-13,34. FoxO proteins are bona fide tumour suppressors, as demonstrated by the phenotype of their conditional deletion in mice8, and as such promote cell cycle arrest, induce apoptosis, contribute to DNA damage repair and suppress oxidative stress by modulating genes involved in these processes30,31.
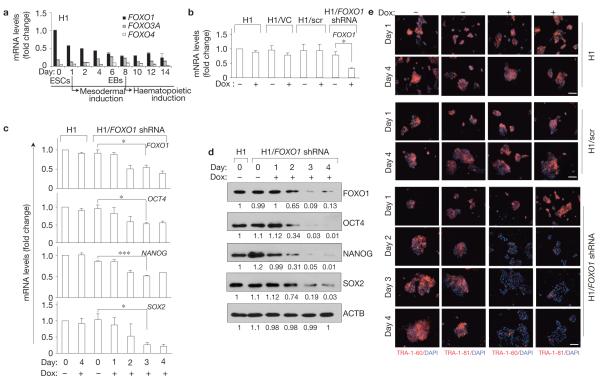
To address the potential function of FoxO proteins in human development, we analysed their expression in hESCs. ESC differentiation recapitulates early events of embryogenesis (reviewed in ref. 35), providing a suitable system for biochemical analyses of developmental processes under tightly controlled in vitro conditions. As previously predicted36, FOXO1 was the most abundant FOXO at the messenger RNA level in undifferentiated pluripotent H1 hESCs (Fig. 1a and Supplementary Fig. S1). Importantly, FOXO1 protein was at least seven times more abundant than FOXO3A and FOXO4 (no FOXO6 mRNA or protein was detectable, Supplementary Fig. S1b–f and data not shown) in these cells. The expression of FOXO1 was markedly downregulated during embryoid body formation and commitment to mesoderm and haematopoietic cells (Fig. 1a). Interestingly, in undifferentiated self-renewing hESCs, most FOXO1 was nuclear (Supplementary Fig. S1g). A similar pattern of FOXO1 distribution was found in a distinct hESC line HES2 (Supplementary Fig. S2a) and was highly conserved during mouse ESC (mESC) differentiation (Supplementary Fig. S2b), collectively indicating a potential role for FOXO1 in regulating ESC fate. To investigate this, we used two distinct FOXO1-targeting short hairpin RNA (shRNA) sequences and a lentiviral vector to deliver tetracycline-inducible shRNAs to inducibly knockdown FOXO1 in hESCs (H1/FOXO1 shRNA, H1/FOXO1 shRNA II). In these cells, the shRNA is driven by a tet-on hybrid promoter where the polymerase (Pol) III promoter H1 is fused to tetracycline operator sequences such that the shRNA expression requires the addition of doxycycline. For a complete description see the Supplementary Information and Supplementary Fig. S2c–j.
Figure 1.
FOXO1 is essential for the expression of hESC pluripotency markers. (a) qRT-PCR analysis of expression of FOXO genes in pluripotent undifferentiated hESCs and during mesodermal induction. The expression levels of FOXO3A and FOXO4 are relative to that of FOXO1 in undifferentiated H1 cells under self-renewal conditions. Note, downregulation of FOXO1 and upregulation of FOXO3A during differentiation of hESCs. FOXO6 expression was not detectable in hESCs. EBs, embryoid bodies. (b) FOXO1 expression was analysed by qRT-PCR in parental H1 hESCs and in H1 cells expressing shRNA targeting FOXO1 (H1/FOXO1 shRNA) cultured in the absence or presence of doxycycline (Dox) for 4 days under undifferentiated self-renewal conditions. H1 cells expressing vector control (H1/VC) or scrambled FOXO1 shRNA (H1/scr) were used as controls. (c) qRT-PCR analysis carried out as in b at the indicated times in cells treated with or without doxycycline. Quantification of the target genes was relative to the endogenous ACTB (-actin) transcript levels. Results are mean s.e.m. of three independent experiments, each carried out in triplicate (a-c); *P < 0.05, *P < 0.01, P < 0.001 (b,c). (d) An aliquot of cells from c was subjected to western blot analysis of the indicated proteins; relative intensities of bands are shown below each panel relative to that measured at time 0 in H1 cells (uncropped scanned gels are shown in Supplementary Fig. S12). (e) hESCs were cultured with or without doxycycline for the indicated times and immunostained for surface markers of pluripotency, TRA-1-60 and TRA-1-81, and counterstained with DAPI. Scale bars, 100 μm.
In the absence of doxycycline, stable expression of shRNA-containing lentiviral vectors did not perturb the normal development of experimental or control hESC-derived lines, which maintained pluripotency under appropriate culture conditions, and preserved full embryoid body formation and commitment to mesoderm and haematopoietic cells in vitro (Fig. 1b-e and Supplementary Figs S3 and S4). Three to four days after the addition of doxycycline, FOXO1 transcript was significantly reduced in both H1/FOXO1- shRNA and H1/FOXO1-shRNA-II cells maintained under pluripotency self-renewing conditions (Fig. 1b,c and Supplementary Fig. S4a). The expression of other FOXO proteins was not significantly altered (Supplementary Figs S4b and S5a,b), nor was their overall nuclear/cytoplasmic distribution (Supplementary Fig. S5c), indicating that the observed phenotype was specifically due to FOXO1 downregulation. Importantly, FOXO1-knockdown cells seemed to conserve their rate of growth and survival, as evidenced by the comparable timing of their confluency and the total number of DAPI (4,6-diamidino-2-phenylindole)-positive cells (data not shown).
Next, we assessed the potential effect of FOXO1 knockdown on hESC pluripotency. Specific inhibition of FOXO1 mRNA using two distinct shRNA sequences resulted in >90% depletion of FOXO1 protein expression within 72 h, which was accompanied by rapid downregulation of pluripotent OCT4, NANOG and SOX2 expression in H1/FOXO1-shRNA and H1/FOXO1-shRNA-II cells (Fig. 1c,d and Supplementary Fig. S4a,b). These alterations were concomitant with a significant loss of surface markers of pluripotency TRA-1-60 and TRA-1-81 (Fig. 1e and Supplementary Figs S3a and S4c). Targeting FOXO1 with a third shRNA led to a similar suppression of pluripotency gene expression, confirming that this phenotype was not the result of an off-target effect of shRNAs (Supplementary Fig. S5d). Furthermore, FOXO1 knockdown in the hESC line HES2 caused a similar downregulation of pluripotency gene expression (Supplementary Fig. S5e). Most hESCs responded to FOXO1 knockdown, as evidenced by immunostaining of cellular distribution of pluripotent proteins, indicating that the phenotype did not result from a minor subset of hESCs (Supplementary Fig. S5f). Together, these results indicate that FOXO1 has a critical function in the regulation of hESC pluripotency.
