Abstract
X-ray repair cross-complementing group 1 (XRCC1) gene Arg194Trp polymorphism has been reported to be associated with risk of lung cancer in many published studies. Nevertheless, the research results were inconclusive and conflicting. To reach conclusive results, several meta-analysis studies were conducted by combining results from literature reports through pooling analysis. However, these previous meta-analysis studies were still not consistent. Hence, we used an updated and cumulative meta-analysis to get a more comprehensive and precise result from 25 case–control studies searching through the PubMed database up to September 1, 2013. The meta-analysis was carried out by the Comprehensive Meta-Analysis software and the odds ratio (OR) with 95 % confidence interval (CI) was used to estimate the pooled effect. The result involving 8,876 lung cancer patients and 11,210 controls revealed that XRCC1 Arg194Trp polymorphism was not associated with lung cancer risk [(OR = 0.97, 95 %CI = 0.92–1.03) for Trp vs. Arg; (OR = 0.92, 95 % CI = 0.85–0.98) for ArgTrp vs. ArgArg; (OR = 1.07, 95 % CI = 0.92–1.23) for TrpTrp vs. ArgArg; (OR = 0.93, 95 % CI = 0.87–1.00) for (TrpTrp + ArgTrp) vs. ArgArg; and (OR = 1.08, 95 % CI = 0.94–1.25) for TrpTrp vs. (ArgTrp + ArgArg)]. The cumulative meta-analysis showed that the results maintained the same, while the ORs with 95 % CI were more stable with the accumulation of case–control studies. The sensitivity and subgroups analyses showed that the results were robust and not affected by any single study with no publication bias. Relevant studies might not be needed for supporting these results.
Keywords: X-ray repair cross-complementing group 1, XRCC1, Lung cancer, Polymorphism, Risk, Meta-analysis
Introduction
X-ray repair cross-complementing group 1 (XRCC1) is involved in base excision repair protein that located on chromosome 19q13.2–13.3 with a length of 33 kb [1–4]. The polymorphisms of XRCC1 gene have been identified as three categories of codons 194(Arg to Trp), 280(Arg to His), and 399 (Arg toGln) [5, 6]. One of them, Arg194Trp polymorphism was first reported in 1998 by Shen and coworkers [7]. In 2001, David-Beabesand coworkers [8] found that Arg194Trp polymorphism might contribute to lung cancer in African-American and Caucasian. Ratnasinghe and coworker [5] found similar results in Chinese during the same year. Later on, many molecular epidemiological studies reported the association of XRCC1 Arg194Trp with lung cancer susceptibility [5, 8–30]. However, these results remain conflicting and inconclusive. To reach conclusive results, several meta-analysis studies were conducted by combining results across studies from literatures through pooling analysis. However, these previous meta-analysis investigations were still not consistent [31–33]. Furthermore, new published research studies were coming out, but the inconclusive results are still a problem to be resolved. Therefore, the association of Arg194Trp with lung cancer susceptibility with lung cancer risk remains unclear.
In order to obtain more comprehensive and precise results, we conducted cumulative meta-analysis [34, 35] to explore the truly association between Arg194Trp polymorphism and lung cancer risk based on 25 case–control studies. The meta-analysis is reported based on preferred reporting items for systematic reviews and meta-analyses (PRISMA) [36] statement.
Material and methods
Inclusion criteria
A study met all of the following inclusion criteria was included: (1) to evaluate the association between XRCC1 Arg194Trp polymorphism and risk of lung cancer; (2) cohort or case–control design and the patients were diagnosed by histology or pathology; (3) the number of genotype distribution in both case and control group were directly reported or calculated from the reported data; and (4) the published language was English or Chinese.
Search strategy
The search terms [(“XRCC1” or “X-ray repair cross-complementing group 1”) and “polymorphism” and (“lung cancer” or “lung carcinoma”)] were used to search the PubMed database up to September 1, 2013. The reference list of the included articles and relevant meta-analyses were manually searched.
