Abstract
Analyzing the dynamics of biofilm formation helps to deepen our understanding of surface colonization in natural environments. While methods for screening biofilm formation in the laboratory are well established, studies in marine environments have so far been based upon destructive analysis of individual samples and provide only discontinuous snapshots of biofilm establishment. In order to explore the development of biofilm over time and under various biotic and abiotic conditions, we applied a recently developed optical biofilm sensor to quasicontinuously analyze marine biofilm dynamics in situ. Using this technique in combination with microscope-assisted imaging, we investigated biofilm formation from its beginning to mature multispecies biofilms. In contrast to laboratory studies on biofilm formation, a smooth transition from initial attachment to colony formation and exponential growth could not be observed in the marine environment. Instead, initial attachment was followed by an adaptation phase of low growth and homogeneously distributed solitary bacterial cells. Moreover, we observed a diurnal variation of biofilm signal intensity, suggesting a transient state of biofilm formation of bacteria. Overall, the biofilm formation dynamics could be modeled by three consecutive development stages attributed to initial bacterial attachment, bacterial growth, and attachment and growth of unicellular eukaryotic microorganisms. Additional experiments showed that the presence of seaweed considerably shortened the adaptation phase in comparison with that on control surfaces but yielded similar growth rates. The outlined examples highlight the advantages of a quasicontinuous in situ detection that enabled, for the first time, the exploration of the initial attachment phase and the diurnal variation during biofilm formation in natural ecosystems.
INTRODUCTION
Colonization of surfaces by microorganisms and formation of biofilms can occur on both living and nonliving surfaces in natural and man-made aquatic environments (1). Depending on composition and location, the effects of biofilm formation are considered positive or negative. For example, biofilm formation on plant roots (2) or human intestine (3) and marine macroalgae (4) are mostly beneficial for their host, while biofilm formation on implants—such as catheters or artificial heart valves—adversely affect human health (5). In aquatic environments, biofilms increase corrosion of metal structures (6); in food processing systems, they may act as a source of contamination (7). Already this short list of beneficial and adverse implications for biological systems as well as human well-being explains the general interest in biofilm formation. As it is known that biofilms are often dominated by bacterial communities, bacterial colonization dynamics of single or mixed bacterial strains have been intensively studied in laboratory experiments. In contrast, little is known about biofilm formation dynamics in natural environments (8–11). In laboratory assays, it has been demonstrated that bacterial biofilm formation is a dynamic process with different phases of development. After biochemical conditioning of the substrate, pioneer bacterial cells from the surrounding water adhere to the surface. This initial attachment phase is followed by a reproduction and accumulation phase, resulting in a monolayer film of bacterial cells. Finally, the bacterial community reaches a pseudostationary phase in which detachment, dispersal, and sloughing are in equilibrium with accumulation and growth (12–14).
In the marine environment, biofilm formation is more complex, and biofilms consist of bacteria, fungi, diatoms, protozoans, larvae, and algal spores (15, 16). However, first colonization of a submersed surface is still initiated by the attachment of bacteria from the surrounding seawater (17), until eukaryotic organisms follow. Depending on the species, eukaryotic settlement may contribute to an enormous growth of marine biofilms (e.g., diatoms [18]) or limit biofilm growth by eukaryotic grazing (19). Of course, biofilm formation is highly variable in marine environments and is affected by physicochemical properties of the surface (20, 21) as well as environmental factors, such as nutrient level, pH, dissolved oxygen concentration, and temperature (22, 23).
Moreover, it has been shown that bacterial community composition and density of the surrounding water (24) play an important role for biofilm establishment. In this context, Dang and Lovell (25, 26) demonstrated that the marine Rhodobacter group not only dominates early stages of biofilm formation on artificial surfaces but also is highly abundant as a potential surface colonizer in coastal seawater all year round (27). So far, to the best of our knowledge, these studies in the marine environment were based solely on destructive methods and the analysis of individual samples after a defined settlement time. These studies provided only snapshots of biofilm formation. Therefore, it was often difficult to attribute the overall effect of selected environmental parameters to a specific growth phase. In particular, short-term effects and/or effects on the initial stages of biofilm formation are often obscured by a limited number of samples or too-long sampling intervals. To overcome this problem and in order to explore in situ the formation of these highly dynamic biofilms under biotic and abiotic parameters in the marine environment, we applied a recently developed optical biofilm sensor in this study (28, 29). The sensor is capable of quasicontinuous monitoring of biofilm development from initial attachment of first bacteria to mature, three-dimensional (3D) multispecies cell clusters. We conducted first experiments to highlight different aspects of biofilm formation in different seasons and habitats and to demonstrate the capability of the biofilm sensor to detect diurnal fluctuations with the finest temporal resolution ever reported for marine biofilms. This technique was used in combination with microscope-assisted imaging to identify known phases of biofilm development that were found to be significantly different from those observed under laboratory conditions.
MATERIALS AND METHODS
Optical biofilm sensor.
For online in situ and nondestructive monitoring of marine biofilms, a novel optical biofilm sensor was employed (28, 29). Basically, the intrinsic fluorescence of the amino acid tryptophan is excited by a UV-LED light source at a wavelength of 280 nm. The emitted fluorescence at 350 nm was collected and guided by 18 fused silica optical fiber bundles that were arranged hemispherically and in two rings around the LED. The optical design of the sensor head was optimized (e.g., the arrangement and inclination angle of the fibers [28]) to detect a fluorescence signal of an extended planar surface without significantly deteriorating the signal by potential background fluorescence from the bulk water. To compensate for the spatially heterogeneous arrangement of bacteria in a typical biofilm with patchy cell clusters several hundred micrometers in diameter (8, 30), the sensor design probed a large substrate surface of approximately 0.5 cm2. The effective penetration depth of the sensor signal was several millimeters (about 50% of the fluorescence signal arose from the first 3.5 mm and about 90% from the first 10 mm), deep enough to measure even thick, mature biofilms.
