Abstract
Objective
To analyse the role of the PTPN22 and CSK genes, previously associated with autoimmunity, in the predisposition and clinical phenotypes of giant cell arteritis (GCA).
Methods
Our study population was composed of 911 patients diagnosed with biopsy-proven GCA and 8,136 unaffected controls from a Spanish discovery cohort and three additional independent replication cohorts from Germany, Norway and the United Kingdom. Two functional PTPN22 polymorphisms (rs2476601/R620W and rs33996649/R263Q) and two variants of the CSK gene (rs1378942 and rs34933034) were genotyped using predesigned TaqMan® assays.
Results
The analysis of the discovery cohort provided evidence of association of PTPN22 rs2476601/R620W with GCA (PFDR=1.06E-04, OR=1.62, CI 95% 1.29-2.04). The association did not appear to follow a specific GCA subphenotype. No statistically significant differences between allele frequencies for the other PTPN22 and CSK genetic variants were evident either in the case/control or in stratified case analysis. To confirm the detected PTPN22 association, three replication cohorts were genotyped, and a consistent association between the PTPN22 rs2476601/R620W variant and GCA was evident in the overall meta-analysis (PMH=2.00E-06, OR= 1.51, CI 95% 1.28-1.79).
Conclusions
Our results suggest that the PTPN22 polymorphism rs2476601/R620W plays an important role in the genetic risk to GCA.
Keywords: GCA, temporal arteritis, SNP, PTPN22, CSK
INTRODUCTION
Giant cell arteritis (GCA) is a chronic vasculitis that shows a complex etiology derived from the interaction between both genetic and environmental factors.[1] Similar to most immune-related disorders, the highest susceptibility signals belong to the human leukocyte antigen (HLA) region. However, different studies have highlighted that genes involved in inflammation pathways may also be implicated in GCA susceptibility.[2] In spite of these findings, the genetic background of this condition is still poorly understood.
Although the etiology of GCA remains unclear, it is well known that innate and adaptive immune responses are involved in its pathogenesis. Several lines of evidence indicate that this vasculitis is a T cell-mediated disease with both Th17 and Th1 cells contributing to inflammation. While Th1 response is associated with chronically persistent vascular lesions, Th17 immunity appears to be more important for acute manifestations, both systemically and in the blood vessels.[3, 4]
The PTPN22/CSK pathway is a master regulator of autoimmunity, with a key role in the negative control of the signaling mediated by the T cell receptor (TCR).[5] Interestingly, several SNPs located within these two genes have been associated with autoimmunity,[6-11] suggesting that this is one of the molecular pathways shared by different autoimmune disorders.
Regarding PTPN22, it has been reported that two autoimmune disease-associated variants, rs2476601 (R620W) and rs33996649 (R263Q) influence the function of the protein.[12, 13] On the other hand, two CSK polymorphisms, rs34933034 and rs1378942, were recently identified as susceptibility factors for systemic sclerosis (SSc)[11] and systemic lupus erythematosus (SLE),[10] respectively. A functional role for the CSK genetic variant rs34933034*A in SLE patients has been proposed in a recent study.[10]
Based on this, we decided to assess the role of the disease associated PTPN22 and CSK polymorphisms in both predisposition to and the clinical phenotypes of GCA.
METHODS
Study population
A total of 911 GCA patients and 8,136 unrelated healthy controls were included in this study. First, we analyzed a discovery cohort of 623 GCA patients and 1,729 healthy controls of Spanish Caucasian ancestry. Subsequently, three independent replication cohorts were analyzed (72 GCA and 937 controls from Germany; 60 GCA and 271 controls from Norway; 156 GCA and 5,199 controls from the United Kingdom). Case and control sets were matched by geographical origin and ethnicity, but not by age (which may represent a limitation of the study). PTPN22 rs2476601 genotype data from the control population of Germany were obtained from Hüffmeier et al.,[14] since this set matched geographically and ethnically our German GCA cohort. More detailed information about the UK controls can be obtained from Morgan et al.[15] Informed written consent from all participants and approval from the local ethical committees were obtained in accordance with the tenets of the Declaration of Helsinki. All patients had a positive temporal artery biopsy (disruption of the internal elastic laminae with infiltration of mononuclear cells into the arterial wall with or without multinucleated giant cells) and fulfilled the 1990 American College of Rheumatology classification criteria for GCA.[16] In the subphenotype analysis, the patients were stratified according to manifestations of polymyalgia rheumatica (PMR) and the presence or absence of visual ischemic manifestations (VIM; if they experienced transient visual loss including amaurosis fugax, permanent visual loss, or diplopia) and irreversible occlusive disease (IOD; if they had at least one of the following features: permanent visual loss, stroke or occlusive disease in the upper extremities or lower extremities).
