Abstract
Surface waters from paired agricultural watersheds under controlled tile drainage (CTD) and uncontrolled tile drainage (UCTD) were monitored over 7 years in order to determine if there was an effect of CTD (imposed during the growing season) on occurrences and loadings of bacterial and viral pathogens, coliphages, and microbial source tracking markers. There were significantly lower occurrences of human, ruminant, and livestock (ruminant plus pig) Bacteroidales markers in the CTD watershed in relation to the UCTD watershed. As for pathogens, there were significantly lower occurrences of Salmonella spp. and Arcobacter spp. in the CTD watershed. There were no instances where there were significantly higher quantitative loadings of any microbial target in the CTD watershed, except for F-specific DNA (F-DNA) and F-RNA coliphages, perhaps as a result of fecal inputs from a hobby farm independent of the drainage practice treatments. There was lower loading of the ruminant marker in the CTD watershed in relation to the UCTD system, and results were significant at the level P = 0.06. The odds of Salmonella spp. occurring increased when a ruminant marker was present relative to when the ruminant marker was absent, yet for Arcobacter spp., the odds of this pathogen occurring significantly decreased when a ruminant marker was present relative to when the ruminant marker was absent (but increased when a wildlife marker was present relative to when the wildlife marker was absent). Interestingly, the odds of norovirus GII (associated with human and swine) occurring in water increased significantly when a ruminant marker was present relative to when a ruminant marker was absent. Overall, this study suggests that fecal pollution from tile-drained fields to stream could be reduced by CTD utilization.
INTRODUCTION
Tile drains or artificial subsurface drainage is commonly used to drain fields in agricultural regions throughout the world to help facilitate crop production. However, it is well documented that conventional tile drainage can serve as an efficient means by which agricultural pollutants from field systems can enter the broader surface water environment (1–4). Fecal pollution in tile drainage as derived from land application of manure or municipal biosolids is well documented (5–11).
Controlled tile drainage (CTD) is a beneficial management practice (BMP) that physically regulates tile discharge from tile-drained fields through the use of water flow control structures (4, 12). Documented benefits of the practice include reduced export of agricultural contaminants from fields to surface water systems (4, 13–15) as well as improved crop yields as a result of the conservation of nutrients and water (16). Controlled tile drainage, which is part of a family of drainage water management practices (17), is a practice that is increasing in use worldwide. Its potential impact on water quality targets can be nontrivial since in many tile-drained landscapes a significant amount of water input to streams comes from tile drainage networks. For instance, Sunohara et al. found that watershed-scale adoption of CTD employed just during the growing season can significantly reduce mass fluxes of water and nutrients (M. D. Sunohara, N. Gottschall, G. Wilkes, E. Craiovan, E. Topp, Z. Que, O. Seidou, S. Frey, and D. R. Lapen, submitted for publication). Notwithstanding these benefits, controlled tile drainage is currently not a practice that is ubiquitous in tile-drained regions throughout the world, and little is known about how this practice, when imposed en masse at a watershed scale, impacts the sources and degree of fecal pollution in surface water. A majority of experimental research on CTD is set at the field/plot scale and has focused primarily on other pollution targets (18–20). However, recently Schmidt et al. (21) found that CTD could potentially increase instantaneous loads and concentrations of fecal indicator bacteria and Campylobacter spp. in watersheds, but at plot scales, Frey et al. (9) found that regulating tile drainage has the potential for significantly reducing bacterial movement to surface water relative to conventional tile drainage following land applications of liquid swine manure. Other studies at field scale that completely shut down tile flow following manure application found a marked reduction in fecal indicator bacteria loads in comparison to free drainage (22). However, fully controlling tile drainage in this way may not be practical to carry out or necessarily beneficial with respect to field trafficking or water ponding potential at the soil surface.
In watersheds, which are open systems, the efficacy of a beneficial management practice (BMP) on microbial water quality will potentially be masked by multiple sources of fecal pollution (23–28). This was underscored in a study by Wilkes et al. (24), whereby occurrences of source-specific Bacteroidales microbial source tracking markers shifted as a result of restricting livestock access to streams. In many tile-drained landscapes, tile drainage can contribute a significant proportion of flow to surface water drainage systems (Sunohara et al., submitted), and it is hypothesized here that CTD imposed en masse on a field-to-field basis in a watershed will impact the sources of fecal contamination and pathogen occurrence in streams by virtue of CTD's considerable control of drainage water from farm field to stream.
A 7-year study was undertaken to examine the effects of CTD, employed during the growing season only, on the loading and/or occurrences of selected Bacteroidales microbial source tracking markers, viruses (pathogens and coliphages), and bacterial pathogens in paired agricultural watersheds in eastern Ontario Canada. The “test” watershed was under CTD intervention (CTD watershed), and the “reference” watershed was under conventional, freely draining conditions (uncontrolled tile drainage [UCTD] watershed).
MATERIALS AND METHODS
Study site and controlled tile drainage.
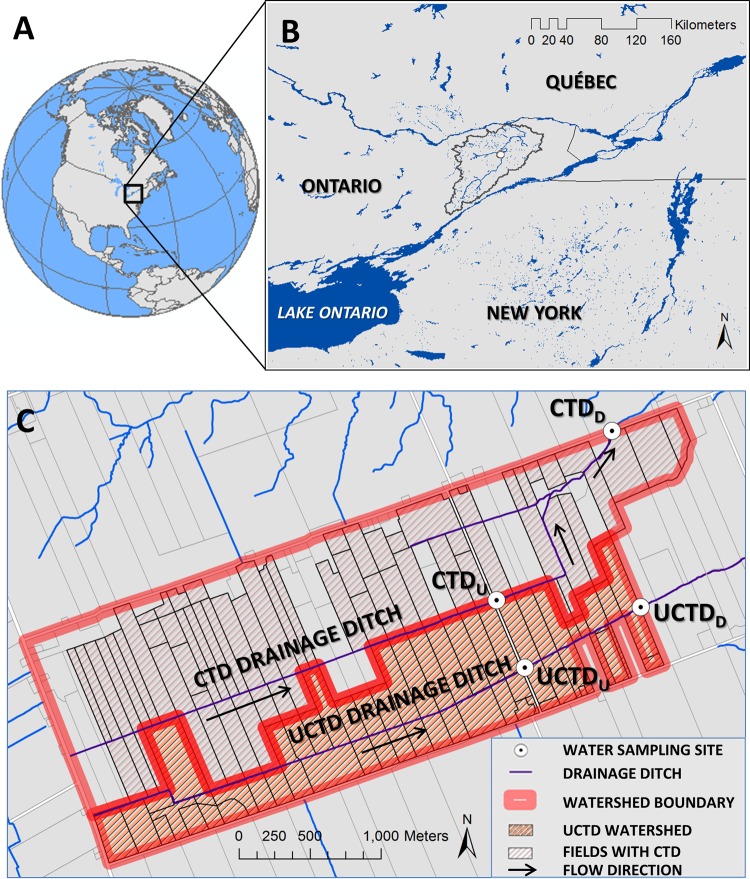
Schmidt et al. (21) and Sunohara et al. (submitted) provide background on the CTD and UCTD watersheds. The paired watersheds situated in eastern Ontario, Canada (Fig. 1A) are located in the South Nation River basin (Fig. 1B). The total surface catchment areas of the CTD and UCTD watersheds are 467 and 250 ha, respectively. The tile drainage-contributing areas (tile shed) for the CTD and UCTD watersheds are 415 and 225 ha, respectively. Due to the very flat topography of the watersheds, and the fact that the drainage ditches are manmade, the tile shed was used in the “load” calculations discussed later in the text. The two watersheds have similar flat topographies, soils, and land uses. Soils are dominated by Bainsville silt loams of the Gleysolic order (29). Mean daily temperatures peaked in July (∼22°C) and were lowest in January to February (∼5°C to −10°C) for the period from 2005 to 2011, whereas total yearly precipitation for 2005 to 2011 ranged between 600 and 1,100 mm (historical climate data for Russell, Ontario, from Environment Canada [http://climate.weather.gc.ca/] [climate ID 6107247], accessed 4 October 2013). The surface water systems are ice covered in winter. Tile drainage from fields is the dominant water input to the streams/ditches described in this study. Sunohara et al. (submitted) estimates that over 73% of stream flow is derived from tile drainage for these watersheds.
FIG 1.
(A) Study location in North America. (B) Study location in Eastern Ontario, Canada, and outline of the South Nation River basin, as well as sample locations (small white dots). (C) Map of the CTD and UCTD watersheds and water sample locations.
There are multiple farms on this watershed that operate independent cash/livestock cropping operations. Based on yearly field surveys of the watersheds made from 2005 to 2012, the CTD watershed was under ∼29 to 50% corn, ∼8 to 37% soybean, and ∼27 to 38% pasture/forage. Similar land cover percentages were found for corn (∼28 to 45%), soybean (∼5 to 22%), and pasture/forage (33 to 50%) on the UCTD watershed. Livestock fecal inputs to streams would be derived exclusively from land applications of dairy cow manure above sites CTDU and UCTDD (Fig. 1C). Land applications of manure (usually surface spread plus incorporation) occur in spring prior to planting (roughly an order of days prior) and in fall after crops have been harvested. Usually liquid bovine manure is applied to fields at application rates of around 47,000 to 75,000 liters ha−1. Within 200 m upstream of site 18, there exists a small hobby farm that consists of penned animals of low and variable seasonal intensity (observed were up to 10 to 20 goats, 1 donkey, and several horses) (31). Wildlife in these watersheds consisted of, at least, raccoons, skunks, muskrats, voles, mice, frogs, fish, turtles, white-tailed deer, and birds (including, but not limited to songbirds, geese, wild turkeys, crows, gulls, and ruffed grouse).
Tile-drained fields occupy roughly 89% and 90% of total catchment areas for the CTD and UCTD watersheds, respectively. The years 2005 and 2006 represented years where CTD intervention in the CTD watershed was insignificant, occupying only 4 to 9% of tile-drained fields in the watershed (pre-CTD intervention). From 2007 on, CTD was installed more intensely, representing ∼79% of the tile-drained fields in the watershed (CTD intervention period). Therefore, for 2007 to 2011, the CTD watershed was under CTD intervention and within the paired-watershed context was considered the “test” watershed. The UCTD watershed is considered a nontreated watershed, or within the paired-watershed context the “reference” watershed (where conventional or free tile drainage is employed exclusively) (Fig. 2A).