At the end of the combined fiber bundles, the collected fluorescence light was spectrally separated and discriminated from the incident and reflected or scattered excitation light by a combination of two interference filters. A photomultiplier tube operating in single-photon-counting mode was employed as detector. A linear sensor signal response between fluorescence intensity and bacterial cell density and coverage area has been demonstrated with two different bacterial strains and in situ with natural bacterial biofilm communities (28). Therefore, the quasicontinuous fluorescence signal of the biofilm sensor can be reliably converted to bacterial cell numbers and microbial coverage area for a monolayer of a biofilm. For the analysis of the field measurements, the respective conversion factors were obtained for a limited number of reference samples analyzed by epifluorescence microscopy. Cell number and surface coverage were determined, and chlorophyll-containing cells were identified. Overall, it turned out that the assumption of a linear response of the sensor was approximately valid even for the complex natural biofilms observed in this work. Typically, the biofilm was exposed to an optical output power of about 600 μW, and a readout cycle consists of five consecutive fluorescence measurements with integration times of 10 ms per reading. Possible DNA damage of the microbial community (31) was excluded by switching off the UV-LED between the measurement cycles. The first measurement, which was taken 10 min after immersion of the sensor (still without any substantial biofilm), served as the background fluorescence intensity of the seawater and was subtracted from all following data.
Experimental setups.
Between August 2010 and November 2011, several biofilm dynamics experiments were conducted by exposing the sensor at a depth of 0.5 m in the Baltic Sea (Kiel Fjord, GEOMAR institute pier; 54°33′N, 10°14′E). UV transparent foil (Lumox film 25; Greiner Bio-One, Germany) was mounted as the settling substrate onto the sensor head. Biofilm formation was quantified by measuring the fluorescence intensity of the substrate using the optical biofilm sensor. Reference settlement substrates (Lumox film 25; Greiner Bio-One, Germany) of 1 cm2 were exposed in close proximity to the biofilm sensor and were sampled at different phases of biofilm formation with 4 replicates per sampling date. These biofilm samples were fixed in 2% formaldehyde to analyze the cell density, cell size, and quantity of chlorophyll-containing cells by epifluorescence microscopy. Samples were stained with a 2 μg/ml DAPI (4′,6-diamidino-2-phenylindole dihydrochloride; Sigma-Aldrich, Germany) solution for 10 min, dip-rinsed in distilled water, and mounted on a glass slide using a coverslip. For quantification of the cell number, surface coverage, cell size, and chlorophyll content of the biofilm, 20 randomly chosen images were captured with an epifluorescence microscope (Axio Scope.A1; Carl Zeiss, Germany) equipped with a color charge-coupled device (CCD) camera (ProgRes CF; Jenoptik, Germany). The epifluorescence microscopy images were analyzed with the free software ImageJ 1.46e (National Institutes of Health, USA) (32). A stack of fluorescence micrographs of different heights of the reference samples were taken when the mature biofilm showed a complex 3D structure using the “Object counter3D” plugin.
During the experiments at Kiel Fjord, the physical seawater parameters temperature (T), pH, dissolved oxygen concentration (O2), and salinity (S) were measured continuously every 10 min (ProfiLux III; GHL, Germany). Water samples were taken weekly for analysis of chlorophyll a concentrations and nutrient levels of silicate [Si(OH4−)], nitrate (NO3−), and phosphate (PO43−) at a 2-m water depth at the institute pier at Kiel Fjord (data courtesy of A. Stuhr [GEOMAR, Kiel, Germany]). A summary of all data is given in in Table S1.1 in the supplemental material.
Initial attachment experiments.
During the first hours of marine bacterial adhesion, the fluorescence intensity of the biofilm sensor was automatically recorded every 15 min in August 2010. In three independent experiments, the bacterial cell density on the settling substrate of the biofilm sensor head was analyzed by epifluorescence microscopy after 6, 12, and 24 h.
Long-term detection of biofilm formation dynamics.
Biofilm formation was continuously analyzed for 18 to 21 days in October 2010, May 2011, and November 2011; here, an hourly recording time was chosen for the sensor, and replicate reference samples (n = 4) were taken daily. To investigate in more detail whether biofilm formation follows a diurnal rhythmic, water samples of 50 ml were taken and stored at −20°C for further analyses of background fluorescence intensity on 6 days at night (1 a.m.) and during the day (12 noon) in October 2010. At the same time, 20-ml seawater samples were fixed in 2% formaldehyde (final concentration) and stored at 4°C. Three aliquots of each sample were used to count the total number of planktonic cells by flow cytometry (FACSCalibur; Becton, Dickinson, USA).
Effect of seaweed on biofilm formation.
In June 2011, the biotic and abiotic effects of the marine macroalgae Fucus vesiculosus on biofilm formation were studied in independent outdoor mesocosms with a volume of 150 liters and a steady exchange with seawater. The biofilm accumulation on the sensor substrate was quantified hourly in close proximity (5 cm) to 10 plants of the macroalgae F. vesiculosus. In parallel, biofilm formation was quantified daily in mesocosms with or without Fucus vesiculosus by epifluorescence microscopy (n = 5). Additionally, biofilm samples for fluorescence in situ hybridization (FISH) analyses were fixed in 2% formaldehyde at 4°C overnight, rinsed twice in 1× phosphate-buffered saline (PBS; pH 7.8), and stored in 1× PBS and 96% ethanol (1:1) at −20°C until further processing. The basic community composition of the bacterial biofilm was determined by FISH using 5′-end fluorochrome Cy3-labeled oligonucleotide probes synthesized by biomers.net (Ulm, Germany). After dehydration, bacteria were hybridized with the oligonucleotide probes EUB338 I to EUB338 III (33), ALF968 (34), CF319a (35), and G Rb (36) and a negative-control probe, NON338 (37), for nonspecific binding. The probes EUB338 I to III stain all Eubacteria, whereas ALF968 stains Alphaproteobacteria and G Rb hybridize with Roseobacter and Rhodobacter. Cytophaga-Flavobacteria was detected with probe CF319a. Probes were hybridized in the dark for 3 h at 46°C in an equilibrated hybridization chamber, incubated in washing buffer at 48°C for 25 min, rinsed with distilled water, and air dried. For details, see reference 38. Samples were mounted in Citifluor (Citifluor, United Kingdom) and analyzed by epifluorescence microscopy. For the quantification of hybridized cells, between 800 and 1,300 DAPI-stained cells per microscope image were counted. All FISH data obtained were corrected by subtracting counts obtained from the negative-control NON338 probe. Bacterial counts of ALF968, CF319a, and G Rb hybridization were normalized to probes EUB338 I to III (39), and a program implemented in the software ImageJ 1.46e (National Institutes of Health, USA) (32) was built to concatenate the different steps automatically.
