Drug resistance to the proteasome inhibitor, bortezomib/VELCADE (Bz) is a significant clinical problem in the treatment of multiple myeloma (MM), an invariably fatal plasma cell malignancy of the bone marrow.1 Despite initial success and wide use in cocktail regimens for MM treatment,2 the majority of patients treated with Bz eventually relapse, many having developed ‘acquired’ Bz-resistant (BzR) disease.1 Thus, the elucidation of mechanisms by which Bz resistance may occur and the identification of novel intervention strategies are important translational aims.
To identify mechanisms of acquired Bz resistance, we previously utilized in vitro cell lines from the Bcl-XL/Myc mouse model of plasma cell malignancy to systematically ascertain differences between Bz-sensitive (BzS) and derived BzR cells.3 We have employed this model system because malignant plasma cell lines isolated from these mice closely resemble human MM based on gene expression, chromosomal abnormalities and progression of disease in the bone marrow.3–5 Perhaps most importantly, from initially drug-sensitive tumor cell populations we are able to select for drug-resistant cells in vitro, adoptively transfer these cells back into syngeneic recipient mice, and recapitulate the drug-sensitive or -resistant phenotype following in vivo Bz treatment of recipient mice.3 Remarkably, the differences in Bz sensitivity in our mouse cell lines strongly correlated with differences in malignant plasma cell migration following adoptive transfer. BzR cells displayed a significantly reduced affinity for the bone marrow compartment, compared with their sensitive counterparts, and instead infiltrated extramedullary tissues more readily,3 suggesting the possibility that BzR plasma cells (PCs) are more likely to reside outside of the bone marrow milieu. Thus, we hypothesized that Bz treatment and acquisition of resistance selects for cells that can survive independent of the bone marrow microenvironment and that this phenotype may be due to the loss of cell-surface proteins that mediate the MM–bone marrow stromal cell (BMSC) interaction.
We recently identified a 23-gene signature that distinguishes between BzS and BzR mouse cell lines in vitro and significantly predicts differences in patient outcomes in a human MM clinical trial that included Bz.3 We explored the gene expression profiling (GEP) data sets associated with these studies further, and of the 23 genes in this model, 5 genes including chemokine (C-X-C motif) receptor 4 (CXCR4), regulator of G-protein signaling 16 (RGS16), lectin, gaglactoside-binding, soluble, 1 (LGALS1), CD93 and cystathionine-beta-synthase (CBS) have been associated with the trafficking of cells within the immune system (Ingenuity Pathway Analysis). However, of these genes, CXCR4 has been most directly associated with PC migration in both humans and mice.6 We observed a loss in Cxcr4 mRNA expression in all four cases of BzR compared with BzS mouse cell lines.3 CXCR4 is known to be highly expressed on the surface of human plasma cells and contributes to bone marrow homing during normal plasma cell maturation through interaction with its ligand SDF-1α that is secreted by bone marrow stromal cells,6 making this gene an attractive candidate for our hypothesis.
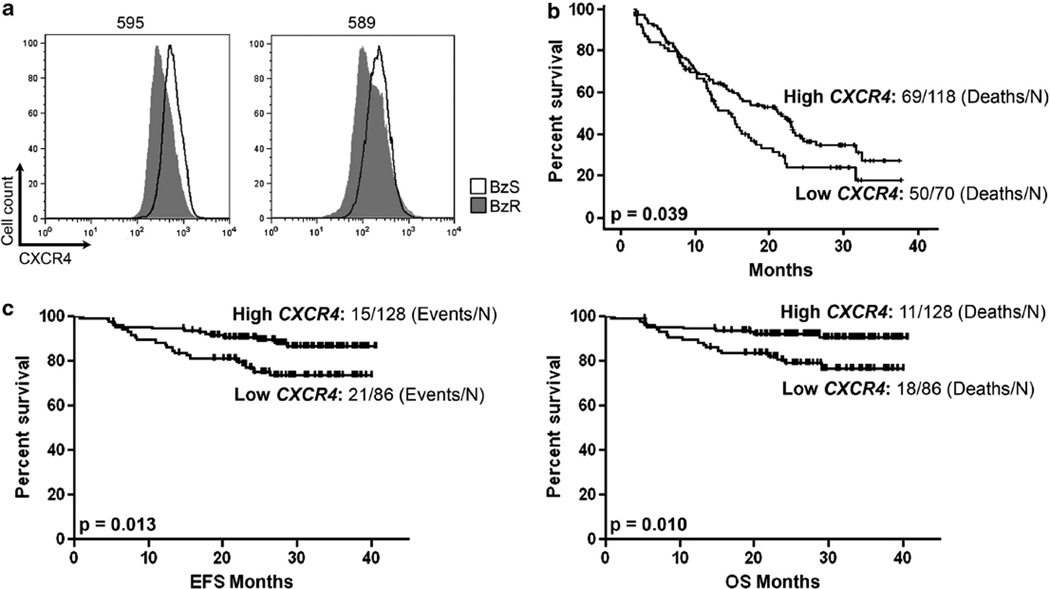
Therefore, we began by validating the protein expression of CXCR4 by flow cytometry in BzS and their derived BzR counterpart mouse cell lines we had isolated as previously described.3 The 2.5-fold average reduction in Cxcr4 mRNA expression that we observed by gene expression microarray3 was further reflected as a 1.5- (cell line 589) to 2 (cell line 595)-fold reduction in total (intracellular and surface) CXCR4 protein in the mouse BzR cell lines in vitro (Figure 1a), suggesting that this could be a functional change in the transcriptional regulation of Cxcr4 or that Bz selects for those cells that express lower levels of CXCR4. However, given that the original mRNA expression data were collected in the absence of cell death over a short-time period3 suggests the possibility that Bz contributes to the reduction of Cxcr4 mRNA expression.