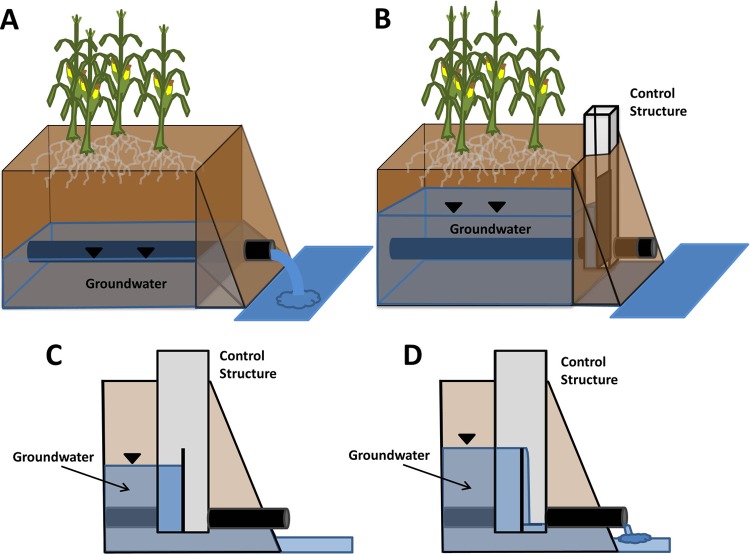
FIG 2.
(A) Tile in field under conventional or free drainage. (B and C) Tile with water flow control structure fully restricting tile flow to adjacent stream. (D) Tile with water flow control structure when water table depth exceeds height of control structure stop logs. Flow to adjacent stream is only partially restricted. Black triangles indicate the groundwater level.
Tile drainage on fields in the CTD watershed was controlled by means of inline water-level control structures (Agri Drain Corporation, Adair, IA) fitted with stop logs (or weir boards) (Fig. 2B to D). Flow control on fields in the CTD watershed was practiced between roughly planting/fertilizing to harvest of each year (i.e., the growing season). Flow control structures in the CTD watershed between roughly harvest and planting (winter season) were set for free drainage. Hence, during this harvest-to-planting time period (winter), the CTD watershed behaved effectively like the UCTD watershed regarding tile drainage. However, often manure applications conducted on the fields were done so during freely draining conditions in order to facilitate field trafficking. During spring, drainage control was nominally imposed on the CTD watershed soon after spring manure applications in timing with planting operations. Manure applied postharvest in the fall would typically be done so on fields that were freely draining, but not always, depending on producer practices and antecedent soil water conditions. During flow control times (approximately from planting/fertilizer application to harvest), the stop logs were set at a depth of ∼0.6 m below the soil surface so that when the water table was less (or shallower) than this depth below the surface, field tile water would overflow the stop logs and drain into the drainage ditch. This CTD attribute helped reduce waterlogging potential during wetter periods and facilitated root-water table interaction for crops. When water levels were below this depth (>0.6 m below surface), tile flow from field to adjacent stream was fully abated.
Stream monitoring and microbiological analyses.
Water sampling/monitoring sites CTDD and UCTDD represent the downstream monitoring sites for the CTD and UCTD watersheds, respectively (Fig. 1C). Water monitoring sites CTDU and UCTDU represent, respectively, upper reach monitoring sites in the CTD and UCTD watershed systems. Site CTDD is the furthest site downstream relative to all other sites and is thereby impacted by a larger area of land uses, including a small hobby farm immediately upstream of the site. For the other monitoring sites, there was no upstream influence from livestock fecal pollution outside bovine manures applied to fields potentially contaminating tile drainage.
From 2005 to 2011, water samples were collected on a biweekly basis (capturing base flow and storm flow events over many seasonal conditions and years) at the water sampling sites (total of 145 sampling occasions) targeting the following: (i) the detection and quantification of total Bacteroidales and host-specific Bacteroidales markers, including human, pig, ruminant, muskrat, and Canada goose markers (from 2005 to 2010); and (ii) detection of bacterial pathogens, including Listeria spp. and Listeria monocytogenes from 2005 to 2006 and Salmonella spp., Campylobacter spp. (Public Health Agency of Canada [PHAC]), and Escherichia coli O157:H7 from 2005 to 2011. Water samples were shipped overnight and processed the next day following the methodologies outlined by Marti et al. (32) for Bacteroidales detection, Lyautey et al. (33, 34) for Listeria and Listeria monocytogenes detection, and Jokinen et al. and Wilkes et al. (26, 35–37) for Salmonella, Campylobacter, and E. coli O157:H7 detection.
Briefly, to detect Bacteroidales source markers, 25 to 300 ml of water was filtered through 0.45-μm-pore-size Nuclepore membrane filters (Whatman, Thermo Fisher Scientific, Ottawa, Ontario, Canada). Filters were placed in a 15-ml Falcon tube containing 0.5 ml of guanidine isothiocyanate (GITC) buffer (5 mol liter−1 guanidine isothiocyanate, 100 mmol liter−1 ethylenediamine tetraacetic acid, 0.5% Sarkosyl) and frozen at −80°C until extraction. DNA was extracted using the DNeasy tissue kit (Qiagen, Mississauga, Ontario, Canada) following the manufacturer's instructions, except that the proteinase K step was omitted (elution volume, 100 μl). The PCR settings, amplification programs, and markers used have been described by Marti et al. (32). Two microliters of template DNA was used for each replicate. Negative controls (no template DNA) were performed in triplicate for each run. The presence/absence of PCR inhibitors was verified on a 10× template DNA dilution by using a TaqMan exogenous internal-positive-control kit (Applied Biosystems, Toronto, Ontario, Canada) following the manufacturer's instructions (46). If inhibitors were present, a 100× dilution of the DNA template proceeded. The number of marker copies per 100 ml was converted to copies day−1 ha−1 using total daily stream discharge (total volume of water discharged per day at the monitoring site) and the catchment area tile-drained fields upstream of the monitoring site. After sample processing for Bacteroidales source markers, ruminant detections were combined with pig detections into a new “livestock” category, and Canada goose plus muskrat markers were combined into a new “wildlife” category. It should be noted that the “total Bacteroidales” parameter is not discussed in depth in this paper because it is nonspecific in regards to source.
In 2008 to 2010, additional biweekly samples were collected targeting the detection of the following viruses: hepatitis A virus, astrovirus, norovirus genogroup I (GI), norovirus GIV, sapovirus, human Torque teno virus, adenovirus 40/41, and general adenovirus. All of these viruses are primarily associated with (a/w) humans. Additionally, norovirus GIII (a/w bovines) and Torque teno sus virus (a/w swine), hepatitis E virus, norovirus GII (a/w swine and human), and rotavirus were analyzed for detection. Samples were processed as described by Wilkes et al. (24, 31). In synchrony with the virus samples, samples were also collected for detection and quantification of F-specific RNA (F-RNA) and F-DNA coliphages as described by Wilkes et al. (24), resulting in F-RNA coliphages of human origin and animal origin.
Campylobacter spp. (Eastern Cereal and Oilseed Research Centre [ECORC]) were quantified by new most probable number (MPN) methods for samples from 2010 to 2011 as described by Schmidt et al. (21). Here, isolation and quantification of Campylobacter cells were performed using an 11 tube (1 of 500 ml, 5 of 100 ml, and 5 of 10 ml) MPN protocol. Each water sample was filtered through a 0.22-μm-pore filter, incubated in enrichment broth, streaked on agar, and reincubated under a microaerophilic condition. The putative Campylobacter colonies selected based on morphology and Gram staining reaction were confirmed by genus- and species-specific PCR assays. The density of Campylobacter cells (MPN, 100 ml−1) was calculated according to the method described by Oblinger and Koburger (38) and subsequently converted to a load identical to the method described previously for the Bacteroidales source markers.
Arcobacter quantification was conducted on samples for 2008 to 2011 according to Whiteduck-Leveillee (39). Water sample volumes of 10, 50, and 100 ml were analyzed by membrane filtration using 0.45-μm-pore filters; enumeration was expressed as CFU 100 ml−1 and subsequently converted to a daily load, as described previously for the quantified water quality endpoints. The filters were aseptically transferred to ASIA (Arcobacter selective isolation agar) medium and incubated under a microaerophilic condition. Putative colonies were further purified on m-AAM agar (modified agarized Arcobacter medium) and then confirmed by PCR assay (39).
Statistical and CART analyses.
Microbial source tracking (MST) marker and pathogen loads were calculated using tile drainage catchment area normalized daily flows (termed “normalized daily loads” for both markers and pathogens). Site and treatment (CTD watershed versus UCTD watershed) comparisons for these calculated loads were made using Mann-Whitney U tests. Occurrence data were also analyzed by treatment, using 2-by-2 contingency tables and Fisher's exact tests for consideration of significance. These analyses were split into preintervention (2005 to 2006) and CTD intervention (2007 to 2011) period data groupings regarding CTD on the CTD test watershed, as previously described. For the Mann-Whitney U and Fisher's exact tests, significance was deemed at P values of <0.05.
Interactions among CTD and UCTD treatments and season (with both drainage treatment and season considered independent variables in analysis) in terms of MST marker copies day−1 ha−1 (dependent variables) were examined by means of classification and regression tree analysis (CART) (Salford Systems, San Diego CA) on CTD intervention period data, following methods described by Wilkes et al. (31, 36) for regression trees. Succinctly, CART is a “machine-driven,” nonparametric and recursive partitioning method that splits dependent data (for, e.g., loads of Bacteroidales source marker copies day−1 ha−1) into homogenous (low-variance) groupings using independent data splitting conditions (for, e.g., season and/or treatment factors, considered potential predictors). CART systematically iterates through a data set and tests all possible split conditions of the dependent data (for, e.g., the aforementioned loads) using all possible independent variable data-dependent variable splitting possibilities. CART then selects the best independent variable split criteria that group (by minimizing variance) the dependent data into what are termed nodes and then repeats the process on these nodes. This process is terminated by the method and/or by user-defined constraints (thereby creating terminal nodes). The result of the process is a tree structure of independent variable splitting rules that identify potentially meaningful groupings of dependent data. The analysis is purely exploratory in the context of this article.