Data analysis.
Specific settling and growth rates for biofilm formation (including bacterial adhesion and growth) were calculated according to reference 40. From the fluorescence signals converted to cell numbers (N), the maximum specific growth rate (μ), defined as the slope at the inflection point (tip) of the exponential growth phase of biofilm formation, was calculated as . Here, A is ln[N(t)/N0], with N0 being the initial cell number and N(t) the cell number after time t.
The seawater samples for quantification of surrounding plankton community and background fluorescence were analyzed by Student's t test (SigmaPlot 11.2, Systat Software Inc., San Jose, CA). All data were normally distributed with homogeneous variances (Shapiro-Wilks test P > 0.65; equal variance test P > 0.194). One-factor analysis of variance (ANOVA) followed by Tukey's honestly significant difference (HSD) test was used to analyze differences in cell size. If some data did not fulfill criteria of normality or homoscedasticity, data were analyzed parametrically at a lower α level of 0.01 (41). For nonlinear curve-fitting using the Levenberg-Marquardt algorithm, Origin 8.0 Pro software (OriginLab, Northampton, MA, USA) was applied.
RESULTS
Initial attachment and biofilm growth.
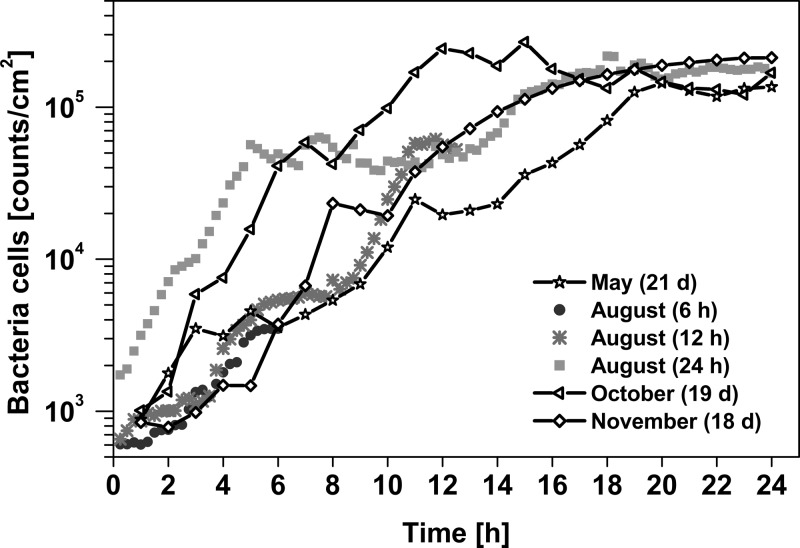
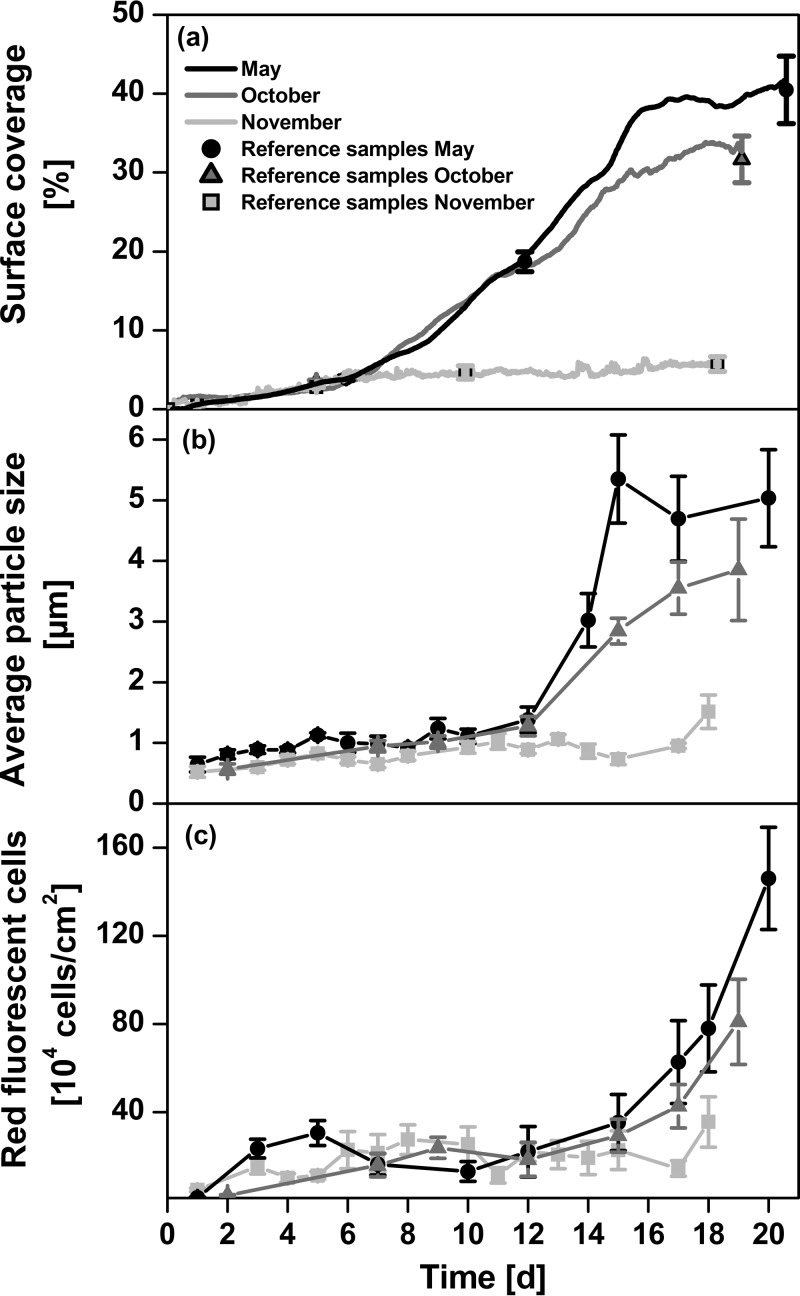
Figure 1 displays the computed bacterial cell numbers during the first hours of bacterial attachment. For the three experiments conducted over a longer period of time, the long-term behavior is illustrated in Fig. 2. Immediately after immersion bacterial colonization started and reached colonization rates (μ [h−1]) that were ≥0.25 and ≤0.49 (μ = 0.27 ± 0.09 h−1; mean ± standard deviation [SD]) in the first hours. In all experiments and independent of the time of year, this rapid initial attachment of bacterial cells to the surface stopped after approximately 18 h when bacterial densities reached 2 × 105 bacterial cells/cm2 on average. The fast initial attachment phase is followed by an adaptation phase, which is characterized by relatively low growth rates of bacterial biofilms (Fig. 2a). In May and October, biofilm growth started to significantly increase again after about 6 days. This exponential accumulation phase of biofilm formation was considerably reduced in November. In May and October, the final surface coverage reached values of 35 to 40%, whereas in November, even after 18 days, the surface coverage stayed at a level of about 5%. Within the first 12 days, biofilms were dominated by bacteria with average cell sizes of 0.6 to 1.2 μm (Fig. 2b). In May, after 12 days (one-way ANOVA; Tukey's HSD P = 0.0001; F = 60), and in October, after 15 days (P = 0.0046; F = 28), a significant increase in cell size to values up to 5 μm was observed. In