Abstract
Members of the genus Bifidobacterium are commonly found in the gastrointestinal tracts of mammals, including humans, where their growth is presumed to be dependent on various diet- and/or host-derived carbohydrates. To understand transcriptional control of bifidobacterial carbohydrate metabolism, we investigated two genetic carbohydrate utilization clusters dedicated to the metabolism of raffinose-type sugars and melezitose. Transcriptomic and gene inactivation approaches revealed that the raffinose utilization system is positively regulated by an activator protein, designated RafR. The gene cluster associated with melezitose metabolism was shown to be subject to direct negative control by a LacI-type transcriptional regulator, designated MelR1, in addition to apparent indirect negative control by means of a second LacI-type regulator, MelR2. In silico analysis, DNA-protein interaction, and primer extension studies revealed the MelR1 and MelR2 operator sequences, each of which is positioned just upstream of or overlapping the correspondingly regulated promoter sequences. Similar analyses identified the RafR binding operator sequence located upstream of the rafB promoter. This study indicates that transcriptional control of gene clusters involved in carbohydrate metabolism in bifidobacteria is subject to conserved regulatory systems, representing either positive or negative control.
INTRODUCTION
Bifidobacteria are Gram-positive, anaerobic bacteria that possess a high GC genome content and belong to the phylum Actinobacteria. Bifidobacteria typically exhibit a forked (bifid) morphology and are commonly found as anaerobic commensals of the gastrointestinal tract (GIT) (1, 2). Bifidobacteria were first isolated as natural inhabitants of the gastrointestinal tracts of humans and mammals by Tissier (3), who was also the first to recognize their probiotic or health-promoting potential (4). Consistent with Tissier's observations, it is known that members of certain bacterial genera, such as bifidobacteria and lactobacilli, provide protection against certain types of gastrointestinal infections (5, 6).
It is also becoming increasingly apparent that a “normal” (or healthy) colonic microbiota may reduce the risk and/or pathology of gastrointestinal diseases and disorders, such as ulcerative colitis, bowel cancer, and pseudomembranous colitis (7–9). Several recent studies have focused on the use of prebiotics, which are nondigestible food ingredients that beneficially affect the host by selectively stimulating growth and/or activity of one or a limited number of beneficial bacteria in the colon (10), thereby improving host health. Certain carbohydrates, including whole-grain wheat, fructo-oligosaccharides, galacto-oligosaccharides, and lactulose, have been shown to exert prebiotic effects (11–14).
Metagenomic analyses of the human gut microbiota have generated knowledge that may allow the rational selection of novel prebiotics that maintain and/or enhance a healthy gut microbiota (15). Integral to the selection of carbohydrates with prebiotic potential is an in-depth understanding of carbohydrate metabolism by members of the gut microbiota. In this respect, bifidobacteria possess a unique hexose metabolism, the bifid shunt, a metabolic pathway that employs the signature enzyme fructose 6-phosphate phosphoketolase (F6PPK), which converts carbohydrates to mainly acetic and lactic acids (16).
Our recent work has established that Bifidobacterium breve UCC2003, an isolate from nursling stool, can metabolize a remarkable range of mono-, di-, oligo-, and polysaccharides (17–23). Bifidobacterial gene clusters associated with carbohydrate metabolism have in several cases been shown to be regulated by LacI-type repressors, representing LacI/GalR family proteins, which typically consist of an N-terminal helix-turn-helix (HTH) DNA-binding motif and a C-terminal domain for oligomerization and effector binding (24, 25). Furthermore, several observations have been made suggesting that such clusters are subject to carbon catabolite control (26).
Recently, we identified a raf gene cluster dedicated to the metabolism of the plant-derived carbohydrates raffinose [α-d-Galp-(1→6)-α-d-Glcp-(1→2)β-d-Fruf], stachyose [α-d-Galp-(1→6)-α-d-Galp-(1→6)-α-d-Glcp-(1→2)α-d-Fruf], and melibiose [α-d-Galp-(1→6)-α-d-Glcp] in B. breve UCC2003 (20). The raf locus in the B. breve UCC2003 genome is located adjacent to the mel gene cluster, which allows the strain to metabolize the trisaccharide melezitose [α-d-Glcp-(1→3)-β-d-Fruf-(2→1)-α-d-Glcp], found in honeydew and manna, which are sugar-rich liquid and solid deposits, respectively, associated with leaves and branches of various trees and shrubs (20, 27). It was initially hypothesized that melezitose is an oligosaccharide that is naturally present in various plants (28). However, it was later found that certain insects are responsible for melezitose production, when they use carbohydrates present in tree sap, by itself devoid of melezitose, to form honeydew (27). In the current work, we investigated the transcriptional regulation of the gene clusters associated with the metabolism of the raffinose family sugars and melezitose.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are detailed in Table 1. Bifidobacteria were routinely cultured in either de Mann Rogosa and Sharpe medium (MRS) (Difco, BD, Le Pont de Claix, France) supplemented with 0.05% cysteine-HCl or reinforced clostridial medium (RCM) (Oxoid Ltd.). Carbohydrate utilization by bifidobacterial strains was examined in modified de Mann Rogosa and Sharpe medium (mMRS) prepared from first principles (29) and to which, prior to inoculation, cysteine-HCl (0.05% [wt/vol]) and a particular carbohydrate source (1% [wt/vol]) had been added. The carbohydrates used were raffinose, stachyose, melibiose, melezitose, and lactose (all purchased from Sigma-Aldrich, Steinheim, Germany). Bifidobacterial cultures were incubated at 37°C under anaerobic conditions, which were maintained using an Anaerocult oxygen-depleting system (Merck, Darmstadt, Germany) in an anaerobic chamber with an atmosphere of 5% CO2, 5% H2, 90% N2. Lactococcus lactis strains were cultivated in M17 broth containing 0.5% glucose (30) at 30°C. Escherichia coli strains were cultured in Luria-Bertani broth (LB) (31) at 37°C with agitation. Where appropriate, growth media contained chloramphenicol (Cm) (5 μg ml−1 for L. lactis, 10 μg ml−1 for E. coli, and 2.5 μg ml−1 for B. breve), erythromycin (Em) (100 μg ml−1 for E. coli), tetracycline (Tet) (10 μg ml−1 for E. coli or B. breve), kanamycin (Km) (50 μg ml−1 for E. coli), or ampicillin (Amp) (100 μg ml−1 for E. coli).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli | ||
| EC101 | Cloning host; repA+ kmr | 44 |
| EC101-pNZ-M.Bbrll+Bbr111 | EC101 harboring a pNZ8048 derivative containing BbrllM and BbrlllM proteins | 40 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| L. lactis | ||
| NZ9000 | MG1363, nisin-inducible overexpression host; pepN::nisRK | 62 |
| NZ9700 | Nisin-producing strain | 62 |
| B. breve | ||
| UCC2003 | Isolate from nursling stool | 19 |
| UCC2003-melR1 | pORI19-tet-mel1 insertion mutant of UCC2003 | This study |
| UCC2003-melR2 | pORI19-tet-melR2 insertion mutant of UCC2003 | This study |
| UCC2003-rafR | pORI19-tet-rafR insertion mutant of UCC2003 | This study |
| Plasmids | ||
| pORI19 | Emr ΔrepA ori+; cloning vector | 44 |
| pORI19-tet-melR1 | Internal 421-bp fragment of melR1 and tetW cloned in pORI19 | This study |
| pORI19-tet-melR2 | Internal 456-bp fragment of melR2 and tetW cloned in pORI19 | This study |
| pORI19-tet-rafR | Internal 331-bp fragment of rafR and tetW cloned in pORI19 | This study |
| pAM5 | pBC1-puC19-Tcr | 45 |
| pNZ8048 | Cmr; nisin-inducible translational-fusion vector | 62 |
| pNZ8150 | Cmr; nisin-inducible translational-fusion vector | 42 |
| PQE30 | Ampr; IPTG-inducible vector | Qiagen |
| pNZ-MelR1-His | melR1 with His tag cloned downstream of nisin-inducible promoter on pNZ8048 | This study |
| pnz-MelR2-His | melR2 with His tag cloned downstream of nisin-inducible promoter on pNZ8150 | This study |
| pQE30-RafR | rafR cloned into the IPTG-inducible vector PQE30 | This study |
In order to determine bacterial growth profiles and final optical densities, 5 ml of freshly prepared mMRS including a particular carbohydrate (see above) was inoculated with 50 μl (1%) of a stationary-phase culture of a particular B. breve strain. Uninoculated mMRS was used as a negative control. The cultures were incubated anaerobically at 37°C for 16 h, and the optical density at 600 nm (OD600) was determined at 30-min intervals using a Powerwave microplate spectrophotometer (BioTek Instruments, Inc., USA) in conjunction with Gen5 Microplate software for Windows.
