Abstract
Background
Canine osteosarcoma is clinically nearly identical to the human disease, but is common and highly heritable, making genetic dissection feasible.
Results
Through genome-wide association analyses in three breeds (greyhounds, Rottweilers, and Irish wolfhounds), we identify 33 inherited risk loci explaining 55% to 85% of phenotype variance in each breed. The greyhound locus exhibiting the strongest association, located 150 kilobases upstream of the genes CDKN2A/B, is also the most rearranged locus in canine osteosarcoma tumors. The top germline candidate variant is found at a >90% frequency in Rottweilers and Irish wolfhounds, and alters an evolutionarily constrained element that we show has strong enhancer activity in human osteosarcoma cells. In all three breeds, osteosarcoma-associated loci and regions of reduced heterozygosity are enriched for genes in pathways connected to bone differentiation and growth. Several pathways, including one of genes regulated by miR124, are also enriched for somatic copy-number changes in tumors.
Conclusions
Mapping a complex cancer in multiple dog breeds reveals a polygenic spectrum of germline risk factors pointing to specific pathways as drivers of disease.
Background
Osteosarcoma (OS) is an aggressive cancer characterized by early metastasis, primary onset in children and adolescents, and high mortality rates (30% to 40%) [1]. Recent work suggests that OS arises when osteoblast differentiation from mesenchymal precursors is disrupted by genetic or epigenetic factors [2]. While no structural variants specific to OS have been identified, somatic alterations in tumors are common and often affect suppressor genes RB1, TP53, and the CDK4 inhibitors CDKN2A/B[2]. Germline mutations in RB1 and p53, two genes essential for OS development, can increase disease risk [2-4]. The only genome-wide association study (GWAS) of OS in humans found two significant associations, one genic (GRM4) and the other in a large gene desert, suggesting inherited variation in regulatory elements underlies susceptibility [5].
OS in dogs is a spontaneously occurring disease with a global tumor gene expression signature indistinguishable from human pediatric tumors [6,7] and, while relative age of onset is higher in dogs, their clinical progression is remarkably similar [8]. Both human and canine OS most commonly arise at the ends of the long bones of the limbs and metastasize readily, usually to the lungs [9]. Unlike human OS, canine OS is a highly heritable disease with some large and giant dog breeds at >10× increased risk, notably greyhounds (mortality from OS = 26%), Rottweilers (17%), and Irish wolfhounds (IWH, 21%) [10-12]. While size and hormonal factors influence risk, variable rates among the larger size breeds suggest breed-specific risk factors [13]. The greyhound American Kennel Club (AKC) registered dogs, a subpopulation that tends to be taller than the racing dogs, has very low rates of OS (G. Couto, unpublished observations).
Mapping disease genes using GWAS in dog breeds, each effectively a genetic isolate only a few hundred years old, requires approximately 10× fewer markers and samples than in human populations [14]. However, population structure, cryptic relatedness, and extensive regions of near fixation in breeds complicate GWAS analysis and to date, few studies have successfully mapped risk factors for complex, multigenic diseases [15]. Here, we use new methods for analyzing breed populations to identify genomic loci associated with OS in the first multibreed association study of a highly polygenic canine disease. We explain the majority of the OS phenotype variance in three high-risk breeds, identify a common regulatory risk factor, and reveal novel genes and pathways underlying this poorly understood disease.
Results
Population genetics of GWAS dog breeds
We genotyped 334 greyhounds, 166 Rottweilers, and 174 IWH on the 170,000 SNP Illumina canine HD arrays, removing SNPs with call rates <95%, and one dog from each pair with the same phenotype and genetic relatedness >0.25, preferentially retaining younger cases and older controls. The final dataset (169,010 SNPs) included 267 racing greyhounds (153 affected (A) + 114 unaffected (U)), 135 Rottweilers (80 A + 55 U), 141 IWH (76 A + 65 U), and 19 AKC greyhounds. The ratio of females to males was approximately equal in cases and controls in the greyhounds (0.72 and 0.75 for A and U) and Rottweilers (1.11 and 1.04), and more skewed in IWH (1.62 and 1.41).
The three breeds are visibly discrete genetic populations; the AKC greyhounds cluster near but distinct from their racing brethren (Figure 1a). The racing greyhound population is the least inbred, likely reflecting a large effective population size, (inbreeding coefficient θ = 0.11 +/− 0.02), followed by the Rottweilers (θ = 0.23 +/− 0.04), IWHs (θ = 0.25 +/− 0.04), and AKC greyhounds (0.30 +/− 0.07) (Figure 1b) [16]. While linkage disequilibrium in all breeds is long, as compared to human populations, it varies markedly by breed, with average r2 dropping below 0.2 at 196 kb in the greyhounds, 632 kb in the Rottweilers, and 1,533 kb in the IWH, suggesting GWAS regions will be shortest in greyhound, facilitating identification of associated functional elements in this breed (Figure 1c).
Figure 1.
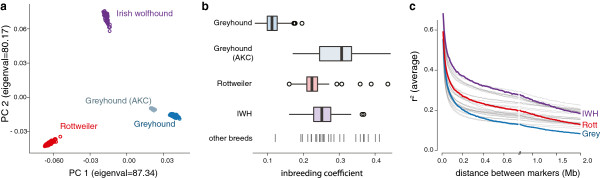
The three GWAS breeds (high risk of OS) and the AKC greyhounds (low risk of OS) are genetically distinct and highly inbred populations. (a) Along the top two principal components of variation, the three breeds (Rottweilers (n = 135, red), Irish wolfhounds (IWH, n = 141, purple), and racing greyhounds (n = 267, dark blue) ) are each tight clusters equidistant apart, with the AKC subpopulation of greyhounds (n = 19, light blue) near but not overlapping the racing dogs. (b) Estimate of the inbreeding coefficient (IC) shows the racing greyhounds as the least inbred, on average, while the other three populations fall within the range seen in an earlier survey of 28 dog breeds (grey bars mark average IC in breed; two outliers at 0.53 and 0.62 not shown) [17]. (c) The extent of linkage disequilibrium, measured as average pair-wise r2, drops below 0.2 at 196 kb, 632 kb, and 1,533 kb in the greyhounds, Rottweilers, and IWH, respectively.
GWAS identifies 33 regions associated with osteosarcoma
We tested for association between OS and germ-line variants with minor allele frequency >0.05 in each of the three breeds independently, controlling for cryptic relatedness and population structure using a mixed model approach with the top principal component as a covariate [18,19]. We identified all SNPs with either significant association (exceeding 95% confidence intervals defined empirically using 1,000 random phenotype permutations) or suggestive association (P<0.0005; Figure 2a-f; Methods) and defined regions of association using linkage disequilibrium (Table 1, Additional file 1: Figure S1). The proportion of very young cases in our IWH dataset allowed us to focus on the more powerful comparison of young cases and older controls (Additional file 1: Figure S2; 28 affected <6 years old and 62 unaffected >6 years old).
Figure 2.
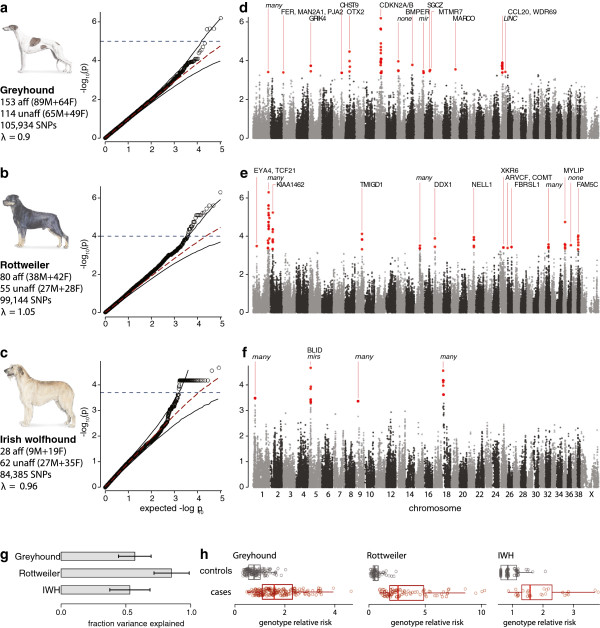
Mixed-model GWAS corrects for population structure and identifies 33 OSA associated loci explaining a large fraction of phenotype variance. In each breed, the QQ plots show no evidence of stratification relative to the expected distribution, identifying nominal significance at -log10p of 3.5 and the 95% empirically determined confidence intervals (dashed grey line) at -log10p of (a) 5 in the greyhounds, (b) 4 in the Rottweilers, and (c) 3.7 in IWH. In the IWH, a plateau of SNPs at P = 6.6 × 10-5 corresponds to a 1.65 Mb haplotype on chromosome 18 peaking at the gene GRB10. (d) In greyhounds, 14 loci have P<0.0005, with one locus, on chromosome 11, exceeding 95% confidence intervals (dashed lines). (e) In Rottweilers, 15 and 6 loci are identified, (f) while only four and two loci are identified in IWHs. (g) For each breed, the phenotype variance explained by the associated loci, broadly defined by SNPs with r2 >0.2 within 5 Mb of the peak SNP, exceeds 50%. In greyhounds, the 14 regions explain 56.9% +/− 12.5%, in Rottweilers, 15 regions explain 85.3 +/− 13.6%, and in IWH, four regions explain 53.1 +/− 15.5%. (h) For each affected dog (red circles) and unaffected dog (black circles), we estimated their relative risk based on the genotypes and odds ratio of the top SNP from each region for the breed (Table 1), showing that even using a small number of SNPs we see clear differences between the cases and the controls.
Table 1.
