Abstract
The in situ metabolic characteristics of the yeasts involved in spontaneous fermentation process of Chinese light-style liquor are poorly understood. The covariation between metabolic profiles and yeast communities in Chinese light-style liquor was modeled using the partial least square (PLS) regression method. The diversity of yeast species was evaluated by sequence analysis of the 26S ribosomal DNA (rDNA) D1/D2 domains of cultivable yeasts, and the volatile compounds in fermented grains were analyzed by gas chromatography (GC)-mass spectrometry (MS). Eight yeast species and 58 volatile compounds were identified, respectively. The modulation of 16 of these volatile compounds was associated with variations in the yeast population (goodness of prediction [Q2] > 20%). The results showed that Pichia anomala was responsible for the characteristic aroma of Chinese liquor, through the regulation of several important volatile compounds, such as ethyl lactate, octanoic acid, and ethyl tetradecanoate. Correspondingly, almost all of the compounds associated with P. anomala were detected in a pure culture of this yeast. In contrast to the PLS regression results, however, ethyl lactate and ethyl isobutyrate were not detected in the same pure culture, which indicated that some metabolites could be generated by P. anomala only when it existed in a community with other yeast species. Furthermore, different yeast communities provided different volatile patterns in the fermented grains, which resulted in distinct flavor profiles in the resulting liquors. This study could help identify the key yeast species involved in spontaneous fermentation and provide a deeper understanding of the role of individual yeast species in the community.
INTRODUCTION
Spontaneous and solid-state fermentation has been used for thousands of years for the preparation of traditional foods such as bread, tempeh, yogurt, and cheese (1–3). Spontaneously fermented foods are generally characterized by a wide variety of tastes and textures, and sensory properties of the foods are generally determined by the involved microflora, besides the raw material (4, 5). With this in mind, it is therefore important to identify the functional microorganisms contributing to the formation of food flavors.
The metabolic characteristics of lactic acid bacteria (LAB) and yeasts have been studied extensively (6–8), and their effects on food flavor have been highlighted through the detection of the corresponding metabolites in pure cultures. Although these studies have provided valuable information regarding the individual species, they take no account of the fact that the level of volatiles produced by a single species may be influenced by other species in a mixed-culture system. The different flavors of foods would be determined to a large extent by the in situ metabolic characteristics of different microorganisms. Several studies have already described the metabolic characteristics of mixed cultures with the aim of identifying the effects of individual strains on the generation of volatile compounds (9–12). For instance, the metabolic footprints of each strain of Saccharomyces in monoculture and mixed cultures were derived during wine fermentation (9). This showed that the volatile compounds produced in the mixed culture could not be predicted based on the volatiles observed in the monocultures of the same yeast species. Recent work indicated that there were obvious differences in the volatile compound production in milk between single species and mixtures of lactic acid bacteria (12). Although these studies have deepened our understanding of the way in which microorganisms affect the generation of aroma compounds, the relationship between the microbial structure and the flavor fingerprint of spontaneous fermented food remains unresolved. The challenge for studying the metabolic traits of a specific microorganism in spontaneous fermentation lies in the determination of the aroma compounds resulting from structural variations in the microbial community. Recently, partial least square (PLS) regression was adopted to establish relationships between two groups of data (14). For example, PLS regression was used to determine which microbes had the biggest influence on host metabolism in the human gut (15), and the results revealed that variations in the Faecalibacterium prausnitzii population were associated with differences in the occurrence of important urinary metabolites. This study highlighted the effectiveness of this method for establishing relationships between the individual microbes present in a system and the metabolic profiles of the samples.
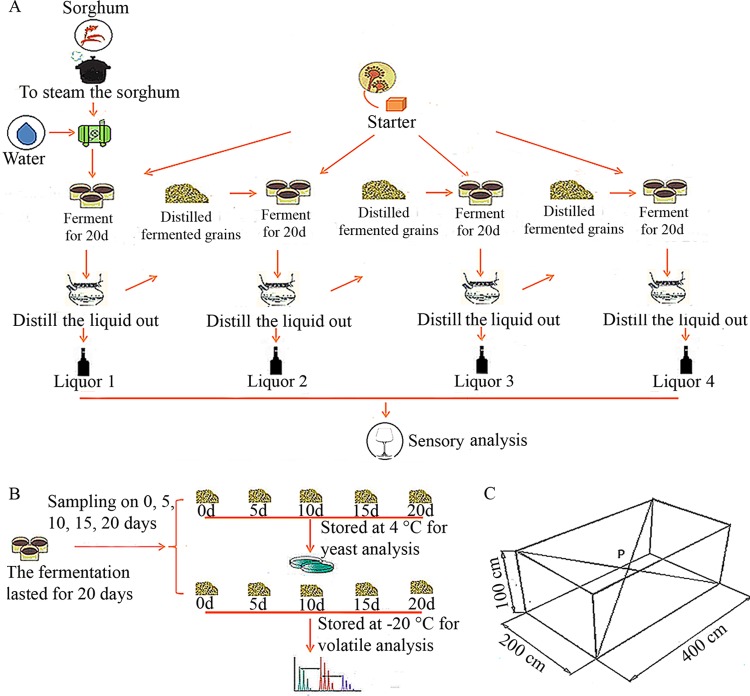
The fermentation of Chinese light-style liquor is a typical spontaneous and solid-state fermentation process, which includes starter making and alcoholic fermentation in pits. The starter used in this study was made from a mixture of barley and peas (6:4), which were stirred together in water (40%). This mixture was then cultivated for 20 to 25 days to allow the growth of crude microorganisms. Following a complex drying process, the starters were then ready to be used. The fermentation process was conducted in a cubic pit in the ground, as shown in Fig. 1C. For the first fermentation batch, a mixture of sorghum and water (1:1.1) was steamed for 30 to 40 min before being mixed with 10% volume of starters. The fermentation lasted for 20 days. Subsequently, the fermented grains were distilled to give a liquid, which was collected. The fermented grains were then mixed with new starters and used for a second fermentation process. In total, the fermentation process was conducted four times, with the same grains being used on all four occasions (Fig. 1A). This kind of fermentation process for Chinese light-style liquor is generally chosen to make full use of the starch. Previous studies have reported that the liquors collected from the first two batches are generally soft and sweet, whereas the liquors collected from the last two batches are generally bitter and pungent (16). It was found that the spontaneous fermentation process accumulated a distinctive community of microorganisms, and of the complex microbiota, yeasts represented the most important group and were primarily responsible for the taste and flavor characteristics of liquor (17, 18). In spite of the importance of yeasts to the fermentation process, knowledge about the influence of different yeast communities on the volatile compound profiles formed during fermentation is still limited.