Odds ratio (OR) estimates and 95% confidence intervals (95% CIs) for the estimates were calculated between pathogens and ruminant and wildlife markers (markers deemed to reflect CTD effect most pertinently). For this study, OR values with 95% confidence values that did not bracket the value of 1 were considered for discussion purposes “proxies” for significance. The Mann-Whitney U test, Fisher's exact tests, CART analyses, and OR estimates between pathogens and markers were applied to all available data in pre-CTD intervention (2005 to 2006) and CTD intervention (2007 to 2011).
RESULTS
Occurrences of pathogens and Bacteroidales markers.
By comparing monitoring site data that had roughly similar temporally collocated support, or in other words, similar total sample numbers during the 2005 to 2006 pre-CTD intervention period, it was found that there were no significant differences among the CTD and UCTD watersheds in terms of the occurrence of any source specific MST marker (with CTDD, CTDU, and UCTDU as the sites considered) (Table 1). This finding was expected given the lack of significant CTD intervention during these years on the CTD watershed (Table 1); albeit only 2 years of pre-CTD intervention monitoring was achieved. For the same sites, and also including UCTDD, there were likewise no significant differences in the occurrences of selected bacterial pathogens among the CTD and UCTD watersheds during this preintervention period (Table 1).
TABLE 1.
The occurrence of microbial source tracking markers and pathogens prior to CTD intervention during years 2005 to 2006 of the study
| Microbial targeta | % of samples positive for microbial targetb |
n |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTDD | CTDU | UCTDD | UCTDU | CTDD − UCTDD | CTDD − UCTDU | CTDU − UCTDD | CTDU − UCTDU | CTDD | CTDU | UCTDD | UCTDU | |
| Total Bacteroidales | 100 | 100 | LD | 88 | LDc | 12 | LD | 12 | 29 | 21 | LD | 25 |
| Bacteroidales markers | ||||||||||||
| Human | 7 | 0 | LD | 4 | LD | 3 | LD | −4 | 29 | 21 | LD | 25 |
| Pig | 3 | 0 | LD | 0 | LD | 3 | LD | 0 | 29 | 21 | LD | 25 |
| Ruminant | 10 | 19 | LD | 12 | LD | −2 | LD | 7 | 29 | 21 | LD | 25 |
| Muskrat | 3 | 14 | LD | 16 | LD | −13 | LD | −2 | 29 | 21 | LD | 25 |
| Canada goose | 7 | 0 | LD | 0 | LD | 7 | LD | 0 | 29 | 21 | LD | 25 |
| Livestock | 14 | 19 | LD | 12 | LD | 2 | LD | 7 | 29 | 21 | LD | 25 |
| Wildlife | 10 | 14 | LD | 16 | LD | −6 | LD | −2 | 29 | 21 | LD | 25 |
| Other bacteria | ||||||||||||
| Listeria spp. | 89 | 100 | 94 | 100 | −5 | −11 | 6 | 0 | 18 | 16 | 17 | 15 |
| Listeria monocytogenes | 33 | 19 | 12 | 13 | 21 | 20 | 7 | 6 | 18 | 16 | 17 | 15 |
| Salmonella spp. | 6 | 9 | 9 | 13 | −3 | −7 | 0 | −4 | 35 | 32 | 33 | 31 |
| Campylobacter spp. | 31 | 31 | 24 | 35 | 7 | −4 | 7 | −4 | 35 | 32 | 33 | 31 |
| E. coli O157:H7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 35 | 32 | 33 | 31 |
Viral pathogens and coliphages were not collected in this period. “Livestock” and “Wildlife” are summary classes: “Livestock” represents pigs and/or ruminants, and “Wildlife” represents muskrats and/or Canada geese.
No significant differences between site pairwise comparisons were observed using Fisher's exact test (significant at P < 0.05). Note that there were percentage point differences for most CTD site − UCTD site percentage point values shown. Negative values indicate the occurrence of a greater percentage in UCTD versus pre-CTD by the indicated percentage point value and site comparison.
LD, limited data.
During the CTD intervention years of 2007 to 2011, for sites CTDU and UCTDU, which had relatively equal numbers of total samples for the aforementioned pairwise comparisons, there were significantly lower occurrences of livestock (pig plus ruminant)-, ruminant-, and human-specific MST markers in the CTD treatment watershed in relation to the UCTD watershed (Table 2). There were also significantly lower occurrences of Salmonella spp. and Arcobacter spp. at the CTDD site in relation to both UCTD sites. Otherwise, outside a significantly higher occurrence of F-DNA coliphage in the CTD watershed, there were no other significant differences in occurrences of other microbial targets among watershed treatments (Table 2).
TABLE 2.
Occurrence of microbial source tracking markers and pathogens within the CTD intervention period 2007 to 2011 on the CTD watershed
| Microbial targeta | % of samples positive for microbial targetb |
n |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTDD | CTDU | UCTDD | UCTDU | CTDD − UCTDD | CTDD − UCTDU | CTDU − UCTDD | CTDU − UCTDU | CTDD | CTDU | UCTDD | UCTDU | |
| Total Bacteroidales | 91 | 96 | LDc | 74 | LD | 17* | LD | 22* | 57 | 57 | LD | 46 |
| Bacteroidales markers | ||||||||||||
| Human | 2 | 0 | LD | 9 | LD | −7 | LD | −9* | 57 | 57 | LD | 46 |
| Pig | 9 | 0 | LD | 0 | LD | 9 | LD | 0 | 57 | 57 | LD | 46 |
| Ruminant | 16 | 0 | LD | 9 | LD | 7 | LD | −9* | 57 | 57 | LD | 46 |
| Muskrat | 5 | 23 | LD | 11 | LD | −6 | LD | 12 | 57 | 57 | LD | 46 |
| Canada goose | 4 | 0 | LD | 0 | LD | 4 | LD | 0 | 57 | 57 | LD | 46 |
| Livestock | 24 | 0 | LD | 9 | LD | 15 | LD | −9* | 54 | 57 | LD | 46 |
| Wildlife | 7 | 23 | LD | 11 | LD | −4 | LD | 12 | 54 | 57 | LD | 46 |
| Coliphages | ||||||||||||
| F-DNA | 34 | 20 | 10 | 23 | 24* | 11 | 10 | −3 | 44 | 46 | 39 | 39 |
| F-RNA a/w humans | 5 | 15 | 13 | 15 | −8 | −10 | 2 | 0 | 44 | 46 | 39 | 39 |
| F-RNA a/w animals | 34 | 15 | 18 | 18 | 16 | 16 | −3 | −3 | 44 | 46 | 39 | 39 |
| Viruses | ||||||||||||
| Hepatitis A virus | 2 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 46 | 47 | 39 | 40 |
| Astrovirus a/w humans | 9 | 9 | 11 | 4 | −2 | 5 | −2 | 5 | 32 | 33 | 27 | 28 |
| Norovirus GI a/w humans | 9 | 9 | 5 | 5 | 4 | 4 | 4 | 4 | 46 | 47 | 39 | 40 |
| Norovirus GIII a/w bovines | 13 | 15 | 11 | 7 | 2 | 6 | 4 | 8 | 32 | 33 | 27 | 28 |
| Norovirus GIV a/w humans | 6 | 12 | 7 | 4 | −1 | 2 | 5 | 8 | 32 | 33 | 27 | 28 |
| Sapovirus a/w humans | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 32 | 32 | 27 | 28 |
| Torque teno virus | 24 | 17 | 23 | 23 | 1 | 1 | −6 | −6 | 45 | 47 | 39 | 40 |
| Torque teno sus virus | 27 | 15 | 15 | 15 | 12 | 12 | 0 | 0 | 44 | 47 | 39 | 40 |
| Adenovirus 40/41 a/w humans | 3 | 6 | 0 | 3 | 3 | 0 | 6 | 3 | 32 | 47 | 39 | 40 |
| General adenovirus a/w humans | 3 | 0 | 3 | 0 | 0 | 3 | −3 | 0 | 32 | 47 | 39 | 40 |
| Pathogenic bacteria | ||||||||||||
| Campylobacter spp. (ECORC) | 70 | 100 | 68 | 73 | 2 | UDSd | UDS | 27 | 27 | 12 | 22 | 11 |
| Arcobacter spp. | 69 | 79 | 78 | 94 | −9 | −25* | 1 | −15 | 58 | 43 | 51 | 34 |
| Salmonella spp. | 0 | 2 | 8 | 5 | −8* | −5 | −6 | −3 | 58 | 45 | 50 | 37 |
| Campylobacter spp. (PHAC) | 57 | 73 | 62 | 76 | −5 | −19 | 11 | −3 | 58 | 45 | 50 | 37 |
| E. coli O157:H7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 58 | 45 | 50 | 37 |
| Other viruses | ||||||||||||
| Hepatitis E virus GIII (swine) | 0 | 4 | 5 | 8 | −5 | −8 | −1 | −4 | 46 | 47 | 39 | 40 |
| Norovirus GII a/w human and swine | 9 | 6 | 3 | 8 | 6 | 1 | 3 | −2 | 46 | 47 | 39 | 40 |
| Rotavirus | 6 | 0 | 0 | 7 | 6 | −1 | 0 | −7 | 32 | 33 | 27 | 28 |
Listeria was not monitored in this period. “Livestock” and “Wildlife” are summary classes: “Livestock” represents pigs and/or ruminants, and “Wildlife” represents muskrats and/or Canada geese.
Note that there were percentage point differences for most of the CTD − UCTD site percentage point values shown. Negative values indicate the occurrence of a percentage greater in UCTD versus CTD by the indicated percentage point value and site comparison. *, significant result by Fisher's exact test (P < 0.05) for pairwise comparison of sites.
LD, limited data.
