Abstract
Genome-wide techniques provide robust and comprehensive identification of lncRNAs in adult mouse neural stem cells and their derivatives, illuminating the functions of these underappreciated transcripts.
Mammalian genomes have unexpectedly few (20,000 or so) protein-encoding genes [1]. Our view of the mammalian genome has additionally been revolutionized with new knowledge that the number of non-protein-coding genes and their product, non-coding RNAs (ncRNAs), has long been underestimated. The finding that more than 70% of the mammalian transcriptome consists of ncRNAs [1] has promoted a search for ncRNA functions. In particular, one category of ncRNAs that has been the target of recent intensive research consists of long ncRNAs (lncRNAs). lncRNAs are defined as transcripts of at least 200 nucleotides that possess little theoretical protein-coding potential. These intriguing RNAs resemble mRNAs in many ways: they are transcribed by RNA polymerase II and capped, they can undergo splicing and polyadenylation, and a small fraction of those exported to the cytoplasm can associate with ribosomes, although with uncertain consequences [2]. However, unlike mRNAs, most lncRNAs are primarily retained within nuclei [3]. Moreover, lncRNAs manifest rates of sequence shifts that throughout evolution have surpassed those of mRNAs [4].
Since lncRNAs generally do not encode proteins, many researchers had relegated them to transcriptional noise. However, increasing lines of evidence suggest that lncRNAs can function to regulate mammalian gene expression at multiple levels, and that they are responsible for a number of key cellular and developmental processes (see Rinn and Chang [5] and the references therein). Rapid advances in high-throughput techniques, especially RNA-seq, have enabled extensive efforts in identifying lncRNAs and the generation of lncRNA databases for various species [3,6]. Nevertheless, since the expression of lncRNAs appears to be more cell type-specific than the expression of mRNAs, and as most lncRNA databases are derived from a mixture of cell types, there is currently a void of reliable lncRNA information for individual cell types.
To fully appreciate the functions of lncRNAs, one key task that remained to be undertaken was to construct accurately annotated cell type-specific lncRNA expression maps for a dynamic, developmental process in vivo. Towards this goal, a recent study by Alexander Ramos and colleagues [7] employed complementary high-throughput methods to identify more than 12,000 lncRNAs expressed during mouse brain development. The authors examined the expression patterns of lncRNAs during subventricular zone (SVZ) neurogenesis in adult mice. They subsequently established an online resource to predict regulatory roles of lncRNAs in the (1) SVZ, which contains neuronal stem cells (NSCs) that can migrate to the olfactory bulb (OB), (2) OB, where NSCs terminally differentiate into interneurons, and (3) dentate gyrus (DG), which harbors a complete neuronal lineage.
Neural lncRNA identification - combining completeness and specificity
To investigate the relationship between lncRNAs and adult mouse-brain development, an issue of emerging interest, Ramos et al. sequenced cDNA libraries from microdissected SVZ, OB and DG [7]. After including RNA-seq data from mouse embryonic stem cells (ESCs) and ESC-derived neural progenitor cells (ESC-NPCs) to increase coverage of potential lncRNAs, the authors used ab initio transcriptome reconstruction to identify 8,992 lncRNAs that derived from 5,731 genomic loci. To incorporate lncRNAs that might have been missed by short-read Illumina-based sequencing, the authors employed long-read RNA CaptureSeq to sequence SVZ cDNAs hybridized to probe libraries that tiled across 100 Mbp of putative lncRNA loci. The additional >3,500 lncRNAs brought the number of lncRNAs identified in neuronal lineages in vivo to an unprecedented >12,000, which is two- to three-fold more than previously known (see Mitchell Guttman et al. [6], for example). The surprising increase in the number of lncRNAs was explained by the focus of previous studies on only one or a combination of a few closely related cell or tissue types. This focus would inherently fail to capture certain sets of lncRNAs, given the finding that lncRNAs exhibit greater spatiotemporal expression specificity than mRNAs ([7], see below). Furthermore, previous studies were limited by the use of relatively insensitive techniques. For instance, custom microarrays do not cover the entire transcriptome, and Illumina-based RNA-seq rarely picks up lower abundance transcripts, many of which are lncRNAs.