November 2011, the average cell size remained small (<2 μm). Along with the increase in cell size, an increase in red fluorescence from chlorophyll-containing cells was observed (Fig. 2c) in the microscope images. These eukaryotic phototrophic organisms were predominantly diatoms, as determined by microscopy. Toward the end of the experimental period, diatoms and bacteriochlorophyll a-containing bacteria increased to a maximum of 104 to 105 cells/cm2. This is 1 to 2 orders of magnitude lower than the total cell counts (see Table S1.1 in the supplemental material).
FIG 1.
Accumulation of bacterial cells over the first 24 h of six independent settlement experiments in the Baltic Sea. The times in parentheses are the maximum experimental observation times of the biofilm.
FIG 2.
Long-term experiments on biofilm formation in the Baltic Sea in different seasons. (a) Compiled quasicontinuous data for surface coverage. A 24-h running mean was applied to smooth out short-term fluctuations. The single data points refer to the surface coverages observed for the DAPI-stained reference samples. These were used to determine the conversion factor between original biofilm sensor signal and surface coverage. (b and c) Average particle size (b) and number of red fluorescent cells (photosynthetic biofilm components, such as diatoms and bacteria) (c) of DAPI-stained biofilm communities. Error bars indicate standard errors of the means (n = 4).
Diurnal variations in biofilm formation.
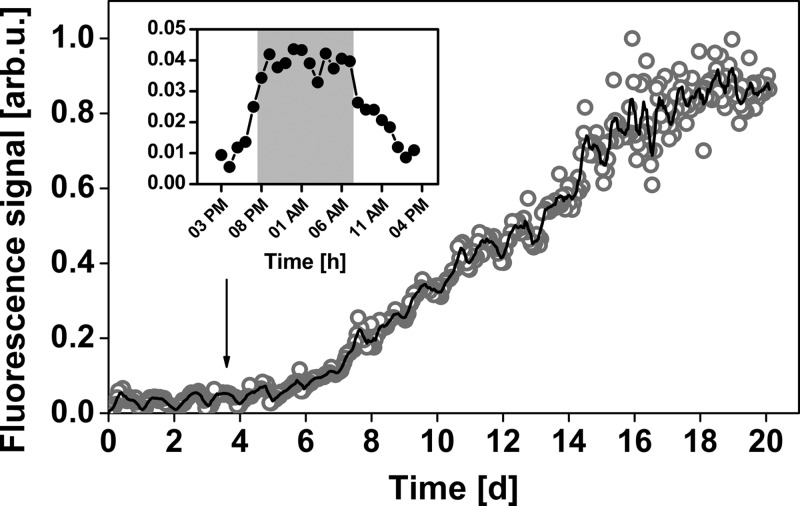
As a representative example of the capability of the biofilm sensor to detect fine-scale temporal fluctuations, Fig. 3 illustrates pronounced diurnal fluctuations of biofilm formation recorded in the Baltic Sea in October 2010. The fluorescence intensity was four times higher during the night than in the morning (Fig. 3, inset), corresponding to approximately 5 × 104 cells/cm2 during the night between days 3 and 4. Quantification of the surrounding plankton community by flow cytometry revealed no significant difference in cell densities between night and day (t test; n = 6; t = −0.634; P = 0.54), with cell densities (average ± standard error) of (1.5 ± 0.1) × 106 cells/ml during the day and (1.7 ± 0.2) × 106 cells/ml during the night. Moreover, there were no significant differences in background fluorescence intensity of the surrounding seawater between day and night (t test; n = 8; t = −0.701; P = 0.49).
FIG 3.
Normalized fluorescence signal of long-term observation of biofilm establishment in the Baltic Sea (open circles) and moving average over 6 h (solid curve). The inset shows a typical day cycle of the fluorescence signal. The gray background marks the hours of darkness. arb.u., arbitrary units.
Impact of the marine seaweed Fucus vesiculosus on biofilm formation.