UDS, data support among sites not considered equal enough to support comparative statistical analyses.
Odds ratio analysis, using all available data, showed that Salmonella spp. had higher odds of occurring when the ruminant marker was present relative to when it was absent (OR, 6.37; 95% CI, 1.00 to 40.67) (Table 3), and the association had a P value of 0.050. Yet for Arcobacter spp., the odds of this pathogen occurring was lower if a ruminant marker was present, relative to when it was absent (OR, 0.28; 95% CI, 0.08 to 0.95). Norovirus GII (a/w human or swine) was found to have a greater odds of occurring in the presence of the ruminant marker, in relation to when it was absent (OR, 4.71; 95% CI, 1.07 to 20.85).
TABLE 3.
Odds ratios and 95% confidence intervals for selected pathogens and ruminant and wildlife MST markers for all years of the study (2005 to 2011)a
| Pathogen | Bacteroidales marker | Lower 95% CI of OR | OR | Upper 95% CI of OR | P valueb |
|---|---|---|---|---|---|
| Salmonella spp. | Ruminant | 1.00 | 6.37 | 40.67 | 0.050 |
| Wildlife | 0.45 | 2.82 | 17.47 | 0.267 | |
| Arcobacter spp. | Ruminant | 0.08 | 0.28 | 0.95 | 0.041* |
| Wildlife | 0.76 | 5.94 | 46.61 | 0.090 | |
| Norovirus GII | Ruminant | 1.07 | 4.71 | 20.85 | 0.041* |
| Adjusted norovirus GIIc | Wildlife | 0.01 | 0.17 | 3.01 | 0.228 |
Note that other pathogens did not have enough data support and/or had confidence intervals that straddled the value of 1 and therefore are not presented here. Only ruminant and wildlife markers were used here since they were deemed more directly influenced by CTD practices in these experimental watersheds.
*, result considered significant due to the confidence interval not bracketing unity and P < 0.05.
Here we added 0.5 within cells of the 2-by-2 matrix to perform OR and OR CI calculation due to a “zero” cell (i.e., there were no positive samples for both norovirus GII and Bacteroidales wildlife markers at the same site and time of sample).
Stream loads of pathogens and Bacteroidales markers.
There were no significant differences among the CTD and UCTD watershed monitoring sites for the pre-CTD intervention period (2005 to 2006) regarding loads of copies day−1 ha−1of source-specific markers (Table 4). For the CTD intervention period (2007 to 2011), the distributions of F-DNA and F-RNA coliphage normalized loads were significantly higher in the CTD watershed relative to the UCTD watershed (Table 5). Of note, the ruminant marker was only marginally insignificant at the 0.06 level for upstream monitoring sites (CTDU and UCTDU), where loads of mean copies day−1 ha−1 were higher in the UCTD watershed in relation to the CTD watershed. Other loads of microbial endpoints, including some bacterial pathogens, were not significantly different among watershed treatments during the CTD intervention period.
TABLE 4.
Mann-Whitney U test results and descriptive statistical summaries for specific Bacteroidales markers for the pre-CTD intervention period (2005 to 2006)a
| Site comparisonb | Bacteroidales microbial markerc | Total copies day−1 ha−1 |
Mean rank sum for: |
n for: |
Mann-Whitney U test P valued | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean |
Median |
CTD site | UCTD site | CTD site | UCTD site | |||||
| CTD site | UCTD site | CTD site | UCTD site | |||||||
| CTDD vs UCTDU | Total | 4.04 × 109 | 2.02 × 1010 | 5.17 × 108 | 6.65 × 109 | 16.3 | 25.3 | 23 | 16 | 0.017* |
| Human | 8.04 × 106 | 0 | 0 | 0 | 20.3 | 19.5 | 23 | 16 | 0.434 | |
| Pig | 0 | 0 | 0 | 0 | 20.0 | 20.0 | 23 | 16 | 0.989 | |
| Ruminant | 5.25 × 106 | 3.76 × 105 | 0 | 0 | 20.6 | 19.2 | 23 | 16 | 0.498 | |
| Muskrat | 1.36 × 104 | 4.42 × 107 | 0 | 0 | 18.8 | 21.8 | 23 | 16 | 0.136 | |
| Canada goose | 6.12 × 104 | 0 | 0 | 0 | 20.3 | 19.5 | 23 | 16 | 0.434 | |
| CTDU vs UCTDU | Total | 4.49 × 109 | 2.02 × 1010 | 1.47 × 109 | 6.65 × 109 | 14.0 | 17.9 | 15 | 16 | 0.244 |
| Human | 0 | 0 | 0 | 0 | 16.0 | 16.0 | 15 | 16 | 0.984 | |
| Pig | 0 | 0 | 0 | 0 | 16.0 | 16.0 | 15 | 16 | 0.984 | |
| Ruminant | 1.86 × 106 | 3.76 × 105 | 0 | 0 | 16.1 | 15.9 | 15 | 16 | 0.963 | |
| Muskrat | 2.43 × 106 | 4.42 × 107 | 0 | 0 | 15.4 | 16.6 | 15 | 16 | 0.579 | |
| Canada goose | 0 | 0 | 0 | 0 | 16.0 | 16.0 | 15 | 16 | 0.984 | |
Arithmetic means are used here for purely descriptive purposes.
UCTDD had limited data, and therefore data are not presented.
Densities of pathogens, viruses, and coliphages were not monitored in this period.
*, significant (P < 0.05).
TABLE 5.
Mann-Whitney U test results and descriptive statistical summaries for specific Bacteroidales markers, coliphages, and pathogens in the CTD intervention period (2007 to 2011)a
| Site comparison | Microbial target | Result for: |
Avg rank sum for: |
n for: |
Mann-Whitney U test P valueb | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean |
Median |
CTD site | UCTD site | CTD site | UCTD site | |||||
| CTD site | UCTD site | CTD site | UCTD site | |||||||
| CTDD vs UCTDU | Coliphages (total PFU day−1 ha−1) | |||||||||
| F-DNA | 4.32 × 106 | 4.26 × 105 | 0 | 0 | 37.8 | 32.0 | 36 | 33 | 0.142 | |
| F-RNA | 5.89 × 106 | 9.62 × 105 | 0 | 0 | 39.1 | 30.6 | 36 | 33 | 0.040* | |
| F-RNA GI (a/w animal) | 5.88 × 106 | 1.02 × 105 | 0 | 0 | 38.1 | 31.6 | 36 | 33 | 0.065 | |
| F-RNA GII (a/w human) | 2.19 × 103 | 8.60 × 105 | 0 | 0 | 33.4 | 36.7 | 36 | 33 | 0.195 | |
| F-RNA GIII (a/w human) | 0 | 0 | 0 | 0 | 35.0 | 35.0 | 36 | 33 | 0.995 | |
| F-RNA GIV (a/w animal) | 0 | 0 | 0 | 0 | 35.0 | 35.0 | 36 | 33 | 0.995 | |
| Bacteroidales (total copies day−1 ha−1) | ||||||||||
| Total | 8.95 × 109 | 7.49 × 1010 | 2.35 × 109 | 4.34 × 108 | 46.3 | 37.9 | 46 | 38 | 0.116 | |
| Human marker | 3.58 × 106 | 1.13 × 107 | 0 | 0 | 41.4 | 43.8 | 46 | 38 | 0.233 | |
| Pig marker | 1.12 × 108 | 0 | 0 | 0 | 44.2 | 40.5 | 46 | 38 | 0.066 | |
| Ruminant marker | 5.77 × 107 | 1.90 × 109 | 0 | 0 | 44.6 | 39.9 | 46 | 38 | 0.152 | |
| Muskrat marker | 2.78 × 105 | 3.15 × 106 | 0 | 0 | 41.8 | 43.3 | 46 | 38 | 0.490 | |
| Canada goose marker | 5.08 × 106 | 0 | 0 | 0 | 42.9 | 42.0 | 46 | 38 | 0.376 | |
| CTDD vs UCTDD | Campylobacter spp. (ECORC) (total MPN day−1 ha−1) | 3.02 × 105 | 1.19 × 105 | 8.40 × 103 | 6.21 × 103 | 22.0 | 21.9 | 24 | 19 | 0.990 |
| Arcobacter spp. (total CFU day−1 ha−1) | 4.93 × 106 | 2.91 × 107 | 4.43 × 105 | 1.57 × 106 | 40.5 | 47.0 | 46 | 40 | 0.222 | |
| Coliphage (total PFU day−1 ha−1) | ||||||||||
| F-DNA | 4.32 × 106 | 7.87 × 105 | 0 | 0 | 38.3 | 29.1 | 36 | 31 | 0.012* | |
| F-RNA | 5.89 × 106 | 3.87 × 105 | 0 | 0 | 37.3 | 30.2 | 36 | 31 | 0.088 | |
| F-RNA GI (a/w animal) | 5.88 × 106 | 3.12 × 105 | 0 | 0 | 36.2 | 31.4 | 36 | 31 | 0.181 | |
| F-RNA GII (a/w human) | 2.19 × 103 | 7.75 × 104 | 0 | 0 | 32.8 | 35.4 | 36 | 31 | 0.292 | |
| F-RNA GIII (a/w human) | 0 | 0 | 0 | 0 | 34.0 | 34.0 | 36 | 31 | 0.995 | |
| F-RNA GIV (a/w animal) | 0 | 0 | 0 | 0 | 34.0 | 34.0 | 36 | 31 | 0.995 | |
| CTDU vs UCTDU | Campylobacter spp. (ECORC) (total MPN day−1 ha−1) | 2.39 × 105 | 1.85 × 105 | 2.21 × 104 | 0 | 11.2 | 8.9 | 9 | 10 | 0.373 |
| Arcobacter spp. (total CFU day−1 ha−1) | 3.40 × 107 | 5.82 × 106 | 4.74 × 105 | 3.29 × 105 | 30.9 | 29.0 | 32 | 27 | 0.673 | |
| Coliphages (total PFU day−1 ha−1) | ||||||||||
| F-DNA | 4.23 × 104 | 4.26 × 105 | 0 | 0 | 34.4 | 36.8 | 37 | 33 | 0.466 | |
| F-RNA | 4.74 × 104 | 9.62 × 105 | 0 | 0 | 35.1 | 35.9 | 37 | 33 | 0.814 | |
| F-RNA GI (a/w animal) | 2.29 × 104 | 1.02 × 105 | 0 | 0 | 35.3 | 35.7 | 37 | 33 | 0.890 | |
| F-RNA GII (a/w human) | 2.38 × 104 | 8.60 × 105 | 0 | 0 | 34.8 | 36.3 | 37 | 33 | 0.599 | |
| F-RNA GIII (a/w human) | 2.31 × 103 | 0 | 0 | 0 | 35.9 | 35.0 | 37 | 33 | 0.360 | |