Data extraction
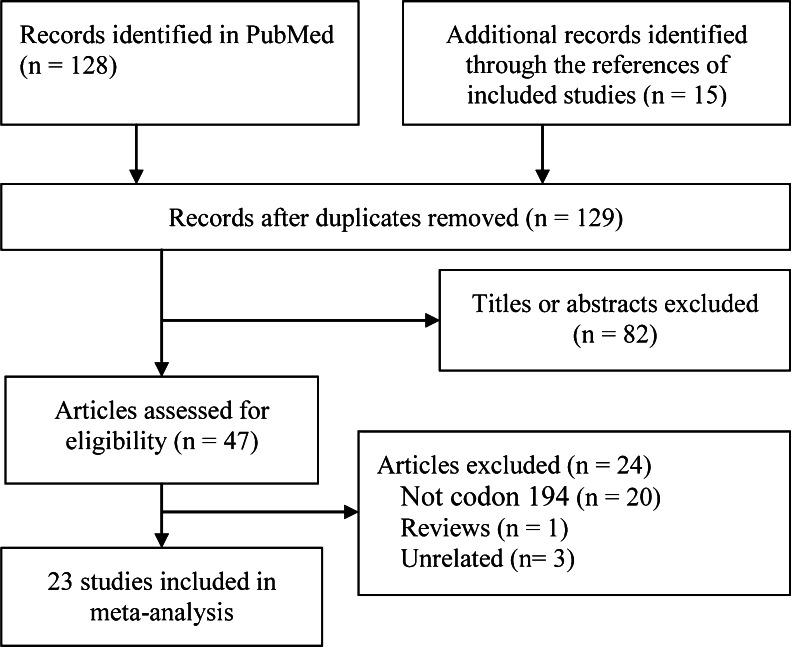
Two authors independently chose 25 case–control studies, which were illustrated in Fig. 1. The data were independently extracted by authors according to the pre-specified table. The following data were extracted: the surname of first author, publication year, country origin and ethnicity, study design, cancer type, source of control, number and genotyping distribution of cases and controls, genotyping method, Hardy–Weinberg equilibrium (HWE) for controls. Disagreements were resolved through discussion with the third author.
Fig. 1.
Flow chart from identification of eligible studies to final inclusion
Statistical analysis
Five genetic models [Trp vs. Arg; ArgTrp vs. ArgArg; TrpTrp vs. ArgArg; (TrpTrp + ArgTrp) vs. ArgArg; and TrpTrp vs. (ArgTrp + ArgArg)] were used to calculate the pooled odds ratio (OR) and its 95 % confidence interval (CI) to present the strength of associations between XRCC1 Arg194Trp polymorphism and risk of lung cancer. The fixed-effects model was used firstly, if heterogeneity among included studies was detected by I 2 statistics (I 2 ≤ 40 %) [37], we shifted to random-effects model. Subgroups analysis were conducted based on the ethnicity, source of controls, cancer types, study design, and HWE for controls.
The influence of sample size on the overall risk estimation was carried out by cumulative meta-analysis [35], and the influence of single study on the overall risk estimation was determined through sensitivity analysis by omitting one study each time. The publication bias was detected by funnel plot analysis. All the analysis was performed using the Comprehensive Meta-Analysis software, version 2.2 (Biostat, Englewood, New Jersey) [38].
Results
Study section and characteristics
The electronic searching yielded 128 studies, and the hand searching yielded 15 studies initially; finally, 23 articles involving 25 case–control studies [5, 8–29] contained 8,876 lung cancer patients and 11,210 controls were included. Figure 1 presents flow chart of study selection. The main characteristics of these eight studies were shown in Table 1. Of them, three were multicenter studies [12, 16, 17], two articles were included two case–control studies [8, 24], and only one study was out of HWE [27].
Table 1.
Characteristics of included studies
| References | Country (ethnicity) | Case | Source of control | Control | Genotyping | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | ArgArg | ArgTrp | TrpTrp | N | ArgArg | ArgTrp | TrpTrp | |||||
| David-Beabes [8] | USA (Caucasian) | 180 | 158 | 22 | 0 | PB | 461 | 407 | 54 | 0 | PCR-RFLP | 0.39 |
| David-Beabes [8] | USA (African-Americans) | 154 | 142 | 10 | 2 | PB | 243 | 205 | 36 | 2 | PCR-RFLP | 0.67 |
| Ratnasinghe [5] | China (Asian) | 108 | 52 | 47 | 9 | PB | 216 | 85 | 104 | 21 | TaqMan | 0.22 |
| Chen [9] | China (Asian) | 109 | 48 | 44 | 11 | PB | 109 | 57 | 40 | 5 | PCR-RFLP | 0.79 |
| Chan [10] | China (Asian) | 75 | 50 | 22 | 3 | HB | 162 | 79 | 67 | 16 | PCR-RFLP | 0.71 |