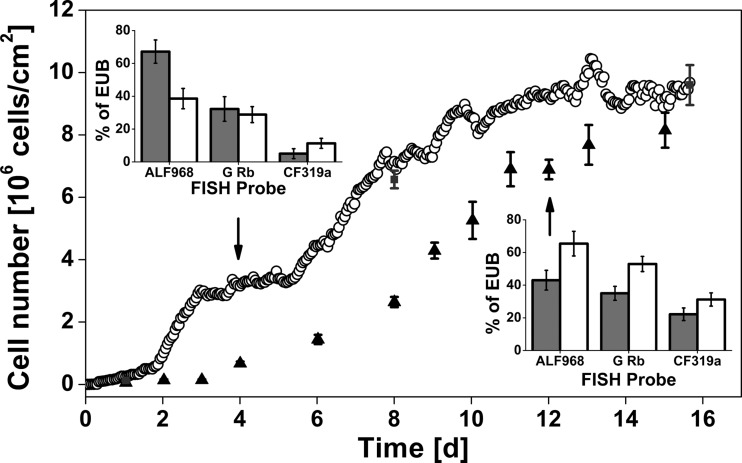
Figure 4 shows the comparison of cell density of a biofilm developed in association with the macroalga Fucus vesiculosus and without. The presence of the marine macroalga Fucus vesiculosus affected bacterial biofilm formation on artificial surfaces at two different stages. First, biofilm establishment under the influence of Fucus vesiculosus was characterized by a reduced induction phase of approximately 2 days compared to approximately 4 days of biofilms grown in the absence of alga. Second, in the presence of alga, biofilms reached their maximum specific growth rate of 0.28 day−1 after 6 days, while the maximum growth rate without alga was 0.32 day−1 and was reached later, after 9 days. In both experimental setups, the total cell density reached similar values, about 8 × 106 to 9 × 106 cells/cm2 after 16 days.
FIG 4.
Numbers of colonizing cells of biofilm growth associated with (open circles, quasicontinuous measurements) and not associated with (triangles, single measurements) the macroalga Fucus vesiculosus in Baltic Sea water. Analyses of cell density were carried out using DAPI-stained reference samples without algae (triangles) and with algae (gray squares). The insets illustrate dominant bacterial groups determined by FISH probes relative to the bacterial counts obtained by using the probe EUB338 after 4 (upper left) and 12 (lower right) days. Gray bars represent a biofilm community established near the macroalgae, and white bars represent one without algae. Error bars indicate standard errors of the mean (n = 4).
Dominating bacterial groups as determined by FISH are illustrated by the insets in Fig. 4. The relative abundance of Cytophaga-like bacteria and Alphaproteobacteria with its genera Rhodobacter and Roseobacter were determined by FISH after 4 and 12 days. Bacterial biofilm communities grown in the presence of Fucus vesiculosus were dominated by Alphaproteobacteria (68% ± 7%) after 4 days. Approximately 50% of these Alphaproteobacteria belonged to the genera Rhodobacter and/or Roseobacter. The relative abundance of Alphaproteobacteria was lower for biofilms grown in the absence of Fucus vesiculosus (39% ± 6%), but the proportion of Rhodobacter and/or Roseobacter cells was relative high and accounted for 70% of all Alphaproteobacteria. Moreover, there were no differences in the relative abundance of Rhodobacter and/or Roseobacter between the two treatments after a colonization time of 4 days. After 12 days, the relative abundance of Alphaproteobacteria decreased from 68% ± 7% to 43% ± 6% in biofilms grown in the presence of Fucus vesiculosus. In contrast, the relative abundance of Alphaproteobacteria increased to 65% ± 8% in biofilms grown without algae. This increase of Alphaproteobacteria was driven mainly by an increase of Rhodobacter and/or Roseobacter, accounting for more than 80% of all Alphaproteobacteria. However, the relative abundance of Rhodobacter and/or Roseobacter did not change between days 4 and 12 in the presence of macroalgae. The abundance of Cytophaga-like bacteria was higher in biofilms that were grown in the absence of algae irrespective of the sampling time. After 12 days, the relative abundance of Cytophaga-like bacteria increased in both treatments.
DISCUSSION
In the present study, an optical biofilm sensor was utilized to quasicontinuously analyze biofilm formation in situ. The sensor enabled the detection of short-term fluctuations, the initial attachment phase, and long-term trends of biofilm behavior. It was shown in a preceding publication (28) that these quantities scale linearly with respect to fluorescence intensity; however, for field measurements, calibration of the overall sensor response by a few reference samples is advised to extract the respective conversion factors and to ensure the linear response. Using the sensor in combination with microscope-assisted imaging, we investigated biofilm formation from its beginning to mature multispecies biofilms. The experiments revealed new insights into biofilm formation.
In all experiments we observed a rapid initial attachment, which was affected by neither season (Fig. 1) nor environment (Fig. 4). Most interesting was that the initial attachment phase was terminated after 18 h, when bacterial densities of approximately 2 × 105 cells/cm2 were reached. In contrast to laboratory studies on biofilm formation of selected strains (42), a smooth transition from initial attachment to colony formation and exponential growth was not observed in the marine environment. Instead, initial attachment was followed by a phase of low accumulation with homogeneously distributed solitary bacterial cells. It can be speculated that further colonization from the surrounding water was inhibited by already-adherent bacteria or that there was a steady state of attachment and detachment of bacteria, similar to the transient state of biofilm formation that has been described for Pseudoalteromonas (43). The first evidence for a transient state of biofilm formation within the first days of settlement is the observation of diurnal variations in biofilm densities. The fluorescence intensity detected by the biofilm sensor shows a clear short-term fluctuation between night and day times in two out of three experiments. One example is shown in Fig. 3. Variations in the fluorescence signal could not be explained by changes in the surrounding seawater or abiotic factors, such as seawater temperature. Bacterial counts in the surrounding water were not different, nor was a clear dependence of the sensor signal on water temperature changes observed (merely a 0.7°C difference between day and night in our experiments). We therefore attribute the observed fluctuation to a change of the established biofilm itself. However, we can only speculate that either differential settlement and detachment of bacterial cells to the surface increased grazing during the day (44) or changes in the metabolic state of bacteria (and hence potential changes of fluorescence quenching) caused or contributed to the observed pattern (45).