| F-RNA GIV (a/w animal) | 0 | 0 | 0 | 0 | 35.5 | 35.5 | 37 | 33 | 0.995 | |
| Bacteroidales (total copies day−1 ha−1) | ||||||||||
| Total | 1.35 × 1010 | 7.49 × 1010 | 1.95 × 109 | 4.34 × 108 | 45.4 | 37.0 | 44 | 38 | 0.107 | |
| Human marker | 0 | 1.13 × 107 | 0 | 0 | 40.0 | 43.2 | 44 | 38 | 0.061 | |
| Pig marker | 0 | 0 | 0 | 0 | 41.5 | 41.5 | 44 | 38 | 0.996 | |
| Ruminant marker | 0 | 1.90 × 109 | 0 | 0 | 40.0 | 43.2 | 44 | 38 | 0.061 | |
| Muskrat marker | 1.54 × 107 | 3.15 × 106 | 0 | 0 | 44.0 | 38.6 | 44 | 38 | 0.102 | |
| Canada goose marker | 0 | 0 | 0 | 0 | 41.5 | 41.5 | 44 | 38 | 0.996 | |
| CTDU vs UCTDD | Arcobacter spp. (total CFU day−1 ha−1) | 3.40 × 107 | 2.91 × 107 | 4.74 × 105 | 1.57 × 106 | 34.5 | 38.1 | 32 | 40 | 0.462 |
| Coliphage (total PFU day−1 ha−1) | ||||||||||
| F-DNA | 4.23 × 104 | 7.87 × 105 | 0 | 0 | 35.3 | 33.6 | 37 | 31 | 0.558 | |
| F-RNA | 4.74 × 104 | 3.87 × 105 | 0 | 0 | 33.8 | 35.3 | 37 | 31 | 0.663 | |
| F-RNA GI (a/w animal) | 2.29 × 104 | 3.12 × 105 | 0 | 0 | 33.6 | 35.5 | 37 | 31 | 0.503 | |
| F-RNA GII (a/w human) | 2.38 × 104 | 7.75 × 104 | 0 | 0 | 34.2 | 34.9 | 37 | 31 | 0.800 | |
| F-RNA GIII (a/w human) | 2.31 × 103 | 0 | 0 | 0 | 34.9 | 34.0 | 37 | 31 | 0.376 | |
| F-RNA GIV (a/w animal) | 0 | 0 | 0 | 0 | 34.5 | 34.5 | 37 | 31 | 0.995 | |
Arithmetic means are presented here for purely descriptive purposes.
*, significant (P < 0.05).
Interactions among season and tile drainage practice for pathogens and Bacteroidales markers.
CART analyses of copies day−1 ha−1 for Bacteroidales MST markers and loads of coliphages and a subset of enumerated pathogens indicated that optimal data split structures were dominated by drainage practice and seasonal interactions (Table 6 and Table 7). Only Arcobacter spp. exhibited a sole seasonal effect (summer > spring and fall) for regression tree growth under the CART frameworks defined previously (Table 6). Drainage treatment was found to be a sole data split variable for only F-RNA coliphage GII (a/w human) (UCTD > CTD) and the muskrat marker (UCTD > CTD). The CART analysis regression trees for other microbial endpoints demonstrated interactions among season and drainage treatment. For specific tree models generated, the groups with the highest mean copies day −1 ha−1 among the source tracking markers associated with the UCTD system, under seasonal interaction, were ruminants (UCTD in spring) and humans (UCTD in fall) (Table 7). For specific CART analysis regression trees produced, the highest groupings in mean copies day−1 ha−1 for the CTD system were found for the pig Bacteroidales marker (CTD in summer [site CTDD versus UCTDU]) and the muskrat Bacteroidales marker (CTD in spring [site CTDU versus UCTDU]). For Campylobacter spp. (ECORC), F-DNA coliphage, F-RNA coliphage, and F-RNA GI coliphage (a/w animal), the highest mean load groups were associated with the CTD watershed, under different seasonal dispositions (coliphages higher in summer and Campylobacter spp. [ECORC] higher in fall).
TABLE 6.
Daily normalized loads of pathogens and coliphages associated with final CART least-square regression tree analysisa
| Microbial target | CART data split criterion/criteria | n | Mean ± SD of terminal node microbial target data |
|---|---|---|---|
| Bacteria | |||
| Campylobacter spp. (total MPN day−1 ha−1) | Spring, summer | 44 | 2.08 × 104 ± 4.12 × 104 |
| Fall and UCTD | 10 | 3.90 × 105 ± 3.52 × 105 | |
| Fall and CTD | 8 | 1.09 × 106 ± 1.05 × 106 | |
| Arcobacter spp. (total CFU day−1 ha−1) | Fall, spring | 71 | 2.92 × 106 ± 5.93 × 106 |
| Summer | 74 | 3.28 × 107 ± 1.38 × 108 | |
| Coliphages (total PFU day−1 ha−1) | |||
| F-DNA | Fall, spring | 66 | 8.13 × 103 ± 3.59 × 104 |
| Summer and UCTD | 33 | 1.16 × 106 ± 3.02 × 106 | |
| Summer and CTD | 38 | 4.12 × 106 ± 1.61 × 107 | |
| F-RNA | Fall, spring | 66 | 3.62 × 105 ± 2.30 × 106 |
| Summer and UCTD | 33 | 6.71 × 105 ± 2.62 × 106 | |
| Summer and CTD | 38 | 5.56 × 106 ± 3.23 × 107 | |
| F-RNA GI (a/w animal) | Fall, spring | 66 | 8.94 × 104 ± 4.20 × 105 |
| Summer and UCTD | 33 | 2.81 × 105 ± 1.53 × 106 | |
| Summer and CTD | 38 | 5.54 × 106 ± 3.23 × 107 | |
| F-RNA GII (a/w human) | CTD | 73 | 1.31 × 104 ± 7.22 × 104 |
| UCTD | 64 | 4.81 × 105 ± 2.49 × 106 | |
| F-RNA coliphage GIII (a/w human) | No tree createdb | ||
| F-RNA coliphage GIV (a/w animal) | No tree createdc |
Shown are the mean (arithmetic) daily normalized loads (and standard deviation [SD]) of pathogens and coliphages associated with final CART least-square regression tree analysis using as input drainage practice (CTD or UCTD) and season (spring, summer, or fall) as independent criteria. The results presented here are limited to the CTD intervention period (2007 to 2011) but include all available site data.
There were only 3 values above zero for this endpoint in this case, limiting the possibility of split combinations available for the program to test for group differences in microbial targets. After running CART on these data, CART lists the following as the classic output for this condition: “No useful split was found. No tree created.”
There were no nonzero data to apply splitting rules to in this case. (All data here were 0.) CART lists the following as the classic output for this condition after running the routine: “No learn sample variance for target.”
TABLE 7.
Daily normalized loads of microbial source tracking endpoints associated with final CART least-square regression tree analysisa
| Site comparison | Bacteroidales microbial target | CART data split criterion/criteria | n | Mean ± SD of terminal node microbial target data (total copies day−1 ha−1) |
|---|---|---|---|---|
| CTDD vs UCTDU | Total | Fall, summer | 54 | 6.29 × 109 ± 1.38 × 1010 |
| Spring and CTD | 16 | 1.17 × 1010 ± 2.24 × 1010 | ||
| Spring and UCTD | 14 | 1.95 × 1011 ± 6.69 × 1011 | ||
| Human marker | CTD | 46 | 3.58 × 106 ± 2.40 × 107 | |
| UCTD and spring, summer | 32 | 6.84 × 106 ± 2.68 × 107 | ||
| UCTD and fall | 6 | 3.53 × 107 ± 7.89 × 107 | ||
| Pig marker | Fall, spring | 44 | 7.12 × 106 ± 4.67 × 107 | |
| Summer and UCTD | 18 | 0 ± 0 | ||
| Summer and CTD | 22 | 2.21 × 108 ± 7.04 × 108 | ||
| Ruminant marker | Fall, summer | 54 | 2.16 × 107 ± 1.15 × 108 | |
| Spring and CTD | 16 | 1.42 × 108 ± 5.16 × 108 | ||
| Spring and UCTD | 14 | 5.11 × 109 ± 1.84 × 1010 | ||
| Muskrat marker | CTD | 46 | 2.78 × 105 ± 1.31 × 106 | |
| UCTD | 38 | 3.15 × 106 ± 1.37 × 107 | ||
| Canada goose marker | No tree createdb | |||
| CTDU vs UCTDU | Total | Fall, summer | 52 | 9.98 × 109 ± 2.79 × 1010 |
| Spring and CTD | 16 | 1.18 × 1010 ± 1.51 × 1010 | ||
| Spring and UCTD | 14 | 1.95 × 1011 ± 6.69 × 1011 | ||
| Human marker | CTD | 44 | 0 ± 0 | |
| UCTD and spring, summer | 32 | 6.84 × 106 ± 2.68 × 107 | ||
| UCTD and fall | 6 | 3.53 × 107 ± 7.89 × 107 | ||
| Pig marker | No tree createdc | |||
| Ruminant marker | Fall, summer | 52 | 1.52 × 107 ± 1.09 × 108 | |
| Spring and CTD | 16 | 0 ± 0 | ||
| Spring and UCTD | 14 | 5.11 × 109 ± 1.84 × 1010 | ||
| Muskrat marker | Fall, summer | 52 | 3.98 × 106 ± 1.69 × 107 | |
| Spring and UCTD | 14 | 3.25 × 106 ± 1.17 × 107 | ||
| Spring and CTD | 16 | 3.41 × 107 ± 7.56 × 107 | ||
| Canada goose marker | No tree createdc |
Shown are the arithmetic mean and standard deviation (SD) of microbial source tracking endpoints associated with final CART least-square regression tree analysis using as input drainage practice (CTD or UCTD) and season (spring, summer, or fall) as independent criteria. The results presented here are limited to the CTD intervention period (2007 to 2011).