FIG 1.
(A) Flow diagram for Chinese light-style liquor production. A complete production process consisted of four fermentation batches. The fermentation of each batch lasted for 20 days. The fermented grains were then distilled, and the liquor was collected. (B) Duplicate samples were collected from each fermentation batch at 5-day intervals, with one sample being used for yeast analysis and the other being used for volatile-compound analysis. The zero point was defined as the time of starter inoculation. (C) Schematic representation of the fermentation tank. P, sampling point.
In this study, the yeast community structures and dynamics during four different fermentation batches of Chinese light-style liquor were analyzed. The volatile compounds in the fermented grains were detected by gas chromatography-mass spectrometry (GC-MS), and the relationship between the volatile compound profile and the yeast community structure was established using the PLS regression method. Multivariate statistical analyses of the metabolism and yeast population dynamics provided new insights into the in situ metabolic characteristics of the different yeast species involved in spontaneous fermentation processes.
MATERIALS AND METHODS
Sampling.
The samples used in this study were obtained from the four successive fermentation batches involved in a complete production process of Chinese light-style liquor (Fig. 1A). All of the fermented grain samples were collected from the center of the pits (Fig. 1C). The samples were collected at five-day intervals in duplicate, with one sample being stored at 4°C for yeast analysis and the other being stored at −20°C for analysis of its volatile-compound composition (Fig. 1B). The collected samples were transported to the laboratory by air and analyzed within 24 h. These sampling procedures were conducted in September and October of 2011 in western China.
Yeast isolation and identification.
For the isolation of the yeasts, fermented grains (5 g) were suspended in 100 ml of sterile physiological saline (0.85% NaCl) and shaken for 15 min at room temperature (RT). For the enumeration of the yeasts, 0.1 ml of each decimal dilution was spread in triplicate on Wallerstein laboratory nutrient agar (WL). Based on colony morphology and frequency, 160 representative colonies were proportionally isolated for further identification. Selected isolates were identified by sequencing the D1/D2 domain of the 26S ribosomal DNA (rDNA) region. Yeast colonies were inoculated into yeast extract-peptone-dextrose (YPD) medium and incubated at 30°C for 24 h. Cultures were centrifuged at 8,000 × g for 1 min at 4°C, and the yeast pellets were washed twice with ice-cold distilled water. The genomic DNA was then extracted according to the method reported previously by Makimura et al. (20). The primers used for amplification of the D1/D2 domain of the 26S rDNA region were NL1 (5′ GCATATCAATAAGCGGAGGAAAAG 3′) and NL4 (5′ GGTCCGTGTTTCAAGACGG 3′) as described previously (21). PCRs were performed as described by Baleiras Couto et al. (22) and Esteve-Zarzoso et al. (23). The resulting sequences were compared with those available in the GenBank database at the National Center for Biotechnology Information (NCBI) using the basic local alignment search tool (BLAST). Sequences with 98% nucleotide identity or higher in the D1/D2 domain of the 26S rDNA region were considered to represent the same species. With the morphology and identification results in hand, we proceeded to count the number of different yeast species on each plate.
Inoculated fermentations on a laboratory scale with selected single strains.
To identify the differences in volatile compounds produced by different yeast species, we carried out liquid and solid-state inoculated fermentations on the laboratory scale with eight selected single strains. Control batches containing the culture medium without inoculation (i.e., one containing the solid culture medium and one containing the liquid culture medium) were also prepared. The volatile compounds from the experimental and the control groups were determined by GC-MS. The results for the control group were subtracted from those of the experimental group, which allowed the volatile compounds produced by the yeasts to be calculated exactly.
(i) Liquid fermentations of signal strains.
The sorghum extract used in the liquid fermentation was prepared according to the methods described below. Two kilograms of milled sorghum powder was added to 8 liters of deionized water. The mixture was steamed for 2 h, followed by saccharification at 60°C for 4 h with the addition of glucoamylase (5 U/liter). The mixture was then filtered through gauze and the filtrate was centrifuged at 8,000 × g for 15 min. The supernatant was collected and used as the liquid fermentation medium. The sugar content was measured on a Leica refractometer (Fisher Scientific, Pittsburg, PA, USA). The original sugar content was determined to be 15 degrees Brix (15°Bx). The mixture was then diluted with water to give a final sugar concentration of 12°Bx (95 ± 5 g/liter reducing sugar) before the sterilization.
The liquid fermentations were carried out in 125 ml of sorghum extract medium in 250-ml Erlenmeyer flasks, which were sealed with Muller valves filled with pure sulfuric acid. The fermentation flasks were inoculated with a 48-h culture grown in the same extract to obtain a cellular population of 1 × 104 viable cells per milliliter. The fermentations were conducted at 30°C without shaking, and the fermentative course was monitored by measuring weight loss, determined by carbon dioxide evolution during the process. At the end of the fermentation, indicated by constant weight of the samples, the fermented medium was centrifuged at 4,000 × g for 5 min, and the supernatant was stored at −20°C until required for analysis. All of the experiments were performed in duplicate.
(ii) Solid-state fermentations of single strains.
Commercial-quality sorghum (particle size, 120 to 160 μm) was obtained from the production and used as the solid substrate. Sorghum (100 g) was added to 100 ml of water, and the mixture was then saccharified at 60°C for 4 h with the addition of glucoamylase (5 U/g). The experiments were carried out in 250-ml conical flasks containing 100 g of saccharified sorghum. Each flask was autoclaved at 121°C for 20 min. Preliminary studies showed that no changes were detected in the moisture content of the substrate after the autoclaving process. After cooling, each flask was inoculated with a 48-h culture grown in the sorghum extract to obtain a cellular population of 1 × 104 cells per 100 g grains. The flask was then sealed and incubated at 30°C for 20 days. After that, the samples were withdrawn and stored at −20°C until required for analysis.
Volatile-compound analysis. (i) Sample pretreatment.
The volatile compounds produced by the different yeasts in the laboratory-scale fermentations and the volatile-compound profiles of the fermented grains were analyzed by GC-MS. The solid samples (i.e., the solid substrate in the laboratory-scale fermentations and the fermented grains from the actual production) were pretreated according to the method described below. The samples (5 g) were added to 20 ml of sterile saline and soaked overnight. The mixture was then subjected to an ultrasonic treatment for 30 min before being centrifuged at 8,000 × g for 10 min at 4°C. The supernatant was then collected and filtered through a 0.22-μm filter and stored at −20°C prior to its analysis.