| Hu [11] | China (Asian) | 710 | 335 | 311 | 64 | HB | 710 | 339 | 308 | 63 | PCR | 0.59 |
| Hung [12] | European (Caucasian) | 2,188 | 1,878 | 259 | 10 | HB | 2,198 | 1,828 | 292 | 12 | PCR | 0.87 |
| Schneider [13] | Germany (Caucasian) | 446 | 389 | 53 | 4 | HB | 622 | 544 | 75 | 3 | PCR | 0.74 |
| Shen [14] | China (Asian) | 118 | 65 | 41 | 12 | HB | 112 | 64 | 40 | 8 | PCR | 0.62 |
| Hao [15] | China (Asian) | 1,024 | 524 | 409 | 91 | PB | 1,118 | 572 | 459 | 87 | PCR | 0.77 |
| Landi [16] | Europe (Caucasian) | 295 | 118 | 143 | 34 | HB | 314 | 123 | 149 | 42 | PCR | 0.96 |
| Matullo [17] | Europe (Caucasian) | 116 | 98 | 16 | 2 | PB | 1,094 | 951 | 141 | 2 | TaqMan | 0.22 |
| Zienolddiny [18] | Norway (Caucasian) | 336 | 309 | 26 | 1 | PB | 405 | 368 | 35 | 2 | TaqMan | 0.23 |
| De Ruyck [19] | Belgium (Caucasian) | 110 | 101 | 8 | 1 | HB | 110 | 93 | 17 | 0 | PCR-RFLP | 0.38 |
| Pachouri [20] | India (Asian) | 103 | 40 | 39 | 24 | PB | 122 | 52 | 47 | 23 | PCR-RFLP | 0.051 |
| Yin [21] | China (Asian) | 241 | 120 | 98 | 23 | HB | 249 | 119 | 109 | 21 | PCR-RFLP | 0.65 |
| Li [23] | China (Asian) | 350 | 184 | 136 | 30 | HB | 350 | 196 | 133 | 21 | PCR-RFLP | 0.89 |
| Improta [22] | Italy (Caucasian) | 94 | 42 | 41 | 11 | HB | 121 | 53 | 61 | 7 | PCR-RFLP | 0.15 |
| Chang [24] | USA (Latinos) | 113 | 89 | 23 | 1 | PB | 299 | 223 | 66 | 10 | Illumina | 0.1 |
| Chang [24] | USA (African–Americans) | 255 | 221 | 34 | 0 | PB | 280 | 248 | 31 | 1 | Illumina | 0.97 |
| Tanaka [25] | Japan (Asian) | 50 | 28 | 15 | 7 | PB | 50 | 25 | 23 | 2 | PCR | 0.47 |
| Janik [26] | Poland (Caucasian) | 88 | 64 | 24 | 0 | HB | 79 | 51 | 28 | 0 | PCR-SSCP | 0.55 |
| Buch [27] | USA (Caucasian) | 720 | 682 | 36 | 2 | HB | 928 | 839 | 83 | 6 | Illumina | 0.03 |
| Wang [28] | China (Asian) | 209 | 105 | 83 | 21 | HB | 256 | 137 | 96 | 23 | PCR-RFLP | 0.59 |
| Guo [29] | China (Asian) | 684 | 314 | 302 | 68 | HB | 602 | 265 | 274 | 63 | PCR-LDR | 0.58 |
N total sample size, PB population-based controls, HB hospital-based controls, HWE Hardy–Weinberg equilibrium, PCR-RFLP polymerase chain reaction-restriction fragment length polymorphism, PCR-LDR polymerase chain reaction-ligase detection reaction, PCR-SSCP polymerase chain reaction-single strand conformation polymorphism
Meta-analysis
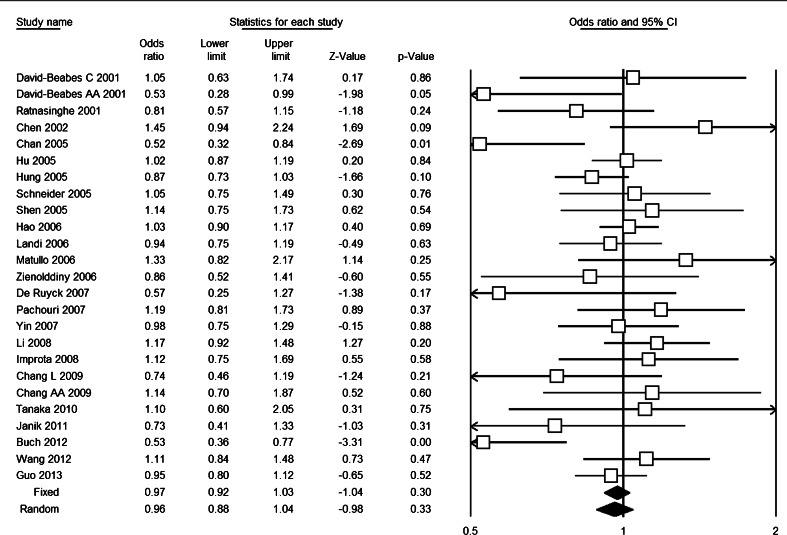
Table 2 presented the overall and subgroups results of XRCC1 Arg194Trp polymorphism and lung cancer risk. Overall, the heterogeneity of all five genetic models were acceptable (I 2 ≤ 40 %), and meta-analysis based on fixed-effects model showed that there was no association of XRCC1 Arg194Trp polymorphism with risk of lung cancer [(OR = 0.97, 95 % CI = 0.92–1.03) for Trp vs. Arg, Fig. 2; (OR = 0.92, 95 % CI = 0.85–0.98) for ArgTrp vs. ArgArg; (OR = 1.07, 95 % CI = 0.92–1.23) for TrpTrp vs. ArgArg; (OR = 0.93, 95 % CI = 0.87–1.00) for (TrpTrp + ArgTrp) vs. ArgArg; and (OR = 1.08, 95 % CI = 0.94–1.25) for TrpTrp vs. (ArgTrp + ArgArg)].