Nucleotide sequence analysis.
Sequence data were obtained from the Artemis-mediated (32) genome annotations of B. breve UCC2003 (33). Database searches were performed using nonredundant sequences accessible at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) using BLAST (34, 35). Sequence verification and analysis were performed using the Seqman and Seqbuilder programs of the DNASTAR software package v10.1.2 (DNASTAR, Madison, WI, USA). Operator sequences for RafR and MelR1/2 in other bifidobacterial species were detected using MEME (36), and consensus sequences were visualized using Weblogo (37). Regions 250 bp upstream and 50 bp downstream of the translation start site of B. breve UCC2003 genes were searched for additional RafR- and LacI-type binding sites using HMMSEARCH (38) with models built from all detected bifidobacterial RafR recognition sites and from known bifidobacterial LacI-type repressor binding motifs (18, 22). Putative binding sites present in the upstream regions of genes in the vicinity of genes coding for predicted LacI-type regulators were extracted and used to build an improved model to find LacI-type repressor binding sites in B. breve UCC2003. Genes coding for LacI-type regulators were searched by screening the encoded proteins for the presence of the PF00356 hidden Markov model (HMM). Distances between binding sites were determined using ClustalW.
DNA manipulations.
Chromosomal DNA was isolated from bifidobacteria as previously described (39). Minipreparation of plasmid DNA from E. coli or L. lactis was achieved using the Qiaprep spin plasmid miniprep kit (Qiagen GmBH, Hilden, Germany). For L. lactis, an initial lysis step was incorporated into the plasmid isolation procedure by resuspending cells in lysis buffer supplemented with lysozyme (30 mg ml−1), followed by incubation at 37°C for 30 min (31). Procedures for DNA manipulations were performed essentially as described previously. Restriction enzymes and T4 DNA ligase were used according to the supplier's instructions (Roche Diagnostics, East Sussex, United Kingdom). The synthetic single-stranded oligonucleotide primers used in this study, detailed in Table S1 in the supplemental material, were synthesized by Eurofins (Ebersberg, Germany). Standard PCRs were performed using TaqPCR mastermix (Qiagen), while B. breve colony PCRs were performed as described previously (40). PCR fragments were purified using the Qiagen PCR purification kit (Qiagen). Electroporation of plasmid DNA into E. coli was performed as previously described (31). Electrotransformation of B. breve UCC2003 (19) and L. lactis (41) was performed according to published protocols. The correct orientation and integrity of all constructs were verified by DNA sequencing, performed by Eurofins (Ebersberg, Germany).
Construction of plasmids pNZ-MelR1-His, pNZ-MelR2-His, and pQE30-RafR.
DNA fragments containing the complete coding regions of Bbr_1864 (here designated melR1) and Bbr_1863 (here designated melR2) were generated by PCR amplification employing chromosomal DNA of B. breve UCC2003 as a template, employing PFU Ultra DNA polymerase (Agilent Technologies) and primer combinations melR1EcorVF and melR1Xba1R, and melR2Nco1F and melR2Xba1R, respectively (see Table S1 in the supplemental material). NcoI or EcoRV and XbaI restriction sites were incorporated at the 5′ ends of each forward and reverse primer combination, respectively, for the melR1- and melR2-encompassing primers (see Table S1 in the supplemental material). In addition, an in-frame His10-encoding sequence was incorporated into each of the forward primers to facilitate downstream protein purification using the Ni-nitrilotriacetic acid (NTA) affinity system (Qiagen). The melR1- and melR2-encompassing amplicons were digested with NcoI/EcoRV and XbaI and ligated into the NcoI/ScaI and XbaI-digested nisin-inducible translational fusion plasmids pNZ8048 and pNZ8150, respectively (42). The ligation mixtures were introduced into L. lactis NZ9000 (Table 1) by electrotransformation, and transformants were then selected based on chloramphenicol resistance (Cmr) (5 μg ml−1 for L. lactis). The plasmid contents of a number of Cmr transformants were screened by restriction analysis, and the integrity of positively identified recombinant melR1- or melR2-containing plasmids was verified by sequencing, resulting in constructs pNZ-MelR1-His and pNZ-MelR2-His, respectively.
The coding region of the predicted repressor open reading frame kinase (ROK)-type regulator-encoding gene Bbr_1868 (here designated rafR) was PCR amplified by employing Taq DNA polymerase and chromosomal DNA of B. breve UCC2003 as a template and the primer combination ROKBglIIF and ROKPst1R, which had BglII and PstI restriction sites incorporated at the 5′ ends of the forward and reverse primers, respectively (see Table S1 in the supplemental material). The rafR-encompassing fragment generated was digested with BglII and PstI and ligated to similarly digested pQE30 (Qiagen), an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible protein expression plasmid, which allows incorporation of a His tag into the N terminus of the expressed protein target. The ligation mixture was introduced into E. coli XL1-Blue (Table 1) by electrotransformation, and transformants were selected based on ampicillin and tetracycline resistance. The plasmid contents of a number of Ampr and Tetr transformants were screened by restriction analysis, and the integrity of a positively identified recombinant plasmid, designated pQE30-RafR, was verified by sequencing.
Protein (over)production and purification.
For the (over)production of MelR1 and MelR2, 25 ml of M17 broth supplemented with 0.5% glucose was inoculated with a 2% inoculum of L. lactis NZ9000 harboring either pNZ-MelR1-His, pNZ-MelR2-His, or the empty vector pNZ8048 (used as a negative control), followed by incubation at 30°C until an OD600 of 0.5 was reached, at which point protein expression was induced by the addition of filter-sterilized, nisin-containing, cell-free supernatant of L. lactis NZ9700, followed by continued incubation at 30°C for 90 min. Cells were harvested, resuspended in 10 mM Tris-HCl buffer (pH 7.0), and disrupted with glass beads in a mini-bead beater (BioSpec Products, Bartlesville, OK). Cellular debris was removed by centrifugation to produce a crude cell extract. Although protein purification of MelR1-His and MelR2-His was achieved using His tag affinity chromatography, the purification procedure appeared to render the proteins inactive in subsequent electrophoretic mobility shift assays (EMSAs) (results not shown). For this reason, crude cell extracts, prepared in a 10 mM Tris-HCl lysis buffer (pH 7.0), were adopted for the EMSAs (see below).
In the case of RafR, a 300-ml volume of LB supplemented with tetracycline and ampicillin was inoculated with 6 ml of an overnight culture of E. coli XL1-Blue cells harboring pQE30-RafR and then incubated at 37°C. At an OD600 of 0.5, expression of RafR was induced by the addition of 1 mM IPTG (Roche Diagnostics Ltd., West Sussex, United Kingdom). Following 2 h of incubation in the presence of IPTG, cells were harvested by centrifugation, and crude cell extracts for protein purification were prepared as described above. Subsequent protein purification was performed using a PrepEase kit for His-tagged protein purification (USB, Germany). Elution fractions were analyzed by SDS-polyacrylamide gel electrophoresis, as described previously (43), on a 12.5% polyacrylamide gel. Following electrophoresis, the gels were fixed and stained with Coomassie brilliant blue to identify fractions containing the purified protein. Prestained Rainbow low-molecular-weight protein marker (New England BioLabs, Herefordshire, United Kingdom) was used to estimate the molecular weight of the expressed and/or purified proteins.
Construction of B. breve insertion mutant strains.