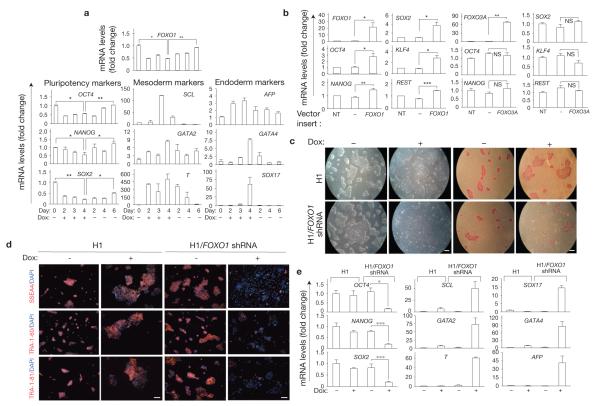
Interestingly, the effect of FOXO1 knockdown on hESC pluripotency was reversible in that doxycycline withdrawal after 96 h resulted in recovery of FOXO1 expression as well as OCT4, NANOG and SOX2 within a few days (Fig. 2a and Supplementary Fig. S6). These results indicate that FOXO1 tightly controls pluripotency genes. FOXO1 knockdown resulted in the spontaneous differentiation of hESCs maintained under pluripotent self-renewal conditions, as shown by the induction of mesoderm (SCL, also known as TAL1; GATA2; and brachyury, also known as T) and endoderm lineage markers (AFP, GATA4 and SOX17; Fig. 2a and Supplementary Fig. S6a–c). As none of these genes is known to be directly regulated by FoxO, these results also indicate that FOXO1 regulation of pluripotency is due in part to its participation in suppressing the mesoderm and endoderm lineage commitment. In this context, FOXO1 knockdown did not impact the expression of markers of ectoderm specification NES, TUBB3 and GFAP (data not shown).
Figure 2.
Reversible effect of loss of FOXO1 on pluripotency and differentiation of hESCs. (a) qRT-PCR analysis of pluripotency (left), mesoderm (middle) and endoderm (right) gene expression in H1/FOXO1-shRNA hESCs. Cells were maintained under pluripotency self-renewing conditions and treated with or without doxycycline for up to 4 days, after which cells were washed extensively and maintained in the absence of doxycycline. Quantification of the genes was relative to the endogenous β-actin transcript levels. Error bars indicate s.e.m. of three independent experiments, each carried out in triplicate. *P <0.05, *P <0.01. (b) In H1 cells, ectopic expression of FOXO1 and not FOXO3A induces expression of pluripotency genes. GFP-positive hESCs lentivirally transduced with an empty control vector or a vector containing FOXO1 or FOXO3A were FACS sorted 72 h after transduction and analysed by qRT-PCR for FOXO1 or FOXO3A and pluripotency marker gene expression; results shown are relative to endogenous ACTB and normalized to untransduced H1 cells under self-renewal conditions. Error bars indicate s.e.m. of three independent experiments each, carried out in triplicate; *P < 0.05, *P < 0.01, *P < 0.001. NT, not transduced. NS, not significant. (c) FOXO1-knockdown-mediated loss of pluripotency and induction of differentiation markers in H1/FOXO1-shRNA cells after several passages. Morphology (left) and alkaline phosphatase staining (right) of hESCs cultured in doxycycline for five passages. (d) Expression of surface markers of pluripotency, SSEA4, TRA-1-60 and TRA-1-81, by immunostaining and DAPI counterstaining of hESCs was analysed after five passages in the absence or presence of doxycycline (Dox). Scale bar, 100 μm. One representative of two independent experiments is shown. (e) qRT-PCR analysis of FOXO1 and pluripotency genes (left), mesoderm (middle) and endoderm (right) markers in H1 cells maintained in pluripotency self-renewal conditions with or without doxycycline for five passages. Results shown are relative to the endogenous ACTB. Error bars indicate s.e.m. (n = 3). *P < 0.05, ***P < 0.001.
In agreement with a critical function of FOXO1 in the regulation of hESC pluripotency, lentiviral-mediated ectopic FOXO1 expression induced a significant upregulation of OCT4, NANOG and SOX2 in two different hESC lines (Fig. 2b and Supplementary Fig. S6d). These effects were specific to FOXO1, because ectopic expression of FOXO3A in these same two hESC lines did not significantly alter the expression of pluripotency genes (Fig. 2b and Supplementary Fig. S6d). Of note, overexpression of FOXO1 in hESCs also resulted in upregulation of KLF4 and REST, genes associated with mESC pluripotency37,38 (Fig. 2b).
Targeting FOXO1 in hESCs undergoing few passages in self-renewing conditions (Fig. 2c–e and Supplementary Fig. S7a, top) resulted in a morphological transition towards an epithelial flattened appearance. These morphological changes were accompanied by a nearly complete loss of alkaline phosphatase activity (Fig. 2c), loss of cell surface markers of pluripotency (Fig. 2d and Supplementary Fig. S7a) and loss of transcripts for OCT4, NANOG and SOX2 (Fig. 2e and Supplementary Fig. S7b), all indicating loss of pluripotency in FOXO1-knockdown cells after few passages. Importantly, these alterations were concomitant with full acquisition of differentiation markers (Fig. 2e and Supplementary Fig. S7c,d). In contrast to its short-term impact (Fig. 2a), the effect of FOXO1 knockdown on pluripotency did not seem to be reversible after few passages, indicating that FOXO1 is critical for the maintenance of pluripotency in hESCs over time (Supplementary Fig. S7e).