Table 2.
Results of overall and subgroup meta-analysis
| No. of studies | Trp vs. Arg | ArgTrp vs. ArgArg | TrpTrp vs. ArgArg | TrpTrp + ArgTrp vs. ArgArg | TrpTrp vs. ArgTrp + ArgArg | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95 % CI) | p for OR | I 2 (%) | OR (95 % CI) | p for OR | I 2 (%) | OR (95 % CI) | p for OR | I 2 (%) | OR (95 % CI) | p for OR | I 2 (%) | OR (95 % CI) | p for OR | I 2 (%) | ||
| Total | 25 | 0.97 (0.92–1.03) | 0.30 | 38.8 | 0.92 (0.85–0.98) | 0.017 | 17.4 | 1.07 (0.92–1.23) | 0.380 | 9.5 | 0.93 (0.87–1.00) | 0.047 | 28.5 | 1.08 (0.94–1.25) | 0.255 | 3.8 |
| Ethnicity | ||||||||||||||||
| Asian | 12 | 1.02 (0.95–1.09) | 0.662 | 28.3 | 0.97 (0.88–1.06) | 0.479 | 0.0 | 1.10 (0.93–1.29) | 0.258 | 7.7 | 0.99 (0.91–1.08) | 0.825 | 13.7 | 1.11 (0.95–1.30) | 0.717 | 0.0 |
| Caucasian | 10 | 0.89 (0.80–0.98) | 0.024 | 37.6 | 0.85 (0.76–0.96) | 0.008 | 16.1 | 1.01 (0.70–1.44) | 0.974 | 29.0 | 0.86 (0.76–0.97) | 0.011 | 24.7 | 1.02 (0.72–1.44) | 0.914 | 32.6 |
| Others | 3 | 0.80 (0.59–1.07) | 0.130 | 46.4 | 0.83 (0.60–1.16) | 0.276 | 66.8 | 0.50 (0.15–1.67) | 0.259 | 0.0 | 0.81 (0.59–1.11) | 0.189 | 59.1 | 0.52 (0.15–1.74) | 0.287 | 0.0 |
| Source of controls | ||||||||||||||||
| HB | 14 | 0.95 (0.88–1.01) | 0.113 | 48.7 | 0.90 (0.83–0.99) | 0.022 | 28.2 | 1.02 (0.85–1.22) | 0.847 | 0.0 | 0.91 (0.84–0.99) | 0.035 | 40.0 | 1.03 (0.87–1.23) | 0.725 | 0.0 |
| PB | 11 | 1.01 (0.92–1.12) | 0.816 | 20.9 | 0.94 (0.83–1.07) | 0.375 | 5.6 | 1.17 (0.91–1.50) | 0.217 | 26.6 | 0.97 (0.86–1.10) | 0.657 | 10.6 | 1.20 (0.94–1.52) | 0.144 | 23.2 |
| HWE | ||||||||||||||||
| Yes | 24 | 0.97 (0.91–1.02) | 0.530 | 20.8 | 0.93 (0.87–1.00) | 0.067 | 0.0 | 1.08 (0.93–1.25) | 0.317 | 8.3 | 0.95 (0.89–1.02) | 0.163 | 8.0 | 1.09 (0.95–1.26) | 0.211 | 2.5 |
| No | 1 | 0.53 (0.36–0.77) | 0.001 | – | 0.53 (0.36–0.80) | 0.002 | – | 0.41 (0.08–2.04) | 0.276 | – | 0.53 (0.35–0.78) | 0.001 | – | 0.43 (0.09–2.13) | 0.300 | – |
OR odds ratio, CI confidence interval, PB population-based controls, HB hospital-based controls, HWE Hardy–Weinberg equilibrium
Fig. 2.