After the initial attachment phase, however, the adaptation phase could last for up to 4 days till the accumulation phase started, with an increased settlement of bacteria leading to rapid biofilm growth. In the presence of macroalgae, the adaptation phase was reduced compared to that on control surfaces from 4 to only 2 days. It can be suggested that a combination of factors (46) contribute to this shortening. Bacterial communities established on the artificial surface in the presence of Fucus vesiculosus were dominated by Alphaproteobacteria at the beginning of the accumulation phase. As the bacterial epibiotic community on the surface of Fucus vesiculosus, with 7.7 × 106 to 1.9 × 108 bacteria/cm2 (47), is dominated by Alphaproteobacteria (48, 49) as well, it can be hypothesized that the initial bacterial community is influenced by the presence of Fucus vesiculosus. Biofilm formation in the absence of seaweed on control surfaces was comparable to the colonization pattern on artificial surfaces in coastal marine environments described by Dang and Lovell (26). In both studies, Alphaproteobacteria contributed approximately 50% of the overall abundance of bacteria and were dominated by Rhodobacter and/or Roseobacter spp. Thus, the abundance of Rhodobacter and/or Roseobacter strains did not change under the influence of F. vesiculosus compared to control surfaces; the increase of Alphaproteobacteria in the presence of seaweed can be explained only by bacteria that are negative for the G Rb FISH probe. It can only be speculated that Sphingomonadales, another bacterial group frequently associated with the surface of F. vesiculosus, may have contributed to the increase of Alphaproteobacteria. In addition, seaweeds provide environments rich in bioavailable nutrients (50) and hence may support the primary colonization of Alphaproteobacteria. However, the impact of seaweed on biofilm formation regarding growth and support of Alphaproteobacteria got lost over a longer period of settlement. In fact, shortening the adaptation time in the presence of seaweed resulted in an earlier decrease of growth rate, leading to similar bacterial densities on treatment and control surfaces after 14 days. This reduction of biofilm growth was accompanied by a reduction of Alphaproteobacterial abundance.
After 6 to 12 days, marine biofilms were still dominated by bacteria and attained high growth rates, depending on environmental conditions and season (Fig. 2). Within the first 12 days of biofilm formation, the abundance of diatoms was relatively low, even though first attachment of diatoms was observed by microscopy after approximately 7 days. After 12 days, a dramatic increase of diatoms was detected and contributed together with bacteria to the development of a 3D biofilm.
Several mathematical models were proposed to describe biofilm formation dynamics that can range from common empirical correlations to complex two- and three-dimensional morphology algorithms (51). Generally, biofilm establishment can be described as a sigmoidal function consisting of three main phases, as defined by Bryers and Characklis (12). Based on empirical data from lab cultures, the Gompertz function turned out to be a suitable fit for bacterial growth (40). However, the observations in this study clearly demonstrate that in natural mixed-species ecosystems, the colonization of a surface does not follow a dynamics that can be described by a single Gompertz function but is better described as a sequence of several accumulation stages. Therefore, we modified Zwietering's description of the Gompertz function (40) to allow for multiple stages of biofilm formation:
Here, gi(t) is ln[N(t)/], representing the sigmoidal growth during the ith stage, with N(t) being the biofilm signal (cell count, coverage area, etc.) at time t and being the asymptotic signal level of the proceeding stage, and Ai is ln(/N0), the asymptote reached after stage i. The growth model contains biologically significant parameters, such as the maximum specific growth rate μi and the conditioning or induction time λi. As we observed three main stages of biofilm formation (initial attachment of pioneer bacteria, bacterial biofilm establishment, and attachment of unicellular microorganisms), the corresponding overall growth function, G(t), becomes
with A0 being equal to 0, since . This empirical marine biofilm model should be considered a basic mathematical description of marine biofilm formation with simplifications and limitations. For example, it does not take into account specific environmental conditions. Three sets of experimental biofilm sensor data were applied to validate the model; one example is shown in Fig. 5. The real-time observation data of the colonization sequence show a strong correlation (R2 = 0.998) to the model of marine biofilm establishment and agrees with previously published articles (11, 52). In our case, seven main phases of biofilm establishment were defined as visualized in Fig. 5. After following a biochemical preconditioning (Fig. 5, phase I), an initial attachment phase (phase II) takes place and commences the adaptation phase (phase III). In the exponential accumulation stage (phase IV), the bacterial biofilm grows continuously, and it reaches maximum bacterial cell density in phase V. Moreover, this phase is characterized by intense settlement of unicellular eukaryotic microorganisms like diatoms. The appearance of both phase VI, which represents the exponential accumulation stage of eukaryotic microorganisms, and the pseudostationary state of the final community structure in phase VII depends on the planktonic community as well as seasonal, local, and biological activity of the seawater. Images of characteristic stages of the aforementioned growth phases are shown in Fig. 5 as insets. Note that in natural marine ecosystems, all specified phases may not necessarily occur and that the emergence of each subsequent phase can overlap the asymptotic state of previous phases. For example, the biofilm formation in November showed only four phases, as the settlement of larger microorganisms and red fluorescence cells such as diatoms was insignificant (Fig. 2). Our biological observations and sensor responses conclude that a model with three distinct growth phases is capable of accurate description of biofilm formation dynamics in marine environments.
FIG 5.