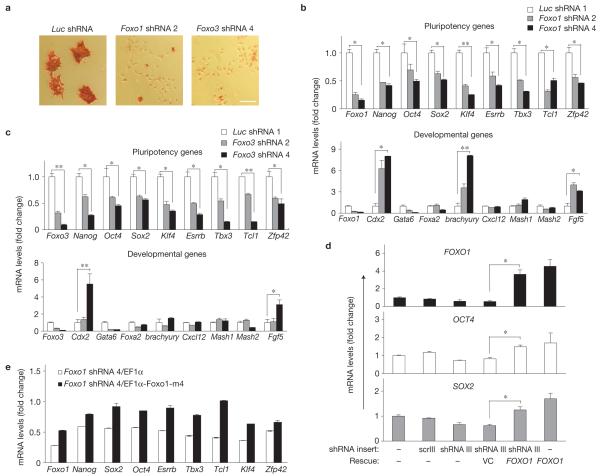
Signalling pathways regulating ESC fate differ between mESCs and hESCs, whereas transcriptional programs of pluripotency are relatively conserved39,40. Importantly, as in hESCs, we found Foxo1 (mouse orthologue of FOXO1) to be critical for the regulation of pluripotency and differentiation of mESCs (Fig. 3), as determined by loss of alkaline phosphatase activity (Fig. 3a), suppression of pluripotency gene expression (Fig. 3b, top) and upregulation of certain developmental genes, such as the mesoderm specific brachyury (T; Fig. 3b, bottom). However, in contrast to hESCs, Foxo1 knockdown resulted in the upregulation of the Cdx2 (caudal type homeobox 2) trophoblastic marker in mESCs, as would have been predicted from OCT4 downregulation41. This result probably reflects differences in responses to OCT4 downmodulation in mESCs versus hESCs that represent discrete pluripotency states developmentally at distinct stages42. Interestingly, in mESCs, although Foxo3 knockdown also resulted in loss of pluripotency (Fig. 3c, top), it did not lead to modulations of brachyury expression as seen with Foxo1 knockdown (Fig. 3c, bottom), indicating a potential function for Foxo3 in upregulation of brachyury gene expression during mesoderm induction.
Figure 3.
Foxo1 and Foxo3 regulate pluripotency of mESCs. (a) Alkaline phosphatase staining in Foxo1-, Foxo3- or control-knockdown mESCs. Scale bar, 50 μm. (b,c) qRT-PCR analysis of pluripotency (top) and developmental genes (bottom) in mESCs expressing one of two distinct shRNA targeting Foxo1 (b) or Foxo3 (c). shRNA-mediated knockdown of luciferase was used as a control. The gene expression levels of four lineages, including trophectoderm (Cdx2 and Mash2, also known as Ascl2), endoderm (Gata6 and Foxa2), ectoderm (Cxcl12, Mash1, also known as Ascl1, and Fgf5) and mesoderm (brachyury) were examined after knockdown of Foxo1 or Foxo3. All graphs show mean ± s.e.m. for n = 3; *P < 0.05, **P < 0.01. P values are from comparing Foxo1 shRNA 4 or Foxo3 shRNA 4 with Luc shRNA 1 (b,c). Ectopic expression of a resistant form of the targeted FoxO1-rescued FoxO1-knockdown-mediated phenotype in both hESCs and mESCs. (d) hESCs were transfected with a lentivirus expressing FOXO1 shRNA III (targeting the 3′ untranslated region of the FOXO1 mRNA); GFP-positive cells were FACS sorted 3 days later (>50-60% GFP positive), transduced with lentiviral vector (pLEIGW-FOXO1) that contains only the FOXO1 coding region and cultured for another 2 days before gene expression analysis. All results are relative to the endogenous ACTB. Error bars indicate s.e.m. (n = 3). *P < 0.05. (e) Endogenous Foxo1 was targeted by Foxo1 shRNA 4 in mESCs and rescued by re-expressing the shRNA-resistant Foxo1-m4 construct (EF1-Foxo1-m4). EF1 promoter empty vector (EF1) was used as a control. Gene expression analysis of Foxo1 and pluripotency genes was carried out by qRT-PCR and all results in Foxo1-shRNA 4 cells are expressed relative to the sample with Luc shRNA and EF1 (set as 1). Error bars indicate s.e.m. (n = 3). P < 0.05 for all genes in rescued samples when compared with controls.
To further confirm the specificity of loss of FoxO1 on pluripotency, we rescued the observed phenotype by overexpression of a resistant form of the targeted FoxO1 using two different strategies in both hESCs (Fig. 3d) and mESCs (Fig. 3e and data not shown). These findings validated unambiguously our results implicating FoxO1 in the regulation of pluripotency in ESCs.
hESCs form teratoma-like masses when introduced into immunodeficient mice43. This generally accepted approach demonstrates the developmental potential of pluripotent hESCs in vivo. We reasoned that if FOXO1 is critical for hESC pluripotency, loss of its activity should prevent hESCs from forming teratomas in vivo. Whereas almost all immunocompromised severe combined immunodeficient (SCID)-Beige mice injected with control cells formed well-encapsulated cystic tumours that contained elements of all three embryonic germ layers, only two of nine mice injected with doxycycline-induced H1/FOXO1 shRNA formed tumours, all of which were relatively small (Supplementary Fig. S8a–m).
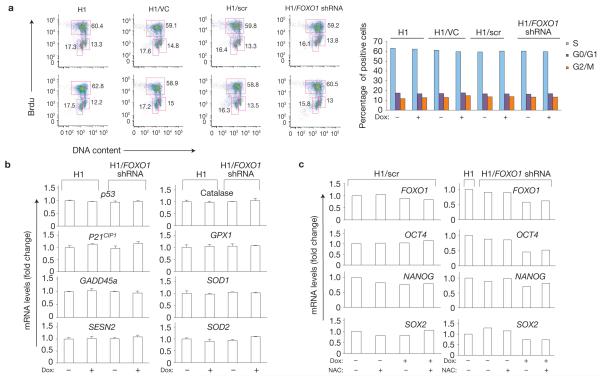
To investigate the mechanism of FOXO1 control of pluripotency, we investigated whether FOXO1 regulates hESC cycling, apoptosis or redox status. FOXO1 knockdown did not significantly modulate hESC proliferation (Fig. 4a and Supplementary Fig. S8n), or apoptosis (Supplementary Fig. S8o). In addition, it did not alter the expression of anti-oxidant enzymes or stress genes that accompany oxidative stress particularly in stem cells9,10 (Fig. 4b). In agreement with these results, reactive oxygen species (ROS) levels were not significantly increased in FOXO1-knockdown hESCs (Supplementary Fig. S8p). Furthermore, the anti-oxidant N-acetyl-cysteine (NAC) treatment did not impact pluripotency genes in these cells (Fig. 4c), altogether strongly arguing against redox modulation in mediating the effect of FOXO1 on pluripotency.