Osteosarcoma associated loci identified by independent GWAS in three dog breeds with high rates of the disease
| SNP | chr | Position | P | Risk allele | OR | f (A) | f (U) | Region start-end | Size (kb) | Genes |
|---|---|---|---|---|---|---|---|---|---|---|
|
Greyhound
|
|
|
|
|
|
|
|
|
|
|
| BICF2P133066 |
11 |
44405676 |
6.4E-07 |
A |
1.26 |
0.84 |
0.65 |
44392734-44414985 |
22 |
None |
| BICF2P1421479 |
8 |
35448126 |
3.4E-05 |
C |
1.36 |
0.12 |
0.03 |
35433142-35454649 |
22 |
OTX2 (50 kb) |
| BICF2S23118341 |
13 |
14588716 |
1.1E-04 |
T |
1.20 |
0.34 |
0.19 |
14549973-14645634 |
96 |
None |
| BICF2S23325120 |
25 |
21912859 |
1.3E-04 |
A |
1.19 |
0.56 |
0.41 |
21831580-21921256 |
90 |
None |
| BICF2P66597 |
14 |
49193217 |
1.6E-04 |
G |
1.19 |
0.37 |
0.23 |
48831824-49203827 |
372 |
BMPER |
| BICF2P1194727 |
5 |
16085937 |
1.8E-04 |
G |
1.23 |
0.28 |
0.14 |
16071171-16152955 |
82 |
GRIK4 |
| BICF2G63051809 |
19 |
34134931 |
2.8E-04 |
T |
1.21 |
0.80 |
0.67 |
33963105-34145310 |
182 |
EN1, MARCO |
| BICF2G630813090 |
16 |
43669044 |
3.0E-04 |
C |
1.16 |
0.64 |
0.48 |
43665149-43737129 |
72 |
MTMR7 |
| BICF2G630418573 |
15 |
63780452 |
3.4E-04 |
A |
1.26 |
0.91 |
0.81 |
63767963-63800415 |
32 |
None |
| TIGRP2P215623 |
16 |
40896559 |
3.5E-04 |
C |
1.36 |
0.97 |
0.89 |
40883517-41081510 |
198 |
SGCZ |
| TIGRP2P331221 |
25 |
43485109 |
3.8E-04 |
G |
1.23 |
0.22 |
0.11 |
43476429-43528145 |
52 |
CCL20 |
| BICF2S23516022 |
1 |
112990983 |
3.8E-04 |
C |
1.21 |
0.82 |
0.69 |
112977233-113081800 |
105 |
CD3EAP, ERCC1, ERCC2, FOSB, PPP1R13L |
| TIGRP2P45171 |
3 |
5564882 |
4.0E-04 |
T |
1.20 |
0.79 |
0.68 |
5162058-6465753 |
1,304 |
FER, MAN2A1, PJA2 |
| BICF2P1090686 |
7 |
64672328 |
4.2E-04 |
C |
1.16 |
0.57 |
0.43 |
64631053-64703475 |
72 |
CHST9 |
|
Rottweiler
|
|
|
|
|
|
|
|
|
|
|
| BICF2P1115364 |
1 |
116524913 |
5.0E-07 |
G |
1.32 |
0.71 |
0.39 |
115582915-116790630 |
1,208 |
Many (35 genes) |
| BICF2P411325 |
2 |
19515571 |
5.8E-06 |
C |
1.43 |
0.91 |
0.73 |
19212450-19542015 |
330 |
KIAA1462 |
| BICF2P1210630 |
1 |
122048812 |
1.1E-05 |
C |
1.30 |
0.73 |
0.46 |
122033806-122051988 |
18 |
C19orf40, CEP89, RHPN2 |
| TIGRP2P407733 |
35 |
18338700 |
1.8E-05 |
A |
1.28 |
0.51 |
0.29 |
18326079-18345318 |
19 |
None |
| BICF2P341331 |
9 |
47659782 |
7.6E-05 |
A |
1.28 |
0.53 |
0.28 |
47647012-47668054 |
21 |
BLMH, TMIGD1 |
| BICF2P1129874 |
38 |
11714169 |
9.4E-05 |
T |
1.24 |
0.49 |
0.26 |
11252518-11739329 |
487 |
FAM5C |
| TIGRP2P286750 |
21 |
46283811 |
1.1E-04 |
C |
1.37 |
0.84 |
0.70 |
46231985-46363479 |
131 |
NELL1 |
| BICF2S23533459 |
17 |
14472761 |
1.3E-04 |
C |
1.39 |
0.18 |
0.01 |
14465884-14482152 |
16 |
None |
| BICF2G630590368 |
32 |
25147661 |
2.7E-04 |
A |
1.35 |
0.95 |
0.81 |
25136302-25156153 |
20 |
EMCN |
| BICF2P92014 |
36 |
29651125 |
3.0E-04 |
A |
1.22 |
0.66 |
0.42 |
29637804-29663408 |
26 |
None |
| TIGRP2P200071 |
15 |
38987072 |
3.1E-04 |
T |
1.27 |
0.82 |
0.59 |
37986345-39974762 |
1,988 |
Many (15 genes) |
| BICF2P1164085 |
1 |
29775073 |
3.3E-04 |
G |
1.24 |
0.46 |
0.23 |
29405587-29914411 |
509 |
EYA4, TCF21 |
| BICF2S23712115 |
26 |
32385934 |
3.7E-04 |
G |
1.35 |
0.91 |
0.76 |
32374093-32428448 |
54 |
ARVCF, C22orf25, COMT |
| BICF2G63095567 |
25 |
29671618 |
3.9E-04 |
G |
1.30 |
0.23 |
0.05 |
29658978-29767164 |
108 |
XKR6 |
| BICF2P841536 |
26 |
3537143 |
4.2E-04 |
A |
1.26 |
0.48 |
0.30 |
3529343-3550075 |
21 |
FBRSL1 |
|
Irish Wolfhound
|
|
|
|
|
|
|
|
|
|
|
| BICF2S23746532 |
5 |
15264066 |
2.1E-05 |
A |
1.40 |
0.45 |
0.16 |
14720254-15466603 |
746 |
BLID |
| BICF2P1466354 |
18 |
4937944 |
2.7E-05 |
C |
1.36 |
0.56 |
0.21 |
4266743-5854451 |
1,588 |
C7orf72, COBL, DDC, FIGNL1, GRB10, IKZF1, VWC2, ZPBP |
| BICF2P1225386 |
1 |
17742179 |
3.2E-04 |
C |
1.31 |
0.46 |
0.19 |
16768869-18150476 |
1,382 |
BCL2, KIAA1468, PHLPP1, PIGN, RNF152, TNFRSF11A, ZCCHC2 |
| BICF2P1125643 | 9 | 19623231 | 4.3E-04 | C | 1.75 | 0.14 | 0.02 | 18896060-19633155 | 737 | ABCA5, KCNJ16, KCNJ2, MAP2K6 |
OR is the conditional odds ratio estimated from beta values calculated by EMMAX.
Within each breed, associated regions (P<0.0005) explain the majority of the phenotype variance: 57% in the greyhound (14 loci), 53% in the IWH (4 loci), and 85% in the Rottweilers (15 loci) (Figure 2g, Additional file 1: Figure S3). The overall genotype relative risk estimated for each dog, based on the risk contributed by each locus, shows a clear separation between cases and controls in each breed (Figure 2h), even at more stringent significance thresholds (Additional file 1: Figure S4). None of the top scoring SNPs was strongly correlated with sex in a genome-wide analysis (Additional file 1: Table S1), and including sex as a covariate revealed no new regions of strong association. Although earlier work on monogenic traits in dogs suggested risk factors often are shared between breeds [20], we see no overlap in regions of association between the breeds and a meta-analysis of the three breeds yielded no significant associations (Additional file 1: Figure S5). Three possible explanations for this observation are: (1) cancer risk factors are abundant in the overall dog population and each breed inherited different subsets; (2) we cannot detect the shared associations in the other breeds at our current sample sizes; and (3) the risk haplotypes are fixed or nearly fixed in the other breeds, and thus undetectable by association.
We tested the top 33 GWAS regions for fixation of the risk allele (frequency >0.95) in the other two breeds. In eight regions, the risk haplotype is fixed in one of the other two breeds, but in only one is it fixed in both: the risk allele tagging the top greyhound locus (A=0.84,U=0.65), on chromosome 11, is fixed in both Rottweilers (0.97) and IWH (0.95) (Table 1). This haplotype occurs at lower frequency in the AKC greyhounds (0.61) and in a panel of 28 other breeds from a previously published dataset (0.51% +/− 0.24) [17]. We sequenced the locus (chr11:43.0-48.9 Mb ) in eight greyhound cases and seven controls, and densely genotyped and imputed 180 greyhound cases and 115 controls (Figure 3a). This narrowed the region of association to a 15 kb non-coding region (chr11:44,390,633-44,406,002) near CDKN2A (encodes p16INK4a and p14ARF), CDKN2B (p15INK4b), and the antisense non-coding gene CDKN2B-AS1/ANRIL (Figure 3a). Both CDKN2A and CDKN2B function as cell cycle regulators and tumor suppressors.
Figure 3.