Genotyping methods
Genomic DNA was extracted from peripheral white blood cells using standard procedures. Two single-nucleotide polymorphisms (SNPs) located within PTPN22, rs2476601/R620W and rs33996649/R263Q, and two SNPs located within CSK, rs1378942 and rs34933034, were genotyped using the TaqMan® allelic discrimination assay technology on a 7900HT Fast Real-Time PCR System, both from Applied Biosystems (Foster City, California, USA). For the United Kingdom samples, rs2476601/R620W was genotyped by direct sequencing.
Statistical analysis
The overall statistical power of the analysis, according to Power Calculator for Genetic Studies 2006 software (http://www.sph.umich.edu/csg/abecasis/CaTS/), is shown in online supplementary table S1.Plink (v1.07) (http://pngu.mgh.harvard.edu/purcell/plink/) and StatsDirect v.2.6.6 (StatsDirect Ltd, Cheshire, UK) were used to perform 2×2 contingency tables and χ2 test and/or Fisher’s exact test. Odds ratios (OR) and 95% confidence intervals (CI) were obtained according to Woolf’s method. The Benjamini & Hochberg (1995) step-up false discovery rate (FDR) control correction for multiple testing[17] was applied to the P-values of the discovery cohort. After correction, P-values lower than 0.05 were considered statistically significant. The allelic combinations were tested using Plink and Haploview (V. 4.2). The analysis of the combined data from all populations was performed using Plink and StatsDirect. Breslow–Day (BD) test method was used to estimate the homogeneity among populations. Pooled analyses were performed by Mantel-Haenszel test under fixed effects.
RESULTS
After genotyping, no divergence from Hardy-Weinberg equilibrium was observed either in controls or cases (P>0.01), and control allelic frequencies were similar to those previously reported in equivalent European Caucasian populations.[8, 10, 11] First, we conducted an association study in a case–control set of Spanish Caucasian origin. As shown in Table 1, when allelic frequencies were compared between cases and controls, a clear association of the PTPN22 rs2476601/R620W*A allele with GCA was observed (PFDR=1.06E-04, OR=1.62, CI 95% 1.29-2.04). Subsequently, to examine whether PTPN22 and CSK polymorphisms might influence the clinical manifestations of the disease, GCA patients were stratified according to the presence of PMR, VIM and IOD (Table 1). Consistently, the subphenotype analysis also reached statistical significance for the rs2476601 polymorphism (PMR+ vs. controls: PFDR=9.02E-04, OR=1.77, CI 95% 1.30-2.40; VIM+ vs. controls: PFDR=4.10E-03, OR=1.82, CI 95% 1.27-2.62; IOD+ vs. controls: PFDR=2.19E-03, OR=2.14, CI 95% 1.38-3.33). However, no statistically significant differences between GCA patients with and without these clinical characteristics were observed (data not shown). No association with any other PTPN22 and CSK genetic variants was evident either in the case/control or subphenotype analysis (Table 1).
Table 1.