Internal fragments of rafR (528 bp, representing codons 88 through 264 out of the 403 codons of the gene), melR1 (380 bp, representing codons 107 through 234 out of the 340 codons of the gene), and melR2 (402 bp, representing codons 93 through 227 out of the 343 codons of the gene) were amplified by PCR using B. breve UCC2003 chromosomal DNA as the template and the oligonucleotide primer combinations rafRfHd3in and rafRrxbaIin, melR1fHd3in and melR1rxba1in, and melR2fHd3in and melR2rxba1in, respectively (see Table S1 in the supplemental material). The PCR products generated were ligated to pORI19, an Ori+ RepA− integration plasmid (44), using HindIII and XbaI restriction sites that were incorporated into the primers for the partial rafR-, melR1-, and melR2-encompassing amplicons, and introduced into E. coli EC101 by electroporation. Recombinant E. coli EC101 derivatives containing pORI19 constructs were selected on LB agar containing Em and supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-d-galactopyranoside; 40 μg ml−1) and 1 mM IPTG.
The expected genetic structures of the recombinant plasmids pORI19-rafR (pORI19 containing an internal 528-bp fragment of the rafR gene), pORI19-melR1 (pORI19 containing an internal 402-bp fragment of the melR1 gene), and pORI19-melR2 (pORI19 containing an internal 380-bp fragment of the melR2 gene) were confirmed by restriction mapping prior to subcloning of the Tet resistance antibiotic cassette, tetW, from pAM5 (45) as a SacI fragment into the unique SacI site present on each of the pORI19 derivatives. The orientation of the tetracycline resistance gene in each of the resulting plasmids, pORI19-tet-rafR, pORI19-tet-melR1, and pORI19-tet-melR2, was determined by restriction analysis. The plasmids were subsequently introduced into E. coli EC101 pNZ-MBbrI-MBbrII (40) (transformants were selected based on Cm and Tet resistance) in order to methylate the plasmid constructs before introduction into B. breve UCC2003.
Methylation of the plasmid complement of the obtained transformants in EC101 pNZ-MBbrI-MBbrII was confirmed by their observed resistance to PstI restriction (40). Plasmid preparations of methylated pORI19-tet-rafR, pORI19-tet-melR1, and pORI19-tet-melR2 were introduced by electroporation into B. breve UCC2003 with subsequent selection on reinforced clostridial agar (RCA) plates supplemented with Tet. Insertion mutants resulting from site-specific homologous recombination were initially confirmed by colony PCR targeting the tetracycline resistance gene, tetW, followed by a second confirmatory PCR adopting a tetW-based primer, either forward or reverse, depending on the orientation of tetW, in combination with a primer specific for each targeted gene to confirm integration at the correct chromosomal position (see Table S1 in the supplemental material). In this case, a product of a particular size would be obtained only if the correct gene disruption had been achieved. In this manner, mutants were obtained carrying chromosomal insertions in either the rafR, melR1, or melR2 gene, resulting in strains B. breve UCC2003-rafR, B. breve UCC2003-melR1, and B. breve UCC2003-melR2, respectively.
Analysis of global gene expression using B. breve DNA microarrays.
Global gene transcription was determined by microarray analysis during growth of B. breve UCC2003-rafR, B. breve UCC2003-melR1, and B. breve UCC2003-melR2 in mMRS supplemented with ribose, and these transcriptomes were compared to that obtained from B. breve UCC2003 wild-type cells that had also been grown on ribose as the sole carbohydrate source. DNA microarrays containing oligonucleotide primers representing each of the 1,864 annotated genes on the genome of B. breve UCC2003 were designed by and obtained from Agilent Technologies (Palo Alto, CA, USA). Cell disruption, RNA isolation, RNA quality control, and cDNA synthesis and labeling were performed as described previously (46). The labeled cDNA was hybridized using the Agilent Gene Expression hybridization kit (part number 5188-5242) as described in the Agilent Two-Color Microarray-Based Gene Expression Analysis v4.0 manual (G4140-90050). Following hybridization, the microarrays were washed in accordance with Agilent's standard procedures and scanned using an Agilent DNA microarray scanner (model G2565A). The generated scans were converted to data files with Agilent's Feature Extraction software (v9.5). The DNA microarray data were processed as previously described (47–49). Differential-expression tests were performed with the Cyber-T implementation of a variant of the t test (50). A gene was considered to exhibit significantly different expression at a P value of <0.001 and an expression ratio of >3 or <0.33 relative to the control.
EMSA.
DNA fragments representing different portions of the promoter regions upstream the rafA, rafB, Bbr_1862, and melA genes were prepared by PCR using IRD800-labeled primer pairs (MWG Biotech) (see Table S2 in the supplemental material). EMSAs were performed essentially as described previously (51). In all cases, the binding reactions were performed in a final volume of 20 μl in the presence of poly(dI-dC) in binding buffer (20 mM Tris-HCl, 5 mM MgCl2, 0.5 mM dithiothreitol [DTT], 1 mM EDTA, 200 mM KCl, 10% glycerol at pH 7.0). Various amounts of a crude cell extract containing either MelR1 or MelR2, or purified RafR, were mixed on ice with a fixed amount of DNA probe (0.1 pmol) and subsequently incubated for 30 min at 37°C. Samples were loaded onto a 6% nondenaturing phosphonoacetic acid (PAA) gel prepared in TAE buffer (40 mM Tris acetate [pH 8.0], 2 mM EDTA) and run in a 0.5× to 2.0× gradient of TAE at 100 V for 90 min in an Atto Mini PAGE system (Atto Bioscience and Biotechnology, Tokyo, Japan). Signals were detected using an Odyssey Infrared Imaging System (Li-Cor Biosciences United Kingdom Ltd., Cambridge, United Kingdom), and images were captured using the supplied Odyssey software v3.0.
Primer extension analysis.
Total RNA was isolated from exponentially growing cells of B. breve UCC2003 or B. breve UCC2003-melR2 cultured in mMRS supplemented with 1% raffinose or melezitose for B. breve UCC2003 or in mMRS supplemented with 1% ribose for B. breve UCC2003-melR2 (52). Primer extension was performed by annealing 1 pmol of an IRD800-labeled synthetic oligonucleotide to 20 μg of RNA, as previously described (53), using primers rafAPERP1, rafAPERP2, rafBPERP1, rafBPERP2, melAPERP1, melAPERP1, 1862PERP1, and 1862PERP1 (see Table S2 in the supplemental material). Sequence ladders of the presumed rafA, rafB, melA, and Bbr_1862 promoter regions were produced using the same primer as in the primer extension reaction and employing a DNA cycle-sequencing kit (Jena Bioscience, Germany) and were run alongside the primer extension products to allow precise alignment of the transcriptional start site with the corresponding DNA sequence. Separation was achieved on a 6.5% Li-Cor Matrix KB Plus acrylamide gel. Signal detection and image capture were performed with a Li-Cor sequencing instrument (Li-Cor Biosciences).
Microarray data accession number.
The microarray data obtained were deposited in NCBI's Gene Expression Omnibus (GEO) and are accessible through GEO series accession number GSE50211.
RESULTS AND DISCUSSION
Transcriptomes of B. breve UCC2003 mutants carrying disruptions in melR1, melR2, or rafR.
In a previous study (20), we investigated the metabolism of the raffinose-related carbohydrates raffinose, stachyose, and melibiose by B. breve UCC2003 and also observed the presence of this cluster in other bifidobacteria. That study identified a gene cluster, comprised of rafA, rafR, rafB, rafC, and rafD, involved in the utilization of raffinose-related sugars, where rafA was shown to encode an α-galactosidase and rafR a putative ROK-type transcriptional regulator, while the adjacent rafBCD genes specified a putative ABC-type sugar uptake system (Fig. 1). The same study also identified a gene cluster in the B. breve UCC2003 genome, located in the immediate vicinity of the raf gene cluster, dedicated to melezitose metabolism and comprised of melA, melB, melC, melD, and melE, with melABC specifying a presumed ABC-type sugar uptake system and melD and melE encoding an α-glucosidase and an α-galactosidase, respectively (Fig. 1), and again also observed the presence of the cluster in other bifidobacteria (20). Located between the aforementioned raf and mel genes are four genes, two of which, designated melR1 and melR2, encode putative LacI-type transcriptional regulators (Fig. 1).
FIG 1.