Figure 1. Low CXCR4 expression is associated with bortezomib-resistance and poor clinical outcomes in patients treated with bortezomib.
(a) Fluorescence-activated cell sorting (FACS) analysis of BzS (solid black line) and BzR (dark gray histogram) mouse cells stained with a CXCR4 antibody for total (intracellular and cell surface) protein. (b) Survival analysis of high and low CXCR4 expressing MM patient groups taken from the APEX drug trial7 treated with either single-agent Bz (left panel) or high-dose dexamethasone (right panel). (c) Event-free (EFS; left panel) and overall survival (OS; right panel) analysis of high and low CXCR4 expressing MM patient groups taken from the total therapy 3 (MMTT3) drug trial.8 P values represent significant differences by one-way analysis of variance (P <0.05 was considered significant). The number of cases analyzed is indicated.
The reduction in CXCR4 expression in BzR cells combined with the in vivo data showing that BzR cells promote aggressive, extramedullary disease resulting in poorer survival3 suggests that CXCR4 could serve as a biomarker for patient survival. We next sought to determine whether reduced CXCR4 expression was associated with poorer survival in MM patients treated with Bz. The APEX drug trial provides initial GEP data from MM patients treated with either single-agent Bz or high-dose dexamethasone.7 We divided the data set into those patients with high versus low CXCR4 expression and assessed overall survival (OS) trends in both groups. As a single biomarker, low CXCR4 expression significantly distinguished those MM patients with poorer OS (P=0.039) in the Bz-treated arm of the clinical trial (Figure 1b), but not the dexamethasone arm (data not shown), suggesting that this could be a novel biomarker associated with poor response to Bz. Because the majority of MM patients are placed on a therapeutic cocktail of drugs that often includes Bz, we next queried the MM total therapy 3 (MMTT3) drug trial that reported GEP data for MM patients treated with VTD-PACE (Bz, thalidomide, dexamethasone, cisplatin, doxorubicin, cyclophosphamide, etoposide) in conjunction with tandem transplantation.8,9 As with the APEX drug trial, low CXCR4 expression significantly distinguished those MM patients with poorer event-free (P=0.013; left panel) and OS (P=0.010; right panel) survival (Figure 1c). Therefore, reduced CXCR4 expression predicts poorer survival in patients treated with Bz.
Using the Bcl-XL/Myc model system of plasma cell malignancy, we identified CXCR4 as a single biomarker whose reduced expression is associated with poorer outcomes in MM patients being treated with Bz. CXCR4 expression is known to have a role in PC homing to the bone marrow.10 When injected back into syngeneic mice, BzR cells, which have reduced CXCR4 expression, display reduced BM homing compared with their BzS counterparts, resulting in a much more severe disease phenotype.3 This finding suggests that reduced CXCR4 expression correlates with increased disease severity and agrees with our analysis of both a single-agent Bz clinical trial (APEX7) and a clinical trial utilizing Bz in cocktail with additional agents (MMTT38), both of which show that low CXCR4 expression is significantly associated with worse outcome compared to patients with high CXCR4 expression.
The biological role of CXCR4 is of particular interest given that studies have shown that the disruption of the interaction between BMSCs and MM cells (for example, by virtue of chemical inhibition of the CXCR4/SDF-1α axis) may increase sensitivity to Bz by mobilizing MM cells from the bone marrow microenvironment. 11 While the full mechanism of reduced CXCR4 expression in BzR cells awaits further investigation, we have evidence to suggest that Bz treatment promotes the reduction of CXCR4 mRNA, which we have previously reported.3 In turn, over time following continuous Bz treatment, reduced Cxcr4 expression may decrease the reliance of the MM cells on the bone marrow microenvironment and thus eventually promote extramedullary disease. Indeed, BzR disease presenting in patients as extramedullary masses has been reported.12 A circulating malignant clonal PC population has already been described by others as highly drug resistant and likely to contribute to disease relapse.13 Whether this population contributes to the proposed tumor-initiating and drug resistant MM ‘side population’,14 which also expresses lower CXCR4 transcript than the bulk tumor population (FZ, unpublished data), will require additional studies.