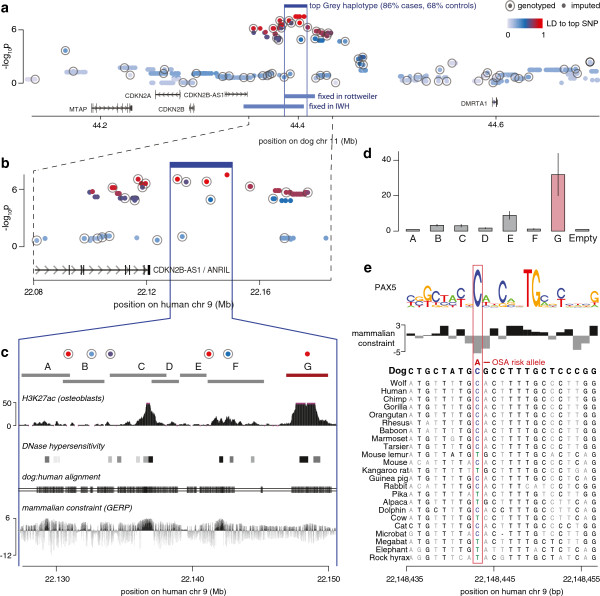
Identification of the top associated variant and functional analysis on chromosome 11. (a) We targeted 2.5 Mb around the greyhound GWAS peak on chromosome 11 for dense sequencing (15 dogs) and finemapping (180 cases and 115 controls). Imputation and association testing of sequenced variants narrowed the peak of association in greyhounds dramatically to a 15 kb risk haplotype (chr11:44390633–44406002), telomeric of the genes CDKN2A and CDKN2B, that is nearly fixed in both the Rottweilers (98% in cases and 96% in controls) and IWH (95% in cases and 92% in controls). (b) The top haplotype (blue vertical lines) maps to a locus downstream of the non-coding gene ANRIL on human chromosome 9 (hg19). (c) We tiled the human chromosome 9 region with luciferase assays and assayed the function in osteosarcoma cell lines compared to renilla. Potential markers of function in the region include H3K27 acetylation in osteoblasts and DNase hypersensitivity clusters (assayed from 125 cell types), most notably in regions that align between the dog and human genomes in a Multiz alignment of 46 species and are constrained across mammals as measured by Genomic Evolutionary Rate Profiling (GERP) [21-24]. (d) Of the seven non-control luciferase assays, four (B, C, E, and G) showed a significant increase compared to empty vector. Construct G showed by far the strongest increase with an approximate 32-fold increased activity suggesting a strong enhancer. (e) This fragment contains one of the top SNPs (Canfam2.0 chr11:44405676) which has a constrained reference allele C corresponding to a predicted transcription factor binding site, while the OS associated allele, A, is not found among 29 mammals or the wolf.
The risk haplotype at 11q16 is syntenic to a non-coding regulatory region on human chromosome 9p21 (Figure 3b), and the alignment between dog and human includes DNase hypersensitivity sites and peaks of H3K27 acetylation characteristic of active enhancers (Figure 3c) [25,26]. The human 9p21 locus is deleted in 5% to 21% of human OS [27], the absence of p16INK4a expression is correlated with decreased survival in pediatric OS patients [28], and increased p16 expression is predictive of better response to chemotherapy [29]. Thus, we hypothesized that the variant(s) carried on the risk haplotype disrupts enhancer elements upstream of the CDKN2A/B locus, thereby altering expression of one or more genes in the region.
We assayed the risk haplotype for regions of enhancer activity using renilla/firefly luciferase assays in the human OS U2OS cell line, tiling the region with seven fragments that were PCR-amplified from human genomic DNA (Figure 3c). Several fragments showed enhancer activity, and one increased luciferase expression >30-fold (Figure 3d). The only greyhound variant identified in this fragment region, a SNP, is also the top associated variant in the greyhound finemapping/imputation dataset (dog chr11:44405676; human chr9:22,148,443, P = 3 × 10-8) (Figure 3a,b) (Additional file 1: Table S2). Additional genotyping validated the imputation (186 dogs genotyped; 98.9% concordance). While this position is strongly constrained to a C or T across mammals, including wolves [30], the OS risk allele, A, is found in 87% of greyhound cases and 68% of greyhounds controls (Figure 3e). We analyzed the locus with two different transcription factor binding motif analysis tools, FIMO [31] and TOMTOM [32]. Just one motif, for PAX5, was significantly detected by both tools and was specific to the non-risk allele (C), suggesting that the risk allele (A) will disrupt binding (Figure 3f, Additional file 1: Figure S6). PAX5 regulates both B-cell and osteoblast differentiation and bone formation [33,34].
We tested for association of chr11:44405676 to osteosarcoma in eight more breeds with high rates of OS. We found the greyhound risk allele at very high frequency in the two other GWAS breeds, Rottweilers (92A + 77U, FA = 0.98, FU = 0.97) and IWH (27A + 31U, FA = 0.93, FU = 0.92), with the risk allele slightly more common in cases, and correlated with OS in Leonbergers (30A + 25U, FA = 0.77, FU = 0.62, P = 0.09) and great Pyrenees (16A + 21U, FA = 0.78, FU = 0.62, P = 0.14). Analyzed together, these four breeds replicate the association seen in the greyhounds (pCMH = 0.03, pCMH = 2 × 10-8 with greyhounds included) and no odds-ratio heterogeneity was seen between breeds (pBreslow-Day = 0.51) (Additional file 1: Table S3) [35]. We found no association with OS risk in mastiffs, Labrador retrievers, great Danes, and golden retrievers. Although small samples limit statistical power, these results suggest the effect of the risk variant may depend on the genetic background in each breed. Pooled sequencing data from an earlier project [30] show that the OS risk allele (A) is never seen in wolves but is common in purebred dogs (approximately 50%). The syntenic human locus at 9p21 has one of the densest known concentrations of regulatory elements in the human genome [36]. Breed-specific variation in nearby functional elements may modulate the effect of the chr11:44405676 variant.
Pathway analysis of osteosarcoma associated regions
While the breeds do not segregate for the same risk variants, their shared predisposition to OS suggests the risk alleles may disrupt the same cellular pathways. All but four of the 33 OS associated regions contain known genes. The majority have just one gene (60%), but a handful have considerably more, including the top Rottweiler region (35 genes) and three out of four IWH regions (4, 7, and 8 genes) (Table 1). We analyzed the GWAS regions using GRAIL, a statistically robust, web-based method that uses published scientific abstracts to find gene relationships connecting distinct genomic regions [37]. We identified significant connectivity between OS associated regions both within and between breeds through key terms including ‘bone’ (13 loci), ‘differentiation’ (13 loci), and ‘development’ (9 loci) (Figure 4a, Additional file 1: Table S4). For example, OTX2, an oncogenic orthodenticle homeobox protein that directly activates cell cycle genes and inhibits differentiation in medulloblastomas [38], is strongly associated with OS in the greyhounds. GRAIL connects OTX2 with genes in six other risk loci (P<0.05): two negative regulators of osteoblast differentiation BMPER and VWC2[39]; EN1, a modulator of osteoblast differentiation and proliferation [40]; DLL3, a notch ligand implicated in human skeletal growth disorders [41]; TCF21, a tumor suppressor that regulates mesenchymal-epithelial cell transitions; and EMCN, a mucin-like anti-adhesion membrane protein and hematopoietic stem cell marker [42]. In a second network, GRAIL connects three genes that regulate bone formation - the osteoblast differentiation enhancer FAM5C[43]; NELL1, a regulator of osteoblast differentiation and ossification [44]; and TNFRSF11A, an essential mediator of osteoclast development [45] - and the pro-apoptotic gene BLID, frequently deleted in human breast, lung, ovarian, and cervical cancers [46].
Figure 4.
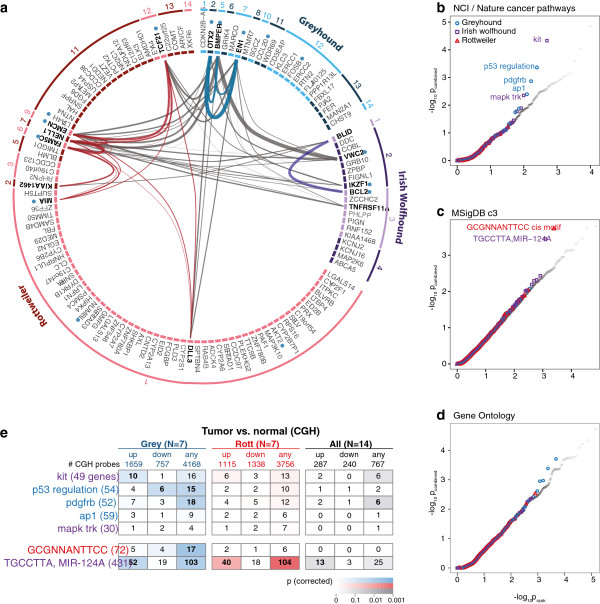
Connectivity and enrichment analysis identifies pathways linked to OS in multiple breeds. (a) Most of the associated regions in each breed contain one or more genes (black text). Genic regions are shown for the greyhounds (blue), IWH (purple), and Rottweilers (red), with hue alternating between light and dark to distinguish regions, numbered as in Table 1. GRAIL [37] analysis identified non-random connectivity (P<0.05) between associated genes (bold text), both within breeds (blue, purple, and red arched lines for greyhounds, IWH, and Rottweilers) and between breeds (grey arched lines). Twelve regions contain genes (blue dots) connected to the key word ‘bone’, one of the top terms identified by GRAIL (Table S4). (b, c, d) When gene set enrichment P values for the associated regions and regions of reduced variability are combined, most sets shows no inflation compared to background (grey circles; RRV P values from 28 other breeds). However a small number are inflated, including (b) five from the NCI pathway interaction database [47], (c) two from the Molecular Signatures database of shared cis-regulatory motif based sets [48], and (d) 0 from the Gene Ontology database. (e) Of the seven gene sets (colors match discovery breed), five are significantly enriched (bold numbers) in regions that are aberrant in all greyhound (blue), all Rottweiler (red), or all dogs (black) for which we compared normal and tumor DNA using comparative genomic hybridization. Boxed numbers show number of genes in gene set overlapping CGH regions; ‘up’ and ‘down’ indicates gain or loss in all samples; ‘any’ indicates all samples are either amplified or deleted.