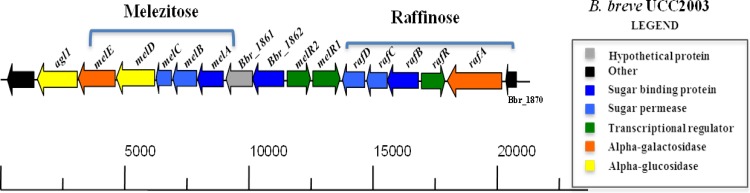
Representation of the raffinose and melezitose utilization operons of B. breve UCC2003.
We previously (20) demonstrated that transcription of the melABCDE and rafABCD genes is induced by the presence of melezitose and raffinose (or raffinose-like sugars), indicative of substrate-specific transcriptional regulation. In order to identify and investigate the regulatory players that control the raffinose and melezitose-induced genes, insertion mutations were made in the melR1, melR2, and rafR genes (see above) (Fig. 1), resulting in strains B. breve UCC2003-melR1, B. breve UCC2003-melR2, and UCC2003-rafR, respectively (see Materials and Methods). It was hypothesized that if any of the putative regulatory genes encoded a repressor, mutation of the gene would lead to increased transcription even in the absence of the inducing carbohydrate.
Altered gene expression patterns were determined by DNA microarray analysis from two independent biological replicates and employing B. breve UCC2003, B. breve UCC2003-melR1, B. breve UCC2003-melR2, or UCC2003-rafR cultures grown to the exponential growth phase in mMRS supplemented with ribose. The transcriptomes obtained revealed that, compared to B. breve UCC2003, the melABCDE genes were significantly upregulated (>5.0-fold change; P < 0.001) in B. breve UCC2003-melR1, while melABCDE, as well as its adjacent genes, Bbr_1861 and Bbr_1862, were upregulated (>5.0-fold change; P < 0.001) in B. breve UCC2003-melR2 (Table 2). However, the insertion mutation in melR2 appears to have a polar effect on the transcription of the downstream gene, melR1, as the gene exhibited a 1.7-fold-lower level of transcription in the B. breve UCC2003-melR2 mutant (compared to UCC2003; P < 0.001), indicating that melR1 and melR2 are cotranscribed. The data obtained are consistent with our prediction that both LacI-type regulators act as transcriptional repressors, since the mutations in B. breve UCC2003-melR1 and UCC2003-melR2 cause transcriptional upregulation of adjacent genes, several of which have been associated with melezitose metabolism. The transcriptome of B. breve UCC2003-rafR did not reveal any statistically significant differences from that of B. breve UCC2003 that could be linked to the metabolism of raffinose-like sugars (results not shown), which is consistent with the notion that RafR is a ROK-type transcriptional activator. ROK regulators were originally described some 20 years ago and include transcriptional repressors, sugar kinases, and as-yet-uncharacterized open reading frames (54). The ROK protein family is characterized by the presence of the PF00480 domain, which belongs to the Actin-ATPase clan in the Pfam database (55).
TABLE 2.
Transcriptome analysis of B. breve UCC2003-melR1 and B. breve UCC2003-melR2 compared to B. breve UCC2003 grown on 1% ribose
| Gene no. | Name | Putative function | Fold upregulationa |
|
|---|---|---|---|---|
| melR2 | melR1 | |||
| Bbr_1862 | Solute binding protein of ABC transporter system for sugars | 148.62 | − | |
| Bbr_1861 | Conserved hypothetical protein | 59.43 | − | |
| Bbr_1860 | melA | Solute binding protein of ABC transporter system for sugars | 77.64 | 80.63 |
| Bbr_1859 | melB | Permease protein of ABC transporter system for sugars | 88.56 | 67.9 |
| Bbr_1858 | melC | Permease protein of ABC transporter system for sugars | 86.8 | 78.1 |
| Bbr_1857 | melD | Alpha-1,4-glucosidase | 10.24 | 8.59 |
| Bbr_1856 | melE | Alpha-galactosidase | 6.6 | 5.51 |
The cutoff point is 5-fold (P < 0.001); values below the cutoff are indicated by a minus.
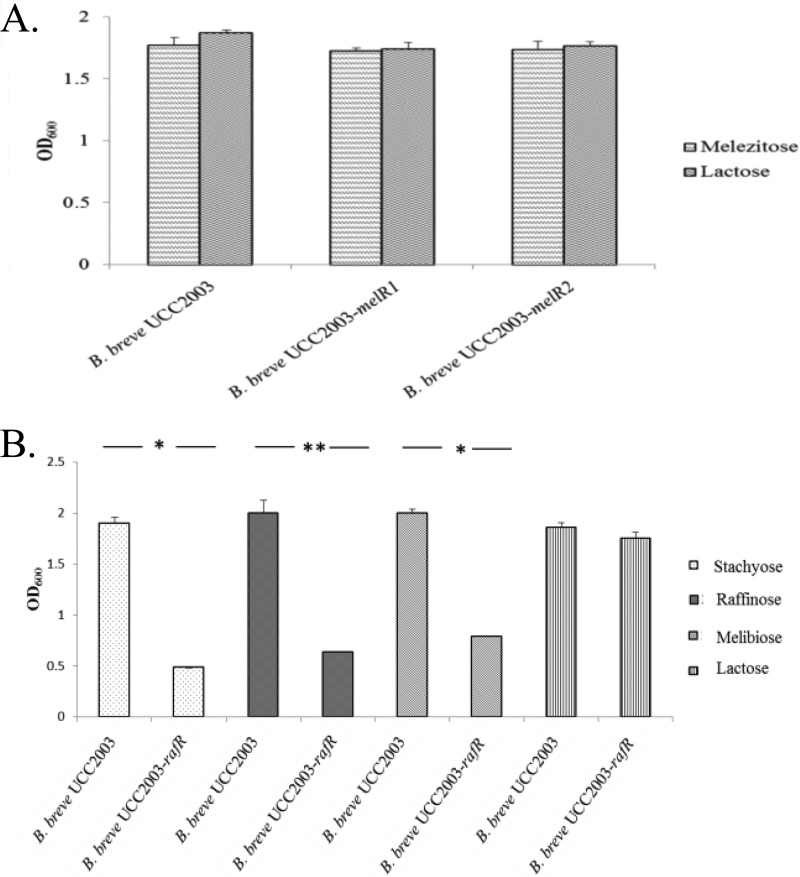
The three mutants were also investigated for the ability to utilize melezitose or raffinose-like sugars. The B. breve UCC2003-melR1 and UCC2003-melR2 mutants both retained the ability to utilize melezitose as the sole carbohydrate source (Fig. 2A). Interestingly, B. breve UCC2003-rafR was incapable of growing to a high optical density in mMRS supplemented with any of the raffinose-like sugars as the sole carbon source (Fig. 2B), while growth of B. breve UCC2003-rafR in mMRS supplemented with 1% lactose was comparable to that observed for the parent strain, B. breve UCC2003. These results demonstrate that rafR is required for growth on raffinose-related carbohydrates and suggest that the ROK-type transcriptional regulator RafR is required for the activation of the raffinose gene cluster.
FIG 2.
(A) Final OD600 values after 16 h of wild-type strain B. breve UCC2003 and insertion mutants B. breve UCC2003-melR1 and B. breve UCC2003-melR2 growth on 1% melezitose or 1% lactose. The results are mean values obtained from three separate experiments. The final optical densities reached by B. breve UCC2003 and insertion mutants B. breve UCC2003-melR1 and B. breve UCC2003-melR2 during growth on lactose or melezitose were not statistically significantly different (P < 0.1). (B) Final OD600 values after 16 h of growth of wild-type strain B. breve UCC2003 and insertion mutant B. breve UCC2003-rafR on 1% stachyose, 1% raffinose, 1% melibiose, or 1% lactose. The final optical densities attained by B. breve UCC2003 compared to insertion mutant B. breve UCC2003-rafR during growth on 1% raffinose, 1% stachyose, or 1% melibiose were statistically significantly different (*, P < 0.001; **, P < 0.01). No significant difference (P < 0.1) was observed in the final optical densities achieved by B. breve UCC2003 and B. breve UCC2003-rafR during growth on 1% lactose. The error bars indicate standard deviations.
MelR1 and MelR2 bind to the melA and Bbr1862 promoter regions, respectively.