Genotype and allele distribution of PTPN22 rs2476601, rs33996649 and CSK rs1378942, rs34933034 in Spanish biopsy-proven GCA patients and healthy controls.
| Genotype, N (%) |
Allele test |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Locus | 1/2 | Subgroup (N) | 1/1 | 1/2 | 2/2 | MAF (%) | P-value* | P FDR ** | OR [CI 95%]*** |
| rs2476601 | PTPN22 | A/G | Controls (n=1729) | 13 (0.75) | 200 (11.57) | 1516 (87.68) | 6.54 | |||
| GCA (n=623) | 4 (0.64) | 119 (19.10) | 500 (80.26) | 10.19 | 2.66E-05 | 1.06E-04 | 1.62 [1.29-2.04] | |||
| PMR+ (n=259) | 2 (0.77) | 53 (20.46) | 204 (78.76) | 11.00 | 2.26E-04 | 9.02E-04 | 1.77 [1.30-2.40] | |||
| VIM+ (n=168) | 1 (0.60) | 36 (21.43) | 131 (77.98) | 11.31 | 1.03E-03 | 4.10E-03 | 1.82 [1.27-2.62] | |||
| IOD+ (n=96) | 2 (2.08) | 21 (21.88) | 73 (76.04) | 13.02 | 5.47E-04 | 2.19E-03 | 2.14 [1.38-3.33] | |||
|
| ||||||||||
| rs33996649 | PTPN22 | T/C | Controls (n=1729) | 4 (0.23) | 110 (6.36) | 1615 (93.41) | 3.41 | |||
| GCA (n=623) | 1 (0.16) | 39 (6.26) | 583 (93.58) | 3.29 | 0.838 | 0.919 | 0.96 [0.67-1.38] | |||
| PMR+ (n=259) | 0 (0.00) | 12 (4.63) | 247 (95.37) | 2.32 | 0.191 | 0.382 | 0.67 [0.37-1.23] | |||
| VIM+ (n=168) | 0 (0.00) | 12 (7.14) | 156 (92.86) | 3.57 | 0.878 | 0.878 | 1.05 [0.57-1.92] | |||
| IOD+ (n=96) | 0 (0.00) | 6 (6.25) | 90 (93.75) | 3.13 | 0.831 | 0.831 | 0.91 [0.40-2.10] | |||
|
| ||||||||||
| rs1378942 | CSK | C/A | Controls (n=1729) | 281 (16.25) | 798 (46.15) | 650 (37.59) | 39.33 | |||
| GCA (n=623) | 129 (20.71) | 230 (36.92) | 264 (42.38) | 39.17 | 0.919 | 0.919 | 0.99 [0.87-1.13] | |||
| PMR+ (n=259) | 55 (21.24) | 99 (38.22) | 105 (40.54) | 40.35 | 0.658 | 0.756 | 1.04 [0.86-1.26] | |||
| VIM+ (n=168) | 34 (20.24) | 69 (41.07) | 65 (38.69) | 40.77 | 0.605 | 0.807 | 1.06 [0.85-1.33] | |||
| IOD+ (n=96) | 20 (20.83) | 39 (40.63) | 37 (38.54) | 41.15 | 0.616 | 0.822 | 1.08 [0.80-1.45] | |||
|
| ||||||||||
| rs34933034 | CSK | A/G | Controls (n=1729) | 72 (4.16) | 537 (31.06) | 1120 (64.78) | 19.69 | |||
| GCA (n=623) | 21 (3.37) | 176 (28.25) | 426 (68.38) | 17.50 | 0.091 | 0.182 | 0.86 [0.73-1.02] | |||
| PMR+ (n=259) | 10 (3.86) | 79 (30.50) | 170 (65.64) | 19.11 | 0.756 | 0.756 | 0.96 [0.76-1.22] | |||
| VIM+ (n=168) | 7 (4.17) | 47 (27.98) | 114 (67.86) | 18.15 | 0.497 | 0.807 | 0.90 [0.68-1.21] | |||
| IOD+ (n=96) | 5 (5.21) | 24 (25.00) | 67 (69.79) | 17.71 | 0.500 | 0.822 | 0.88 [0.60-1.28] | |||
All P-values have been calculated for the allelic model.
Benjamini and Hochberg step-up false discovery rate control.
Odds ratio for the minor allele.