Fixed and selected loci in breeds contribute to disease risk
It is likely that alleles that are fixed or at high frequency in breeds, and thus undetectable by GWAS, contribute to OS risk, as seen at the CDKNA2/B locus. In each breed a substantial portion of the genome is comprised of fixed regions (minor allele frequency <0.05) longer than 250 kb: 2.8% of the autosomal genome in the greyhounds, 2.9% in the Rottweilers, and 7.6% in IWH (Additional file 1: Figure S7a; 25.9%, 30.5%, and 31.5% for chromosome X). These fixed regions overlap genes involved in bone development and OS, including RB1 (IWH), FOS (Rottweilers), RUNX2 (Rottweilers), CCNB1 (IWH), COL11A2 (greyhounds), and POSTN (IWH and greyhounds) [49]. We tested genomic regions fixed in all three breeds for gene set enrichment using INRICH, empirically measuring significance through 100,000 permutations matched for region size, SNP density, and gene number [50]. Among eight sets of microRNAs implicated in OS pathobiology [51-55], we found significant enrichment for one associated with pathogenesis and progression of OS (5/27 genes, P = 0.017, Pcorrected = 0.041, Additional file 1: Table S6) [51].
We next analyzed the fraction of the genome that shows exceptionally low variation in OS breeds compared to 28 other breeds, including regions of incomplete fixation. We defined these regions of reduced relative variability (RRVs) by comparing each OS breed to up to 28 other dog breeds and focusing on the 1% least variable 150 kb regions [17]. RRVs totaled 2.9% (277 regions), 2.9% (344 regions), and 3.1% (387 regions) of the genome in the greyhounds, Rottweilers, and IWH, respectively (Additional file 1: Figure S7b). We tested the RRVs for gene set enrichment using INRICH and combined these P values with those from a matched analysis of the GWAS regions. We hypothesized that, if genes in RRVs contribute to OS risk, we should see the same gene pathways enriched in the two analyses. While, as expected, the vast majority of gene sets in the studied breeds showed no increase in significance compared to the background distribution, seven gene sets are markedly inflated. This includes three pathways (KIT, p53, and PDGFRB) in a list curated by the National Cancer Institute and Nature Publishing Group (Figure 4b) and, from the Molecular Signatures Database, two gene sets defined by cis-regulatory motifs - targets of MIR - 124A (TGCCTTA) and a highly conserved motif with no known transcription factor match (Figure 4c) [47]. We found only weak P value inflation in the Gene Ontology analysis (Figure 4d).
GWAS pathways enriched for somatic mutations in OS tumors
Our analysis of the OS breeds demonstrates that inherited variants are major factors for determining whether a dog develops OS. As somatic changes in the tumor also contribute to progression of the disease, we hypothesize that genes affected by these changes will be enriched in the same pathways as the inherited variants. We investigated the frequency and distribution of somatic DNA copy number aberrations (CNAs) in 22 OS tumor samples (12 greyhounds, 10 Rottweilers) using 26 kb-resolution genome-wide array-based comparative genomic hybridization analysis (array-CGH).
While the CGH profiles exhibit the extensive karyotypic instability characteristic of OS [56], they are remarkably conserved between the two breeds, with no significant regional differences in DNA copy number status (defined as corrected P<0.05; Additional file 1: Figure S8a,b,c). Moreover, the genome-wide CGH profiles of dog and human OS are broadly consistent, both in the frequency and relative distribution of CNAs (Additional file 1: Figure S8d), including genes associated with OS pathogenesis [56], such as MYC gain (60% in dog/67% in human), RB1 loss (36%/33%), RUNX2 gain (45%/67%), and CDKN2A/B loss (73%/67%).
Using the GISTIC algorithm [57], we identified discrete regions that had statistically high CNA frequency in canine tumors relative to the globally chaotic genomic background of OS, suggestive of specific gene targets strongly associated with disease pathogenesis. The most significant was a 2.5 Mb region at 11q16 (chr11:43,615,205-46,137,412) encompassing the CDKN2A and CDKN2B genes, with the strongest signal (Pcorrected = 1.5 × 10-12) at chr11:44,304,860-44,308,340 between CDKN2B and CDKN2A-AS1, approximately 100 kb from the top greyhound GWAS SNP. This region was deleted in 73% of tumors (9/12 greyhounds, 7/10 Rottweilers), of which 59% were consistent with homozygous deletion (7/12 greyhounds, 6/10 Rottweilers). No other regions identified by GISTIC overlapped GWAS loci, reinforcing the fundamental role of the CDKN2A/B region in disease pathogenesis.
We tested the subset of the CGH samples (7 greyhounds and 7 Rottweilers) also included in the GWAS analysis and found that five of the top 29 GWAS SNPs are moderately associated with tumor gain or loss, most significantly at the gene BLMH, a candidate tumor suppressor gene for hepatocellular carcinoma [58] (Additional file 1: Table S5). Furthermore, certain probes were enriched for genomic imbalance; the fraction of probes gained or lost in all Rottweilers (n = 8,087, 4.95%), all greyhounds (n = 8,781, 5.35%), or all 14 dogs (n = 1,603, 0.98%) was much higher than expected by random chance (Pbinomial = 2.71%, 1.3%, and 0.04%, respectively). We found putative human OS driver genes deleted in human tumors [59] are among those lost in all greyhounds (ARHGAP22, ARID5B, RCBTB1; INRICH enrichment P = 0.0004), all Rottweilers (LHFP; P = 0.13), and all dogs (AIFM2, TSC22D1; P = 0.024).
Of the seven gene sets enriched in the GWAS + RRV analysis, five are also enriched in the CGH regions in one or both breeds (Figure 4e). In particular, genes with a MIR-124A cis-regulatory motif, identified in IWH, showed significant enrichment in both Rottweiler and greyhound tumors. MIR-124A has diverse regulatory functions: it is upregulated during chondrogenesis [60], is a potential silencer of CDK6 when downregulated in leukemia [61], and regulates NFκB [62]. Except for MIR-124A, gene sets tend to have stronger enrichment in the greyhounds than in the Rottweilers. While this could suggest breed-specific OS pathways, it may also reflect greater tumor heterogeneity in the Rottweilers diluting the enrichment results.
Discussion
Through a parallel multibreed canine association study, we found 33 genomic regions associated with OS, and identified genes and pathways potentially causing this complex, polygenic, and poorly understood disease. Altogether, the 33 loci identified by GWAS account for 50% to 80% of the disease risk within each of these three breeds, demonstrating that inherited factors are the predominant cause. In addition, regions of unusually low variability, reflective of the small effective population sizes and strong artificial selection used to create dog breeds, are also likely to contribute to an overall increased risk for these breeds.
None of the OS GWAS loci overlapped between breeds, a strikingly different genetic architecture from the shared variants previously found by mapping Mendelian traits in multiple dog breeds. Potentially this could reflect the difference between: (1) a monogenic trait, where a single variant causes a trait, which, if desirable, is then deliberately bred for; and (2) a complex trait, caused by a random assortment of many low frequency risk factors that rise in frequency through population bottlenecks and selection. This latter scenario is more similar to the genetics in human populations, where many rare risk factors may underlie OS, and may increase in frequency by random drift or moderate natural selection. In dog breeds, tighter bottlenecks and stronger selective forces push risk allele frequencies up, making them easier to detect. Thus, dogs bring added value to the study of OS.
The relatively large number of OS associated genes identified in this study facilitated pathway analysis with two different methods. First, the GRAIL software, given only the human genomic regions syntenic to the dog GWAS loci as input (and no information on phenotype) mined the abstracts of all previously published literature and found significant connections between associated genes related to growth, osteoblast differentiation and proliferation, and tumor suppression. Furthermore, a combined gene set enrichment analysis of the associated, fixed, and somatically altered loci identified common pathways affected by inherited and somatically acquired variation, suggesting they may interact to cause tumor initiation and progression. A fraction of the genes and pathways identified have previously been reported to be involved in human OS as either inherited or somatic changes, demonstrating the relevance of the canine model. Potentially more interesting, we also identified novel pathways, including two related to poorly characterized cis-regulatory motifs.
The GWAS loci implicated, several of which contain no known genes, show that canine OS has a complex genetic architecture with key advantages over artificially induced mouse models of the human disease. This is well illustrated by our functional analysis of the top greyhound locus. The strong effect of the variant and low genetic diversity of the breed populations allowed us to move rapidly from GWAS region to candidate functional variants with limited additional sequencing and genotyping. The top candidate causal variant is in a non-coding regulatory element upstream of the CDKNA2A/B locus, near, but not overlapping, a region associated with canine histiocytic sarcoma [63]. Pinpointing functional regulatory variants is harder than finding functional coding variation, but such regulatory variation is likely more representative of the genomic variants underlying common human diseases [64].
The CDKNA2A/B locus in dogs is syntenic to the human 9p21 locus, one of the most complex regulatory loci in the human genome [36]. SNPs in this region are significantly and independently associated with diseases including coronary artery disease, myocardial infarction, type 2 diabetes, melanoma, basal cell carcinoma, glioma, acute lymphoblastic leukemia, and breast cancer [64]. Deciphering the cellular mechanisms disrupted by OS associated regulatory variation in dogs may elucidate mechanisms underlying diverse human diseases.
We hypothesize that the top canine OS risk variant at chr11:44405676 alters regulation of CDKN2A/ARF. The OS associated variant at dog chr11:44405676 disrupts a highly constrained position in a genomic locus that we show, using a luciferase assay, has strong enhancer activity in a human osteosarcoma cell line. CDKN2A/ARF encodes the INK4 family of cyclin-dependent kinase inhibitor proteins (including p16INK4a, p15INK4b, and p14ARF). These proteins control G1-progression by inactivation of D-cyclins, inducing senescence via the RB and p53 pathways [65-67]. Altered levels of CDKN2A, a master regulator of tissue development, are linked to hematopoietic stem cell senescence and development, key feature of malignancies including OS [68,69]. SNPs that disrupt enhancer element binding can change transcriptional activity across the human 9p21 locus, including at CDKN2A[36]. Germline variants affecting regulation of CDKN2A may alter the balance between proliferation and senescence in specific tissues, thereby leading to an increased risk of developing OS and potentially also other cancers in adolescence and adulthood.