(ii) GC-MS analysis of the volatile compounds.
Volatile compounds were determined by GC-MS. The sampling conditions were as follows: 8 ml of sample and 10 μl of 4-methyl-2-pentanol (internal standard) at 4,035 mg/liter were placed into a 20-ml headspace vial with 3 g of NaCl. The vial was hermetically sealed with a polytetrafluoroethylene (PTFE)-faced silicone septum. The sample was then equilibrated at 50°C for 5 min and extracted for 45 min at the same temperature under stirring in a multipurpose sampler with solid-phase microextraction (SPME) capability (MPS 2; Gerstel, Germany).
GC-MS analysis was conducted on an Agilent 6890N GC system coupled to an Agilent 5975 mass selective detector (MSD; Agilent, CA, USA). The concentrate (1 μl) was analyzed on a DB-Wax column (30-m by 0.25-mm inside diameter [i.d.], 0.25-μm film thickness; J&W Scientific, CA, USA). The oven and injector temperatures were set as described previously, with some minor modifications (24). The injector temperature was set at 250°C, and the splitless mode was used. The oven temperature was held at 50°C for 2 min and then raised to 230°C at a rate of 4°C/min, before being held at 230°C for 15 min. Helium was used as the column carrier gas at a constant flow rate of 2 ml/min. The electron impact energy and ion source temperature were set at 70 eV and 230°C, respectively. The mass spectra of unknown compounds were compared with those in the Wiley 275.L (Agilent) database. Retention indices (RIs) of unknown compounds were calculated in accordance with a modified Kovats method (25).
Sensory analysis.
The liquors collected from the four fermentation batches were submitted for taste testing to 10 trained panelists (5 men and 5 women) using a quantitative descriptive analysis. Each sensorial attribute was evaluated on a continuous linear scale ranging from 0 (no perception of the descriptor) to 10 (very intense). The sensory vocabulary used to characterize each liquor sample comprised 10 terms for flavor, including irritation, spirits, sour, sweet, bitter, peppery, astringent, full-bodied, congruous, and aftertaste; seven terms for taste, including spicy, strong, smooth, soft, mellow, elegant, and pungent; four terms for aroma, including rancid, ester flavor, caramel, and faint fruit scent; and five terms for appearance, including color, transparency, muddy, suspension, and sediment. These terms were selected by the panel during their preliminary tasting sessions to describe the samples. Tests were conducted in individual cubicles, with 15-ml liquor samples being presented in transparent glasses covered with a plastic dish.
Statistical analysis.
Principal component analysis (PCA; SPSS Statistics v19.0 win 32) was carried out on the volatile compounds as well as the microbial variables to represent the structuring of these variables. Partial least square (PLS; SIMCA-P-11.5) regression was used to establish a relationship between the metabolites and the microbial variables. Analysis of variance was conducted using an analysis of variance (ANOVA) Tukey test to determine significant differences between the samples. The statistical level of significance was set at a P value of ≤0.05.
Nucleotide sequence accession numbers.
The species-specific partial 26S rDNA region sequences were submitted to the GenBank database under the following accession numbers: Issatchenkia orientalis (Pichia kudriavzevii), KJ469959; Pichia membranifaciens, KJ469960; Pichia scaptomyzae, KJ469961; Saccharomyces cerevisiae, KJ469962; Saccharomyces servazzii (Kazachstania servazzii), KJ469963; Saccharomyces uvarum, KJ469964; Torulaspora delbrueckii, KJ469965; and Pichia anomala (Wickerhamomyces anomalus), KJ469966.
RESULTS
Yeast community structures in the four fermentation batches.
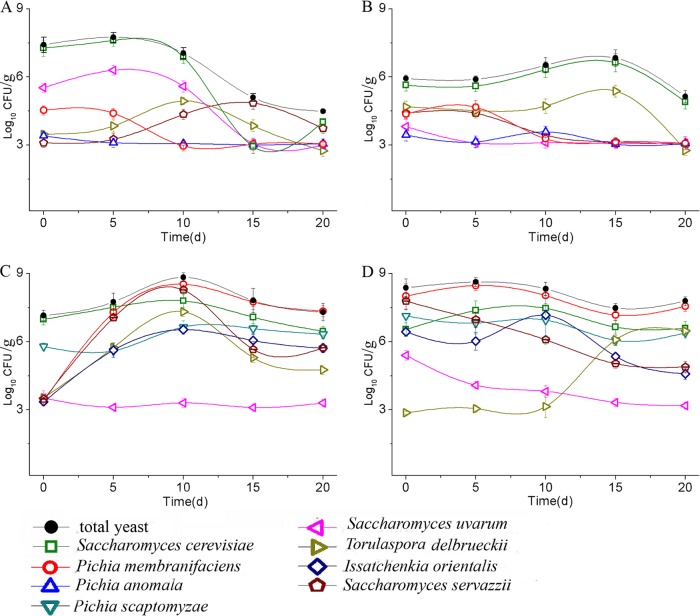
To investigate the yeast community structures and dynamics in the four fermentation batches of Chinese light-style liquor, we collected samples at five time points (i.e., 0, 5, 10, 15, and 20 days) during the fermentations. We evaluated the total yeast count and the non-Saccharomyces and Saccharomyces populations by the spread plate method on WL medium. Based on colony morphology and color (19), 16 representative colonies (K1 to K16) were observed on the plates (see Fig. S1 in the supplemental material). All 160 selected yeast strains were assigned to eight species (Table 1). The results of yeast microflora analyses showed that the four fermentation batches contained a diverse range of yeast communities (Fig. 2). The yeast compositions of the first and second batches were similar (Fig. 2A and B), and in both, S. cerevisiae, S. uvarum, P. membranifaciens, T. delbrueckii, P. anomala, and S. servazzii were detected. Two new species, including I. orientalis and P. scaptomyzae, were identified in the third and fourth batches, but P. anomala was not detected (Fig. 2C and D). In terms of the total number of yeast, the yeast population reached its peak level of 4.2 × 107 CFU/g on the fifth day and then decreased significantly after the tenth day in the first batch (Fig. 2A). The number of yeasts continued to increase slowly from 0 to 15 days (the maximum was 4.6 × 106 CFU/g) and then decreased in the second batch (Fig. 2B). In the third batch, the quantity of yeast reached its maximum (5.4 × 108 CFU/g) on the tenth day and then decreased to about 107 CFU/g by the fifth day (Fig. 2C). In the fourth batch, the quantity of yeast remained stable up until the tenth day and then dropped from 109 to 108 CFU/g (Fig. 2D). These results indicated that in all four batches, the total number of yeasts increased at first and then decreased, and S. cerevisiae was detected as the dominant yeast species. In addition, S. uvarum was found to be the dominant species until the tenth day in the first batch, while P. membranifaciens was identified as one of the dominant species (>108 CFU/g) in the third and fourth batches.