Forest plot based on Trp vs. Arg genetic model
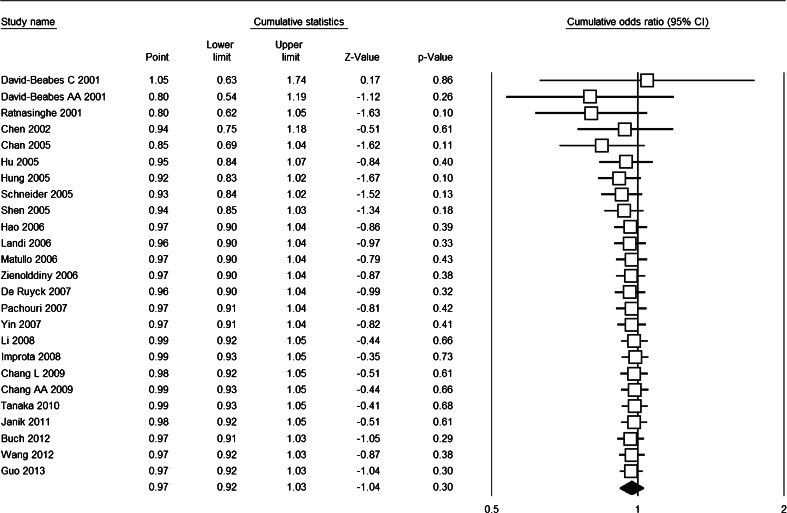
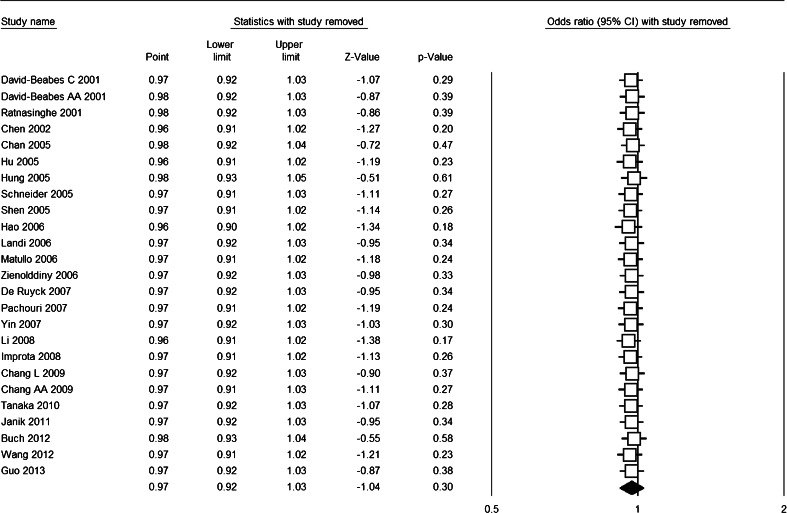
The cumulative meta-analysis accumulated the studies according to the publication year and showed that there was no significant association between XRCC1 Arg194Trp polymorphism and lung cancer risk (Fig. 3). The sensitivity analysis showed that the results were robust and were not influenced by any single study (Fig. 4), with ORs in the range of 0.96–0.98 and 95 % CIs in the range of 0.90–1.05. Subgroup analysis upon source of control, ethnicity, and HWE also revealed similar results (Table 2).
Fig. 3.
Forest plot for cumulative meta-analysis based on Trp vs. Arg genetic model
Fig. 4.