Canine and human OS are remarkably similar diseases, both clinically [8] and in their tumor gene expression profiles [6,7]. The recent publication of the first GWAS of osteosarcoma in humans offers a new opportunity to identify common risk factors shared between the two species and thus of particular etiological interest. The human OS GWAS compared 941 patients with osteosarcoma to 3,291 unaffected adults across 699,000 SNPs and found two genome-wide significant loci, one at the glutamate receptor gene GRM4 and the other in a gene desert [5]. The much larger dataset required for GWAS in human patients illustrates the power offered by mapping in genetically isolated dog breed populations. While we see no association in the dog GWAS at the two loci found in the human GWAS, another glutamate receptor gene (GRIK4) is associated with OS in greyhounds. Although glutamate signaling in primary bone cancers is not well understood, both GRM4 and GRIK4 are expressed in normal bone, and glutamate signaling has been shown to regulate bone formation and resorption [70]. Furthermore, inhibition of glutamate receptors limits cell growth in many cell lines.
The association of glutamate receptors with OS in both dogs and humans suggests glutamate signaling as a potential therapeutic target, although the diverse physiological functions of this pathway could make this difficult. Glutamatergic signaling is critical for learning and memory [71] and is implicated in a range of neuropsychiatric diseases in both humans [72-74] and dogs [75]. Fixation of variants in glutamate related genes, as seen near GRM4 in Rottweilers (Additional file 1: Figure S9), may suggest that selection on behavioral, as well morphological, traits helped drive OS to the exceptionally high rates seen in the breeds studied here.
Conclusions
Detailed characterization of the genetic risk factors involved in initiation and progression of canine OS will give us a comprehensive understanding of the genes involved and the different genetic interactions sufficient to cause the disease. We hypothesize that, by integrating genetic etiology into canine clinical trials, we can identify molecular subtypes of OS and correlate them with treatment efficacy and survival outcomes. In the future, we anticipate that the insights gained from the canine model will help us develop more personalized and effective treatments for human OS.
Methods
Sample collection and genotyping
DNA samples used were collected from pet dogs with the owner’s consent. Osteosarcoma was diagnosed by a qualified veterinarian using X-ray and/or tumor histopathology. Histological subtype was not included in GWAS analysis because of the difficulty of ascertaining it consistently across diverse collection sites. Osteosarcoma can be reliably diagnosed without histopathology in breeds with exceptionally high rates of the disease. Dogs classified as unaffected had no history of cancer. Phenotype included age at disease onset for most (79%) of affected dogs. For almost all (98%) of unaffected dogs, phenotype included age last confirmed OS free. Samples were collected with the appropriate consent and animal care protocols (U Minn 0802A27363, 1101A94713, and MIT 0910-074-13). Genomic DNA was isolated from whole blood (QIAamp DNA Midi kit) and was genotyped for approximately 170,000 SNPs using the Illumina 170 K canine HD array [17].
Data analysis
SNPs and individuals with genotyping rate <0.95 were removed with PLINK [35]. Genetic relatedness and principal component (PC) analysis was done with GCTA [76]. To minimize population structure in the GWA analysis within each breed, we removed one dog from each concordant phenotype pair with genetic relatedness >0.25, preferentially retaining the younger case and older control. We measured phenotype-genotype association with the mixed model method implemented in EMMAX [77], which fits a standard linear regression model and tests the significance of the slope coefficient by the standard t test [77]. We included the top PC as a covariate to control for remaining cryptic relatedness [18].
We defined genome-wide significance in the GWAS using empirical 95% confidence intervals (CIs) rather than using a Bonferroni correction. As previously noted, a Bonferroni correction for the number of SNPs tested is excessively stringent in dog breeds, where extensive LD means that each SNP is not an independent test [14]. Of the approximately 100,000 SNPs used in the GWAS, the majority (98%) are in LD (r2 >0.5) with at least one other SNP within 1 Mb, and most (60%) are in LD with 10 or more SNPs. While an alternative is to correct for the number of independent haplotype blocks (if haplotype blocks are 1 Mb [14], the 2.4 Gb genome is comprised of roughly 2,400 independent haplotype blocks, corresponding to a corrected genome-wide significance threshold of 2 × 10-5), accurately determining the number of independent genomic regions in a particular breed population is difficult. We defined genome-wide significance using 95% CIs calculated from the empirical distribution of P values observed in the absence of real association. We determined this distribution by rerunning the GWAS with randomly permuted phenotypes 1,000 times. Conceptually, the empirical CIs bound the area in which 95% of the permutation QQ plots fall. We note that in dog breeds, the empirical CIs set more conservative thresholds than the CIs for a uniform score distribution used in, for example, human GWAS studies [78] (Additional file 1: Figure S10). We defined genome-wide significance as associations exceeding the 97.5% upper empirical CI, a threshold that varies by breed (1 × 10-5 in Greyhounds, 1 × 10-4 in Rottweilers, and 2 × 10-4 in IWH; Figure 2).
We defined genome-wide significant associations as those exceeding the 97.5% upper CI. To facilitate gene set and pathway analysis, we also identified a larger set of candidate OS regions with P<0.0005. The laxer threshold encompasses associations exceeding the theoretical CIs but not necessarily the stricter empirical CIs. We note that the inflated P values suggest that these regions are enriched for true OS associations. Although some of the regions included may not be true associations, this would most likely weaken rather than strengthen the gene set and pathway analyses, leading to false negatives rather than false positives.
We used linkage disequilibrium clumping in PLINK to define regions of association in two stages, first defining wide regions of SNPs in weak LD (r2 >0.2 within 5 Mb of top SNP) and then narrowing the association to a single peak of SNPs in strong LD (r2 >0.8 within 1 Mb of top SNP). While potentially masking a small number of true disease associated variants, this conservative approach ensured we identified only the strongest peaks and not regions with association reflecting proximity to a stronger peak. For each breed, we used GCTA to estimate the phenotype variance explained by the associated loci, which we defined broadly as SNPs with r2 >0.2 within 5 Mb of the peak SNP, totally 36.4 Mb for greyhound, 58.5 Mb for Rottweilers, and 18 Mb for IWH.
Sequencing of top locus
We resequenced 5.9 Mb (chr11:43,000,000-48,900,000) in eight greyhound cases and seven controls using a Roche Nimblegen sequence capture array comprised of 385,000 probes covering approximately 95% of the target region. After library preparation (Illumina Paired end kit) DNA was hybridized to the array following the manufacturer’s protocol and the enriched samples sequenced using paired-end sequencing (2 × 74 bp) on the Illumina Genome analyzer II. Sequence reads were aligned to the CanFam2 genome reference using BWA [79]. Picard [80] was used to identify and mark duplicate reads (PCR artifacts) and the Genome analysis toolkit [81,82] to recalibrate quality scores, local realignment around indels, and call SNPs and indels. Variants were filtered based on sequence depth, quality of alignments, SNP clusters, and strand bias. In total, we identified 16,475 high quality variants; the Ts/Tv ratio was 2.06. The variants were annotated for cross-species conservation using SEQscoring [83,84], annotated and analyzed for predicted effect by using snpEff [85], and were visually examined by IGV [86].
We genotyped variants discovered in the sequence data using the Sequenom iPLEX Massarray system, tiling the center of the associated region (chr11:44.35 - 44.47 Mb) most densely. We genotyped 124 SNPs in greyhounds (172 A + 110 U) and Rottweilers (64 A + 32 U) and 60 SNPs in IWH (22 A + 30 U). SNPs were prioritized for genotyping if they were located in a protein coding sequence as defined by SnpEff [84] or in a conserved element compared to 29 other mammals [87]. We used Beagle 3.3.2 [88] to imputed genotype for all sequenced variants in LD with a genotyped variant (r2 >0.75) in the greyhounds and tested for association using PLINK [35]. We analyzed 41 bases around the top candidate SNP in greyhound for transcription factor binding motifs using FIMO [31] (JASPAR, UniPROBE, ENCODE-motifs databases, P value <1e-4) and TOMTOM [32] (JASPAR and UniPROBE databases, E-value <10) testing both the OS risk and non-risk alleles.
Enhancer assay
For the enhancer assay, potential enhancer regions were PCR amplified from human gDNA and placed in front of minimal promoter driven luciferase reporter gene (pGL4.26, Promega). HTB-96 U-2 OS osteosarcoma cells were obtained from American Type Culture Collection (ATCC). These cells have been used within 6 months of purchase (February 2013). All cells were authenticated by standard ATCC procedures [89]. U-2 OS cells were seeded in 96 well plates (25,000 cells/well) and grown for 20 to 26 h before transfection. Each well was transfected with 0.1 μg reporter construct and 0.01 μg renilla luciferase driven by CMV promoter to control for cell density, using 0.4 μL/well FuGENE (Promega) according to the manufacturer’s instructions. Twenty-four hours after transfection, luciferase activity was measured sequentially using the Dual-Glo Luciferase System (Promega) using a Synergy H4 hybrid reader (BioTek). At least three independent experiments were performed, each with eight technical replicates of every construct.
Pathway analysis
GRAIL analysis, using the PubMed Text (Aug2012) data source, was run on the OS regions (Table 1) lifted over to human genome hg18 coordinates (genome.ucsc.edu/cgi-bin/hgLiftOver) with 50 kb flanks added to start and end and gene size correction turned on. Gene set enrichment testing was done with INRICH, empirically measuring significance through 100,000 permutations matched for region size, SNP density, and gene number [50]. INRICH reports the significance for each gene set (P), and the experiment-wide significance correcting for the number of gene sets tested (Pcorr); thus, the correction varies depending on the number of sets included in the analysis. We considered Pcorr <0.05 to be significant. We tested gene sets between 10 and 2,000 genes from five catalogs: three MSigDB collections (c2, c3, and c4 with 2593, 819, and 836 gene sets, respectively from [90]); the Gene Ontology ([91]; 1,582 sets); and the NCI/Nature pathway interaction database ([92]; 166 sets). We also made a custom catalog of putative osteosarcoma driver genes based on a recent publication [59]. We ran INRICH on canFam2 using a map file of 17,665 genes lifted over from the hg19 RefSeqGene catalog (UCSC Genome Brower) [93]. We made a custom catalog of 10 microRNA sets associated with OS in recent papers [51-55] and ran INRICH with a canFam2 map file of 532 human microRNAs mapped to dog with liftOver (Additional file 1: Table S6).