Figure 4.
FOXO1 knockdown does not impact hESC proliferation or redox status. (a) Left, H1, H1/VC, H1/scr and H1/FOXO1-shRNA cells maintained in pluripotency self-renewing conditions were cultured with (top) or without (bottom) doxycycline (Dox) for 96 h, after which cell proliferation was analysed by BrdU staining. BrdU incorporation versus DNA content were used to detect percentage of cells that are in G1, S or G2/M phase of cell cycle. Right, quantification of results of two independent experiments is shown. (b) qRT-PCR analysis of anti-oxidant enzymes and anti-oxidant response genes in parental H1 and in H1/FOXO1-shRNA cells maintained with or without doxycycline for 96 h. Quantification of the target genes is relative to the endogenous ACTB transcript levels and normalized to untreated H1 cells under self-renewal conditions. Results are shown as mean ± s.e.m. of triplicate experiments; one representative of two independent experiments is shown. (c) Anti-oxidant treatment does not impact the expression of pluripotency genes in hESCs. qRT-PCR analysis of FOXO1 and pluripotency genes in H1, H1/scr and H1/FOXO1-shRNA cells maintained under pluripotency conditions in the presence or absence of NAC (100 μM) and treated or not with doxycycline for 96 h. Results shown are relative to the endogenous ACTB and normalized to untreated H1/scr or H1 cells under self-renewal conditions. n = 2.
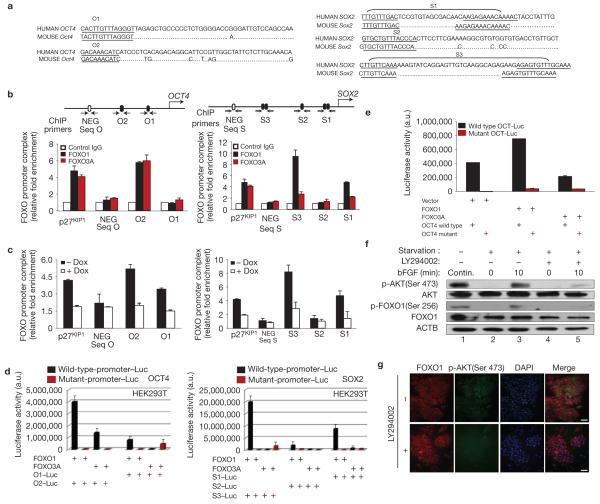
These results led us to investigate whether FOXO1 directly regulates pluripotency gene expression. Using rVista tools, multi-LAGAN alignment and the TRANSFAC database, we analysed 5-kb regions upstream of pluripotency genes. These analyses identified highly conserved sequences containing putative FoxO binding sites44 within the regulatory regions of OCT4 (O1, O2) and SOX2 (S1, S2, S3) genes (S1 and S3 each containing two clusters each) but not those of the NANOG gene (Fig. 5a).
Figure 5.
FOXO1 activates the expression of OCT4 and SOX2 pluripotency genes in hESCs by binding directly to their regulatory regions. (a) Sequence alignment of human OCT4 and SOX2 regulatory regions containing putative FoxO-binding sites. (b) Endogenous FOXO occupation of sites shown within arrows was analysed by ChIP in H1 cells. DNA co-immunoprecipitated with either anti-FOXO1 (H-128), anti-FOXO3A (N-15) or a control pre-immune immunoglobulin was amplified by qPCR. Binding of FOXO1 to p27KIP1 promoter and binding of FOXO1 to a conserved upstream region with no FOXO-binding sequences (in 2.7 kb of human OCT4 (NEG Seq O) and in -4 kb of human SOX2 (NEG - Seq S)) were used, respectively, as positive and negative controls. Results of three independent experiments (mean ± s.e.m.) are shown as relative fold enrichment when compared with control immunoglobulin after normalization to the values obtained from the input samples. (c) ChIP analysis of FOXO1 in undifferentiated H1/FOXO1-shRNA hESCs treated or not with doxycycline (Dox) for 3 days. Results of three independent experiments (mean ± s.e.m.) are shown. (d) pcDNA3, pcDNA3-FOXO1 or pcDNA-FOXO3A were co-transfected into HEK293T cells with a pGL3 containing the O1 or O2 site of human OCT4, or the S1, S2 or S3 site of SOX2 or their mutants driving the luciferase gene; luciferase activity was measured 48 h later. Results are from three independent experiments (mean ± s.e.m.). (e) Luciferase activity measured 48 h after co-transfection of hESCs (H1) with a lentiviral vector control or containing FOXO1 or FOXO3A and a human OCT4 reporter plasmid containing a wild-type or mutant O2 sequence. Results are the mean of two independent experiments. (f) H1 hESCs were maintained continuously (Contin.) in bFGF for 72 h (lane 1) or starved overnight and stimulated or not with bFGF (40 ng ml−1 for 10 min) and/or PI(3)K inhibitor LY294002 (10 μM) for 45 min before stimulation, and preparing the whole-cell lysate (lanes 2-5). One representative immunoblot of two experiments is shown. (g) H1 cells maintained in undifferentiated conditions and treated or not with LY294002 (10 μM) for 2 h before double immunostaining of FOXO1 and phospho-AKT (Ser 473) and counterstaining with DAPI. One representative of three independent experiments is shown (uncropped scanned gels are shown in Supplementary Fig. S12).