TABLE 1.
Yeast species identified by sequence analysis of the D1/D2 domain of the 26S rDNA region
| Species | Colony(ies) | GenBank accession no. | Sequence similarity to type strain (%) | Proportion (%) |
|---|---|---|---|---|
| S. cerevisiae | K5 | KJ469962 | 99 | 6.25 |
| P. anomala | K3, K6, K16 | KJ469966 | 99 | 18.7 |
| I. orientalis | K7, K15 | KJ469959 | 100 | 12.5 |
| S. uvarum | K1 | KJ469964 | 99 | 6.25 |
| S. servazzii | K14 | KJ469963 | 99 | 6.25 |
| P. membranifaciens | K8, K10, K11, K13 | KJ469960 | 99 | 25.0 |
| T. delbrueckii | K4, K2 | KJ469965 | 99 | 12.5 |
| P. scaptomyzae | K9, K12 | KJ469961 | 99 | 12.5 |
FIG 2.
Dynamics of the total and different yeast species established by culture/counting and D1/D2 PCR during the first batch (A), the second batch (B), the third batch (C), and the fourth batch (D). Values are presented as log values.
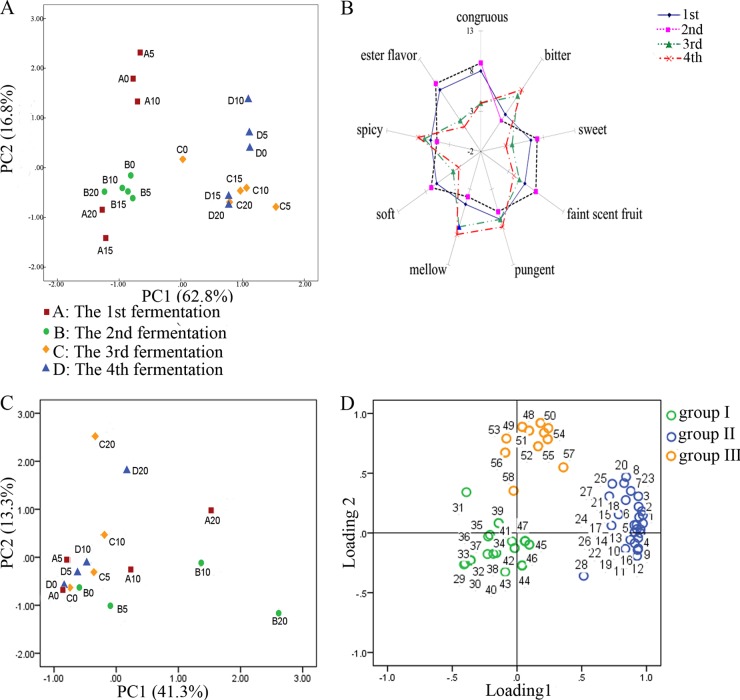
Changes in the yeast community structures of the four fermentation batches were visualized by PCA (Fig. 3A; also, see Fig. S2 in the supplemental material). Each point was labeled with the corresponding fermentation batch (A, B, C, and D represent the first, second, third, and fourth batches, respectively) followed by the fermentation time in days (0, 5, 10, 15, and 20). The yeast community profiles from the early stages of the first batch were most distantly located from the profiles of the other fermentation batches (Fig. 3A). This seems to be mainly affected by the unique dominance of S. uvarum in the early stages of the first batch (Fig. 2A). A big jump, i.e., a relatively more significant change in community structure, between two consecutive time points on the plot was relevant to a remarkable rise or fall in the target yeast populations. For example, in the third batch (C), the jump from the day 0 sample (C0) to the day 5 sample (C5) paralleled the rapid increase in P. membranifaciens, P. scaptomyzae, and I. orientalis during the corresponding period (Fig. 2C and 3A). The jumps from A10 to A15 and from D10 to D15 in the first and fourth batches also coincided with the sudden rise of T. delbrueckii (Fig. 2 and 3A). All of the samples collected in the second batch were grouped together on the plot (Fig. 3A). This grouping of the samples was consistent with a relatively stable population structure for the yeast community in this batch (Fig. 2B). These results suggested that the quantitative shifts in yeast community structures were closely reflected in the PCA results. The PCA results also revealed that the yeast communities of the four fermentation batches had all evolved in different ways.
FIG 3.
(A) Principal component analyses of the yeast community analysis data, showing a plot of principal axes 1 and 2. A, B, C, and D represent the first, second, third, and fourth batches, respectively. For fermented grains, 0-, 5-, 10-, 15- and 20-day points are represented. (B) Sensory profiles of the liquors collected from the four fermentation batches. (C) Principal component analyses of the volatile-compound patterns in the fermented grains collected from different fermentation stages of the four batches. (D) Loading plot for all of the volatile compounds. Compounds in group I were characteristic of the early-stage samples of the four batches. The compounds in group II were present in high concentrations in samples A20 and B20. Compounds in group III were characteristic compounds of samples C20 and D20. The numbers 1 to 58 represent the volatile compounds listed in Table 2.
Sensory analysis.
Sensory analysis was conducted to determine the differences in the aromas of the liquors collected from the four fermentation batches. Figure 3B shows a spiderweb graph of the results obtained in quantitative descriptive analysis (QDA). Attributes that did not achieve scores higher than 1 in any sample were not represented. The liquors collected from the first and second batches had a similar flavor profile (Fig. 3B), and they both reached higher scores for the attributes “ester flavor” and “congruous.” In contrast, the liquors obtained in the third and fourth batches had a similar flavor profile, and they both had higher scores for the attributes “bitter” and “mellow.”
Variation metabolic profiling of yeast community.