For each gene set we calculated the combined enrichment P value from the INRICH empirical P values for the GWAS and RRV regions using Fisher’s combined probability test. We first calculated the scores for each GWAS breed (that is, greyhound GWAS enrichment P combined with greyhound RRV enrichment P). We then determined the expected background distribution by doing the same using RRV enrichment test for each of a panel of 28 reference breeds, and then combining the GWAS enrichment P from each of the three GWAS breeds with the RRV enrichment P from each of the non-GWAS breeds [17].
CGH
CGH analysis of primary canine OS was performed as described previously [94] using an approximate 180,000 feature Agilent canine oligonucleotide CGH array with approximately 26 kb resolution within the canFam 2.0 genome sequence assembly. An equimolar pool of constitutional DNA from 50 female dogs of mixed breed was used as the common reference for all OS cases. Arrays were scanned at 3 μm resolution using an Agilent G2565CA scanner and image data were processed using Feature Extraction version 10.10 and Genomic Workbench version 7.0 (Agilent Technologies) to exclude probes exhibiting non-uniform hybridization or signal saturation. Recurrent CNAs within each tumor were defined using the FASST2 segmentation algorithm in Nexus Copy Number version 6.1 (Biodiscovery Inc.) based on a minimum of three consecutive probes with log2 tumor:reference values ≥0.2 (copy number gain) or ≤ −0.2 (copy number loss). We compared genotypes at the top GWAS SNPs in Greyhound and Rottweiler (Table 1) to the CGH data from tumors for seven case greyhounds and seven case Rottweilers by first defining, for each probe, each dog’s phenotype (gain = log2 tumor:reference values ≥0.2 and loss = log2 tumor:reference values ≤0.2) and then associating phenotype with genotype using the Cochran-Mantel-Haenszel test in PLINK to control for breed clusters.
Data availability
The data presented in this publication are available through the Gene Expression Omnibus (accession number GSE52147) and ArrayExpress (accession numbers E-MTAB-1984 and E-MTAB-1986). Datasets analyzed in the paper are also available at [95].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KLT, KC, and EKK conceived the study and together with JFM, MB, and MPS designed the experiment. KC, SS, GC, WK, MB, RT, JFM, MPS, NT, RS, SF, TB, NA MP, MS, CCC, LY, CAL, and SM coordinated, collected, prepared samples, made/confirmed diagnoses, and characterized samples for the study. SS, EI, MP, RS SF, TB, and NA performed genotyping, sequencing, and other genetic lab work. IE and JW performed the luciferase assays and other functional work. RT and MB generated and analyzed CGH data. EKK designed analysis methods. EKK, SS, EI, CH, SLR, ESL, and KLT performed and interpreted GWAS analysis, sequencing and association analysis, pathways analysis and risk assessment, motif prediction, and germline to somatic mutation analysis. EKK, SS, EI, and KLT wrote the paper with input from the other authors. All authors read and approved the final manuscript.
Supplementary Material
Regions of association defined with LD clumping. Figure S2. Ages of affected and unaffected samples. Figure S3. Phenotype variance explained at different association thresholds. Figure S4. The difference in genotype relative risk between cases and controls persists at more stringent significance thresholds. Figure S5. Cross-breed meta-analysis of OS GWAS datasets. Figure S6. Transcription factor motif analysis of top candidate variant. Figure S7. Size distribution of fixed and RRV regions by breed. Figure S8. CGH penetrance plots of dog and human OS. Figure S9. Dog GWAS results in human OS associated regions. Figure S10. Confidence interval comparison. Table S1. Association at top SNPs when sex is included as a covariate in the EMMAX genomewide analysis. Table S2. Top associated variants in the greyhound finemapping and imputation analysis. Table S3. Meta-analysis of nine breeds at the top candidate variant. Table S4. GRAIL keywords over-represented among genes in more than one osteosarcoma GWAS region. Table S5. Association between top GWAS SNPs and CGH gain/loss in matched germline tumor samples, controlling for breed clusters using Cochran-Mantel-Haenszel (CMH) test. Table S6. Osteosarcoma related microRNA sets curated from literature for INRICH testing.
Contributor Information
Elinor K Karlsson, Email: elinor@broadinstitute.org.
Snaevar Sigurdsson, Email: snaevar.sigurdsson@decode.is.
Emma Ivansson, Email: emma.ivansson@imbim.uu.se.
Rachael Thomas, Email: rthomas3@ncsu.edu.
Ingegerd Elvers, Email: ielvers@broadinstitute.org.
Jason Wright, Email: jwright@broadinstitute.org.
Cedric Howald, Email: chowald@broadinstitute.org.
Noriko Tonomura, Email: tonomura@broadinstitute.org.
Michele Perloski, Email: perloski@broadinstitute.org.
Ross Swofford, Email: swofford@broadinstitute.org.
Tara Biagi, Email: tarabiagi@gmail.com.
Sarah Fryc, Email: smfryc@gmail.com.
Nathan Anderson, Email: ndanderson23@gmail.com.
Celine Courtay-Cahen, Email: celinecc4@gmail.com.
Lisa Youell, Email: woodimp77@gmail.com.
Sally L Ricketts, Email: sally.ricketts@aht.org.uk.
Sarah Mandlebaum, Email: sarah.mandlebaum@quantis-intl.com.
Patricio Rivera, Email: patricio.rivera@djursjukhusen.se.
Henrik von Euler, Email: henrik.von.euler@slu.se.
William C Kisseberth, Email: william.kisseberth@cvm.osu.edu.
Cheryl A London, Email: cheryl.london@cvm.osu.edu.
Eric S Lander, Email: lander@broadinstitute.org.
Guillermo Couto, Email: coutovetconsultants@gmail.com.
Kenine Comstock, Email: keninec@yahoo.com.
Mike P Starkey, Email: mike.starkey@aht.org.uk.
Jaime F Modiano, Email: modiano@umn.edu.
Matthew Breen, Email: matthew_breen@ncsu.edu.
Kerstin Lindblad-Toh, Email: kersli@broadinstitute.org.
Acknowledgments
We thank pet owners and their dogs and all the breed clubs for their participation, which made this research possible. We thank Anna Blom, Susanne Björnefeldt, Susan Fosmire, Cristan Jubala, Lori Gardner, Mitzi Lewellen, Catherine Gavin, Milcah Scott, and the SciLifeLab/SLU Canine Biobank for help with sample collection. We thank Abhirami Ratnakumar, Erik Axelsson, and Matthew T. Webster for assistance with data analysis, as well as Jill Mesirov, Subbaya Subramanian, Colm O’Dushlaine, Aaron Sarver, the Broad Cancer Program, and the Sabeti group for helpful discussions. We thank the Broad Genomics Platform as well as the SNP & Seq Platform at SciLifeLab for help with sequencing and genotyping. Funding is gratefully acknowledged from AKC/CFH (2254, 947, 373A, 757, and 1317), Irish Wolfhound Foundation (USA), Irish Wolfhound Club (UK), Irish Wolfhound Society (UK), Irish Wolfhound Association of New England, Leonberger Club of America, Leonberger Health Foundation, 2 Million Dogs, Uppsala University, Swedish Medical Research Council, Research Council FORMAS, the European Commission (FP7-LUPA, GA-201370), and the Morris Animal Foundation. EI is supported by fellowships from Barncancerfonden and the Swedish Society for Medical Research, EKK by fellowships from the American Cancer Society and the Charles A. King Trust, IE by the Swedish Research Council, CH by the Swiss National Research Foundation and KLT by a EURYI from the ESF and Consolidator Award from the ERC.