In summary, these results support the use of CXCR4 as a valuable diagnostic biomarker that predicts clinical outcome in MM patients treated with Bz. The positive impact of cell-surface markers that may predict Bz response as a diagnostic tool in patient care could mean the early detection of BzR disease and improved OS through individualized medical treatment and new therapeutic targets (Supplementary Information).
Supplementary Material
Acknowledgments
BVN receives research support from Millennium Pharmaceuticals Inc., Cambridge, MA.
Footnotes
CONFLICT OF INTEREST
The other authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
REFERENCES
- 1.Richardson PG, Barlogie B, Berenson J, Singhal S, Jagannath S, Irwin D, et al. A phase 2 study of bortezomib in relapsed, refractory myeloma. N Engl J Med. 2003;348:2609–2617. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- 2.Shah JJ, Orlowski RZ. Proteasome inhibitors in the treatment of multiple myeloma. Leukemia. 2009;23:1964–1979. doi: 10.1038/leu.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stessman HA, Baughn LB, Sarver A, Xia T, Deshpande R, Mansoor A, et al. Profiling bortezomib resistance identifies secondary therapies in a mouse myeloma model. Mol Cancer Ther. 2013 doi: 10.1158/1535-7163.MCT-12-1151. e-pub ahead of print 27 March 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung WC, Kim JS, Linden M, Peng L, Van Ness B, Polakiewicz RD, et al. Novel targeted deregulation of c-Myc cooperates with Bcl-X(L) to cause plasma cell neoplasms in mice. J Clin Invest. 2004;113:1763–1773. doi: 10.1172/JCI20369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boylan KL, Gosse MA, Staggs SE, Janz S, Grindle S, Kansas GS, et al. A transgenic mouse model of plasma cell malignancy shows phenotypic, cytogenetic, and gene expression heterogeneity similar to human multiple myeloma. Cancer Res. 2007;67:4069–4078. doi: 10.1158/0008-5472.CAN-06-3699. [DOI] [PubMed] [Google Scholar]
- 6.Alsayed Y, Ngo H, Runnels J, Leleu X, Singha UK, Pitsillides CM, et al. Mechanisms of regulation of CXCR4/SDF-1 (CXCL12)-dependent migration and homing in multiple myeloma. Blood. 2007;109:2708–2717. doi: 10.1182/blood-2006-07-035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulligan G, Mitsiades C, Bryant B, Zhan F, Chng WJ, Roels S, et al. Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor bortezomib. Blood. 2007;109:3177–3188. doi: 10.1182/blood-2006-09-044974. [DOI] [PubMed] [Google Scholar]
- 8.Shaughnessy JD, Jr, Qu P, Usmani S, Heuck CJ, Zhang Q, Zhou Y, et al. Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myeloma treated with total therapy 3. Blood. 2011;118:3512–3524. doi: 10.1182/blood-2010-12-328252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Usmani SZ, Crowley J, Hoering A, Mitchell A, Waheed S, Nair B, et al. Improvement in long-term outcomes with successive Total Therapy trials for multiple myeloma: are patients now being cured? Leukemia. 2013;27:226–232. doi: 10.1038/leu.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunkel EJ, Butcher EC. Plasma-cell homing. Nat Rev Immunol. 2003;3:822–829. doi: 10.1038/nri1203. [DOI] [PubMed] [Google Scholar]
- 11.Azab AK, Runnels JM, Pitsillides C, Moreau AS, Azab F, Leleu X, et al. CXCR4 inhibitor AMD3100 disrupts the interaction of multiple myeloma cells with the bone marrow microenvironment and enhances their sensitivity to therapy. Blood. 2009;113:4341–4351. doi: 10.1182/blood-2008-10-186668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moriuchi M, Ohmachi K, Kojima M, Tsuboi K, Ogawa Y, Nakamura N, et al. Three cases of bortezomib-resistant multiple myeloma with extramedullary masses. Tokai J Exp Clin Med. 2010;35:17–20. [PubMed] [Google Scholar]
- 13.Peceliunas V, Janiulioniene A, Matuzeviciene R, Zvirblis T, Griskevicius L. Circulating plasma cells predict the outcome of relapsed or refractory multiple myeloma. Leuk Lymphoma. 2012;53:641–647. doi: 10.3109/10428194.2011.627481. [DOI] [PubMed] [Google Scholar]
- 14.Mo SL, Li J, Loh YS, Brown RD, Smith AL, Chen Y, et al. Factors influencing the abundance of the side population in a human myeloma cell line. Bone Marrow Res. 2011;2011:524845. doi: 10.1155/2011/524845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.