GC-MS was performed to evaluate the metabolic profiles of the four fermentation batches. Fifty-eight aroma compounds were identified in all the samples (Table 2). PCA showed that the samples collected in the early stages (before the fifth day) from all four batches formed a group in the negative quadrant of PC1 and PC2 (Fig. 3C), which indicated that they had similar volatile compound profiles. However, the volatile compound profiles of the four batches became totally different after 10 days (Fig. 3C). The evolution of the volatile compound profiles showed the rise or fall of these 58 aroma compounds in different batches (Fig. 3D). These compounds could be divided into three groups according to their loadings on the axes (Table 2). Group I contained 19 aroma compounds, including acids, alcohols, and esters (Table 2). These compounds were characteristic of the samples collected during the early stages (before the fifth day) of the four batches (Fig. 3C and D). Twenty-eight compounds were found in group II, which had high loadings in the positive section of PC 1 (Table 2; Fig. 3D). Most of these aroma compounds were ethyl esters and were present in high concentrations in the samples collected from the late stages of the first two batches (Fig. 3C and D). Most of the aroma compounds in group III were alcohols (Table 2) and had high loadings in the positive section of PC2 (Fig. 3D). These compounds were present in high concentrations in the samples collected from the late stages of the last two batches (Fig. 3D). These results suggested that different yeast communities led to distinct volatile compound profiles during the latter stages of the fermentation process.
TABLE 2.
Volatile compounds detected in the fermented grains and their factor loadings obtained by PCA
| Group and no. | Aroma compounds | Factor loading |
|
|---|---|---|---|
| Loading 1 | Loading 2 | ||
| Group IIb | |||
| 1 | Ethyl tetradecanoate | 0.97 | 0.15 |
| 2 | Acetic acid | 0.97 | 0.08 |
| 3 | Ethyl hexadecanoate | 0.96 | 0.18 |
| 4 | Octanoic acid | 0.95 | 0.00 |
| 5 | Isoamyl lactate | 0.95 | 0.03 |
| 6 | Ethyl 3-phenylpropionate | 0.94 | 0.13 |
| 7 | Ethyl 9-hexadecenoate | 0.94 | 0.22 |
| 8 | Phenylethanol | 0.94 | 0.31 |
| 9 | 2-Methoxy-4-methylphenol | 0.93 | −0.13 |
| 10 | Pentadecanoic acid ethyl ester | 0.93 | −0.04 |
| 11 | 1-Octen-3-ol | 0.93 | −0.14 |
| 12 | n-Decanoic acid | 0.93 | −0.19 |
| 13 | 2,3-Butanediol | 0.93 | 0.03 |
| 14 | Ethyl phenylacetate | 0.92 | 0.07 |
| 15 | Isopentyl acetate | 0.91 | 0.01 |
| 16 | 3-Methylpentanoic acid | 0.91 | −0.09 |
| 17 | 2-Furanmethanol | 0.91 | 0.07 |
| 18 | 2-Phenylethyl acetate | 0.88 | 0.35 |
| 19 | Hexanoic acid | 0.88 | −0.06 |
| 20 | Ethyl caprate | 0.84 | 0.47 |
| 21 | Ethyl hexanoate | 0.84 | 0.28 |
| 22 | Furfural | 0.84 | −0.13 |
| 23 | Ethyl acetate | 0.83 | 0.42 |
| 24 | Ethyl lactate | 0.79 | 0.16 |
| 25 | Ethyl octanoate | 0.74 | 0.41 |
| 26 | Isoamyl alcohol | 0.73 | 0.06 |
| 27 | Geranyl acetone | 0.71 | 0.31 |
| 28 | Ethyl isobutyrate | 0.52 | −0.36 |
| Group Ia | |||
| 29 | 3-Hydroxy-butanoic acid ethyl ester | −0.41 | −0.26 |
| 30 | Propanoic acid | −0.40 | −0.26 |
| 31 | 1-Nonanol | −0.39 | 0.34 |
| 32 | 5-Heptyldihydro-2-(3H)-furanone | −0.36 | −0.23 |
| 33 | Isooctanol | −0.36 | −0.23 |
| 34 | 4-Ethyl guaiacol | −0.23 | −0.18 |
| 35 | Ethyl butyrate | −0.22 | −0.03 |
| 36 | Ethyl propanoate | −0.21 | −0.02 |
| 37 | Butanoic acid | −0.18 | −0.18 |
| 38 | 1-Nonen-3-ol | −0.16 | −0.17 |
| 39 | 4-Dodecanolide | −0.14 | 0.08 |
| 40 | Nonanoic acid | −0.09 | −0.33 |
| 41 | Isobutyl acetate | −0.04 | −0.07 |
| 42 | 2-Heptanol | −0.02 | −0.13 |
| 43 | 5-Methyl-2-hexanol | 0.04 | −0.27 |
| 44 | Dodecanoic acid | 0.04 | −0.27 |
| 45 | Butanedioic acid diethyl ester | 0.10 | −0.10 |
| 46 | Ethyl heptanoate | 0.06 | −0.07 |
| 47 | 2,6-Dimethyl-4-heptanol | 0.06 | −0.07 |
| Group IIIc | |||
| 48 | 4-Vinylguaiacol | 0.18 | 0.92 |
| 49 | 1-Octanol | 0.04 | 0.89 |
| 50 | 1-Pentanol | 0.24 | 0.88 |
| 51 | Ethyl dodecanoate | 0.09 | 0.86 |
| 52 | 2-Propenyl phenylacetate | 0.21 | 0.84 |
| 53 | 3-Hydroxy-2-butanone | −0.08 | 0.79 |
| 54 | γ-Nonalactone | 0.24 | 0.79 |
| 55 | 2-Methyl-1-propanol | 0.16 | 0.73 |
| 56 | Ethyl nonanoate | −0.09 | 0.67 |
| 57 | 2-Methyl-1-butanol | 0.36 | 0.55 |
| 58 | Hexanal | −0.03 | 0.35 |
Group I contained 19 aroma compounds. These compounds were characteristic of the samples collected during the early stages of the four batches.
Group II consisted of aroma compounds which had high loadings in the positive section of PC1. These compounds were present at high concentrations in the samples A20 and B20.
Group III consisted of aroma compounds which had high loadings in the positive section of PC2. These compounds were present at high concentrations in the samples C20 and D20.
Covariation analysis of yeast community profiling and metabolic profiling.