The transcriptome data obtained for the B. breve UCC2003-melR1 and UCC2003-melR2 mutants (see above) suggest that melR1 and melR2 encode LacI-type regulators that control various genes of the mel gene cluster. In order to establish if the MelR1 or MelR2 protein directly and specifically interacts with presumed operator sequences within the promoter region(s) of the mel gene cluster, i.e., the Bbr_1862 and/or melA promoter region (see below), EMSAs were performed. For this purpose, we first cloned melR1 and melR2 in the nisin-inducible vectors pNZ8048 and pNZ8150, respectively, with the introduction of an N-terminal His tag-encoding sequence to facilitate protein expression and purification in L. lactis NZ9000 (generating plasmids pNZ-MelR1-His and pNZ-MelR2-His, respectively).
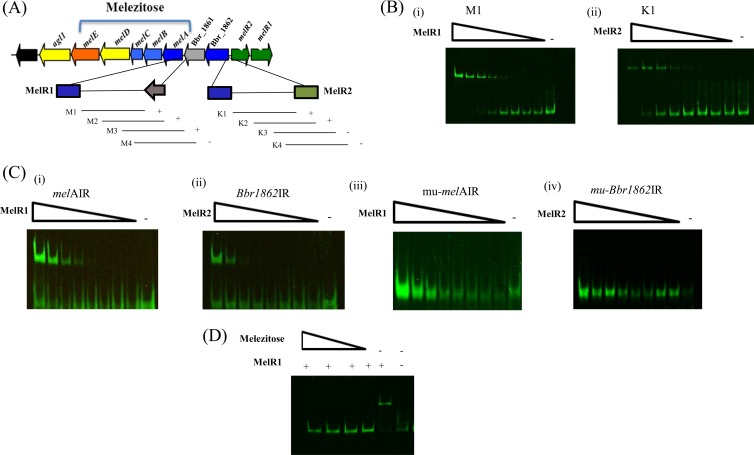
Although MelR1 and MelR2 could be obtained as purified proteins, they proved unable to form DNA-protein complexes under the conditions used, which had been noted previously for LacI-type regulators in bifidobacteria (18). For this reason, instead of the purified proteins, crude cell extracts of (nisin-induced) L. lactis NZ9000 pNZ-MelR1-His or L. lactis NZ9000 pNZ-MelR2-His were employed to perform EMSAs. As a negative control for the EMSAs, we used crude cell extract of L. lactis NZ9000(pNZ8048) (empty vector) incubated with various DNA fragments, as mentioned below, which, as expected, all failed to alter the electrophoretic behavior of these DNA fragments. The results obtained with a crude cell extract containing MelR1 clearly demonstrate that the protein binds to the IRD800-labeled DNA fragment M1, which encompasses the 444-bp promoter-containing (see below) region preceding melA, while MelR1 failed to bind to the IRD800-labeled DNA fragment K1, encompassing the Bbr_1862 promoter region, or the rafA or rafB promoter region [Fig. 3A and B(i) and results not shown].
FIG 3.
(A) Representation of the melezitose utilization cluster of B. breve UCC2003 and DNA fragments used in EMSAs for the melA and Bbr_1862 promoter regions. The plus and minus signs indicate ability or inability of MelR1 or MelR2 to bind to the melA or Bbr_1862 DNA promoter-encompassing fragments, respectively. (B) EMSAs showing MelR1 interaction with DNA fragments encompassing fragment M1 (i) and MelR2 interaction with DNA fragments encompassing fragment K1 (ii). (C) EMSAs illustrating MelR1 interaction with melAIR (i) and mutated derivative mu-melAIR (ii) and MelR2 interaction with Bbr1862IR (iii) and mutated derivative mu-Bbr1862IR (iv). The minus signs indicate lanes with binding reactions to which no protein was added, while the remaining lanes represent binding reactions with the respective DNA probes incubated with increasing amounts of protein at concentrations ranging from 0.04 nM to 0.01 μM. Each successive lane, from right to left, corresponds to a doubling in the amount of protein used for the EMSA. (D) EMSAs showing MelR1 interaction with the DNA fragment M1 with the addition of melezitose at concentrations ranging from 2.5 to 20 mM.
Further dissection of the melA promoter region showed that MelR1 binding required an 84-bp DNA segment that is present within fragments M1 to M3 [Fig. 3B(i); see Fig. S1(i) to (iii) in the supplemental material] and that contains an 18-bp imperfect inverted repeat, 5′-TGCATAAGC><GCTTAGCAA-3′ (with “><” indicating the center of the inverted repeat), representing the putative MelR1-specific operator sequence. This was further validated by EMSAs using a 46-bp synthetic DNA fragment that contained this MelR1 binding (melAIR) [Fig. 3C(i)]. Furthermore, introduction of two point mutations in this presumed MelR1 operator sequence (a G-to-T and a C-to-A mutation at positions 8 and 9 of the sequence, as indicated above) were shown to prevent binding of MelR1 (mu-melAIR) [Fig. 3C(iii)]. Other mutations in bases on either side of this motif did not affect the binding of MelR1 (results not shown) (for all wild-type and mutated primer sequences, see Table S3 in the supplemental material). To investigate whether MelR1 interaction with its target DNA sequence is influenced by a carbohydrate effector molecule, as is known to be the case for other LacI-type regulators (reviewed in references 56 and 57), several carbohydrates were tested for their effects on MelR1-DNA complex formation. The results obtained show that the ability of MelR1 to bind to the M1 fragment of the melA promoter region is lost in the presence of melezitose at concentrations of >2.5 mM (Fig. 3D), whereas under the same experimental conditions, galactose or lactose did not affect MelR1 binding to its target [see Fig. S2(i) and (ii) in the supplemental material].
When EMSAs were performed using a nisin-induced L. lactis NZ9000 pNZ-MelR2-His cell extract, MelR2 was shown to bind to IRD800-labeled DNA fragment K1, which encompasses the 310-bp promoter-containing (see below) region preceding Bbr_1862, but not with similarly labeled DNA fragments containing the promoter regions upstream of melA (fragment M1), rafA, or rafB [Fig. 3A and B(ii) and results not shown]. Further EMSAs showed that MelR2 binding required an 85-bp DNA segment present within fragments K1 and K2 and absent in fragments K3 and K4 of the Bbr_1862 promoter region [Fig. 3B(ii); see Fig. S3 in the supplemental material]. Inspection of this 85-bp fragment revealed the presence of a 18-bp imperfect inverted repeat, 5′-TGCGTAATC><GATATCGCA-3′, representing a putative MelR2 operator sequence. This was validated by EMSAs using a synthetic 50-bp DNA fragment that contained this predicted operator sequence (Bbr1862IR) [Fig. 3C(ii)]. Introduction of two point mutations in the putative 18-bp MelR2 binding motif (an A-to-C and a T-to-G mutation at positions 7 and 8 of the motif) was shown to prevent MelR2 binding (see Table S2 in the supplemental material) for IRD800 primer sequences (mu-Bbr1862IR) [Fig. 3C(iv)]. Other mutations were also made in bases surrounding this motif, but none of the mutations was shown to affect binding under the conditions tested (results not shown) (see Table S4 in the supplemental material for the relevant primer sequences used). To investigate if MelR2 interaction with its target DNA sequence is influenced by a carbohydrate effector molecule, as shown for MelR1, several carbohydrates were tested for their effects on MelR2-DNA complex formation. However, none of the carbohydrates tested (melezitose, turanose, sucrose, glucose, fructose, α-1,3-galactobiose, or α-1,4-galactobiose) was shown to prevent MelR2 binding to its DNA target [see Fig. S4(i) to (vii) in the supplemental material], suggesting that MelR2 binding is abrogated by an as-yet-unknown molecule (20).