References
- Ta HT, Dass CR, Choong PF, Dunstan DE. Osteosarcoma treatment: state of the art. Cancer Metastasis Rev. 2009;14:247–263. doi: 10.1007/s10555-009-9186-7. [DOI] [PubMed] [Google Scholar]
- Tang N, Song WX, Luo J, Haydon RC, He TC. Osteosarcoma development and stem cell differentiation. Clin Orthop Relat Res. 2008;14:2114–2130. doi: 10.1007/s11999-008-0335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman SD, Calo E, Landman AS, Danielian PS, Miller ES, West JC, Fonhoue BD, Caron A, Bronson R, Bouxsein ML, Mukherjee S, Lees JA. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci U S A. 2008;14:11851–11856. doi: 10.1073/pnas.0805462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, Rodda SJ, Snay E, Dunning P, Fahey FH, Alt FW, McMahon AP, Orkin SH. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;14:1662–1676. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage SA, Mirabello L, Wang Z, Gastier-Foster JM, Gorlick R, Khanna C, Flanagan AM, Tirabosco R, Andrulis IL, Wunder JS, Gokgoz N, Patino-Garcia A, Sierrasesumaga L, Lecanda F, Kurucu N, Ilhan IE, Sari N, Serra M, Hattinger C, Picci P, Spector LG, Barkauskas DA, Marina N, de Toledo SR, Petrilli AS, Amary MF, Halai D, Thomas DM, Douglass C, Meltzer PS. et al. Genome-wide association study identifies two susceptibility loci for osteosarcoma. Nat Genet. 2013;14:799–803. doi: 10.1038/ng.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoloni M, Davis S, Lana S, Withrow S, Sangiorgi L, Picci P, Hewitt S, Triche T, Meltzer P, Khanna C. Canine tumor cross-species genomics uncovers targets linked to osteosarcoma progression. BMC Genomics. 2009;14:625. doi: 10.1186/1471-2164-10-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MC, Sarver AL, Gavin KJ, Thayanithy V, Getzy DM, Newman RA, Cutter GR, Lindblad-Toh K, Kisseberth WC, Hunter LE, Subramanian S, Breen M, Modiano JF. Molecular subtypes of osteosarcoma identified by reducing tumor heterogeneity through an interspecies comparative approach. Bone. 2011;14:356–367. doi: 10.1016/j.bone.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withrow SJ, Powers BE, Straw RC, Wilkins RM. Comparative aspects of osteosarcoma. Dog versus man. Clin Orthop Relat Res. 1991. pp. 159–168. [PubMed]
- Mueller F, Fuchs B, Kaser-Hotz B. Comparative biology of human and canine osteosarcoma. Anticancer Res. 2007;14:155–164. [PubMed] [Google Scholar]
- Slatter M. Rottweiler Health Foundation Health Survey Results. Deerfield Beach, FL: Rottweiler Health Foundation; 2000. http://www.rottweilerhealth.org/RHF_surveyresults.html. [Google Scholar]
- Urfer SR. Lifespan and causes of death in the Irish Wolfhound: medical, genetical and ethical aspects. Suedwestdeutscher Verlag fuer Hochschulschriften: Saarbrucken; 2009. [Google Scholar]
- Lord LK, Yaissle JE, Marin L, Couto CG. Results of a web-based health survey of retired racing greyhounds. J Vet Intern Med. 2007;14:1243–1250. doi: 10.1111/j.1939-1676.2007.tb01945.x. [DOI] [PubMed] [Google Scholar]
- Ru G, Terracini B, Glickman LT. Host related risk factors for canine osteosarcoma. Vet J. 1998;14:31–39. doi: 10.1016/S1090-0233(98)80059-2. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ 3rd, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, DeJong PJ, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin CW, Cook A, Cuff J, Daly MJ, DeCaprio D, Gnerre S. et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;14:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Wilbe M, Jokinen P, Truve K, Seppala EH, Karlsson EK, Biagi T, Hughes A, Bannasch D, Andersson G, Hansson-Hamlin H, Lohi H, Lindblad-Toh K. Genome-wide association mapping identifies multiple loci for a canine SLE-related disease complex. Nat Genet. 2010;14:250–254. doi: 10.1038/ng.525. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;14:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaysse A, Ratnakumar A, Derrien T, Axelsson E, Rosengren Pielberg G, Sigurdsson S, Fall T, Seppala EH, Hansen MS, Lawley CT, Karlsson EK, LUPA Consortium, Bannasch D, Vila C, Lohi H, Galibert F, Fredholm M, Haggstrom J, Hedhammar A, Andre C, Lindblad-Toh K, Hitte C, Webster MT. Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 2011;14:e1002316. doi: 10.1371/journal.pgen.1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;14:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;14:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, Salmon Hillbertz NH, Zody MC, Anderson N, Biagi TM, Patterson N, Pielberg GR, Kulbokas EJ 3rd, Comstock KE, Keller ET, Mesirov JP, von Euler H, Kampe O, Hedhammar A, Lander ES, Andersson G, Andersson L, Lindblad-Toh K. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 2007;14:1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;14:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;14:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, Raney BJ, Pohl A, Malladi VS, Li CH, Lee BT, Learned K, Kirkup V, Hsu F, Heitner S, Harte RA, Haeussler M, Guruvadoo L, Goldman M, Giardine BM, Fujita PA, Dreszer TR, Diekhans M, Cline MS, Clawson H, Barber GP. et al. The UCSC genome browser database: extensions and updates 2013. Nucleic Acids Res. 2013;14:D64–D69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbloom KR, Dreszer TR, Long JC, Malladi VS, Sloan CA, Raney BJ, Cline MS, Karolchik D, Barber GP, Clawson H, Diekhans M, Fujita PA, Goldman M, Gravell RC, Harte RA, Hinrichs AS, Kirkup VM, Kuhn RM, Learned K, Maddren M, Meyer LR, Pohl A, Rhead B, Wong MC, Zweig AS, Haussler D, Kent WJ. ENCODE whole-genome data in the UCSC genome browser: update 2012. Nucleic Acids Res. 2012;14:D912–D917. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;14:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee K, London D, Lotakis D, Neph S. et al. The accessible chromatin landscape of the human genome. Nature. 2012;14:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JW, Squire JA, Zielenska M. The genetics of osteosarcoma. Sarcoma. 2012;14:627254. doi: 10.1155/2012/627254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra A, Roberts H, Weinberg AG, Geradts J. Loss of p16(INK4a) expression correlates with decreased survival in pediatric osteosarcomas. Int J Cancer. 2001;14:34–38. doi: 10.1002/1097-0215(20010120)95:1<34::AID-IJC1006>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Borys D, Canter RJ, Hoch B, Martinez SR, Tamurian RM, Murphy B, Bishop JW, Horvai A. P16 expression predicts necrotic response among patients with osteosarcoma receiving neoadjuvant chemotherapy. Hum Pathol. 2012;14:1948–1954. doi: 10.1016/j.humpath.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Axelsson E, Ratnakumar A, Arendt ML, Maqbool K, Webster MT, Perloski M, Liberg O, Arnemo JM, Hedhammar A, Lindblad-Toh K. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;14:360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;14:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;14:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz MC, Fretz JA, Lorenzo JA. How B cells influence bone biology in health and disease. Bone. 2010;14:472–479. doi: 10.1016/j.bone.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoi E, Nakatani E, Yamamoto T, Iezaki T, Takahata Y, Fujita H, Ishiura R, Takamori M, Yoneda Y. The transcription factor paired box-5 promotes osteoblastogenesis through direct induction of Osterix and Osteocalcin. J Bone Miner Res. 2012;14:2526–2534. doi: 10.1002/jbmr.1708. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;14:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, Frazer KA. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;14:264–268. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, Sklar P, Scolnick EM, Xavier RJ, Altshuler D, Daly MJ. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;14:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunt J, Hasselt NE, Zwijnenburg DA, Hamdi M, Koster J, Versteeg R, Kool M. OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells. Int J Cancer. 2012;14:E21–E32. doi: 10.1002/ijc.26474. [DOI] [PubMed] [Google Scholar]
- Koike N, Kassai Y, Kouta Y, Miwa H, Konishi M, Itoh N. Brorin, a novel secreted bone morphogenetic protein antagonist, promotes neurogenesis in mouse neural precursor cells. J Biol Chem. 2007;14:15843–15850. doi: 10.1074/jbc.M701570200. [DOI] [PubMed] [Google Scholar]
- Deckelbaum RA, Majithia A, Booker T, Henderson JE, Loomis CA. The homeoprotein engrailed 1 has pleiotropic functions in calvarial intramembranous bone formation and remodeling. Development. 2006;14:63–74. doi: 10.1242/dev.02171. [DOI] [PubMed] [Google Scholar]
- Bulman MP, Kusumi K, Frayling TM, McKeown C, Garrett C, Lander ES, Krumlauf R, Hattersley AT, Ellard S, Turnpenny PD. Mutations in the human delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nat Genet. 2000;14:438–441. doi: 10.1038/74307. [DOI] [PubMed] [Google Scholar]
- Matsubara A, Iwama A, Yamazaki S, Furuta C, Hirasawa R, Morita Y, Osawa M, Motohashi T, Eto K, Ema H, Kitamura T, Vestweber D, Nakauchi H. Endomucin, a CD34-like sialomucin, marks hematopoietic stem cells throughout development. J Exp Med. 2005;14:1483–1492. doi: 10.1084/jem.20051325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Matsumoto E, Higashimaki Y, Sugimoto T, Seino S, Kaji H. FAM5C is a soluble osteoblast differentiation factor linking muscle to bone. Biochem Biophys Res Commun. 2012;14:134–139. doi: 10.1016/j.bbrc.2011.12.147. [DOI] [PubMed] [Google Scholar]
- Zou X, Shen J, Chen F, Ting K, Zheng Z, Pang S, Zara JN, Adams JS, Soo C, Zhang X. NELL-1 binds to APR3 affecting human osteoblast proliferation and differentiation. FEBS Lett. 2011;14:2410–2418. doi: 10.1016/j.febslet.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa N, Kinosaki M, Yamaguchi K, Shima N, Yasuda H, Yano K, Morinaga T, Higashio K. RANK is the essential signaling receptor for osteoclast differentiation factor in osteoclastogenesis. Biochem Biophys Res Commun. 1998;14:395–400. doi: 10.1006/bbrc.1998.9788. [DOI] [PubMed] [Google Scholar]
- Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;14:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the pathway interaction database. Nucleic Acids Res. 2009;14:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;14:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TM, Kahler RA, Li X, Westendorf JJ. Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J Biol Chem. 2004;14:41998–42007. doi: 10.1074/jbc.M403702200. [DOI] [PubMed] [Google Scholar]