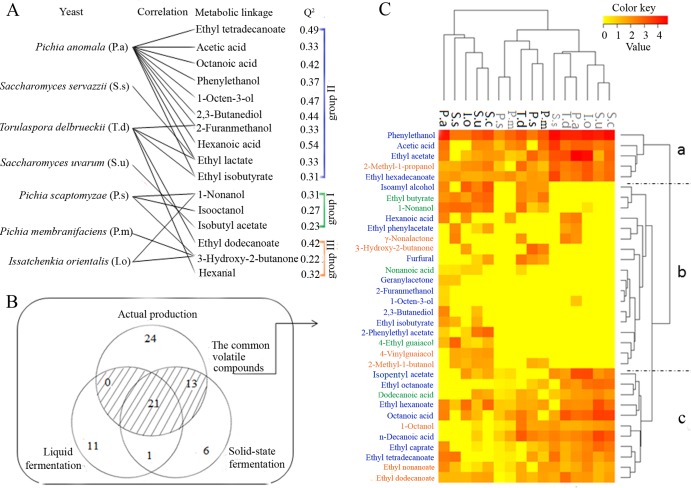
To determine which aroma compounds were most closely associated with structural variations in the yeast community during the fermentation process, we modeled the covariation between the metabolic profile and the yeast community using PLS regression. The predictive power of the model was assessed by 5-fold cross-validation and the Q2 value (goodness of prediction). All of the volatile compounds whose variance could be explained by the yeast population (Q2 > 20%) are shown in Fig. 4A. The percentage (Q2) of variance explained by the model ranged from 22% to 54%. Almost all of the yeasts had a positive association with the metabolites except for S. cerevisiae. In the volatiles that could be well predicted by the yeasts, three compounds came from group I, 10 came from group II, and the remaining three came from group III (Table 2). Some of the yeasts revealed only single-metabolite association, for example, the connection between S. uvarum and isobutyl acetate. P. anomala appeared to be the most important yeast, because it was statistically relevant to the presence of 10 compounds in group II, including ethyl tetradecanoate, acetic acid, octanoic acid, phenylethanol, 1-octen-3-ol, 2,3-butanediol, 2-furanmethanol, hexanoic acid, ethyl lactate, and ethyl isobutyrate. These compounds are all important aroma compounds that make a significant contribution to Chinese liquor flavor (25). According to the results of the PLS regression, most of the volatile compounds in group II could be well predicted and had a positive connection with P. anomala.
FIG 4.
(A) Volatile compounds that could be well predicted (Q2 > 0.2) on the basis of the yeast dynamics in the four fermentation batches by PLS regression. Associations with the volatile compounds are shown for each species of yeast that had a positive effect on the compounds. Groups I to III, labeled on the right, correspond to the groups used to illustrate the characteristic volatile compounds in the fermented grains generated by PCA (Fig. 3D). (B) Venn diagram outlining volatile compounds detected in the single-strain fermentations on a laboratory scale. (C) Comparisons of the volatile compounds produced by the different yeast species during the liquid (black) and solid-state (gray) fermentations. The Z score was used for data standardization by SPSS. The volatile compounds in group I, group II, and group III are in blue, green, and orange, respectively.
Volatile compounds produced in fermentations on a laboratory scale.
Single-strain fermentations were carried out on a laboratory scale to verify the PLS regression model, and the results are shown in Fig. 4B and C. Thirty-three volatile compounds were detected in the liquid fermentations, and 51 were detected in the solid-state fermentations (Fig. 4B). Of the 58 volatiles detected in the fermented grains, 34 were detected during the single-strain fermentations (Fig. 4B). The heat map shown in Fig. 4C was used to visualize the details of these 34 compounds. More compounds in group II and group III could be detected (Fig. 4C). The hierarchical clustering results showed that all 34 compounds were classified into 3 main groups (i.e., groups a, b, and c). The yeasts produced relatively high levels of the compounds belonging to group b during the solid-state fermentation process (Fig. 4C). Of the 10 aroma compounds that had a positive connection with P. anomala, only two were not detected, including ethyl lactate and ethyl isobutyrate. The aroma compounds 2,3-butanediol and 2-furanmethanol were detected only in the sorghum fermented by P. anomala. Hexanoic acid and 1-octen-3-ol were determined to be at their highest concentrations in the sorghum fermented by P. anomala. However, large quantities of ethyl tetradecanoate, octanoic acid, acetic acid, and phenylethanol were also detected in the sorghum fermented by other yeasts. These results demonstrated the credibility of the PLS regression method and also showed that some metabolites could be generated by P. anomala only when the functional yeast was involved in the yeast community.
DISCUSSION
Studying the in situ metabolic characteristics of the yeasts in Chinese liquor could help to identify the key yeasts contributing to the formation of liquor flavor and develop a deeper understanding of the fermentative mechanisms involved in spontaneous and solid-state fermentations. In the current study, eight species of yeast were detected in the fermentation process of Chinese light-style liquor. Of the eight species of yeast observed in this work, however, only five species, including S. cerevisiae, I. orientalis, P. membranifaciens, P. anomala, and T. delbrueckii, have been reported previously (18, 26, 27). To the best of our knowledge, there are few reports about the detection of S. uvarum, S. servazzii, or P. scaptomyzae. A combination of the bar-coded pyrosequencing of internal transcribed spacer region 1 (ITS1) and quantitative real-time PCR (qPCR) was used to investigate microbial diversity throughout the fermentation process of Chinese light-style liquor (18). The results of this study revealed that the yeast species involved in the fermentation process were Saccharomyces bulderi, P. anomala, Saccharomyces castellii, S. cerevisiae, T. delbrueckii, and I. orientalis. The results of the current study were therefore consistent with those of previously reported works using the uncultured method.
In this work, 38 volatile compounds were detected in the fermented grains collected on the first day from all four fermentation batches (see Table S1 in the supplemental material). ANOVA analysis showed that there was no significant difference in the concentration of these 38 compounds among the samples (95% LSD). These results indicated that the fermented grains possessed similar volatile-compound profiles at the beginning of the fermentation process. Different yeast communities in the four fermentation batches, however, led to dissimilar compound patterns in the fermented grains (Fig. 3A and C), which consequently led to the distinct liquor flavor profiles (Fig. 3B). Considering that the material used in each successive fermentation process came from the previous fermentation, it is likely that variations in the fermentable sugar available in the raw material may have influenced the yeast communities, which led to the observed differences in the liquor flavors. In the current study, we found that the samples collected on the last day from the first two batches were characterized by 28 aroma compounds, 11 of which were ethyl esters (Fig. 3C; Table 2). Recent work has shown that the aroma of Chinese light-style liquor is mainly attributable to esters (16). Among these 11 esters, ethyl acetate, ethyl lactate, ethyl octanoate, and ethyl hexanoate were found to be the most important esters in terms of their contributions to the overall flavor based on their Osme values (16). Most of the characteristic aroma compounds in the samples collected on the last day from the last two batches were higher alcohols (Fig. 3C; Table 2), including 1-octanol, 2-methyl-1-propanol, and 2-methyl-1-butanol. The high concentrations of these compounds made the liquor taste spicy and bitter (28).