Forest plot for sensitivity analysis based on Trp vs. Arg genetic model
Publication bias
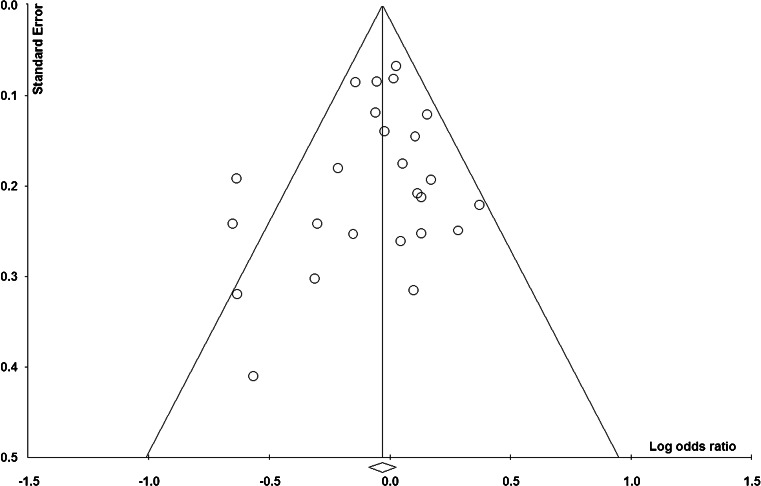
Figure 5 shows the funnel plot of based on Trp vs. Arg genetic model. The relatively symmetric distribution indicated that there was no publication bias, which was confirmed by Egger’s test [(p = 0.33 for Trp vs. Arg; p = 0.12 for ArgTrp vs. ArgArg; p = 0.65 for TrpTrp vs. ArgArg; p = 0.25 for (TrpTrp + ArgTrp) vs. ArgArg; and p = 0.50 for TrpTrp vs. (ArgTrp + ArgArg))].
Fig. 5.
Funnel plot based on Trp vs. Arg genetic model
Discussion
Meta-analysis is a statistical method of combining results across studies from literatures to resolve discrepancy in genetic association studies [39]. The meta-analysis of 25 case–control studies indicated that XRCC1 Arg194Trp polymorphism is not associated with lung cancer risk within human populations, and subgroup analysis upon source of controls, ethnicity, and HWE for controls is consistent with this result, which was also supported by cumulative meta-analysis and sensitivity analysis.
Compared to previously meta-analyses [31–33], the included studies of our analysis are most precise and comprehensive attributing to the largest sample size and accumulative meta-analysis method. Hence, the results are more precise and comprehensive. In addition, cumulative meta-analysis was performed to investigate the tendency of results by accumulating single study year by year. This analysis could be used to determine whether new relevant studies are needed or not. Indeed, we found that the results remained the same when studies were accumulated. Coincidentally, the sensitivity analysis indicated that the results were not influenced by any single study. Hence, our results were more precise and useful for appropriate care in lung cancer.
Obviously, there were potential to moderate level heterogeneity. From the subgroups analysis, we found that ethnicity and source of control might not be the source of heterogeneity (Table 2). When we deleted the study reported by Buch et al.[27], which was not according to HWE any more, the heterogeneity of all genetic models were decreased and the results of all five genetic models were of no significance (Table 2). This further indicated that violations and deviations in HWE might be one source of heterogeneity and do largely influence the results [40].
There were some limitations of our meta-analysis. First, there was heterogeneity among included studies. Although the heterogeneity was probably from the study reported by Buch et al. [27], we could not conclude whether the heterogeneity came from ethnicity or inconsistent results. Obviously, the homogeneity of Asians and Caucasian was good, but only the one combined with mixed ethnicities was significant. Second, although no obvious publication bias was detected; the funnel plot was not very symmetry. Our meta-analysis is limited to language and database restrictions. The PubMed database is the only search source and included published studies were either in English or Chinese [28]. Third, this meta-analysis was based on unadjusted data, lacking of detailed genotype information stratified by main confounding variables from original studies. Therefore, gene-gene and gene-environment interactions remain unclear.
In conclusion, this meta-analysis suggests that XRCC1 Arg194Trp polymorphism is not associated with lung cancer risk, either in Asians or Caucasians, either the controls were sourced with or without HWE. These results were not influenced by any single study, and relevant studies are not needed for supporting this result. Due to the limitations of this meta-analysis, current results should be viewed with caution and future studies should be conducted in gene-gene and gene-environment interactions.
Acknowledgment
We specially thank Ru Wen (who is currently working in the Department of Chemistry, University of Georgia) for her help in revising the grammar issue of our manuscript.