The above-mentioned binding studies, combined with the transcriptome analyses, indicate that MelR1 is the melezitose-responsive repressor of the melABCDE gene cluster, while MelR2 is a LacI-type repressor controlling the transcription of Bbr_1861 and Bbr_1862. The observation that the melR2 mutation leads to an increase in melABCDE transcription is most likely an indirect effect due to its negative polar effect on melR1 transcription, leading to reduced MelR1 expression and, consequently, derepression of the melABCDE gene cluster. Motif searches using a LacI operator consensus model, based on sequences recognized by bifidobacterial LacI-type repressors as determined in this work and reported previously (17, 18, 22), reveal the presence of other LacI-type recognition sites in the genome of B. breve UCC2003. These putative LacI-type binding sites are located in intergenic regions upstream of genes involved in carbohydrate metabolism, consistent with experimentally verified LacI-type binding sites, although validation will require EMSAs involving such DNA regions and their cognate LacI-type proteins (see Fig. S5 and Table S5 in the supplemental material). The sequence differences between these putative LacI-type binding motifs are believed to be responsible for target specificity, a finding that has been observed previously in Lactobacillus plantarum (58). Interestingly, analysis of the genomes of Bifidobacterium longum KACC 91563 and B. longum ATCC 55813, which contain homologs of the melezitose locus (20), including two LacI-type regulator-encoding genes, shows the conservation of the deduced MelR1 and MelR2 operator sites (data not shown).
RafR binds to the rafB promoter region.
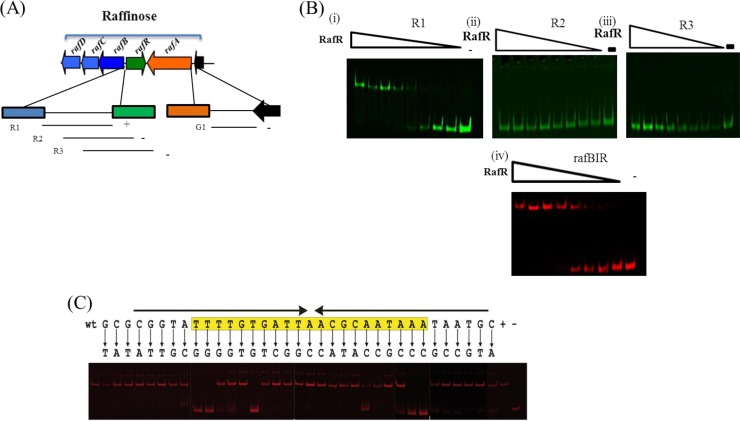
The presence of rafR, encoding a putative ROK-type regulator within the raffinose gene cluster, suggests that the gene plays a role in the transcriptional regulation of the raf gene cluster, a notion that was supported by the inability of a rafR mutant to support proper growth on raffinose-type sugars (see above). In order to establish if RafR is capable of interacting with specific sequences within the raffinose-induced (20) promoter regions upstream of rafA and rafB (see below), we overexpressed and purified RafR as a His-tagged protein (see Materials and Methods). RafR was then used to perform EMSAs, the results of which clearly demonstrate that RafR causes a mobility shift of IRD800-labeled DNA fragment R1, which includes the 241-bp promoter region upstream of rafB [Fig. 4A and B(i)]. In contrast, under the conditions used, RafR was unable to bind to IRD800-labeled DNA fragment G1, which includes the 223-bp promoter region upstream of rafA (results not shown). This suggests that rafA is regulated by an as-yet-unknown regulator or molecule.
FIG 4.
(A) Representation of the raffinose utilization cluster of B. breve UCC2003 and DNA fragments used in EMSAs for the rafB and rafA promoter regions. The plus and minus signs indicate the ability or inability, respectively, of RafA to bind to the rafB or rafA DNA fragments. (B) EMSAs showing RafR interaction with DNA fragments encompassing fragments R1 (i), R2 (ii), and R3 (iii) and the annealed oligonucleotides representing rafBIR (iv). The minus signs indicate lanes with binding reactions to which no protein was added, while the remaining lanes represent binding reactions with the respective DNA probes incubated with increasing amounts of protein at concentrations ranging from 0.04 nM to 0.01 μM. Each successive lane, from right to left, corresponds to a doubling in the amount of protein used for the EMSA. (C) EMSAs showing RafR interaction with the mutated operator motif of the rafB promoter region. DNA fragments were obtained by PCR using IRD700-labeled primers. The operator sequence and incorporated mutations are shown above the image. wt and +, original promoter sequence; −, no added protein. The conserved ROK motif is highlighted in yellow. The arrows indicate the inverted-repeat structure of the motif.
Further EMSAs involving RafR and fragments R2 and R3 [Fig. 4B(ii) and (iii)] of the rafB promoter region showed that RafR binding requires an 81-bp DNA segment that contains an imperfect inverted repeat, 5′-TTTATTGCGTT>A<ATCACAAAATA-3′, thus representing the putative RafR operator sequence. This was validated by EMSAs using a 38-bp DNA fragment that contained this sequence (rafBIR) [Fig. 4B(iv)]. Introduction of point mutations in a 35-bp sequence encompassing the putative RafR binding motif revealed various positions within the operator sequence that are crucial for RafR binding (Fig. 4C; for the primer sequences used, see Table S6 in the supplemental material). This operator binding sequence is very similar to the consensus operator sequence determined for two other members of the ROK family proteins, Mlc and NagC, in E. coli (54, 59). Analysis of sequenced bifidobacterial genomes, many of which contain a homolog of the B. breve raf locus, reveals the presence of a single conserved RafR operator site in the rafB promoter region, whereas no other sites could be detected. Bases important for binding (as determined above) are conserved between bifidobacterial species (see Fig. S6 in the supplemental material).
RafR interaction with its target DNA sequence was not affected by the presence of raffinose, stachyose, melibiose, galactose, glucose, sucrose, α-1,3-galactobiose, and α-1,4-galactobiose at concentrations ranging from 2.5 mM to 20 mM [see Fig. S7(i) to (viii) in the supplemental material]. Binding of RafR to the rafA promoter region was also tested in the presence of these sugars; however, this did not result in binding of RafR to the rafA promoter region (results not shown). A summary of all protein interactions is presented in Table S7 in the supplemental material.
Identification of the transcription start sites of rafA, rafB, melA, and Bbr_1862.
From the genetic organization (Fig. 1) and the expression pattern of the raf locus (20) it was deduced that the raf cluster contained two raffinose/stachyose/melibiose-inducible promoters, one in front of the rafA gene and one in front of the rafB gene. In relation to the mel cluster, potential promoter regions were identified based on in silico analysis of the mel locus (Fig. 1) coupled with transcriptomic data obtained during growth on melezitose (20), as well as the transcriptomes of UCC2003-melR1and UCC2003-melR2 (see above). It was reasoned that one melezitose-inducible, MelR1-dependent promoter was located directly upstream of the melA coding sequence and one promoter (transcription of which is subject to MelR2 and an as-yet-unknown inducer molecule) preceded the Bbr_1862 coding sequence.
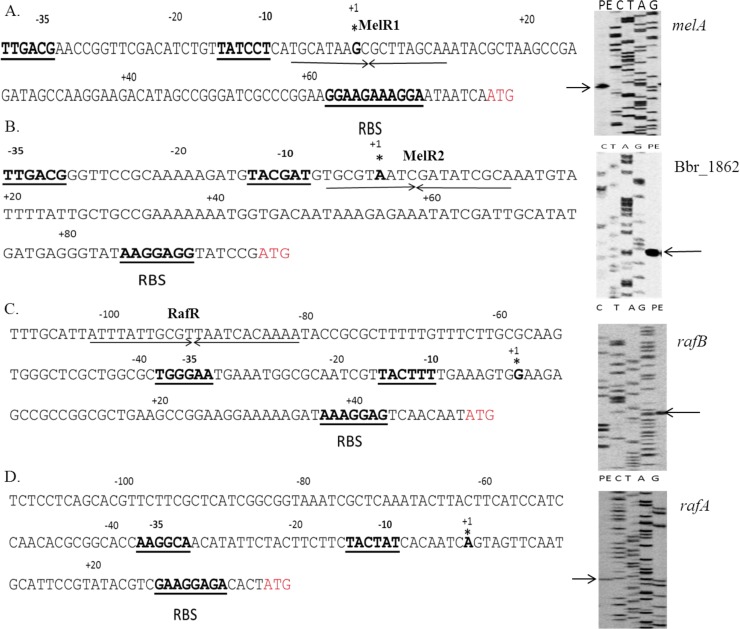
In order to determine the transcription start sites of these presumed promoters, primer extension analysis was performed using RNA extracted from B. breve UCC2003 grown in mMRS containing either 1% melezitose or 1% raffinose, as appropriate. To determine the transcription start site of Bbr_1862, B. breve UCC2003-melR2 (whose transcriptome revealed constitutive transcription of Bbr_1862) was grown in mMRS supplemented with ribose as the sole carbohydrate source. Single extension products were identified upstream of melA, the Bbr_1862 gene, rafB, and rafA, and in all cases, potential promoter recognition sequences resembling consensus −10 and −35 hexamers could be identified upstream of these transcription start points (Fig. 5A to D). In the cases of melA- and Bbr_1862-associated promoters, the MelR1 and MelR2 binding operator sequences overlap the respective −10 sequences, which supports our findings that MelR1 and MelR2 are LacI-type transcriptional repressors. For the rafB promoter region, the RafR operator sequence is located upstream of the −35 and −10 promoter recognition sequences, a location that is consistent with the notion that RafR functions as a transcriptional activator.