By correlating the yeast communities with the volatile compound profiles, it was possible to visualize the covariance of yeast community structures and volatile-compound profiles (Fig. 4A). A total of 16 compounds could be well predicted (Q2 > 0.2), and 10 of these compounds came from group II and had a positive connection with P. anomala. A comparison of the PLS regression results (Fig. 4A) with the single strain fermentations (Fig. 4C) revealed that the metabolic characteristics were different between the yeast community and the individual yeast species. P. anomala had a positive connection with 10 aroma compounds and eight of these volatiles could be detected in sorghum fermented by P. anomala. Of these eight compounds, four of them, including 1-octen-3-ol, 2-furanmethanol, hexanoic acid, and 2, 3-butanediol, could be detected only in or had their highest concentration in the sorghum fermented by P. anomala, whereas the other four compounds, including octanoic acid, acetic acid, phenylethanol, and ethyl tetradecanoate, were also detected in large quantities in the sorghum samples fermented by other yeasts. For example, ethyl tetradecanoate was associated with P. anomala, but S. servazzii could also produce this compound at high levels (Fig. 4C). Thus, the lower production of ethyl tetradecanoate may be due to the lower growth of S. servazzii in the community. Ethyl lactate also had a close relationship with P. anomala but could not be detected in the sorghum fermented by P. anomala. Recent work has shown that the mixed fermentation process involving S. cerevisiae and P. anomala led to the production of more esters than the individual species in isolation (29). Some of the compounds produced by P. anomala could react with other compounds in the yeast community and this could explain why the generation of ethyl lactate was related to P. anomala. Alternatively, P. anomala could promote the growth of some other strains capable of producing ethyl lactate in the community. We also noticed that all of the yeasts produced more volatile compounds in the solid-state fermentation than in the liquid fermentation process. Solid-state fermentation resembles the natural habitat of a microorganism and is, therefore, the preferred choice for microorganisms to grow and produce more aroma compounds (11).
The results of the current study also revealed that S. cerevisiae can produce many volatile compounds during a single-strain fermentation process (Fig. 4C). The PLS regression model, however, revealed that S. cerevisiae had no positive connection with any of these compounds in the fermentation of Chinese light-style liquor (Fig. 4A). A recent study showed that there is a significant polymorphism among different strains of S. cerevisiae (30) and that these strains vary considerably in terms of their volatiles profiles (31). It is therefore likely that the spontaneous fermentation of Chinese liquor could involve different strains of S. cerevisiae. With this in mind, it would be difficult to establish a relationship between S. cerevisiae and the various volatile profiles. We did notice, however, that while S. cerevisiae played a dominant role during the early stages of the four fermentation batches, its level declined during the later stages, when the volatile compound profiles changed. It is therefore likely that S. cerevisiae produced aroma compounds during the early stages of the fermentation process, and its contribution to the liquor flavor would have been reduced during the latter stages.
In conclusion, we have demonstrated that the in situ metabolic characteristics of yeasts in a mixed system are different from those of the individual yeast species in single cultures. The PLS regression method allowed the identification of potentially important relationships between the different yeast present in the fermentation process and the volatile compound profiles of the liquors. P. anomala appeared to be the most important yeast in Chinese light-style liquor and was statistically associated with the presence of 10 important aroma compounds that make a significant contribution to the liquor flavor. This work represents a step forward in our understanding of how the activity of a yeast community translates into liquor flavor and may therefore provide a rationale for the selection of functional yeasts for liquor fermentation.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Menhui Zhang for technical assistance.
This work was supported by the National High Technology Research and Development Program of China (2012AA021301 and 2013AA102108), National Natural Science Foundation of China (31000806, 31371822, and 31271921), Cooperation Project of Jiangsu Province among Industries, Universities and Institutes (BY2010116), and Program of Introducing Talents of Discipline to Universities (111 Project) (111-2-06).
Footnotes
Published ahead of print 11 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.04219-13.
REFERENCES
- 1.Beresford TP, Fitzsimons NA, Brennan NL, Cogan TM. 2001. Recent advances in cheese microbiology. Int. Dairy J. 11:259–274. 10.1016/S0958-6946(01)00056-5 [DOI] [Google Scholar]
- 2.De Vuyst L, Vancanneyt M. 2007. Biodiversity and identification of sourdough lactic acid bacteria. Food Microbiol. 24:120–127. 10.1016/j.fm.2006.07.005 [DOI] [PubMed] [Google Scholar]
- 3.Holzapfel W. 2002. Appropriate starter culture technologies for small-scale fermentation in developing countries. Int. J. Food Microbiol. 75:197–212. 10.1016/S0168-1605(01)00707-3 [DOI] [PubMed] [Google Scholar]
- 4.Caplice E, Fitzgerald GF. 1999. Food fermentations: role of microorganisms in food production and preservation. Int. J. Food Microbiol. 50:131–149. 10.1016/S0168-1605(99)00082-3 [DOI] [PubMed] [Google Scholar]
- 5.Nisiotou A, Chorianopoulos N, Nychas GJ, Panagou E. 2010. Yeast heterogeneity during spontaneous fermentation of black Conservolea olives in different brine solutions. J. Appl. Microbiol. 108:396–405. 10.1111/j.1365-2672.2009.04424.x [DOI] [PubMed] [Google Scholar]