In pluripotent self-renewing hESCs, endogenous FOXO1 bound specifically to sequences within regulatory regions of OCT4 (O2) and SOX2 (S1, S3) genes, as revealed by chromatin immunoprecipitation (ChIP; Fig. 5b and Supplementary Fig. S9a,b). In these experiments the p 27KIP1 promoter, a known FOXO target, served as a positive control, whereas a conserved upstream region of human OCT4 (NEG Seq O) and human SOX2 (NEG Seq S) lacking FOXO-binding sequences served as negative controls. Importantly, FOXO1 binding to these sites was significantly reduced in doxycycline-treated cells when compared with controls (Fig. 5c), further validating the specificity of FOXO1 binding. These in vivo findings were corroborated by in vitro gel mobility shift assays using nuclear extracts of HEK293T cells ectopically expressing FOXO1 (Supplementary Figs. S9c,d). In agreement with these results, reporter assays showed that FOXO1 activates OCT4 and SOX2 transcription through regulation of O2 in OCT4, and the S1 and S3 regions in SOX2 (Fig. 5d and Supplementary Fig. S10a–c). These experiments also revealed that FOXO3A binds to the O2 sequence of OCT4 in hESCs and activates to some extent OCT4 luciferase reporter in HEK293T cells (Fig. 5b,d). To further evaluate this, we examined the activation of the full-length (3 kb) human OCT4 promoter by overexpressed FOXO proteins in hESCs. As shown in Fig. 5e, ectopically expressed FOXO1 highly activates (four times more than FOXO3A) human OCT4 in hESCs. Importantly, mutation of the O2 sequence abolishes the activation, indicating that O2 is the principal mediator of FOXO1 induction of OCT4 in hESCs.
In contrast to FOXO1, and consistent with overexpression studies in hESCs (Fig. 2b and Supplementary Fig. S6d), FOXO3A did not bind to SOX2 regulatory regions in vivo, nor did it have any impact on SOX2 transcription (Fig. 5b,d and Supplementary Fig. S10b), despite binding to the SOX2 promoter in gel mobility shift assays in vitro (Supplementary Fig. S10e).
The differential impact of FOXO1 versus FOXO3A on pluripotency genes may be explained by the differential regulation of FOXO proteins, and specifically their nuclear localization, in hESCs (see Supplementary Figs S1g and S5c). Indeed, the function of FoxO proteins is determined by the integrated balance of competing stimuli30,31. Notable among negative regulators of FOXO proteins is AKT, which is critical for ESC self-renewal45,46. AKT-dependent phosphorylation of FOXO proteins triggers their rapid nuclear export by multiple means to ensure the inhibition of their transcriptional activity30,31. As anticipated, in self-renewing hESCs the PI(3)K/AKT signalling pathway is activated in response to basic fibroblast growth factor (bFGF) stimulation resulting in AKT phosphorylation of FOXO1 (Fig. 5f, lanes 2–5). Interestingly, FOXO1 (but not FOXO3A) is mostly nuclear despite abundant pAKT in hESCs cultured in bFGF (Fig. 5f, lane 1, Fig. 5g, top and Supplementary Fig. S11).
Inhibition of AKT by the PI(3)K inhibitor LY294002 leads to FOXO1 dephosphorylation and increased nuclear FOXO1 intensity (Fig. 5f, lane5; Fig. 5g). These results indicate that in hESCs, FOXO1 is nuclear and transcriptionally active despite its phosphorylation by AKT, presumably because its inhibition by pAKT is overriden by other signalling pathways28. These findings are reminiscent of regulation of Foxo3 in adult stem cells10,47 and are consistent with the notion that the output of competing signalling pathways (and not AKT alone) ultimately determines the nuclear localization and activity of FOXO proteins30,31.
The present study demonstrates an essential function for FOXO1 in the regulation of hESC fate. Thus, the longevity FoxO proteins emerge as critical non-redundant regulators of both somatic and embryonic stem cell activity. Specifically, Foxo3 is essential for the maintenance of haematopoietic, neural and leukaemic stem cells9-13, whereas FOXO1 is critical for regulating hESC pluripotency. Similarly to certain other pluripotency gene-deleted mice such as Sox2−/−, Tcl1−/− or Tbx3−/− (refs 7,48,49), Foxo1−/− mice do not exhibit an early (pre-gastrulation) developmental defect33. This may be due to the regulation of pluripotency by both Foxo1 and Foxo3 in mESCs in contrast to hESCs (mESCs being at an earlier developmental stage than hESCs)42, indicating that perhaps during early stages of mouse development Foxo1 and Foxo3 are redundant in vivo, as supported by our findings in mESCs (Fig. 3). Alternatively, lack of early lethality of Foxo1−/− mice may be due to species-specific differences or, as in the case of Sox2−/− (ref. 7), to the potential presence of long-lived maternal Foxo1 protein at the early stages of Foxo1−/− embryonic development. Given OCT4 binding to FOXO1 promoter in hESCs14, these findings indicate that FOXO1 is a component of the circuitry of hESC pluripotency and support the notion that activation of FOXO1 may be used for improving somatic cell reprogramming15-17.
METHODS
Maintenance and differentiation of ESCs and flow cytometry
hESC lines H1 (NIH code WA01, WiCell Research Institute) and HES2 (NIH code ES02, ES Cell International (ESI)) were maintained on Matrigel (Becton Dickenson)-treated six-well plates using mTeSR1 medium (StemCell Technologies). hESC differentiation and haematopoietic colony-forming assays were carried out as previously described50. mESC maintenance and differentiation were carried out as previously described46.
Cells were stained with anti-TRA-1-81 (Santa Cruz), anti-KDR-PE (R&D Systems), anti-CD34-PE-cy7 (BD Pharmingen), anti-PECAM-1 (CD31)–FITC (BD Pharmingen) and/or anti-CD117–APC (Invitrogen) and analysed with a fluorescence-activated cell sorting (FACS) LSRII flow cytometer (Becton Dickinson).
Plasmids, generation of H1TetR cells, lentiviral production, ESC transduction and PCR reactions
H1 hESCs (2 × 106) were transfected with 5 μg of FspI-linearized tetracycline repressor protein (TetR)-expressing plasmid pcDNA6/TR (Invitrogen) using nucleofection (Amaxa kit VPH-1001 & Nucleofector). Blasticidin-S-resistant (5 μg ml−1, Invitrogen) hESC colonies were picked two weeks later, expanded and maintained in the presence of 10 μg ml−1 blasticidin S. One of positive colonies was used in subsequent experiments.