MAF, minor allele frequency; GCA, giant cell arteritis; PMR, polymyalgia rheumatica; VIM, visual ischemic manifestations; IOD, irreversible occlusive disease
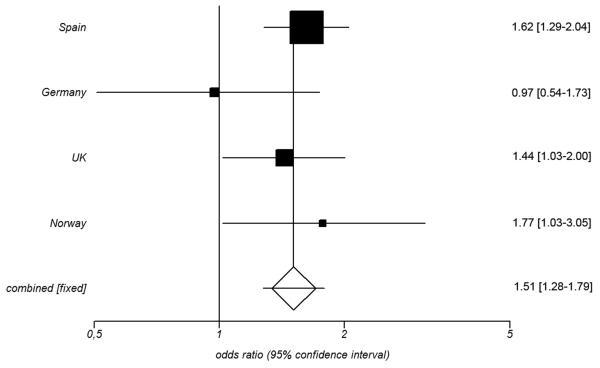
To follow up the positive finding of an association between PTPN22 rs2476601/R620W and GCA in the Spanish population, we attempted to confirm the detected association in a replication set of three independent cohorts of Caucasian ancestry. No heterogeneity between the ORs from the three replication cohorts was evident by BD test (P=0.05), and therefore a combined meta-analysis was performed (Table 2 and online supplementary table S2). Statistically significant differences were observed for the PTPN22 rs2476601*A allele in the pooled analysis (PMH=0.0154, OR= 1.38, CI 95% 1.07-1.77) (Table 2). Subsequently, the overall meta-analysis including both the discovery and the three replication cohorts showed a consistent association between the PTPN22 rs2476601*A variant and GCA (PMH=2.00E-06, OR= 1.51, CI 95% 1.28-1.79; Figure 1). Again, no significant differences were found when GCA patients with and without specific clinical features were compared (data not shown).
Table 2.
Replication and pooled analysis of the PTPN22 rs2476601 variant in Caucasian biopsy-proven GCA patients and controls.
| Genotype, N (%) |
Allele test |
||||||
|---|---|---|---|---|---|---|---|
| Population | Subgroup (N) | 1/1 | 1/2 | 2/2 | MAF (%) | P-value* | OR [CI 95%]** |
| Germany | Controls (n=937) | 9 (0.98) | 164 (17.94) | 741 (81.07) | 9.96 | ||
| GCA (n=72) | 0 (0.00) | 14 (19.44) | 58 (80.56) | 9.72 | 0.9280 | 0.97 [0.54-1.73] | |
|
| |||||||
| Norway | Controls (n=271) | 1 (0.37) | 56 (20.66) | 214 (78.97) | 10.70 | ||
| GCA (n=60) | 1 (1.67) | 19 (31.67) | 40 (66.67) | 17.50 | 0.0376 | 1.77 [1.03-3.05] | |
|
| |||||||
| UK | Controls (n=5199) | 42 (0.80) | 933 (17.95) | 4224 (81.25) | 9.78 | ||
| GCA (n=156) | 3 (1.92) | 36 (23.08) | 117 (75.00) | 13.46 | 0.0319 | 1.44 [1.03-2.00] | |
|
| |||||||
| Replication meta-analysis*** | Controls (n=6384) | 52 (0.81) | 1153 (18.06) | 5179 (81.12) | 9.84 | ||
| GCA (n=288) | 4 (1.39) | 69 (23.96) | 215 (74.65) | 13.37 | 0.0154 | 1.38 [1.07-1.77] | |
|
| |||||||
| Overall meta-analysis**** | Controls (n=8113) | 65 (0.80) | 1353 (16.68) | 6695 (82.52) | 9.14 | ||
| GCA (n=911) | 8 (0.88) | 188 (20.64) | 715 (78.49) | 11.20 | 2.00E-06 | 1.51 [1.28-1.79] | |
All P-values have been calculated for the allelic model.
Odds ratio for the minor allele.
Including independent cohorts from Germany, Norway and UK
Including independent cohorts from Spain, Germany, Norway and UK
Figure 1.
Forest plot showing the odds ratios (OR) and confidence intervals (CI) of the PTPN22 rs2476601 association in the discovery and replication cohorts. OR and CI were calculated under the fixed effect model.
The comparisons of the different detected allelic combinations between cases and controls did not yield additional information (data not shown).