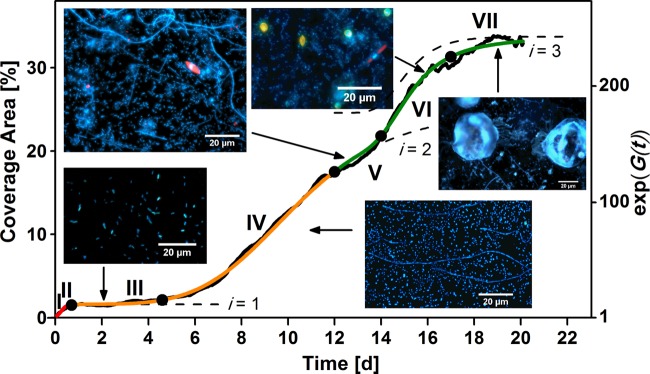
Model of marine biofilm growth in the Baltic Sea fitted by three consecutive Gompertz functions (i = 1 to 3) with respect to coverage area. It is compared with data from the biofilm sensor recorded in October (black line). The colored lines (red, orange, and green) represent the three main stages of biofilm establishment, whereas the numbers indicate the seven main phases (I to VII) of marine biofilm formation dynamics. The dashed curves depict the contributions of the three biofilm formation stages, exp(gi), where i is 1 to 3. The insets show images of biofilm microorganisms stained with DAPI at five stages of establishment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Annett Klemm (University of York), Thomas F. Krauss (University of York), and Tilmann Harder (University of New South Wales) for helpful discussions on the manuscript. In particular, we acknowledge the continued scientific support of Martin Wahl (GEOMAR), including valuable suggestions regarding the experimental design.
Financial support by the German Science Foundation (DFGEC 80) in the framework of the cluster of excellence “The Future Ocean” is gratefully acknowledged. The work of Tim Lachnit was supported by the German Science Foundation (DFG-LA 297 8/1-1). We also acknowledge the EPSRC of the UK (grant no. EP/J01771X/1, “Structured Light”) for financial support.
Footnotes
Published ahead of print 11 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00298-14.
REFERENCES
- 1.Flemming HC. 2009. Why microorganisms live in biofilms and the problem of biofouling. Mar. Ind. Biofouling 4:3–12. 10.1007/978-3-540-69796-1_1 [DOI] [Google Scholar]
- 2.Danhorn T, Fuqua C. 2007. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 61:401–422. 10.1146/annurev.micro.61.080706.093316 [DOI] [PubMed] [Google Scholar]
- 3.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920. 10.1126/science.1104816 [DOI] [PubMed] [Google Scholar]
- 4.Lachnit T, Meske D, Wahl M, Harder T, Schmitz R. 2011. Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 13:655–665. 10.1111/j.1462-2920.2010.02371.x [DOI] [PubMed] [Google Scholar]
- 5.Donlan RM. 2002. Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8:881–890. 10.3201/eid0809.020063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beech IB, Sunner J. 2004. Biocorrosion: towards understanding interactions between biofilms and metals. Curr. Opin. Biotechnol. 15:181–186. 10.1016/j.copbio.2004.05.001 [DOI] [PubMed] [Google Scholar]
- 7.Van Houdt R, Michiels CW. 2010. Biofilm formation and the food industry, a focus on the bacterial outer surface. J. Appl. Microbiol. 109:1117–1131. 10.1111/j.1365-2672.2010.04756.x [DOI] [PubMed] [Google Scholar]
- 8.Stoodley P, Wilson S, Hall-Stoodley L, Boyle JD, Lappin-Scott HM, Costerton JW. 2001. Growth and detachment of cell clusters from mature mixed-species biofilms. Appl. Environ. Microbiol. 67:5608–5613. 10.1128/AEM.67.12.5608-5613.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao D, Webb JS, Kjelleberg S. 2005. Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 71:1729–1736. 10.1128/AEM.71.4.1729-1736.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doiron K, Linossier I, Fay F, Yong J, Abd Wahid E, Hadjiev D, Bourgougnon N. 2012. Dynamic approaches of mixed species biofilm formation using modern technologies. Mar. Environ. Res. 78:40-47. 10.1016/j.marenvres.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 11.O'Toole G, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49–79. 10.1146/annurev.micro.54.1.49 [DOI] [PubMed] [Google Scholar]
- 12.Bryers JD, Characklis WG. 1982. Processes governing primary biofilm formation. Biotechnol. Bioeng. 24:2451–2476. 10.1002/bit.260241111 [DOI] [PubMed] [Google Scholar]
- 13.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95–108. 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- 14.Nadell CD, Xavier JB, Foster KR. 2009. The sociobiology of biofilms. FEMS Microbiol. Rev. 33:206–224. 10.1111/j.1574-6976.2008.00150.x [DOI] [PubMed] [Google Scholar]
- 15.Marshall KC, Stout R, Mitchell R. 1971. Mechanism of the initial events in the sorption of marine bacteria to surfaces. J. Gen. Microbiol. 68:337–348. 10.1099/00221287-68-3-337 [DOI] [Google Scholar]
- 16.Railkin A. 2004. Marine biofouling: colonization processes and defenses, p. 25–31 CRC Press, Boca Raton, FL [Google Scholar]
- 17.Wahl M. 1989. Marine epibiosis. I. Fouling and antifouling: some basic aspects. Mar. Ecol. Prog. Ser. 58:175–189 [Google Scholar]
- 18.Mieszkin S, Callow ME, Callow JA. 2013. Interactions between microbial biofilms and marine fouling algae: a mini review. Biofouling 29:1097–1113. 10.1080/08927014.2013.828712 [DOI] [PubMed] [Google Scholar]
- 19.Huws SA, McBain AJ, Gilbert P. 2005. Protozoan grazing and its impact upon population dynamics in biofilm communities. J. Appl. Microbiol. 98:238–244. 10.1111/j.1365-2672.2004.02449.x [DOI] [PubMed] [Google Scholar]
- 20.Dang H, Li T, Chen M, Huang G. 2008. Cross-ocean distribution of Rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl. Environ. Microbiol. 74:52–60. 10.1128/AEM.01400-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fletcher M, Loeb GI. 1979. Influence of substratum characteristics on the attachment of a marine pseudomonad to solid surfaces. Appl. Environ. Microbiol. 37:67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiu JMY, Thiyagarajan V, Tsoi MMY, Qian PY. 2005. Qualitative and quantitative changes in marine biofilms as a function of temperature and salinity in summer and winter. Biofilms 2:183–195. 10.1017/S147905050500195X [DOI] [Google Scholar]
- 23.Costerton JW. 1995. Overview of microbial biofilms. J. Ind. Microbiol. 15:137–140. 10.1007/BF01569816 [DOI] [PubMed] [Google Scholar]