All shRNAs (Supplementary Table S1) used in hESCs were cloned into EcoRI/XhoI sites of the pL4–H1/TetO4 lentiviral vector. FOXO1 shRNA III targets the 3′ untranslated region of the FOXO1 mRNA. shRNAs used in mESCs were inserted into the lentivirus-based pLKO.bsd vector. FOXO1 and FOXO3A complementary DNAs were cloned into the HpaI site of the pLEIGW vector. The 3-kb human OCT4 promoter from phOCT4–Luc (plasmid # 17221, Addgene)16, amplified by PCR (see Supplementary Table S1), and the regulatory regions containing putative FOXO-binding sites of human OCT4 or SOX2 were inserted between KpnI and BglII sites of the pGL3 vector containing a TATA box51. shRNA-resistant Foxo1 (Foxo1-m4), generated by site-directed mutagenesis without altering the amino acid sequence, was inserted into the pSIN-EF1 α-IRES-Puro lentiviral vector.
Lentiviral vectors were packaged by co-transfection of pLP1, pLP2 and pLPVSVG plasmids (all from Invitrogen) into HEK293T cells, concentrated and stored at −80 °C. Cells were transduced with viral particles in media containing polybrene (Sigma; hESC, 4 μg ml−1; mESC cell line CCE, 8 μg ml−1). Zeocin (Invitrogen, 1–2.5 μg ml−1) and blasticidin S (5 μg ml−1) were added to H1-derived cells. Resistant and GFP–positive hESC colonies were picked 2-3 weeks later and expanded. mESCs were selected four days later in ESC media supplemented with puromycin (1 μg ml−1) and blasticidin S (5 μg ml−1) added for an additional four days. PCR with reverse transcription (RT–PCR) and quantitative PCR with reverse transcription (qRT–PCR) analyses were carried out as previously described13 (see Supplementary Table S1 for primers).
GST-fusion protein and western blot analysis
Glutathione S-transferase (GST)–FOXO1 was generated by inserting FOXO1 amino acids 471–598 into pGEX–4T1 and bacterially expressed in DH5α induced with 0.5 mM isopropyl-β-D-thiogalactoside (IPTG) at 37 °C for 2 h. GST-FOXO3A (Addgene plasmid 1790, provided by M. Greenberg, Harvard University, USA) and GST–FOXO4 (provided by L. Zhi-Ping, UTSW, USA, PMID: 17242183) were induced in Rosetta 2(DE3) with 0.5 mM IPTG overnight. All GST fusions were purified over glutathione affinity resins, and the predominant bands were quantified in Coomassie-stained gels by comparison with an albumin standard (Thermo Scientific). The immunogens were amino acids 471–598 of FOXO1 (Santa Cruz Cat. No. 11350), regions surrounding amino acid 50 of FOXO3A (Cell Signalling Cat. No. 2497) and the amino terminus of FOXO4 (Santa Cruz cat. No. 5221).
The nuclear and cytoplasmic extracts were prepared using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce) following the manufacturer’s instructions. Cell lysates were prepared in Laemmeli buffer (50 mM Tris-Cl at pH 6.8, 2% SDS, 10% glycerol, 0.1% β-mercaptoethanol and 0.1% bromo phenol blue), resolved by SDS–polyacrylamide gel electrophoresis (PAGE), transferred onto polyvinylidene fluoride membranes and probed with the following primary antibodies: goat anti-FOXO1 (N-18; 1:250), rabbit anti-FOXO1 (H-128; 1:250), goat anti-FOXO3A (N-15; 1:500), goat anti-FOXO4 (N-19; 1:500), mouse anti-OCT4 (1:500), rabbit anti-NANOG (1:500), rabbit anti-SOX2 (1:300) and goat anti-actin (1:1,000), all from Santa Cruz biotechnology, and rabbit anti-TetR (Sigma, T-0951; 1:500). Rabbit anti-phosphoFOXO1 (Ser 256), anti-AKT and anti-phosphoAKT (Ser 473), all from Cell Signaling, were used at 1:1,000. The bound antibodies were detected by by horseradish peroxidase (HRP)-conjugated secondary antibodies (1:2,000, Pierce or Santa Cruz).
ChIP assay
ChIP was carried out as previously reported52,53. Briefly, 1 × 107 H1 hESCs were crosslinked at room temperature with formaldehyde (1% for FOXO1 antibodies, 0.4% for FOXO3A antibody). Cells were sonicated in SDS-lysis buffer (50 mM Tris–HCl at pH 8.0, 10 mM EDTA, 1% SDS), after which protein lysate was diluted fivefold in ChIP dilution buffer (20 mM Tris-HCl at pH 8.0, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl and 0.01% SDS). An aliquot of precleared lysates (100 μl) was used as a control for the amount of input DNA. The rest was immunoprecipitated with 5 μg anti-FOXO1 (N-18 or H-128, Santa Cruz Biotechnology), or anti-FOXO3A (N-15, Santa Cruz Biotechnology or 07-702, Millipore) antibodies, overnight at 4 °C. As a negative control, anti-goat or anti-rabbit IgG (Cell Signaling) was used. The antibody–FoxO–DNA complexes were reverse crosslinked for DNA isolation and quantitative PCR (qPCR) analysis. qPCR was carried out in duplicate on an ABI PRISM 7500 Sequence Detection system (Applied Biosystems) using SYBR Premix Ex Taq (Takara). Primer sequences are shown in Supplementary Table S1.
Sequence alignment and identification of FOXO-binding sites in OCT4 and SOX2 regulatory regions
To identify the putative FOXO-binding sites in OCT4 and SOX2 regulatory regions, the most recent annotated versions of the 5-kb genomic sequences upstream of the transcription start site of the OCT4 and SOX2 locus in mouse, rat and human were obtained from the ENSEMBL Genome Browser (http://www.ensembl.org). The three-way alignment was created using the Multi-LAGAN tool (http://genome.lbl.gov/vista/phylovista/index.shtml). The rVista tool (http://rvista.dcode.org) was run to identify putative FoxO binding sites. Conserved binding sites are defined in rVista as sequence fragments conserved between at least two species at the level of over 80% over a 24-base-pair window. For comparison, visualization of the alignment and FOXO-binding site searches within the alignment were repeated using the TRANSFAC database (http://www.generegulation.com/pub/databases.html).