There is only 1 positive value for this case, limiting the possibility of split combinations available for the program to test for group differences in microbial targets. CART lists the following as the classic output for this particular CART analysis routine: “No useful split was found. No tree created.”
There were no nonzero data to apply splitting rules to in this case. (All data here were 0.) CART lists the following as the classic output for this condition after running the CART routine: “No learn sample variance for target.”
DISCUSSION
Significant differences in frequencies of occurrence of host-specific DNA markers and selected pathogens were not found for the pre-CTD intervention years of 2005 to 2006. This is consistent with initial expectations because, first, there was no significant CTD intervention to control water discharge from fields to the water courses during this time, and second, the environmental and land use affinities among the two watersheds were effectively similar. For the CTD intervention period of 2007 to 2011, when on average during these years ∼79% of the tile-drained fields were tile drain managed in the CTD watershed, significant differences associated with CTD and UCTD treatment effects for microbial source tracking host-specific DNA marker and pathogen occurrences were found. Overall, there were significantly lower occurrences in human-, ruminant-, and livestock (pig and ruminant)-specific Bacteroidales markers in the CTD watershed, by approximately 9 percentage points each. The finding of modestly lower ruminant marker occurrences associated with the upstream monitoring sites (upstream of the influence of the small hobby farm in the CTD watershed and therefore more purely influenced by tile drainage management practice) makes biophysical sense since CTD can physically control the degree of fecal pollution being transported from fields, where manure is applied and wildlife interact, to adjacent surface water. In these watersheds, a prevailing driver of input of livestock fecal material into streams is tile drainage. Sunohara et al. (submitted) estimated that ∼73% of stream discharge is derived from tile drainage in these watersheds. The predominant ruminants in the watershed are dairy cattle (Holstein), and dairy cow manure is always applied to various fields in this watershed in both spring and fall. Fecal pollution by other ruminants in this agricultural watershed is considerably rescinded to livestock sources. Frequent surveillance of livestock and wildlife activity in these watersheds supports such contentions. Nevertheless, since dairy operations dominate the livestock activities in these experimental watersheds, the ruminant findings are promising in terms of the potential for CTD to reduce fecal contamination of surface waters from fields that receive manure applications. The relatively higher occurrences of the human Bacteroidales marker in the upstream UCTD monitoring site cannot be easily explained on the basis of land use activities, since there was no known location upstream of CTDU and UCTDU where human fecal inputs were known to have systematically occurred.
In terms of loading processes associated with fecal markers, the lack of significant differences among the host-specific Bacteroidales markers for the watershed treatments during 2005 to 2006 (pre-CTD intervention) is consistent with the explanations already provided. During the CTD intervention years, however, no significant differences in source-specific Bacteroidales marker copies day−1 ha−1 were observed. However, it is important to note though that the differences in ruminant marker copies day−1 ha−1 between site CTDU in relation to site UCTDU were almost significant (P = 0.06 versus the significance threshold of 0.05) with lower copies day−1 ha−1 associated with site CTDU (Table 5). These marginally insignificant findings complement the significant ruminant marker occurrence results and support contentions of CTD being a beneficial management practice. It is speculated that the inline water-level control structures limit the transport of fecal pollution from fields where manure is applied and wildlife interact to adjacent surface water; as the upstream differences in marker endpoints observed here were not confounded by downstream land uses. Such downstream-confounding land uses include the hobby farm located just upstream of CTDD. The significantly higher loading of F-RNA and F-DNA coliphages associated with site CTDD in relation to the UCTD sample sites suggests some impact from the small hobby farm since comparisons among sites upstream of the farm on the CTD watershed (CTDu) and UCTD sites revealed no significant differences. Why these differences would have manifested themselves for coliphages and not the other microbial endpoints is unclear. However, it may have something to do with persistence of these organisms in environmental matrices, and moreover, they can be significantly shed by both animals and humans alike (40).
It should be noted again that controlled tile drainage was imposed roughly between planting and harvest (growing season), since farmers most often require free tile drainage outside this time to some degree in order to traffic their fields. Manure applications on fields typically occur just prior to planting and after harvest, when tile drainage control in the CTD watershed is not imposed. Therefore, the abatement potential of CTD regarding ruminant fecal pollution would manifest itself most strongly following spring manure applications at a minimum, since the period between manure applications on fields and conversion of free drainage on the CTD watershed to CTD was on the order of days. Thus, given the use of CTD during the growing season only, these results are especially promising as a basis to support CTD-based mitigation strategies of fecal pollution of surface waters. Such beneficial impacts of CTD on MST endpoints were further demonstrated in the CART analysis employed in this study.
In this study, CART analyses uncovered seasonal interactions with watershed tile drainage treatment for a majority of the microbial water quality targets quantified as a “load.” The groups with the greatest mean ruminant marker copies day−1 ha−1 were delineated for the UCTD watershed in spring when manure is applied to fields. Moreover, in middle to late spring, tile flow is controlled in the CTD watershed, accounting for lower flows in the CTD watershed in relation to the UCTD watershed (21; Sunohara et al, submitted). Also in spring, tile flows are usually higher in relation to summer tile flow (Sunohara et al., submitted). Controlled tile drainage would indeed have a maximal effect on ruminant loading during these times given that dominant sources of ruminant fecal material would be derived from livestock manure applications and that CTD controls contaminated tile water discharge from field to stream on a field-to-field basis in this experimental watershed setting. This maximal effect was observable and evidenced at site CTDU in spring, which had no observable ruminant detection during the treatment period. However, there were also seasonal-drainage treatment interactions that identified higher relative loads in the CTD watershed during the CTD intervention period for Campylobacter spp. (ECORC), F-RNA coliphage, F-DNA coliphage, and F-RNA coliphage GI (a/w animal). This was the only time coliphage level was determined, as it was not done in 2005 to 2006 prior to CTD intervention. Reasons for these differences are not entirely clear, but since there were at least animal affinities with the coliphage data and affinities among Campylobacter spp. and avian source-classed Cryptosporidium identified in other studies (26), it is plausible that reduction of field-based water inputs to a stream via CTD reduces flushing and dilution of fecal material in the stream system, thereby increasing concentrations (see reference 21 for a similar discussion). This consideration is supported by the fact that (i) summer is a time that farmers who grow crops do not apply manure to their fields, (ii) tile flow control is strongest during the growing season (15; Sunohara et al., submitted), (iii) stream flow is lowest during this period to begin with, which may have been especially critical in terms of exposing streambed sediments to rainfall erosion (more so in the flow-controlled CTD watershed), (iv) wildlife frequently interact and produce fecal droppings in the stream or near the stream corridor in the summer months (31), (v) the hobby farm animals are present in open pens and pasture in the summer, and (vi) summer had a higher coliphage load in the stream in relation to the other seasons.
It was found that occurrences of Salmonella spp. and Arcobacter spp. were significantly lower at CTD monitoring sites in relation to the UCTD monitoring sites during the CTD intervention period (by 8 and 25 percentage points, respectively). Salmonella spp. was found to be associated with livestock-source-classed Cryptosporidium (26) and ruminant Bacteroidales markers (32) in surface waters in the study region. Wilkes et al. (36) indicated that relatively significant discharge-generating events may need to transpire in order for the detection of Salmonella spp. to occur in surface waters in the area (i.e., runoff, erosion, and drainage events). These results are supporting evidence of Salmonella spp. originating directly from on-farm sources or sediment/soil receptors that are influenced by agricultural activities (S. K. Frey, N. Gottschall, G. Wilkes, D. Gregoire, E. Topp, K. D. M. Pintar, M. Sunohara, and D. R. Lapen, submitted for publication). The link between Salmonella spp. and livestock sources is underscored by the odds ratio results where the odds of Salmonella spp. occurring is higher (∼6.4) when the ruminant marker occurs in water than when the marker does not occur in water. Thus, controlling tile water flow from fields where manures are applied could be a means to control at least some movement of Salmonella spp. to surface water bodies. Frey et al. (9) also found that controlling tile drainage in fall after manure application significantly reduced fecal indicator bacterial loads in surface water, supporting the above contention. In addition, Frey et al. (41) found strong links among Salmonella detection and ruminant markers in a nearby watershed in the study area. Arcobacter spp. can exist in the gastrointestinal tracts of healthy cattle (42) and therefore could enter surface water in this watershed via cattle manure leaching into tile drains, as would be the case for any other manure-derived pathogen. However, unlike Salmonella spp., odds ratios among Arcobacter spp. and the ruminant marker indicated that the odds of this pathogen occurring in water decreases when the ruminant marker occurs, relative to when the marker is absent (OR, ∼0.3). However, the odds ratios among Arcobacter spp. occurrence and wildlife markers were nearly significant in the context of 95% confidence intervals bracketing 1, with a lower 95% confidence value of 0.76 and an OR value of ∼5.9. These findings are worth noting on their own since they are opposite from the trends that occur among the Salmonella spp. and the ruminant marker. Similarly, Campylobacter spp. versus ruminant ORs, although having confidence intervals bracketing 1, were lower than the ORs for Campylobacter spp. versus wildlife (data not shown). Thus, there may be an association between wildlife fecal pollution and Arcobacter spp. and the closely related Campylobacter spp. in these watersheds. The finding of statistically lower occurrences of Arcobacter spp. in the CTD watershed is in support of potential wildlife pollution sources (for, e.g., avian [43]) on fields where crops are grown. The resulting fecal pollution from these sources appears, at least statistically speaking, to be partially mitigated by tile drainage control.