Conflicts of interest
None
References
- 1.Xue H, Ni P, Lin B, Xu H, Huang G. X-ray repair cross-complementing group 1 (XRCC1) genetic polymorphisms and gastric cancer risk: a huge review and meta-analysis. Am J Epidemiol. 2011;173:363–75. [DOI] [PubMed]
- 2.Duell EJ, Holly EA, Bracci PM, Wiencke JK, Kelsey KT. A population-based study of the arg399gln polymorphism in x-ray repair cross-complementing group 1 (XRCC1) and risk of pancreatic adenocarcinoma. Cancer Res. 2002;62:4630–6. [PubMed]
- 3.Kubota Y, Nash RA, Klungland A, Schar P, Barnes DE, Lindahl T. Reconstitution of DNA base excision-repair with purified human proteins: Interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–70. [PMC free article] [PubMed]
- 4.Duell EJ, Millikan RC, Pittman GS, Winkel S, Lunn RM, Chiu-Kit JT, et al. Polymorphisms in the DNA repair gene XRCC1 and breast cancer. Cancer Epidemiol Biomark Prev. 2001;10:217–22. [PubMed]
- 5.Ratnasinghe D, Yao SX, Tangrea JA, Qiao YL, Andersen MR, Barrett MJ, et al. Polymorphisms of the DNA repair gene XRCCL and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10:119–23. [PubMed]
- 6.Park JY, Lee SY, Jeon HS, Bae NC, Chae SC, Joo S, et al. Polymorphism of the DNA repair gene XRCCL and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2002;11:23–7. [PubMed]
- 7.Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–8. [PubMed]
- 8.David-Beabes GL, London SJ. Genetic polymorphism of XRCC1 and lung cancer risk among African-Americans and Caucasians. Lung Cancer. 2001;34:333–9. [DOI] [PubMed]
- 9.Chen S, Tang D, Xue K, Xu L, Ma G, Hsu Y, et al. DNA repair gene XRCC1 and XPD polymorphisms and risk of lung cancer in a Chinese population. Carcinogenesis. 2002;23:1321–5. [DOI] [PubMed]
- 10.Chan EC, Lam SY, Fu KH, Kwong YL. Polymorphisms of the GSTM1, GSTP1, MPO, XRCC1, and NQO1 genes in Chinese patients with non-small cell lung cancers: Relationship with aberrant promoter methylation of the cdkn2a and rarb genes. Cancer Genet Cytogenet. 2005;162:10–20. [DOI] [PubMed]
- 11.Hu Z, Ma H, Lu D, Zhou J, Chen Y, Xu L, et al. A promoter polymorphism (−77 T >C) of DNA repair gene XRCC1 is associated with risk of lung cancer in relation to tobacco smoking. Pharmacogenet Genomics. 2005;15:457–63. [DOI] [PubMed]
- 12.Hung RJ, Brennan P, Canzian F, Szeszenia-Dabrowska N, Zaridze D, Lissowska J, et al. Large-scale investigation of base excision repair genetic polymorphisms and lung cancer risk in a multicenter study. J Natl Cancer Inst. 2005;97:567–76. doi: 10.1093/jnci/dji101. [DOI] [PubMed] [Google Scholar]
- 13.Schneider J, Classen V, Bernges U, Philipp M. Xrcc1 polymorphism and lung cancer risk in relation to tobacco smoking. Int J Mol Med. 2005;16:709–16. [PubMed] [Google Scholar]
- 14.Shen M, Berndt SI, Rothman N, Mumford JL, He X, Yeager M, et al. Polymorphisms in the DNA base excision repair genes APEX1 and XRCC1 and lung cancer risk in Xuan Wei, China. Anticancer Res. 2005;25:537–42. [PubMed]
- 15.Hao B, Miao X, Li Y, Zhang X, Sun T, Liang G, et al. A novel T-77C polymorphism in DNA repair gene xrcc1 contributes to diminished promoter activity and increased risk of non-small cell lung cancer. Oncogene. 2006;25:3613–20. [DOI] [PubMed]
- 16.Landi S, Gemignani F, Canzian F, Gaborieau V, Barale R, Landi D, et al. DNA repair and cell cycle control genes and the risk of young-onset lung cancer. Cancer Res. 2006;66:11062–9. doi: 10.1158/0008-5472.CAN-06-1039. [DOI] [PubMed] [Google Scholar]
- 17.Matullo G, Dunning AM, Guarrera S, Baynes C, Polidoro S, Garte S, et al. DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis. 2006;27:997–1007. doi: 10.1093/carcin/bgi280. [DOI] [PubMed] [Google Scholar]
- 18.Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland L, et al. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27:560–7. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]
- 19.De Ruyck K, Szaumkessel M, De Rudder I, Dehoorne A, Vral A, Claes K, et al. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat Res. 2007;631:101–10. doi: 10.1016/j.mrgentox.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Pachouri SS, Sobti RC, Kaur P, Singh J. Contrasting impact of DNA repair gene XRCC1 polymorphisms Arg399Gln and Arg194Trp on the risk of lung cancer in the north-Indian population. DNA Cell Biol. 2007;26:186–91. [DOI] [PubMed]