Considering that lncRNA expression is highly specific to cell type and strictly regulated during development, those lncRNAs identified by Ramos et al. [7] are anticipated to be only part of the mouse lncRNA repertoire. Thus, it is likely that the number of lncRNAs in other organisms has been underestimated as well, since no other thorough genome- and developmental-wide analysis has been performed. It follows that existing underestimates of lncRNA numbers are accompanied by an under-appreciation of lncRNA functions, some of which have been conserved through evolution from zebrafish to humans [8].
The dynamics of lncRNA expression during neurogenesis
The finding that lncRNAs exhibit greater spatiotemporal expression specificity than do mRNAs - a finding that derived in part from published RNA-seq data from different regions of the mouse brain and during different stages of mouse brain development - indicated that lncRNAs have specific spatiotemporal roles. Thus, equally as important as identifying lncRNAs is determining lncRNA expression patterns. To map the expression patterns of lncRNAs in distinct cell types in vivo, Ramos et al. [7] used specific markers and fluorescence-activated cell sorting (FACS) to sort SVZ-derived cells that represent the three main neurogenic cell types - namely, activated NSCs, transit-amplifying cells and migratory neuroblasts - and they did likewise for niche astrocytes. The authors then interrogated the cDNAs generated from these cells using a microarray of probes corresponding to the lncRNAs that they had previously identified. They found a unique lncRNA expression pattern for each of the three stages of neurogenesis analyzed that can be distinguished from the expression pattern in niche astrocytes. Thus, the differential expression of lncRNAs at different stages of the same lineage likely contributes to the specification of these stages.
In addition, Ramos and co-workers [7] found that lncRNAs are transcriptionally regulated in a manner analogous to mRNAs. Using ChIP-seq, they showed that, as for mRNAs dynamically regulated in neurogenesis, the transcription start sites (TSSs) of many of the identified lncRNAs had both an activating and a repressive histone mark (H3K4me3 and H3K27me3, respectively) in NSCs. With this bivalent mark, their promoters are held inactive but poised for either activation or repression upon differentiation via loss of either one of the histone modifications. The presence of these marks was consistent with the expression patterns of the particular lncRNAs as determined using microarrays. These findings enable the prediction of lncRNAs that may function in NSC maintenance and/or differentiation. The authors have incorporated their annotations of putative lncRNAs that derive from RNA-seq and RNA CaptureSeq, along with lncRNA expression patterns determined using microarray analyses, into an online database [7]. This lncRNA identification and expression analysis pipeline constitutes an important resource for future analyses of lncRNA function in the mouse brain and during SVZ neurogenesis.
lncRNAs: a hidden clue to cures?
Ramos et al. [7] used their newly established pipeline to predict lncRNAs that may function in SVZ neurogenesis. One of these lncRNAs, Six3os, was expressed specifically in NSCs but not in subsequently differentiated cells. Downregulating Six3os using a short-hairpin RNA reduced by two-fold the number of NSCs that after differentiation stained positive for the neuron-specific class III beta-tubulin (TUJ1), indicating a Six3os short hairpin RNA (shRNA)-mediated defect in neurogenesis.
While only a few identified lncRNAs were functionally validated, results demonstrate the utility of the authors' workflow to predict with high confidence lncRNAs that function in the neurogenic process. Notably, when the authors generated different transcript modules, consisting of lncRNAs and known protein-coding transcripts whose variation in expression typify a brain region or brain developmental stage, they found that some modules are closely related to human neurodegenerative diseases such as Huntington's disease, Alzheimer's disease and so on. This suggests that lncRNAs classified into such modules are potentially associated with these diseases. For example, 88 lncRNAs were found in a module that correlates with a gene expression set that is misregulated in mouse models of Huntington's disease, implying potential roles for these lncRNAs in this neurodegenerative condition. More than solving the argument of whether these lncRNAs have functional relevance, it now becomes imperative to understand lncRNA function in order to understand how they contribute to neurodegenerative disorders.
Future directions
The study by Ramos et al. [7] has provided a generalizable way to comprehensively identify novel functional transcripts (Figure 1a). This is of special significance given that lncRNAs are playing more roles in cellular functions than originally anticipated (Figure 1b). These roles pertain not only to neurogenesis but also to embryogenesis [9], myogenesis [10] and likely many other processes. With such workflows, researchers can begin constructing more complete functional genomics maps for different cell types and from various developmental stages. These maps will help to unravel mammalian gene expression networks and provide a basis for the study of the largely uninvestigated but clearly diverse roles of lncRNAs in normal and disease-associated cellular metabolism.