An interesting and unexpected finding is the odds ratio associated with norovirus GII (a/w human and swine) and the ruminant marker (Table 3). The OR indicates the odds of this virus occurring increases when a ruminant marker occurs, relative to when the marker is not present. For now, the presence of norovirus GII (a/w human and swine) in bovine fecal material has not been reported. However, Mattison (44) identified partial GII.4 norovirus genomic sequences for the first time in cattle feces from a Canadian farm. Recombination, which is common in noroviruses, can occur when a host is infected with 2 different strains of viruses and has been recognized as partially responsible for the genetic diversity and continuing emergence of new noroviruses (45). Sequencing as well as other confirmatory evaluations regarding norovirus GII (a/w human and swine) links with bovine sources was not conducted in this study. It should be noted that there are reports in many rural areas in North America of human septage being put into manure storage systems. If that kind of material is applied to land as a manure amendment on farms in this study, it could account for the ruminant marker-norovirus GII (a/w human and swine) relationships seen in this study. Although the data were not presented, we did not find odds ratios among norovirus GII (a/w human and swine) and the human Bacteroidales marker that had confidence intervals that did not bracket the value of 1.
Some important conclusions can be drawn from this work. They include the following. (i) There were no differences in occurrences of pathogens and microbial source tracking markers and stream loading of microbial endpoints among the tile drain managed (CTD) and uncontrolled or conventional/free tile drained (UCTD) watersheds during the 2 years (2005 to 2006) before CTD was implemented broadly in the CTD watershed (P ≥ 0.05).
(ii) During the CTD intervention period (2007 to 2011) when the CTD watershed had widespread tile drainage control (with 79% of tile-drained fields controlled during the growing season during these years), lower occurrences of the human, ruminant, and livestock (ruminant plus pig) markers were found for the CTD watershed in relation to the UCTD watershed (P < 0.05). As for pathogens and other microbial water quality parameters, there were only lower occurrences of Salmonella spp. and Arcobacter spp. in the CTD watershed (P < 0.05). There were no instances, other than for F-DNA coliphage, where there were significantly higher (at P < 0.05) occurrences of any marker or microbial target in the CTD watershed. This supports the general contention that water flow control was generally effective at reducing inputs of fecal material from fields to streams during the growing season.
(iii) It was found that the odds of Salmonella spp. occurring increased (OR, 6.37) when a ruminant marker was present (relative to marker absence), yet for Arcobacter spp., the opposite was true (OR, 0.28). Moreover, the odds of Arcobacter presence increased when a wildlife marker was present (OR, 5.94), relative to when the wildlife marker(s) was absent. Additionally the odds of norovirus GII (a/w human and swine) occurring in water increased when a ruminant marker was present relative to when a ruminant marker was absent (OR, 4.71). The mechanisms for this particular association are not entirely clear. From these findings we conclude, in combination with factors identified in conclusion ii above, that CTD reduces transport of some pathogens from tile-drained fields to streams.
(iv) Regarding loads of microbial source tracking markers, pathogens, and viral indicators, there were only significantly higher loads of F-RNA and F-DNA coliphages from the CTD watershed during the CTD intervention period (P < 0.05). Coliphage loading was higher in the CTD watershed associated with a downstream monitoring site, possibly as a result of fecal material derived from a small hobby farm located upstream of the site. Yet it is also worthy of note that there was lower loading of the ruminant marker in the CTD watershed, but results were only marginally insignificant at the P = 0.06 level (in relation to the significance threshold of 0.05).
(v) Data mining by means of classification and regression tree (CART) (Salford Systems, San Diego, CA) analyses delineated a vast array of seasonal and drainage control interactions among microbial load targets of importance. For example, it was found for the ruminant marker that highest loads existed during spring in the UCTD watershed, which is consistent with reduced manure inputs into the stream system via tile drainage control in the CTD watershed. F-RNA coliphage loads associated with animals were highest in summer in the CTD watershed which may have been primarily of wildlife origin and the hobby farm as noted above. Irrespective, during this period, stream flushing was likely constrained and dilution inhibited in the CTD watershed.
(vi) While overall, this study suggests that CTD imposed at the watershed scale has a beneficial impact on occurrences of some pathogens and markers of ruminant origin, it should be noted that in this region, free tile drainage (to reduce soil water contents) is usually used around harvest time and immediately prior to planting to allow for field traffic activities such as tillage, planting, and manure/fertilizer application to land. Unfortunately, during these earlier spring and fall periods, manure is applied to fields, and thus abatement of fecal pollution potential by CTD would be reduced during these periods, and the CTD watershed would be expected to behave more in line of a freely draining watershed (UCTD watershed). Controlled tile drainage could be employed as a temporary manure management practice as described by Frey et al. (9). However, to reduce manure borne fecal contamination to streams in more temperate regions of the world where tile drainage is utilized, CTD is often imposed more readily during winter or “fallow” periods of the year, and therefore, we would expect greater potential for CTD to mitigate field-based fecal pollution of streams on a yearly basis. Nevertheless, as documented in this study, there are significant environmental benefits that can be garnered via the use of CTD during just the growing season.
ACKNOWLEDGMENTS
This study was funded by Agriculture and Agri-Food Canada through the Sustainable Agriculture Environmental Systems (SAGES) program, the Watershed Evaluation of Beneficial Management Practice (WEBs) program, the National Water Quality Surveillance Research Initiative through an agreement with Health Canada, the National Agri-Environmental Standards Initiative (NAESI) program of Environment Canada, and the Alberta Water Research Institute.
We thank Diane Medeiros (Health Canada) and South Nation Conservation Authority for helping to facilitate this research.
Footnotes
Published ahead of print 11 April 2014
REFERENCES
- 1.Kladivko EJ, Van Scoyoc GE, Monke EJ, Oates KM, Pask W. 1991. Pesticide and nutrient movement into subsurface tile drains on a silt loam soil in Indiana. J. Environ. Qual. 20:264–270. 10.2134/jeq1991.00472425002000010043x [DOI] [Google Scholar]
- 2.Skaggs RW, Breve MA, Gilliam JW. 1994. Hydrologic and water quality impacts of agricultural drainage. Crit. Rev. Environ. Sci. Technol. 24:1–32. 10.1080/10643389409388459 [DOI] [Google Scholar]
- 3.Blann KL, Anderson JL, Sands GR, Vondracek B. 2009. Effects of agricultural drainage on aquatic ecosystems: a review. Crit. Rev. Environ. Sci. Technol. 39:909–1001. 10.1080/10643380801977966 [DOI] [Google Scholar]
- 4.Evans RO, Skaggs RW, Gilliam JW. 1995. Controlled versus conventional drainage effects on water quality. J. Irrig. Drain. Eng. 121:271–276. 10.1061/(ASCE)0733-9437(1995)121:4(271) [DOI] [Google Scholar]
- 5.Thornley S, Bos AW. 1985. Effects of livestock wastes and agricultural drainage on water quality: an Ontario case study. J. Soil Water Conserv. 40:173–175 [Google Scholar]
- 6.Scott CA, Geohring LD, Walter MF. 1998. Water quality impacts of tile drains in shallow, sloping, structured soils as affected by manure application. Appl. Eng. Agric. 14:599–603. 10.13031/2013.19428 [DOI] [Google Scholar]
- 7.Lapen DR, Topp E, Edwards M, Sabourin L, Curnoe W, Gottschall N, Bolton P, Rahman S, Ball-Coelho B, Payne M, Kleywegt S, McLaughlin N. 2008. Effect of liquid municipal biosolid application method on tile and groundwater quality. J. Environ. Qual. 37:925–936. 10.2134/jeq2006.0486 [DOI] [PubMed] [Google Scholar]
- 8.Pappas EA, Kanwar RS, Baker JL, Lorimor JC, Mickelson SK. 2008. Fecal indicator bacteria in subsurface drain water following swine manure application. Trans. ASABE 51:1567. 10.13031/2013.25313 [DOI] [Google Scholar]
- 9.Frey SK, Topp E, Ball BR, Edwards M, Gottschall N, Sunohara M, Zoski E, Lapen DR. 2013. Tile drainage management influences on surface-water and groundwater quality following liquid manure application. J. Environ. Qual. 42:881–892. 10.2134/jeq2012.0261 [DOI] [PubMed] [Google Scholar]
- 10.Gottschall N, Edwards M, Topp E, Bolton P, Payne M, Curnoe W, Ball-Coelho B, Lapen DR. 2009. Nitrogen, phosphorus, and bacteria tile and groundwater quality following direct injection of dewatered municipal biosolids into soil. J. Environ. Qual. 38:1066–1075. 10.2134/jeq2008.0085 [DOI] [PubMed] [Google Scholar]
- 11.Geohring LD, Wright PE, Steenhuis TS. 1998. Preferential flow of liquid manure to subsurface drains, p 1–8 In Brown L. (ed), Drainage in the 21st century: food production and the environment. ASAE, St. Joseph, MI [Google Scholar]