- Lee PH, O'Dushlaine C, Thomas B, Purcell SM. INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;14:1797–1799. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KB, Salah Z, Del Mare S, Galasso M, Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, Volinia S, Stein GS, Croce CM, Lian JB, Ageilan RI. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;14:1865–1877. doi: 10.1158/0008-5472.CAN-11-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lulla RR, Costa FF, Bischof JM, Chou PM, de Bonaldo FM, Vanin EF, Soares MB. Identification of differentially expressed microRNAs in Osteosarcoma. Sarcoma. 2011;14:732690. doi: 10.1155/2011/732690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maire G, Martin JW, Yoshimoto M, Chilton-MacNeill S, Zielenska M, Squire JA. Analysis of miRNA-gene expression-genomic profiles reveals complex mechanisms of microRNA deregulation in osteosarcoma. Cancer Genet. 2011;14:138–146. doi: 10.1016/j.cancergen.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Sarver AL, Phalak R, Thayanithy V, Subramanian S. S-MED: sarcoma microRNA expression database. Lab Invest. 2010;14:753–761. doi: 10.1038/labinvest.2010.53. [DOI] [PubMed] [Google Scholar]
- Thayanithy V, Sarver AL, Kartha RV, Li L, Angstadt AY, Breen M, Steer CJ, Modiano JF, Subramanian S. Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma. Bone. 2012;14:171–181. doi: 10.1016/j.bone.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angstadt AY, Thayanithy V, Subramanian S, Modiano JF, Breen M. A genome-wide approach to comparative oncology: high-resolution oligonucleotide aCGH of canine and human osteosarcoma pinpoints shared microaberrations. Cancer Genet. 2012;14:572–587. doi: 10.1016/j.cancergen.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du J, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff IK. et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;14:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura Y, Nomoto S, Hayashi M, Hishida M, Nishikawa Y, Yamada S, Fujii T, Sugimoto H, Takeda S, Kodera Y, Nakao A. Identification of the bleomycin hydrolase gene as a methylated tumor suppressor gene in hepatocellular carcinoma using a novel triple-combination array method. Cancer Lett. 2011;14:150–157. doi: 10.1016/j.canlet.2011.07.028. [DOI] [PubMed] [Google Scholar]
- Kuijjer ML, Rydbeck H, Kresse SH, Buddingh EP, Lid AB, Roelofs H, Burger H, Myklebost O, Hogendoorn PC, Meza-Zepeda LA, Cleton-Jansen AM. Identification of osteosarcoma driver genes by integrative analysis of copy number and gene expression data. Genes Chromosomes Cancer. 2012;14:696–706. doi: 10.1002/gcc.21956. [DOI] [PubMed] [Google Scholar]
- Suomi S, Taipaleenmaki H, Seppanen A, Ripatti T, Vaananen K, Hentunen T, Saamanen AM, Laitala-Leinonen T. MicroRNAs regulate osteogenesis and chondrogenesis of mouse bone marrow stromal cells. Gene Regul Syst Bio. 2008;14:177–191. doi: 10.4137/grsb.s662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agirre X, Vilas-Zornoza A, Jimenez-Velasco A, Martin-Subero JI, Cordeu L, Garate L, San Jose-Eneriz E, Abizanda G, Rodriguez-Otero P, Fortes P, Rifon J, Bandres E, Calasanz MJ, Martin V, Heiniger A, Torres A, Siebert R, Roman-Gomez J, Prosper F. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009;14:4443–4453. doi: 10.1158/0008-5472.CAN-08-4025. [DOI] [PubMed] [Google Scholar]
- Lindenblatt C, Schulze-Osthoff K, Totzke G. IkappaBzeta expression is regulated by miR-124a. Cell Cycle. 2009;14:2019–2023. doi: 10.4161/cc.8.13.8816. [DOI] [PubMed] [Google Scholar]
- Shearin AL, Hedan B, Cadieu E, Erich SA, Schmidt EV, Faden DL, Cullen J, Abadie J, Kwon EM, Grone A, Devauchelle P, Rimbault M, Karyadi DM, Lynch M, Galibert F, Breen M, Rutteman GR, Andre C, Parker HG, Ostrander EA. The MTAP-CDKN2A locus confers susceptibility to a naturally occurring canine cancer. Cancer Epidemiol Biomarkers Prev. 2012;14:1019–1027. doi: 10.1158/1055-9965.EPI-12-0190-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;14:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas Z, Vojtesek B, Anton M, Picksley SM, Kubalek V. Overexpression of p53 and MDM2 proteins in cervical neoplasia. Folia Biol (Praha) 1995;14:197–199. [PubMed] [Google Scholar]
- Qi Y, Gregory MA, Li Z, Brousal JP, West K, Hann SR. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature. 2004;14:712–717. doi: 10.1038/nature02958. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Ramsey MR, Balasubramanian P, Castrillon DH, DePinho RA. The differential impact of p16(INK4a) or p19(ARF) deficiency on cell growth and tumorigenesis. Oncogene. 2004;14:379–385. doi: 10.1038/sj.onc.1207074. [DOI] [PubMed] [Google Scholar]
- Hidalgo I, Herrera-Merchan A, Ligos JM, Carramolino L, Nunez J, Martinez F, Dominguez O, Torres M, Gonzalez S. Ezh1 is required for hematopoietic stem cell maintenance and prevents senescence-like cell cycle arrest. Cell Stem Cell. 2012;14:649–662. doi: 10.1016/j.stem.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;14:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan RW, Seidlitz EP, Singh G. Glutamate signaling in healthy and diseased bone. Front Endocrinol (Lausanne) 2012;14:89. doi: 10.3389/fendo.2012.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel G, Platt B, Micheau J. Glutamate receptor function in learning and memory. Behav Brain Res. 2003;14:1–47. doi: 10.1016/S0166-4328(02)00272-3. [DOI] [PubMed] [Google Scholar]
- Elia J, Sackett J, Turner T, Schardt M, Tang SC, Kurtz N, Dunfey M, McFarlane NA, Susi A, Danish D, Li A, Nissley-Tsiopinis J, Borgmann-Winter K. Attention-deficit/hyperactivity disorder genomics: update for clinicians. Curr Psychiatry Rep. 2012;14:579–589. doi: 10.1007/s11920-012-0309-4. [DOI] [PubMed] [Google Scholar]
- Lee PH, Perlis RH, Jung JY, Byrne EM, Rueckert E, Siburian R, Haddad S, Mayerfeld CE, Heath AC, Pergadia ML, Madden PA, Boomsma DI, Penninx BW, Sklar P, Martin NG, Wray NR, Purcell SM, Smoller JW. Multi-locus genome-wide association analysis supports the role of glutamatergic synaptic transmission in the etiology of major depressive disorder. Transl Psychiatry. 2012;14:e184. doi: 10.1038/tp.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SE, Yu D, Scharf JM, Neale BM, Fagerness JA, Mathews CA, Arnold PD, Evans PD, Gamazon ER, Davis LK, Osiecki L, McGrath L, Haddad S, Crane J, Hezel D, Illman C, Mayerfield C, Konkashbaev A, Liu C, Pluzhnikov A, Tikhomirov A, Edlund CK, Rauch SL, Moessner R, Falkai P, Maier W, Ruhrmann S, Grabe HJ, Lennertz L, Wagner M. et al. Genome-wide association study of obsessive-compulsive disorder. Mol Psychiatry. 2013;14:788–798. doi: 10.1038/mp.2012.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodman NH, Karlsson EK, Moon-Fanelli A, Galdzicka M, Perloski M, Shuster L, Lindblad-Toh K, Ginns EI. A canine chromosome 7 locus confers compulsive disorder susceptibility. Mol Psychiatry. 2010;14:8–10. doi: 10.1038/mp.2009.111. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee S, Goddard M, Visscher P. GCTA: a tool for genome-wide complex trait analysis. Am J Human Gen. 2010;14:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Sul J, Service S, Zaitlen N, Kong S-Y, Freimer N, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;14:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;14:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;14:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard. http://picard.sourceforge.net.
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;14:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;14:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellay J. Host genetics influences on HIV type-1 disease. Antivir Ther. 2009;14:731–738. doi: 10.3851/IMP1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truve K, Eriksson O, Norling N, Wilbe M, Mauceli E, Lindblad-Toh K, Bongcam-Rudloff E. SEQscoring: a tool to facilitate the interpretation of data generated with next generation sequencing technologies. EMBnet journal. 2011;14:38–45. [Google Scholar]
- Cingolani P, Platts A, Wang Le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;14:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2012;14:2012. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, Ward LD, Lowe CB, Holloway AK, Clamp M, Gnerre S, Alfoldi J, Beal K, Chang J, Clawson H, Cuff J, Di Palma F, Fitzgerald S, Flicek P, Guttman M, Hubisz MJ, Jaffe DB, Jungreis I, Kent WJ, Kostka D, Lara M. et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;14:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;14:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://www.atcc.org/Standards/Standards_Programs/ATCC_Standards_Development_Organization.aspx.
- Gene Set Enrichment Analysis. http://www.broadinstitute.org/gsea/msigdb.
- Ensembl. http://www.ensembl.org/biomart.
- Pathway Interaction Database. http://pid.nci.nih.gov.
- Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI reference sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;14:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Seiser EL, Motsinger-Reif A, Borst L, Valli VE, Kelley K, Suter SE, Argyle D, Burgess K, Bell J, Lindblad-Toh K, Modiano JF, Breen M. Refining tumor-associated aneuploidy through ‘genomic recoding’ of recurrent DNA copy number aberrations in 150 canine non-Hodgkin lymphomas. Leuk Lymphoma. 2011;14:1321–1335. doi: 10.3109/10428194.2011.559802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://www.broadinstitute.org/ftp/pub/vgb/dog/OSA_GenomeBiology2013paper.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Regions of association defined with LD clumping. Figure S2. Ages of affected and unaffected samples. Figure S3. Phenotype variance explained at different association thresholds. Figure S4. The difference in genotype relative risk between cases and controls persists at more stringent significance thresholds. Figure S5. Cross-breed meta-analysis of OS GWAS datasets. Figure S6. Transcription factor motif analysis of top candidate variant. Figure S7. Size distribution of fixed and RRV regions by breed. Figure S8. CGH penetrance plots of dog and human OS. Figure S9. Dog GWAS results in human OS associated regions. Figure S10. Confidence interval comparison. Table S1. Association at top SNPs when sex is included as a covariate in the EMMAX genomewide analysis. Table S2. Top associated variants in the greyhound finemapping and imputation analysis. Table S3. Meta-analysis of nine breeds at the top candidate variant. Table S4. GRAIL keywords over-represented among genes in more than one osteosarcoma GWAS region. Table S5. Association between top GWAS SNPs and CGH gain/loss in matched germline tumor samples, controlling for breed clusters using Cochran-Mantel-Haenszel (CMH) test. Table S6. Osteosarcoma related microRNA sets curated from literature for INRICH testing.
Data Availability Statement
The data presented in this publication are available through the Gene Expression Omnibus (accession number GSE52147) and ArrayExpress (accession numbers E-MTAB-1984 and E-MTAB-1986). Datasets analyzed in the paper are also available at [95].