Reporter gene assay
HEK293T (1.5 × 105 cells per well) were co-transfected with 0.5 μg pcDNA3–FOXO1, pcDNA3–FOXO3A or an empty control vector and 0.5 μg of the Firefly luciferase reporter gene along with 50 ng pRL–TK (Promega). hESCs (1 × 106 H1 cells) were co-transfected with 2.5 μg pLEIGW–FOXO1, pLEIGW–FOXO3A or pLEIGW empty vector and 2.5 μg of the luciferase reporter vector along with 250 ng pRL-TK (Promega) using nucleofection. Firefly luciferase activity was normalized to the corresponding Renilla luciferase activity.
Measurement of intracellular ROS levels
ROS levels were measured as previously described10,13,34. hESCs (5 × 105) were probed with 2,3,4,5,6-pentafluorodihydrotetramethylfosamine (RedoxSensor Red CC-1; Molecular Probes-Invitrogen, 3 μM), and hESCs incubated with H2O2 (200 μM) were used as positive controls.
Electrophoretic mobility shift assay
Nuclear extracts of HEK293T cells were isolated in two steps using hypotonic buffer (20 mM HEPES, 10 Mm KCl, 1 mM MgCl2, 0.5 mM dithiothreitol, 0.1% Triton X-100 and 20% glycerol) containing protease inhibitors (Roche) and nuclear extraction buffer (hypotonic buffer with 420 mM NaCl) and stored at −80 °C. The 2× DNA–protein binding reaction buffer was composed of 100 mM KCl, 20 mM MgCl2 protein, 0.1 mM dithiothreitol, 1% NP-40 and 5% glycerol in 20 mM Tris-HCl at pH 7.5, supplemented with 0.1 μg ml−1 polydeoxyinosinic-deoxycytidylic acid. Total nuclear extracts (5 μg) were used in each binding reaction in a total volume of 20 μl at room temperature. A total of 100 ng of each probe (S1, S3, O2 wild type and mutant, and insulin response sequence as a positive control)19 was labelled with [γ-32P]ATP (PerkinElmer) using the T4 Polynucleotide Kinase. In competition experiments, 200 molar excess of unlabelled probe was added to the binding reaction 15 min before the 32P-labelled probe. For supershift experiments, nuclear extracts were pre-incubated with 2 μg of anti-FOXO1 (N-18) or anti-HA antibodies (Santa Cruz Biotechnology) for 45 min at 4 °C.
Cell proliferation and apoptosis
BrdU (5-bromodeoxyuridine) labelling and annexin V-binding assays were carried out as previously described10,13. Ki-67 staining for flow cytometry was carried out according to the manufacturer’s recommendations.
Histochemistry and immunocytochemistry staining
Alkaline phosphatase activity was detected using Alkaline Phosphatase Substrate kit I (Vector Laboratories) following the manufacturer’s protocol.
Cells were fixed with 4% paraformaldehyde, then permeablized with 0.3% Triton X-100 for 30 min followed by washing and blocking with 4% goat serum in PBS. Cells were then incubated with the following antibodies: mouse anti-SSEA-4, mouse anti-TRA-1-60, mouse anti-TRA-1-81 monoclonal and rabbit anti-FOXO3A (07-702, Millipore) antibodies (1:50), mouse anti-OCT4 (C-10) and rabbit anti-SOX2 (H-65) antibodies (1:50), goat anti-FOXO1 (N-18; 1:50), goat anti-FOXO3A (N-15; 1:200), goat anti-FOXO4 (N-19; 1:200), all from Santa Cruz, and goat anti-brachyury (T, AF-2085, R & D systems) antibody (1:50). The Alexa Fluor 594-conjugated secondary antibodies (Invitrogen) were used at 1:500 dilution (1:100 for brachyury staining). Double immunostaining was carried out using anti-FOXO1 (N-18; Santa Cruz) and anti phospho-AKT (Ser 473; Cell Signalling) at 1:50 and 1:25 dilution, respectively. After washing three times, slides were mounted using Vectashield with DAPI (Vector Laboratories) and visualized by a Leica DMRA2 fluorescence microscope (Leica).
Southern blot analysis
Genomic DNA was isolated using a DNeasy Tissue kit (QIAGEN), digested with EcoRI and KpnI, separated by 1% agarose gel electrophoresis and transferred to a nylon membrane (Amersham Bioscience) in 10× SSC (1.5 M NaCl and 0.15 M sodium citrate) overnight. PCR-amplified TetR and enhanced fluorescent green protein (eGFP) gene fragments were used as probes for hybridization. The TetR probe detects a 1.5-kb band and the eGFP probe a 1.7-kb band.
Teratoma formation
Doxycycline-treated or non-treated hESCs (1 × 106) were resuspended in 20 μl hESC media and injected under the kidney capsule of four-week-old male SCID-Beige mice. Four to eight weeks later, tumours (>5 mm in diameter) were surgically removed, fixed in 4% paraformaldehyde overnight and embedded in paraffin. Sections were stained with haematoxylin and eosin.
Statistical analysis
Data represent mean ± s.e.m. of the number (n) of independent experiments unless indicated otherwise. Statistical significance has been calculated by a two-tailed Student t-test between the indicated groups.
Supplementary Material
ACKNOWLEDGEMENTS
We thank F. Lohmann for advice on the ChIP assay, J. Bieker (Mount Sinai School of Medicine) and G. Blobel (University of Pennsylvania) for critical reading of the manuscript, I. George and M. Grisotto for cell sorting, and the Flow Cytometry Shared Research Facility of Mount Sinai School of Medicine. This work was supported in part by a National Institutes of Health grant RO1 DK077174, an American Cancer Society Research Scholarship (RSG LIB-110480), a Career Enhancement Award (K18 HL76510-01), a Black Family Stem Cell Institute Exploratory Research Award, a New York State Stem Cell Science (NYSTEM) award (CO24408), an Irma Hirschl/Weill-Caulier Trust Research Award and a Roche Foundation for Anemia Research (RoFAR) Award to S.G., and an NIH P20 GM75019 (S.G. CoPI).