Figure 1.
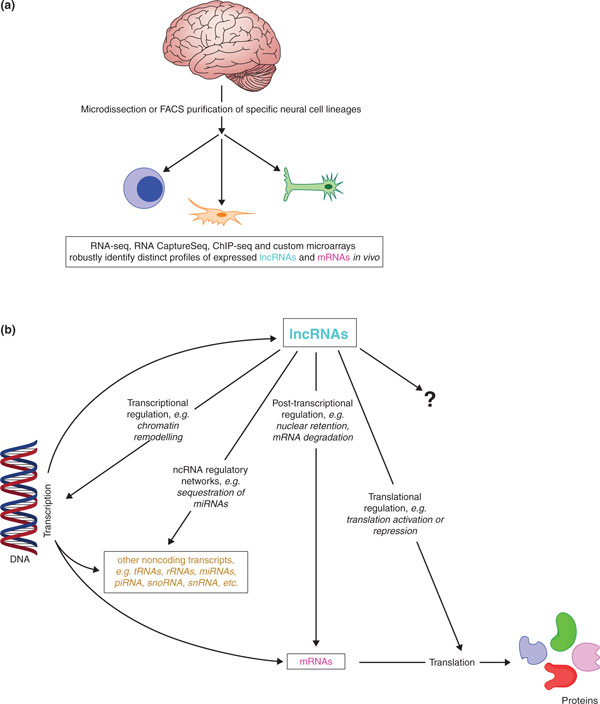
Characterizing lncRNA profiles and functions in specific neural cell lineages. (a) Genome-wide techniques provide a robust and comprehensive way of identifying lncRNAs [7]. (b) lncRNAs can regulate gene expression at multiple levels during mouse neurogenesis, and likely other developmental processes [5,7]. miRNA, microRNA; piRNA, piwi-interacting RNA; rRNA, ribosomal RNA; snoRNA, small nucleolar RNA; snRNA, small nuclear RNA; tRNA, transfer RNA.
Abbreviations
cDNA: complementary DNA; ChIP-seq: chromatin immunoprecipitation followed by high-throughput DNA sequencing; DG: dentate gyrus; ESC: embryonic stem cell; ESC-NPC: ESC-derived neural progenitor cell; FACS: fluorescence-activated cell sorting; H3K4me3 or H3K27me3: histone containing a trimethylated lysine at position 4 or 27; lncRNA: long non-coding RNA; Mbp: megabase pair; ncRNA: non-coding RNA; NSC: neuronal stem cell; OB: olfactory bulb; RNA CaptureSeq: a combination of cDNA capture on tiling arrays that enriches for particular transcripts or genomic regions followed by 454 sequencing-to-saturation to mine the depth of a particular part of the transcriptome; RNA-seq: high-throughput whole-transcriptome shotgun sequencing of cellular cDNAs; SVZ: subventricular zone; TSS: transcription start site; TUJ1: neuron-specific class III beta-tubulin.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Jiashi Wang, Email: Jiashi_Wang@URMC.Rochester.edu.
Bronwyn A Lucas, Email: Bronwyn_Lucas@URMC.Rochester.edu.
Lynne E Maquat, Email: lynne_maquat@urmc.rochester.edu.
Acknowledgements
We thank Max Popp for comments on the manuscript. Work on lncRNAs in the Maquat laboratory is supported by R01 grant GM074593 from the NIH to LEM.
References
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F. Landscape of transcription in human cells. Nature. 2012;14:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;14:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;14:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, Ponting CP, Odom DT, Marques AC. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 2012;14:e1002841. doi: 10.1371/journal.pgen.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Ann Rev Biochem. 2012;14:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell Guttman IA, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;14:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos AD, Diaz A, Nellore A, Delgado RN, Park K-Y, Gonzales-Roybal G, Oldham MC, Song JS, Lim DA. Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell. 2013;14:616–628. doi: 10.1016/j.stem.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;14:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;14:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Gong C, Maquat LE. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 2013;14:793–804. doi: 10.1101/gad.212639.112. [DOI] [PMC free article] [PubMed] [Google Scholar]