- 12.Gilliam JW, Skaggs RW, Weed SB. 1979. Drainage control to diminish nitrate loss from agricultural fields. J. Environ. Qual. 8:137–142. 10.2134/jeq1979.00472425000800010030x [DOI] [Google Scholar]
- 13.Adeuya R, Utt N, Frankenberger J, Bowling L, Kladivko E, Brouder S, Carter B. 2012. Impacts of drainage water management on subsurface drain flow, nitrate concentration, and nitrate loads in Indiana. J. Soil Water Conserv. 67:474–484. 10.2489/jswc.67.6.474 [DOI] [Google Scholar]
- 14.Helmers M, Christianson R, Brenneman G, Lockett D, Pederson C. 2012. Water table, drainage, and yield response to drainage water management in southeast Iowa. J. Soil Water Conserv. 67:495–501. 10.2489/jswc.67.6.495 [DOI] [Google Scholar]
- 15.Sunohara M, Craiovan E, Topp E, Gottschall N, Drury CF, Lapen DR. 2014. Comprehensive nitrogen budgets for controlled tile drainage fields in eastern Ontario, Canada. J. Environ. Qual. 10.2134/jeq2013.04.0117 [DOI] [PubMed] [Google Scholar]
- 16.Ghane E, Fausey N, Shedekar V, Piepho H-P, Shang Y, Brown L. 2012. Crop yield evaluation under controlled drainage in Ohio, United States. J. Soil Water Conserv. 67:465–473. 10.2489/jswc.67.6.465 [DOI] [Google Scholar]
- 17.Frankenberger J, Kladivko E, Sands G, Jaynes D, Fausey N, Helmers M, Cooke R, Strock J, Nelson K, Brown L. 2006. Drainage water management for the Midwest questions and answers about drainage water management for the Midwest. Publication no. WQ-44-W. Purdue Agriculture, Purdue University, West Lafayette, IN [Google Scholar]
- 18.Gaynor JD, Tan CS, Drury CF, Welacky TW, Ng HYF, Reynolds WD. 2002. Runoff and drainage losses of atrazine, metribuzin, and metolachlor in three water management systems. J. Environ. Qual. 31:300–308. 10.2134/jeq2002.3000 [DOI] [PubMed] [Google Scholar]
- 19.Wesström I, Messing I, Linnér H, Lindström J. 2001. Controlled drainage—effects on drain outflow and water quality. Agric. Water Manag. 47:85–100. 10.1016/S0378-3774(00)00104-9 [DOI] [Google Scholar]
- 20.Skaggs RW, Fausey NR, Evans RO. 2012. Drainage water management. J. Soil Water Conserv. 67:167A–172A. 10.2489/jswc.67.6.167A [DOI] [Google Scholar]
- 21.Schmidt PJ, Pintar KD, Fazil AM, Flemming CA, Lanthier M, Laprade N, Sunohara MD, Simhon A, Thomas JL, Topp E, Wilkes G, Lapen DR. 2013. Using Campylobacter spp. and Escherichia coli data and Bayesian microbial risk assessment to examine public health risks in agricultural watersheds under tile drainage management. Water Res. 47:3255–3272. 10.1016/j.watres.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 22.McLellan JE, Fleming RJ, Bradshaw SH. 1993. Reducing manure output to streams from subsurface drainage systems. American Society of Agricultural Engineers, St. Joseph, MI [Google Scholar]
- 23.Drozd M, Merrick NN, Sanad YM, Dick LK, Dick WA, Rajashekara G. 2013. Evaluating the occurrence of host-specific Bacteroidales, general fecal indicators, and bacterial pathogens in a mixed-use watershed. J. Environ. Qual. 42:713–725. 10.2134/jeq2012.0359 [DOI] [PubMed] [Google Scholar]
- 24.Wilkes G, Brassard J, Edge T, Gannon V, Jokinen C, Jones TH, Marti R, Neumann N, Ruecker N, Sunohara M, Topp E, Lapen DR. 2013. Coherence among different microbial source tracking markers in a small agricultural stream with and without livestock exclusion practices. Appl. Environ. Microbiol. 79:6207–6219. 10.1128/AEM.01626-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gentry RW, Layton AC, McKay LD, McCarthy JF, Williams DE, Koirala SR, Sayler GS. 2007. Efficacy of Bacteroides measurements for reducing the statistical uncertainty associated with hydrologic flow and fecal loads in a mixed use watershed. J. Environ. Qual. 36:1324–1330. 10.2134/jeq2006.0496 [DOI] [PubMed] [Google Scholar]
- 26.Wilkes G, Ruecker NJ, Neumann NF, Gannon VP, Jokinen C, Sunohara M, Topp E, Pintar KD, Edge TA, Lapen DR. 2013. Spatiotemporal analysis of Cryptosporidium species/genotypes and relationships with other zoonotic pathogens in surface water from mixed-use watersheds. Appl. Environ. Microbiol. 79:434–448. 10.1128/AEM.01924-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogel J, Stoeckel DM, Lamendella R, Zelt RB, Santo Domingo W, Walker SR, Oerther DB. 2007. Identifying fecal sources in a selected catchment reach using multiple source tracking tools. J. Environ. Qual. 36:718–729. 10.2134/jeq2006.0246 [DOI] [PubMed] [Google Scholar]
- 28.Fremaux B, Gritzfeld J, Boa T, Yost CK. 2009. Evaluation of host-specific Bacteroidales 16S rRNA gene markers as a complementary tool for detecting fecal pollution in a prairie watershed. Water Res. 43:4838–4849. 10.1016/j.watres.2009.06.045 [DOI] [PubMed] [Google Scholar]
- 29.Wicklund RE, Richards NR. 1962. Soil survey of Russell and Prescott counties. Report no. 33 of the Ontario Soil Survey. Research Branch, Canada Department of Agriculture, Ottawa, Ontario, Canada [Google Scholar]
- 30. Reference deleted.
- 31.Wilkes G, Brassard J, Edge T, Gannon V, Jokinen CC, Jones TH, Neumann N, Pintar KDM, Ruecker N, Schmidt PJ, Sunohara M, Topp E, Lapen DR. 2013. Bacteria, viruses, and parasites in an intermittent stream protected from and exposed to pasturing cattle: prevalence, densities, and quantitative microbial risk assessment. Water Res. 47:6244–6257. 10.1016/j.watres.2013.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marti R, Gannon VP, Jokinen C, Lanthier M, Lapen DR, Neumann NF, Ruecker NJ, Scott A, Wilkes G, Zhang Y, Topp E. 2013. Quantitative multi-year elucidation of fecal sources of waterborne pathogen contamination in the South Nation River basin using Bacteroidales microbial source tracking markers. Water Res. 47:2315–2324. 10.1016/j.watres.2013.02.009 [DOI] [PubMed] [Google Scholar]
- 33.Lyautey E, Hartmann A, Pagotto F, Tyler K, Lapen DR, Wilkes G, Piveteau P, Rieu A, Robertson WJ, Medeiros DT, Edge TA, Gannon V, Topp E. 2007. Characteristics and frequency of detection of fecal Listeria monocytogenes shed by livestock, wildlife, and humans. Can. J. Microbiol. 53:1158–1167. 10.1139/W07-084 [DOI] [PubMed] [Google Scholar]
- 34.Lyautey E, Lapen DR, Wilkes G, McCleary K, Pagotto F, Tyler K, Hartmann A, Piveteau P, Rieu A, Robertson WJ, Medeiros DT, Edge TA, Gannon V, Topp E. 2007. Distribution and characteristics of Listeria monocytogenes isolates from surface waters of the South Nation River watershed, Ontario, Canada. Appl. Environ. Microbiol. 73:5401–5410. 10.1128/AEM.00354-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkes G, Edge T, Gannon V, Jokinen C, Lyautey E, Medeiros D, Neumann N, Ruecker N, Topp E, Lapen DR. 2009. Seasonal relationships among indicator bacteria, pathogenic bacteria, Cryptosporidium oocysts, Giardia cysts, and hydrological indices for surface waters within an agricultural landscape. Water Res. 43:2209–2223. 10.1016/j.watres.2009.01.033 [DOI] [PubMed] [Google Scholar]
- 36.Wilkes G, Edge TA, Gannon VP, Jokinen C, Lyautey E, Neumann NF, Ruecker N, Scott A, Sunohara M, Topp E, Lapen DR. 2011. Associations among pathogenic bacteria, parasites, and environmental and land use factors in multiple mixed-use watersheds. Water Res. 45:5807–5825. 10.1016/j.watres.2011.06.021 [DOI] [PubMed] [Google Scholar]
- 37.Jokinen CC, Edge TA, Koning W, Laing CR, Lapen DR, Miller J, Mutschall S, Scott A, Taboada EN, Thomas JE, Topp E, Wilkes G, Gannon VP. 2012. Spatial and temporal drivers of zoonotic pathogen contamination of an agricultural watershed. J. Environ. Qual. 41:242–252. 10.2134/jeq2011.0203 [DOI] [PubMed] [Google Scholar]
- 38.Oblinger JL, Koburger JA. 1975. Understanding and teaching the most probable number technique. J. Milk Food Technol. 38:540–545 [Google Scholar]
- 39.Whiteduck-Leveillee K. 2014. Characterization of two novel Arcobacter sp. isolated from fecal matter. M.S. thesis Centre INRS-Institut Armand-Frappier Laval, Quebec, Canada [Google Scholar]
- 40.Cole D, Long SC, Sobsey MD. 2003. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl. Environ. Microbiol. 69:6507–6514. 10.1128/AEM.69.11.6507-6514.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frey SK, Topp E, Edge TA, Fall C, Gannon VPJ, Jokinen CC, Marti R, Neumann NF, Ruecker NJ, Wilkes G, Lapen DR. 2013. Using SWAT, Bacteroidales microbial source tracking markers, and fecal indicator bacteria to predict waterborne pathogen occurrence in an agricultural watershed. Water Res. 47:6326–6337. 10.1016/j.watres.2013.08.010 [DOI] [PubMed] [Google Scholar]
- 42.Wesley I, Wells S, Harmon K, Green A, Schroeder-Tucker L, Glover M, Siddique I. 2000. Fecal shedding of Campylobacter and Arcobacter spp. in dairy cattle. Appl. Environ. Microbiol. 66:1994–2000. 10.1128/AEM.66.5.1994-2000.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pacha R, Clark GW, Williams EA, Carter AM. 1988. Migratory birds of central Washington as reservoirs of Campylobacter jejuni. Can. J. Microbiol. 34:80–82. 10.1139/m88-015 [DOI] [PubMed] [Google Scholar]
- 44.Mattison K, Shukla A, Cook A, Pollari F, Friendship R, Kelton D, Bidawid S, Farber JM. 2007. Human noroviruses in swine and cattle. Emerg. Infect. Dis. 13:1184–1188. 10.3201/eid1308.070005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bull RA, Tanaka MM, White PA. 2007. Norovirus recombination. J. Gen. Virol. 88:3347–3359. 10.1099/vir.0.83321-0 [DOI] [PubMed] [Google Scholar]
- 46.Marti R, Zhang Y, Tien Y-C, Lapen DR, Topp E. 2013. Assessment of a new Bacteroidales marker targeting North American beaver (Castor canadensis) fecal pollution by real-time PCR. J. Microbiol. Methods 95:201–206. 10.1016/j.mimet.2013.08.016 [DOI] [PubMed] [Google Scholar]