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website
AUTHOR CONTRIBUTIONS
X.Z. and S.G. designed experiments and analysed data; X.Z. carried out most of the experiments, with significant help from S.Y. and some assistance from S-M.L., S.K.M. and P.R.; M.K. helped with the set-up of some critical techniques; R.S. analysed data; D-F.L. and J.S. designed and carried out experiments involving mESCs; N-W.C. designed and carried out antibody calibration experiments with the help of T-Y.J.Y.; M.L. and T.T. contributed key reagents; I.L. and G.K. provided valuable reagents and advice; S.G. conceived the project and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr. Opin. Genet. Dev. 2006;16:455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 2.Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- 3.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nichols J, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- 5.Chambers I, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- 6.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 7.Avilion AA, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paik JH, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyamoto K, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Yalcin S, et al. Foxo3 is essential for the regulation of Ataxia telangiectasia mutated and oxidative stress-mediated homeostasis of hematopoietic stem cells. J. Biol. Chem. 2008;283:25692–25705. doi: 10.1074/jbc.M800517200. [DOI] [PubMed] [Google Scholar]
- 11.Renault VM, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naka K, et al. TGF-β-FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature. 463:676–680. doi: 10.1038/nature08734. [DOI] [PubMed] [Google Scholar]
- 13.Yalcin S, et al. ROS-mediated amplification of AKT/mTOR signalling pathway leads to myeloproliferative syndrome in Foxo3(-/-) mice. EMBO J. 2010;29:4118–4131. doi: 10.1038/emboj.2010.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 18.Ogg S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 19.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 20.Kops GJ, et al. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 21.Essers MA, et al. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 2004;23:4802–4812. doi: 10.1038/sj.emboj.7600476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu MC, et al. I B kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 23.Yang W, Dolloff NG, El-Deiry WS. ERK and MDM2 prey on FOXO3a. Nat. Cell Biol. 2008;10:125–126. doi: 10.1038/ncb0208-125. [DOI] [PubMed] [Google Scholar]
- 24.Mahmud DL, et al. Phosphorylation of forkhead transcription factors by erythropoietin and stem cell factor prevents acetylation and their interaction with coactivator p300 in erythroid progenitor cells. Oncogene. 2002;21:1556–1562. doi: 10.1038/sj.onc.1205230. [DOI] [PubMed] [Google Scholar]
- 25.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 26.Daitoku H, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc. Natl Acad. Sci. USA. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H, et al. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl Acad. Sci. USA. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamagata K, et al. Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol. Cell. 2008;32:221–231. doi: 10.1016/j.molcel.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 29.Dansen TB, et al. Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat. Chem. Biol. 2009;5:664–672. doi: 10.1038/nchembio.194. [DOI] [PubMed] [Google Scholar]
- 30.van den Berg MC, Burgering BM. Integrating opposing signals toward Forkhead box O. Antioxid Redox Signal. 2011;14:607–621. doi: 10.1089/ars.2010.3415. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Rielland M, Yalcin S, Ghaffari S. Regulation and function of FoxO transcription factors in normal and cancer stem cells: what have we learned? Curr. Drug Targets. 2011;12 doi: 10.2174/138945011796150325. [DOI] [PubMed] [Google Scholar]
- 32.Castrillon DH, Miao L, Kollipara R, Horner JW, DePinho RA. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301:215–218. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]
- 33.Hosaka T, et al. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl Acad. Sci. USA. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marinkovic D, et al. Foxo3 is required for the regulation of oxidative stress in erythropoiesis. J. Clin. Invest. 2007;117:2133–2144. doi: 10.1172/JCI31807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 36.Brandenberger R, et al. Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat. Biotechnol. 2004;22:707–716. doi: 10.1038/nbt971. [DOI] [PubMed] [Google Scholar]
- 37.Jiang J, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 2008;10:353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 38.Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453:223–227. doi: 10.1038/nature06863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei CL, et al. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells. 2005;23:166–185. doi: 10.1634/stemcells.2004-0162. [DOI] [PubMed] [Google Scholar]
- 40.Boiani M, Scholer HR. Regulatory networks in embryo-derived pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 2005;6:872–884. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- 41.Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 42.Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 43.Adewumi O, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 44.Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 2000;349:629–634. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe S, et al. Activation of Akt signaling is sufficient to maintain pluripotency in mouse and primate embryonic stem cells. Oncogene. 2006;25:2697–2707. doi: 10.1038/sj.onc.1209307. [DOI] [PubMed] [Google Scholar]
- 46.Ivanova N, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 47.Lee JY, et al. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell. 2010;7:593–605. doi: 10.1016/j.stem.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davenport TG, Jerome-Majewska LA, Papaioannou VE. Mammary gland, limb and yolk sac defects in mice lacking Tbx3, the gene mutated in human ulnar mammary syndrome. Development. 2003;130:2263–2273. doi: 10.1242/dev.00431. [DOI] [PubMed] [Google Scholar]
- 49.Narducci MG, et al. TCL1 participates in early embryonic development and is overexpressed in human seminomas. Proc. Natl Acad. Sci. USA. 2002;99:11712–11717. doi: 10.1073/pnas.182412399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kennedy M, D’Souza SL, Lynch-Kattman M, Schwantz S, Keller G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood. 2007;109:2679–2687. doi: 10.1182/blood-2006-09-047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hua X, Miller ZA, Wu G, Shi Y, Lodish HF. Specificity in transforming growth factor β-induced transcription of the plasminogen activator inhibitor-1 gene: interactions of promoter DNA, transcription factor muE3, and Smad proteins. Proc. Natl Acad. Sci. USA. 1999;96:13130–13135. doi: 10.1073/pnas.96.23.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Im H, et al. Measurement of protein-DNA interactions in vivo by chromatin immunoprecipitation. Methods Mol. Biol. 2004;284:129–146. doi: 10.1385/1-59259-816-1:129. [DOI] [PubMed] [Google Scholar]
- 53.Lohmann F, Bieker JJ. Activation of Eklf expression during hematopoiesis by Gata2 and Smad5 prior to erythroid commitment. Development. 2008;135:2071–2082. doi: 10.1242/dev.018200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.