DISCUSSION
Our data indicate, for the first time, an important role for PTPN22 in the genetic susceptibility of GCA. The combined analysis of the four independent cohorts showed a strong association between the PTPN22 rs2476601/R620W variant and this disease. The effect size detected in our study (OR=1.51) is similar to that described for other autoimmune conditions, such as rheumatoid arthritis (OR=1.45), SLE (OR=1.45) or type 1 diabetes mellitus (OR>1.80),[6, 7] and, interestingly, for other vasculitides, such as Behçet’s disease (OR>2.0) or anti-neutrophil cytoplasmic antibodies (ANCA)-associated vasculitis (OR>1.90).[18, 19] Despite this, a previous study failed to show association between PTPN22 rs2476601 and GCA;[20] however, it should be noted that the statistical power of this study was compromised because of the small sample size included in this report (96 GCA cases and 229 controls). In the subphenotype analysis, no specific association with any analyzed clinical feature was observed, indicating that this variant may represent a risk factor for the global disease. Nevertheless, this should be taken with caution, because of the low statistical power, which was a limitation of this stratified analysis.
Regarding CSK, our analysis had enough statistical power to detect a possible weak signal (power > 80% to detect an OR>1.25 in the discovery cohort); therefore, it is unlikely that CSK may play an important role in GCA susceptibility. Since an association between PTPN22 and GCA was observed, it makes sense that its interacting partner CSK may also play a role in this pathology, but in most of the diseases in which an involvement of PTPN22 has been described, an association with CSK has not been reported. Nevertheless, an effect of other CSK polymorphisms, showing low linkage disequilibrium with those analyzed in our study, in GCA susceptibility cannot be discarded.
Initially, the PTPN22 allele rs2476601*A, located within a protein–protein interaction domain, was reported as a gain-of-function allele that cause a decrease in TCR signaling.[12] However, a recent study has reported that this variant is a loss-of-function allele, leading to an accelerated degradation of lyp that results in enhanced signaling in several immune cell types.[13] Although the mechanisms underlying the role of the PTPN22 rs2476601 genetic variant in autoimmunity remain unclear, the association of this SNP with GCA suggests that a deregulation of TCR signaling is involved in the pathophysiological mechanisms of this vasculitis.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Sofía Vargas, Sonia Rodríguez and Gema Robledo for their excellent technical assistance, and all the patients and healthy controls for kindly accepting their essential collaboration. Banco Nacional de ADN (University of Salamanca, Spain) is thanked for supplying part of the control material. The Norwegian Systemic Vasculitis and Connective Tissue Disease Registry (NOSVAR) at Oslo University Hospital is acknowledged for providing data on the Norwegian patients and the Norwegian Bone Marrow Donor Registry is acknowledged for providing the Norwegian controls. This study was supported by ‘Fondo de Investigaciones Sanitarias’ through grants PI06-0024 and PS09/00748 (Spain), and partially supported by RETICS Program RD08/0075 (RIER) from ‘Instituto de Salud Carlos III’ (ISCIII). FDC was supported by Consejo Superior de Investigaciones Científicas (CSIC) through the program JAE-DOC. MCC and JH-R were granted by SAF 11/30073. TW was granted by DFG WI 1031/6.1. SLM was supported by a Clinical Lectureship from the National Institute for Health Research, UK, and by a grant from the Wellcome Trust/Academy of Medical Sciences.