FIG 5.
Schematic representation of the melA (A), Bbr_1862 (B), rafB (C), and rafA (D) promoter regions. Boldface type and underlining indicate the −10 and −35 hexamers deduced from the primer extension results; deduced ribosomal binding sites (RBS) and experimentally determined transcriptional start sites (TSS) are indicated by asterisks. The arrows under sequences with names in boldface indicate the inverted-repeat MelR1, MelR2, and RafR binding sequences displayed above the arrows. The arrows in the right panels indicate the primer extension products.
Concluding remarks.
The data assembled in this study provide significant information on the ability of B. breve UCC2003 to regulate two adjacent genetic loci dedicated to the utilization of raffinose-containing carbohydrates and melezitose (20). We conclude that MelR1 is a LacI-type regulator that acts as a melezitose-responsive repressor of the melABCDE gene cluster, while MelR2 is also a LacI-type regulator that acts as a repressor of the Bbr_1861 and Bbr_1862 genes, the latter encoding a predicted carbohydrate binding protein of an ABC-type sugar transport system. Because of its vicinity and the finding that melE encodes an α-galactosidase, we speculate that MelR2 responds to an oligosaccharide containing a melezitose backbone to which one or more α-galactose moieties are attached. This α-galactose-containing oligosaccharide is bound by the product of Bbr_1862 and internalized by the melABC uptake system, after which the α-galactose moiety (or moieties) is removed by the action of MelE, followed by further degradation by MelD. Unfortunately, no such substrates are currently commercially available, and this hypothesis therefore remains to be verified.
In a classic example discussed by Irwin and Ptashne (60), it was observed that mutating the catabolite activator protein (CAP) resulted in lack of transcriptional activation of the gal promoter. Similarly, in C. glutamicum 13032, a deletion in the gene encoding the ROK-type protein CysR rendered the strain incapable of utilizing sulfate or sulfonates as the sole source of sulfur, leading the authors to speculate that CysR functions as a transcriptional activator (61). Based on our experimental findings, we conclude that RafR also acts as a transcriptional activator of the rafBCD cluster.
Somewhat to our surprise, RafR did not bind to the rafA promoter, at least not under the conditions used here, although the promoter is transcriptionally induced when B. breve UCC2003 is grown in raffinose-like sugars (20). It may be that RafR requires an effector for binding to the promoter, and future research will further examine this.
Supplementary Material
ACKNOWLEDGMENTS
This report is based on research conducted with the financial support of Science Foundation Ireland (SFI) under grants 07/CE/B1368, 08/SRC/B13404, and SFI/12/RC/2273. K.J.O. was supported by a postgraduate fellowship funded through the Tomar Trust, while M.O.M. is the recipient of an HRB postdoctoral fellowship (grant no. PDTM/20011/9).
We sincerely thank Stephen Cunningham, Marian Keane, and Lokesh Joshi of the Alimentary Glycoscience Research Cluster, NUIG, Ireland, for kindly supplying α-1,4- and α-1,3-galactobiose. We also acknowledge Kieran James for technical assistance.
Footnotes
Published ahead of print 4 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00130-14.
REFERENCES
- 1.Miyake T, Watanabe K, Watanabe T, Oyaizu H. 1998. Phylogenetic analysis of the genus Bifidobacterium and related genera based on 16S rDNA sequences. Microbiol. Immunol. 42:661–667 [DOI] [PubMed] [Google Scholar]
- 2.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. 2007. Genomics of actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71:495–548. 10.1128/MMBR.00005-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tissier H. 1900. Recherchers sur la flora intestinale normale pathologique du nourisson. Thesis, University of Paris, Paris, France [Google Scholar]
- 4.Tissier H. 1906. The treatment of intestinal infections by the method of transformation of the bacterial intestinal flora. C. R. Soc. Biol. 60:359–361 [Google Scholar]
- 5.Erdogan O, Tanyeri B, Torun E, Gonullu E, Arslan H, Erenberk U, Oktem F. 2012. The comparition of the efficacy of two different probiotics in rotavirus gastroenteritis in children. J. Trop. Med. 2012:787240. 10.1155/2012/787240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parker MW, Schaffzin JK, Lo Vecchio A, Yau C, Vonderhaar K, Guiot A, Brinkman WB, White CM, Simmons JM, Gerhardt WE, Kotagal UR, Conway PH. 2013. Rapid adoption of Lactobacillus rhamnosus GG for acute gastroenteritis. Pediatrics 131(Suppl. 1):S96–S102. 10.1542/peds.2012-1427l [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rohlke F, Stollman N. 2012. Fecal microbiota transplantation in relapsing Clostridium difficile infection. Therap. Adv. Gastroenterol. 5:403–420. 10.1177/1756283X12453637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Q, Gao R, Wu W, Qin H. 2013. The role of gut microbiota in the pathogenesis of colorectal cancer. Tumour Biol. 34:1285–1300. 10.1007/s13277-013-0684-4 [DOI] [PubMed] [Google Scholar]
- 9.Gough E, Shaikh H, Manges AR. 2011. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin. Infect. Dis. 53:994–1002. 10.1093/cid/cir632 [DOI] [PubMed] [Google Scholar]
- 10.Gibson GR, Roberfroid MB. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401–1412 [DOI] [PubMed] [Google Scholar]
- 11.Costabile A, Klinder A, Fava F, Napolitano A, Fogliano V, Leonard C, Gibson GR, Tuohy KM. 2008. Whole-grain wheat breakfast cereal has a prebiotic effect on the human gut microbiota: a double-blind, placebo-controlled, crossover study. Br. J. Nutr. 99:110–120. 10.1017/S0007114507793923 [DOI] [PubMed] [Google Scholar]
- 12.Bouhnik Y, Vahedi K, Achour L, Attar A, Salfati J, Pochart P, Marteau P, Flourie B, Bornet F, Rambaud JC. 1999. Short-chain fructo-oligosaccharide administration dose-dependently increases fecal bifidobacteria in healthy humans. J. Nutr. 129:113–116 [DOI] [PubMed] [Google Scholar]
- 13.Macfarlane GT, Steed H, Macfarlane S. 2008. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J. Appl. Microbiol. 104:305–344. 10.1111/j.1365-2672.2007.03520.x [DOI] [PubMed] [Google Scholar]
- 14.Bouhnik Y, Attar A, Joly FA, Riottot M, Dyard F, Flourie B. 2004. Lactulose ingestion increases faecal bifidobacterial counts: a randomised double-blind study in healthy humans. Eur. J. Clin. Nutr. 58:462–466. 10.1038/sj.ejcn.1601829 [DOI] [PubMed] [Google Scholar]
- 15.Cecchini DA, Laville E, Laguerre S, Robe P, Leclerc M, Doré J, Henrissat B, Remaud-Siméon M, Monsan P, Potocki-Véronèse G. 2013. Functional metagenomics reveals novel pathways of prebiotic breakdown by human gut bacteria. PLoS One 8:e72766. 10.1371/journal.pone.0072766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scardovi V, Trovatelli ID. 1965. The fructose-6-phosphate shunt as a peculiar pattern of hexose degradation in the genus Bifidobacterium. Ann. Microbiol. 15:19–29 [Google Scholar]