- 6.Van der Meulen R, Scheirlinck I, Van-Schoor A, Huys G, Vancanneyt M, Vandamme P, Vuyst LD. 2007. Population dynamics and metabolite target analysis of lactic acid bacteria during laboratory fermentations of wheat and spelt sourdoughs. Appl. Environ. Microbiol. 73:4741–4750. 10.1128/AEM.00315-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravyts F, De Vuyst L. 2011. Prevalence and impact of single-strain starter cultures of lactic acid bacteria on metabolite formation in sourdough. Food Microbiol. 28:1129–1139. 10.1016/j.fm.2011.03.004 [DOI] [PubMed] [Google Scholar]
- 8.Gobbetti M, Angelis MD, Corsetti A, Cagno RD. 2005. Biochemistry and physiology of sourdough lactic acid bacteria. Trends Food Sci. Technol. 16:57–69. 10.1016/j.tifs.2004.02.013 [DOI] [Google Scholar]
- 9.Guerzoni M, Vernocchi P, Ndagijimana M, Gianotti A, Lanciotti R. 2007. Generation of aroma compounds in sourdough: effects of stress exposure and Lactobacilli-yeasts interactions. Food Microbiol. 24:139–148. 10.1016/j.fm.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 10.Howell KS, Cozzolino D, Bartowsky EJ, Fleet GH, Henschke PA. 2006. Metabolic profiling as a tool for revealing Saccharomyces interactions during wine fermentation. FEMS Yeast Res. 6:91–101. 10.1111/j.1567-1364.2005.00010.x [DOI] [PubMed] [Google Scholar]
- 11.Singhania RR, Patel AK, Soccol CR, Pandey A. 2009. Recent advances in solid-state fermentation. Biochem. Eng. J. 44:13–18. 10.1016/j.bej.2008.10.019 [DOI] [Google Scholar]
- 12.de Bok FA, Janssen PW, Bayjanov JR, Sieuwerts S, Lommen A, Johan ET, van Hylckama V, Molenaar D. 2011. Volatile compound fingerprinting of mixed-culture fermentations. Appl. Environ. Microbiol. 77:6233–6239. 10.1128/AEM.00352-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talbot G, Topp E, Palin M, Masse D. 2008. Evaluation of molecular methods used for establishing the interactions and functions of microorganisms in anaerobic bioreactors. Water Res. 42:513–537. 10.1016/j.watres.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 14.Duthoit F, Callon C, Tessier L, Montel MC. 2005. Relationships between sensorial characteristics and microbial dynamics in “Registered Designation of Origin” Salers cheese. Int. J. Food Microbiol. 103:259–270. 10.1016/j.ijfoodmicro.2004.11.040 [DOI] [PubMed] [Google Scholar]
- 15.Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, Zhang Y, Shen J, Pang X, Zhang M. 2008. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. U. S. A. 105:2117–2122. 10.1073/pnas.0712038105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo JH, Yan X, Fan WL. 2012. Aroma compounds of fresh distillates from different runs of fermented grains in Chinese light aroma type liquor. Sci. Technol. Food Ind. 33:52–55 (In Chinese.) http://www.cnki.com.cn/Article/CJFDTotal-SPKJ201213004.htm [Google Scholar]
- 17.Han S, Lei ZH, Li Q, Lv LH, Zhao LQ. 2009. Study on the cultured microbial community and the metabolism regulation during the brewing process of the fen liquor. Sci. Technol. Food Ind. 35:9–13 (In Chinese.) http://www.cnki.com.cn/Article/CJFDTotal-SPFX200901006.htm [Google Scholar]
- 18.Li XR, Ma EB, Yan LZ, Meng H, Du XW, Zhang SW, Quan ZX. 2011. Bacterial and fungal diversity in the traditional Chinese liquor fermentation process. Int. J. Food Microbiol. 146:31–37. 10.1016/j.ijfoodmicro.2011.01.030 [DOI] [PubMed] [Google Scholar]
- 19.Pallmann C, Brown JA, Olineka TL, Cocolin L, Mills D, Bisson L. 2001. Use of WL medium to profile native flora fermentations. Am. J. Enol. Vitic. 52:198–203 [Google Scholar]
- 20.Makimura K, Murayama SY, Yamaguchi H. 1994. Detection of a wide range of medically important fungi by the polymerase chain reaction. J. Med. Microbiol. 40:358–364. 10.1099/00222615-40-5-358 [DOI] [PubMed] [Google Scholar]
- 21.Riethmüller A, Voglmayr H, Göker M, Weiß M, Oberwinkler F. 2002. Phylogenetic relationships of the downy mildews (Peronosporales) and related groups based on nuclear large subunit ribosomal DNA sequences. Mycologia 94:834–849. 10.2307/3761698 [DOI] [PubMed] [Google Scholar]
- 22.Baleiras Couto M, Reizinho R, Duarte F. 2005. Partial 26S rDNA restriction analysis as a tool to characterise non-Saccharomyces yeasts present during red wine fermentations. Int. J. Food Microbiol. 102:49–56. 10.1016/j.ijfoodmicro.2005.01.005 [DOI] [PubMed] [Google Scholar]
- 23.Esteve-Zarzoso B, Belloch C, Uruburu F, Querol A. 1999. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol. 49:329–337. 10.1099/00207713-49-1-329 [DOI] [PubMed] [Google Scholar]
- 24.Fan WL, Xu Y, Zhang Y. 2007. Characterization of pyrazines in some Chinese liquors and their approximate concentrations. J. Agric. Food Chem. 55:9956–9962. 10.1021/jf071357q [DOI] [PubMed] [Google Scholar]
- 25.Fan WL, Qian MC. 2006. Identification of aroma compounds in Chinese ‘Yanghe Daqu' liquor by normal phase chromatography fractionation followed by gas chromatography/olfactometry. Flavour Fragr. J. 21:333–342. 10.1002/ffj.1621 [DOI] [Google Scholar]
- 26.Zhang WX, Qiao ZW, Tang YQ, Hu C, Sun Q, Morimura S, Kida K. 2007. Analysis of the fungal community in zaopei during the production of Chinese luzhou-flavour liquor. J. Inst. Brew. 113:21–27. 10.1002/j.2050-0416.2007.tb00251.x [DOI] [Google Scholar]
- 27.Wu Q, Xu Y, Chen LQ. 2012. Diversity of yeast species during fermentative process contributing to Chinese maotai-flavour liquor making. Lett. Appl. Microbiol. 55:301–307. 10.1111/j.1472-765X.2012.03294.x [DOI] [PubMed] [Google Scholar]
- 28.Verginer M, Leitner E, Berg G. 2010. Production of volatile metabolites by grape-associated microorganisms. J. Agric. Food Chem. 58:8344–8350. 10.1021/jf100393w [DOI] [PubMed] [Google Scholar]
- 29.Tang J, Wang HY, Xu Y. 2012. Effect of mixed culture of Saccharomyces cerevisiae and Pichia anomala on fermentation efficiency and flavor compounds in Chinese liquor. Microbiol. China 39:921–930 (In Chinese.) http://www.cnki.com.cn/Article/CJFDTotal-WSWT201207003.htm [Google Scholar]
- 30.Rosanna T, Sandra T, Clemencia CL, Maria M, Antonello P, Giovanna S. 2007. A survey of Saccharomyces populations associated with wine fermentations from the Apulia region (south Italy). Ann. Microbiol. 57:545–552. 10.1007/BF03175353 [DOI] [Google Scholar]
- 31.Patel S, Shibamoto T. 2002. Effect of different strains of Saccharomyces cerevisiae on production of volatiles in Napa Gamay wine and Petite Sirah wine. J. Agric. Food Chem. 50:5649–5653. 10.1021/jf020337f [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.