REFERENCES
- 1.Gonzalez-Gay MA, Vazquez-Rodriguez TR, Lopez-Diaz MJ, et al. Epidemiology of giant cell arteritis and polymyalgia rheumatica. Arthritis Rheum. 2009;61(10):1454–1461. doi: 10.1002/art.24459. [DOI] [PubMed] [Google Scholar]
- 2.Ly KH, Regent A, Tamby MC, et al. Pathogenesis of giant cell arteritis: More than just an inflammatory condition? Autoimmun Rev. 2010;9:635–645. doi: 10.1016/j.autrev.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Deng J, Younge BR, Olshen RA, et al. Th17 and Th1 T-cell responses in giant cell arteritis. Circulation. 2010;121:906–915. doi: 10.1161/CIRCULATIONAHA.109.872903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espigol-Frigole G, Corbera-Bellalta M, Planas-Rigol E, et al. Increased IL-17A expression in temporal artery lesions is a predictor of sustained response to glucocorticoid treatment in patients with giant-cell arteritis. Ann Rheum Dis. 2012 doi: 10.1136/annrheumdis-2012-201836. [DOI] [PubMed] [Google Scholar]
- 5.Levinson NM, Seeliger MA, Cole PA, et al. Structural basis for the recognition of c-Src by its inactivator Csk. Cell. 2008;134:124–134. doi: 10.1016/j.cell.2008.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bottini N, Musumeci L, Alonso A, et al. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 7.Orozco G, Sanchez E, Gonzalez-Gay MA, et al. Association of a functional single-nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with rheumatoid arthritis and systemic lupus erythematosus. Arthritis Rheum. 2005;52(1):219–224. doi: 10.1002/art.20771. [DOI] [PubMed] [Google Scholar]
- 8.Diaz-Gallo LM, Gourh P, Broen J, et al. Analysis of the influence of PTPN22 gene polymorphisms in systemic sclerosis. Ann Rheum Dis. 2011;70:454–462. doi: 10.1136/ard.2010.130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez-Rodriguez L, Taib WR, Topless R, et al. The PTPN22 R263Q polymorphism is a risk factor for rheumatoid arthritis in Caucasian case-control samples. Arthritis Rheum. 2011;63:365–372. doi: 10.1002/art.30145. [DOI] [PubMed] [Google Scholar]
- 10.Manjarrez-Orduno N, Marasco E, Chung SA, et al. CSK regulatory polymorphism is associated with systemic lupus erythematosus and influences B-cell signaling and activation. Nat Genet. 2012;44:1227–1230. doi: 10.1038/ng.2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin JE, Broen JC, Carmona FD, et al. Identification of CSK as a systemic sclerosis genetic risk factor through Genome Wide Association Study follow-up. Hum Mol Genet. 2012;21:2825–2835. doi: 10.1093/hmg/dds099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vang T, Congia M, Macis MD, et al. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet. 2005;37:1317–1319. doi: 10.1038/ng1673. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Zahir N, Jiang Q, et al. The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness. Nat Genet. 2011;43:902–907. doi: 10.1038/ng.904. [DOI] [PubMed] [Google Scholar]
- 14.Huffmeier U, Steffens M, Burkhardt H, et al. Evidence for susceptibility determinant(s) to psoriasis vulgaris in or near PTPN22 in German patients. J Med Genet. 2006;43:517–522. doi: 10.1136/jmg.2005.037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan AW, Thomson W, Martin SG, et al. Reevaluation of the interaction between HLA-DRB1 shared epitope alleles, PTPN22, and smoking in determining susceptibility to autoantibody-positive and autoantibody-negative rheumatoid arthritis in a large UK Caucasian population. Arthritis Rheum. 2009;60(9):2565–2576. doi: 10.1002/art.24752. [DOI] [PubMed] [Google Scholar]
- 16.Hunder GG, Bloch DA, Michel BA, et al. The American College of Rheumatology 1990 criteria for the classification of giant cell arteritis. Arthritis Rheum. 1990;33:1122–1128. doi: 10.1002/art.1780330810. [DOI] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 18.Baranathan V, Stanford MR, Vaughan RW, et al. The association of the PTPN22 620W polymorphism with Behcet’s disease. Ann Rheum Dis. 2007;66:1531–1533. doi: 10.1136/ard.2007.073866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martorana D, Maritati F, Malerba G, et al. PTPN22 R620W polymorphism in the ANCA-associated vasculitides. Rheumatology (Oxford) 2012;51:805–812. doi: 10.1093/rheumatology/ker446. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez-Gay MA, Oliver J, Orozco G, et al. Lack of association of a functional single nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with susceptibility to biopsy-proven giant cell arteritis. J Rheumatol. 2005;32:1510–1512. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.