- 24.Lee J-W, Nam J-H, Kim Y-H, Lee K-H, Lee D-H. 2008. Bacterial communities in the initial stage of marine biofilm formation on artificial surfaces. J. Microbiol. 46:174–182. 10.1007/s12275-008-0032-3 [DOI] [PubMed] [Google Scholar]
- 25.Dang H, Lovell CR. 2000. Bacterial primary colonization and early succession on surfaces in marine waters as determined by amplified rRNA gene restriction analysis and sequence analysis of. Appl. Environ. Microbiol. 66:467–475. 10.1128/AEM.66.2.467-475.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang H, Lovell CR. 2002. Numerical dominance and phylotype diversity of marine Rhodobacter species during early colonization of submerged surfaces in coastal marine waters as determined by 16S ribosomal DNA sequence. Appl. Environ. Microbiol. 68:496–504. 10.1128/AEM.68.2.496-504.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dang H, Lovell CR. 2002. Seasonal dynamics of particle-associated and free-living marine Proteobacteria in a salt marsh tidal creek as determined using fluorescence in situ hybridization. Environ. Microbiol. 4:287–295. 10.1046/j.1462-2920.2002.00295.x [DOI] [PubMed] [Google Scholar]
- 28.Fischer M, Wahl M, Friedrichs G. 2012. Design and field application of a UV-LED based optical fiber biofilm sensor. Biosens. Bioelectron. 33:172–178. 10.1016/j.bios.2011.12.048 [DOI] [PubMed] [Google Scholar]
- 29.Fischer M, Wahl M, Friedrichs G. 2013. Field sensor for in situ detection of marine bacterial biofilms. Sea Technol. 54:49–52 [Google Scholar]
- 30.Dalton HM, Goodman AE, Marshall KC. 1996. Diversity in surface colonization behavior in marine bacteria. J. Ind. Microbiol. Biotechnol. 17:228–234. 10.1007/BF01574697 [DOI] [Google Scholar]
- 31.Elasri MO, Miller RV. 1999. Study of the response of a biofilm bacterial community to UV radiation. Appl. Environ. Microbiol. 65:2025–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amann RI, Krumholz L, Stahl DA. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glöckner FO, Fuchs BM, Amann R. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 142:1097–1106. 10.1099/13500872-142-5-1097 [DOI] [PubMed] [Google Scholar]
- 36.Eilers H, Pernthaler J, Glöckner FO, Amann R. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044–3051. 10.1128/AEM.66.7.3044-3051.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallner G, Amann R, Beisker W. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136–143. 10.1002/cyto.990140205 [DOI] [PubMed] [Google Scholar]
- 38.Loy A, Maixner F, Wagner M, Horn M. 2007. probeBase—an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Res. 35:800–804. 10.1093/nar/gkl856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouvier T, Del Giorgio PA. 2003. Factors influencing the detection of bacterial cells using fluorescence in situ hybridization (FISH): a quantitative review of published reports. FEMS Microbiol. Ecol. 44:3–15. 10.1016/S0168-6496(02)00461-0 [DOI] [PubMed] [Google Scholar]
- 40.Zwietering MH, Jongenburger I, Rombouts FM, van 't Riet K. 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56:1875–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Underwood AJ. 1997. Experiments in ecology: their logical design and interpretation using analysis of variance. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 42.Thormann KM, Saville RM, Shukla S, Pelletier DA, Spormann AM. 2004. Initial phases of biofilm formation in Shewanella oneidensis MR-1. J. Bacteriol. 186:8096–8104. 10.1128/JB.186.23.8096-8104.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140–1154. 10.1128/jb.184.4.1140-1154.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huws SA, McBain AJ, Gilbert P. 2005. Protozoan grazing and its impact upon population dynamics in biofilm communities. J. Appl. Microbiol. 98:238–244. 10.1111/j.1365-2672.2004.02449.x [DOI] [PubMed] [Google Scholar]
- 45.Vivian JT, Callis PR. 2001. Mechanisms of tryptophan fluorescence shifts in proteins. Biophys. J. 80:2093–2109. 10.1016/S0006-3495(01)76183-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lam C, Harder T. 2007. Marine macroalgae affect abundance and community richness of bacterioplankton in close proximity 1. J. Phycol. 43:874–881. 10.1111/j.1529-8817.2007.00385.x [DOI] [Google Scholar]
- 47.Wahl M, Shahnaz L, Dobretsov S, Saha M, Symanowski F, David K, Lachnit T, Vasel M, Weinberger F. 2010. Ecology of antifouling resistance in the bladder wrack Fucus vesiculosus: patterns of microfouling and antimicrobial protection. Mar. Ecol. Prog. Ser. 411:33–48. 10.3354/meps08644 [DOI] [Google Scholar]
- 48.Lachnit T, Fischer M, Künzel S, Baines JF, Harder T. 2013. Compounds associated with algal surfaces mediate epiphytic colonization of the marine macroalga Fucus vesiculosus. FEMS Microbiol. Ecol. 84:411–420. 10.1111/1574-6941.12071 [DOI] [PubMed] [Google Scholar]
- 49.Lachnit T, Wahl M, Harder T. 2010. Isolated thallus-associated compounds from the macroalga Fucus vesiculosus mediate bacterial surface colonization in the field similar to that on the natural alga. Biofouling 26:247–255. 10.1080/08927010903474189 [DOI] [PubMed] [Google Scholar]
- 50.Delille D, Marty G, Cansemi-Soullard M, Frankignoulle M. 1997. Influence of subantarctic Macrocystis bed metabolism in diel changes of marine bacterioplankton and CO2 fluxes. J. Plankton Res. 19:1251–1264. 10.1093/plankt/19.9.1251 [DOI] [Google Scholar]
- 51.Monds RD, O'Toole GA. 2009. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 17:73–87. 10.1016/j.tim.2008.11.001 [DOI] [PubMed] [Google Scholar]
- 52.Dobretsov S. 2010. Marine biofilms, p 123–133 In Dürr S, Thomason JC. (ed), Biofouling. Wiley-Blackwell, Oxford, United Kingdom [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.