- 21.Yin J, Vogel U, Ma Y, Qi R, Sun Z, Wang H. The DNA repair gene XRCC1 and genetic susceptibility of lung cancer in a northeastern Chinese population. Lung Cancer. 2007;56:153–60. [DOI] [PubMed]
- 22.Improta G, Sgambato A, Bianchino G, Zupa A, Grieco V, La Torre G, et al. Polymorphisms of the DNA repair genes XRCC1 and XRCC3 and risk of lung and colorectal cancer: a case–control study in a southern Italian population. Anticancer Res. 2008;28:2941–6. [PubMed]
- 23.Li M, Yin Z, Guan P, Li X, Cui Z, Zhang J, et al. XRCC1 polymorphisms, cooking oil fume and lung cancer in Chinese women nonsmokers. Lung Cancer. 2008;62:145–51. [DOI] [PubMed]
- 24.Chang JS, Wrensch MR, Hansen HM, Sison JD, Aldrich MC, Quesenberry CP, Jr, et al. Base excision repair genes and risk of lung cancer among san francisco bay area latinos and African-Americans. Carcinogenesis. 2009;30:78–87. doi: 10.1093/carcin/bgn261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka Y, Maniwa Y, Bermudez VP, Doi T, Nishio W, Ohbayashi C, et al. Nonsynonymous single nucleotide polymorphisms in DNA damage repair pathways and lung cancer risk. Cancer. 2010;116:896–902. doi: 10.1002/cncr.24850. [DOI] [PubMed] [Google Scholar]
- 26.Janik J, Swoboda M, Janowska B, Ciesla JM, Gackowski D, Kowalewski J, et al. 8-oxoguanine incision activity is impaired in lung tissues of NSCLC patients with the polymorphism of OGG1 and XRCC1 genes. Mutat Res. 2011;709–710:21–31. [DOI] [PubMed]
- 27.Buch SC, Diergaarde B, Nukui T, Day RS, Siegfried JM, Romkes M, et al. Genetic variability in DNA repair and cell cycle control pathway genes and risk of smoking-related lung cancer. Mol Carcinog. 2012;51(Suppl 1):E11–20. doi: 10.1002/mc.20858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang N, Wu Y, Zhou X. Association between genetic polymorphism of metabolizing enzymes and DNA repairing enzymes and the susceptibility of lung cancer in henan population. Wei Sheng Yan Jiu. 2012;41:251–6. [PubMed] [Google Scholar]
- 29.Guo S, Li X, Gao M, Li Y, Song B, Niu W. The relationship between XRCC1 and XRCC3 gene polymorphisms and lung cancer risk in northeastern Chinese. PLoS One. 2013;8:e56213. [DOI] [PMC free article] [PubMed]
- 30.Butkiewicz D, Rusin M, Enewold L, Shields PG, Chorazy M, Harris CC. Genetic polymorphisms in DNA repair genes and risk of lung cancer. Carcinogenesis. 2001;22:593–7. doi: 10.1093/carcin/22.4.593. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Yang H, Li H, Li L, Wang H, Liu C, et al. Association between x-ray repair cross complementing group 1 codon 399 and 194 polymorphisms and lung cancer risk: a meta-analysis. Cancer Lett. 2009;285:134–40. doi: 10.1016/j.canlet.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 32.Dai L, Duan F, Wang P, Song C, Wang K, Zhang J. XRCC1 gene polymorphisms and lung cancer susceptibility: a meta-analysis of 44 case–control studies. Mol Biol Rep. 2012;39:9535–47. [DOI] [PubMed]
- 33.33 Wu T, Xu YH, Ye XL: X-ray repair cross-complementing group 1 Arg194Trp polymorphism is associated with increased risk of lung cancer in Chinese han population. Tumour Biol 2013 [DOI] [PubMed]
- 34.Pabalan NA. Meta-analysis in cancer genetics. Asian Pac J Cancer Prev. 2010;11:33–8. [PubMed] [Google Scholar]
- 35.Rotondi MA, Bull SB. Cumulative meta-analysis for genetic association: When is a new study worthwhile? Hum Hered. 2012;74:61–70. doi: 10.1159/000345604. [DOI] [PubMed] [Google Scholar]
- 36.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the prisma statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huedo-Medina TB, Sanchez-Meca J, Marin-Martinez F, Botella J. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods. 2006;11:193–206. [DOI] [PubMed]
- 38.Borenstein M, Hedges L, Rothstein H. Comprehensive meta-analysis. Englewood, New Jersey: Biostat; 2005. [Google Scholar]
- 39.Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20:439–44. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 40.Trikalinos TA, Salanti G, Khoury MJ, Ioannidis JP. Impact of violations and deviations in hardy-weinberg equilibrium on postulated gene-disease associations. Am J Epidemiol. 2006;163:300–9. doi: 10.1093/aje/kwj046. [DOI] [PubMed] [Google Scholar]