- 17.Pokusaeva K, Neves AR, Zomer A, O'Connell-Motherway M, MacSharry J, Curley P, Fitzgerald GF, van Sinderen D. 2010. Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb. Biotechnol. 3:311–323. 10.1111/j.1751-7915.2009.00152.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pokusaeva K, O'Connell-Motherway M, Zomer A, Macsharry J, Fitzgerald GF, van Sinderen D. 2011. Cellodextrin utilization by Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 77:1681–1690. 10.1128/AEM.01786-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maze A, O'Connell-Motherway M, Fitzgerald GF, Deutscher J, van Sinderen D. 2007. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73:545–553. 10.1128/AEM.01496-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Connell KJ, O'Connell Motherway M, O'Callaghan J, Fitzgerald GF, Ross RP, Ventura M, Stanton C, van Sinderen D. 2013. Metabolism of four α-glycosidic linkage-containing oligosaccharides by Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 79:6280–6292. 10.1128/AEM.01775-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryan SM, Fitzgerald GF, van Sinderen D. 2005. Transcriptional regulation and characterization of a novel β-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 71:3475–3482. 10.1128/AEM.71.7.3475-3482.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Connell Motherway M, Fitzgerald GF, van Sinderen D. 2011. Metabolism of a plant derived galactose-containing polysaccharide by Bifidobacterium breve UCC2003. Microb. Biotechnol. 4:403–416. 10.1111/j.1751-7915.2010.00218.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Connell Motherway M, Fitzgerald GF, Neirynck S, Ryan S, Steidler L, van Sinderen D. 2008. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 74:6271–6279. 10.1128/AEM.01169-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukami-Kobayashi K, Tateno Y, Nishikawa K. 2003. Parallel evolution of ligand specificity between LacI/GalR family repressors and periplasmic sugar-binding proteins. Mol. Biol. Evol. 20:267–277. 10.1093/molbev/msg038 [DOI] [PubMed] [Google Scholar]
- 25.Weickert MJ, Adhya S. 1992. A family of bacterial regulators homologous to Gal and Lac repressors. J. Biol. Chem. 267:15869–15874 [PubMed] [Google Scholar]
- 26.Parche S, Beleut M, Rezzonico E, Jacobs D, Arigoni F, Titgemeyer F, Jankovic I. 2006. Lactose-over-glucose preference in Bifidobacterium longum NCC2705: glcP, encoding a glucose transporter, is subject to lactose repression. J. Bacteriol. 188:1260–1265. 10.1128/JB.188.4.1260-1265.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bacon JS, Dickinson B. 1957. The origin of melezitose: a biochemical relationship between the lime tree (Tilia spp.) and an aphis (Eucallipterus tiliae L.). Biochem. J. 66:289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hudson CS. 1946. Melezitose and turanose, p 1–36 In Pigman MLW, Stanley P. (ed), Advances in carbohydrate chemistry, vol 2 Academic Press, San Diego, CA [Google Scholar]
- 29.De Man JC, Rogosa M, Sharpe ME. 1960. A medium for the cultivation of lactobacilli. J. Appl. Microbiol. 23:130–135 [Google Scholar]
- 30.Terzaghi BE, Sandine WE. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- 33.O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JA, Kearney B, Houston AM, O'Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O'Toole PW, van Sinderen D. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U. S. A. 108:11217–11222. 10.1073/pnas.1105380108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 35.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37:W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Riordan K, Fitzgerald GF. 1999. Molecular characterisation of a 5.75-kb cryptic plasmid from Bifidobacterium breve NCFB 2258 and determination of mode of replication. FEMS Microbiol. Lett. 174:285–294 [DOI] [PubMed] [Google Scholar]
- 40.O'Connell Motherway M, O'Driscoll J, Fitzgerald GF, Van Sinderen D. 2009. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb. Biotechnol. 2:321–332. 10.1111/j.1751-7915.2008.00071.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wells JM, Wilson PW, Le Page RW. 1993. Improved cloning vectors and transformation procedure for Lactococcus lactis. J. Appl. Bacteriol. 74:629–636 [DOI] [PubMed] [Google Scholar]
- 42.Mierau I, Leij P, van Swam I, Blommestein B, Floris E, Mond J, Smid E. 2005. Industrial-scale production and purification of a heterologous protein in Lactococcus lactis using the nisin-controlled gene expression system NICE: the case of lysostaphin. Microb. Cell Fact. 4:15. 10.1186/1475-2859-4-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 44.Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alvarez-Martin P, O'Connell-Motherway M, van Sinderen D, Mayo B. 2007. Functional analysis of the pBC1 replicon from Bifidobacterium catenulatum L48. Appl. Microbiol. Biotechnol. 76:1395–1402. 10.1007/s00253-007-1115-5 [DOI] [PubMed] [Google Scholar]
- 46.Zomer A, Fernandez M, Kearney B, Fitzgerald GF, Ventura M, van Sinderen D. 2009. An interactive regulatory network controls stress response in Bifidobacterium breve UCC2003. J. Bacteriol. 191:7039–7049. 10.1128/JB.00897-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia De La Nava J, Santaella DF, Alba JC, Carazo JM, Trelles O, Pascual-Montano A. 2003. Engene: the processing and exploratory analysis of gene expression data. Bioinformatics 19:657–658. 10.1093/bioinformatics/btg028 [DOI] [PubMed] [Google Scholar]
- 48.van Hijum SAFT, Garcia De La Nava J, Trelles O, Kok J, Kuipers OP. 2003. MicroPreP: a cDNA microarray data pre-processing framework. Appl. Bioinformatics 2:241–244 [PubMed] [Google Scholar]
- 49.van Hijum SAFT, De Jong A, Baerends RJ, Karsens HA, Kramer NE, Larsen R, den Hengst CD, Albers CJ, Kok J, Kuipers OP. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. 10.1186/1471-2164-6-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Long AD, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937–19944. 10.1074/jbc.M010192200 [DOI] [PubMed] [Google Scholar]
- 51.Hellman LM, Fried MG. 2007. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2:1849–1861. 10.1038/nprot.2007.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuipers OP, Beerthuyzen MM, Siezen RJ, de Vos WM. 1993. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 216:281–291 [DOI] [PubMed] [Google Scholar]
- 53.Ventura M, Zink R, Fitzgerald GF, van Sinderen D. 2005. Gene structure and transcriptional organization of the dnaK operon of Bifidobacterium breve UCC 2003 and application of the operon in bifidobacterial tracing. Appl. Environ. Microbiol. 71:487–500. 10.1128/AEM.71.1.487-500.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Titgemeyer F, Reizer J, Reizer A, Saier MH. 1994. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 140:2349–2354 [DOI] [PubMed] [Google Scholar]
- 55.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson CJ, Zhan H, Swint-Kruse L, Matthews KS. 2007. The lactose repressor system: paradigms for regulation, allosteric behavior and protein folding. Cell. Mol. Life Sci. 64:3–16. 10.1007/s00018-006-6296-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swint-Kruse L, Matthews KS. 2009. Allostery in the LacI/GalR family: variations on a theme. Curr. Opin. Microbiol. 12:129–137. 10.1016/j.mib.2009.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Francke C, Kerkhoven R, Wels M, Siezen RJ. 2008. A generic approach to identify transcription factor-specific operator motifs; inferences for LacI-family mediated regulation in Lactobacillus plantarum WCFS1. BMC Genomics 9:145. 10.1186/1471-2164-9-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plumbridge J. 2001. Regulation of PTS gene expression by the homologous transcriptional regulators, Mlc and NagC, in Escherichia coli (or how two similar repressors can behave differently). J. Mol. Microbiol. Biotechnol. 3:371–380 [PubMed] [Google Scholar]
- 60.Irwin N, Ptashne M. 1987. Mutants of the catabolite activator protein of Escherichia coli that are specifically deficient in the gene-activation function. Proc. Natl. Acad. Sci. U. S. A. 84:8315–8319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruckert C, Milse J, Albersmeier A, Koch D, Puhler A, Kalinowski J. 2008. The dual transcriptional regulator CysR in Corynebacterium glutamicum ATCC 13032 controls a subset of genes of the McbR regulon in response to the availability of sulphide acceptor molecules. BMC Genomics 9:483. 10.1186/1471-2164-9-483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Ruyter PG, Kuipers OP, Beerthuyzen MM, van Alen-Boerrigter I, de Vos WM. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.