Abstract
The anoxic and freezing brine that permeates Lake Vida's perennial ice below 16 m contains an abundance of very small (≤0.2-μm) particles mixed with a less abundant population of microbial cells ranging from >0.2 to 1.5 μm in length. Fluorescent DNA staining, electron microscopy (EM) observations, elemental analysis, and extraction of high-molecular-weight genomic DNA indicated that a significant portion of these ultrasmall particles are cells. A continuous electron-dense layer surrounding a less electron-dense region was observed by EM, indicating the presence of a biological membrane surrounding a cytoplasm. The ultrasmall cells are 0.192 ± 0.065 μm, with morphology characteristic of coccoid and diplococcic bacterial cells, often surrounded by iron-rich capsular structures. EM observations also detected the presence of smaller unidentified nanoparticles of 0.020 to 0.140 μm among the brine cells. A 16S rRNA gene clone library from the brine 0.1- to 0.2-μm-size fraction revealed a relatively low-diversity assemblage of Bacteria sequences distinct from the previously reported >0.2-μm-cell-size Lake Vida brine assemblage. The brine 0.1- to 0.2-μm-size fraction was dominated by the Proteobacteria-affiliated genera Herbaspirillum, Pseudoalteromonas, and Marinobacter. Cultivation efforts of the 0.1- to 0.2-μm-size fraction led to the isolation of Actinobacteria-affiliated genera Microbacterium and Kocuria. Based on phylogenetic relatedness and microscopic observations, we hypothesize that the ultrasmall cells in Lake Vida brine are ultramicrocells that are likely in a reduced size state as a result of environmental stress or life cycle-related conditions.
INTRODUCTION
One of the coldest stable liquid cryoenvironments known on Earth is the recently described Lake Vida brine (1, 2). Lake Vida, situated in Victoria Valley, East Antarctica, is one of the largest lakes in the McMurdo Dry Valleys. Most of the dry valley lakes are perennially covered with three to six meters of ice, which allows for limited atmospheric exchange and light penetration. However, the thick (>27-m) perennial ice of Lake Vida is void of sunlight below ∼6 m due to gas bubbles with hoar frost and sediment layers (1) and creates a sealed brine reservoir below 16 m. This brine has not exchanged gases with the atmosphere for an estimated 2,800 years (1). The anoxic brine (with a salinity range from 188 to 210) is a unique ecosystem with a temperature of −13.4°C, pH of 6.2, high solute concentrations (e.g., NH4+, 3.8 mM; Fe, >300 μM; NO3−, 905 μM), and high levels of dissolved organic carbon (580 mg liter−1) (2).
Lake Vida brine microbial assemblage is dominated by Bacteria (∼107 cells ml−1) (2). Eight bacterial phyla were identified from a 16S rRNA gene clone library from brine collected by filtration with 0.2-μm-pore-size filters: Proteobacteria (classes Gammaproteobacteria, Deltaproteobacteria, and Epsilonproteobacteria), Lentisphaera, Firmicutes, Spirochaeta, Bacteroidetes, Actinobacteria, Verrucomicrobia, and candidate division TM7 (2). According to the authors, the brine microbial assemblage is composed of two cell size classes. One class consists of typical aquatic bacteria with sizes of >0.2 to 1.0 μm. The second cell size class, with 100 times the abundance of the larger cells, consists of ultrasmall cell-like particles of ∼0.2 μm.
Ultrasmall microorganisms (organisms small enough to pass through 0.2-μm-pore-size filters) have been observed inhabiting different environments as parasites, symbionts, and free-living cells. Phylogenetic, morphological, and (meta)genomic characterizations of cultivated and uncultivated ultrasmall microbial cells from glacial ice (3–5), seawater (6–8), permafrost (9–11), soil (12–14), subsurface water and deep groundwater (15), freshwater (16), acid mine drainage (17, 18), deep-sea hydrothermal vents (19–21), and insect guts (22) show how prevalent these tiny microorganisms are on our planet.
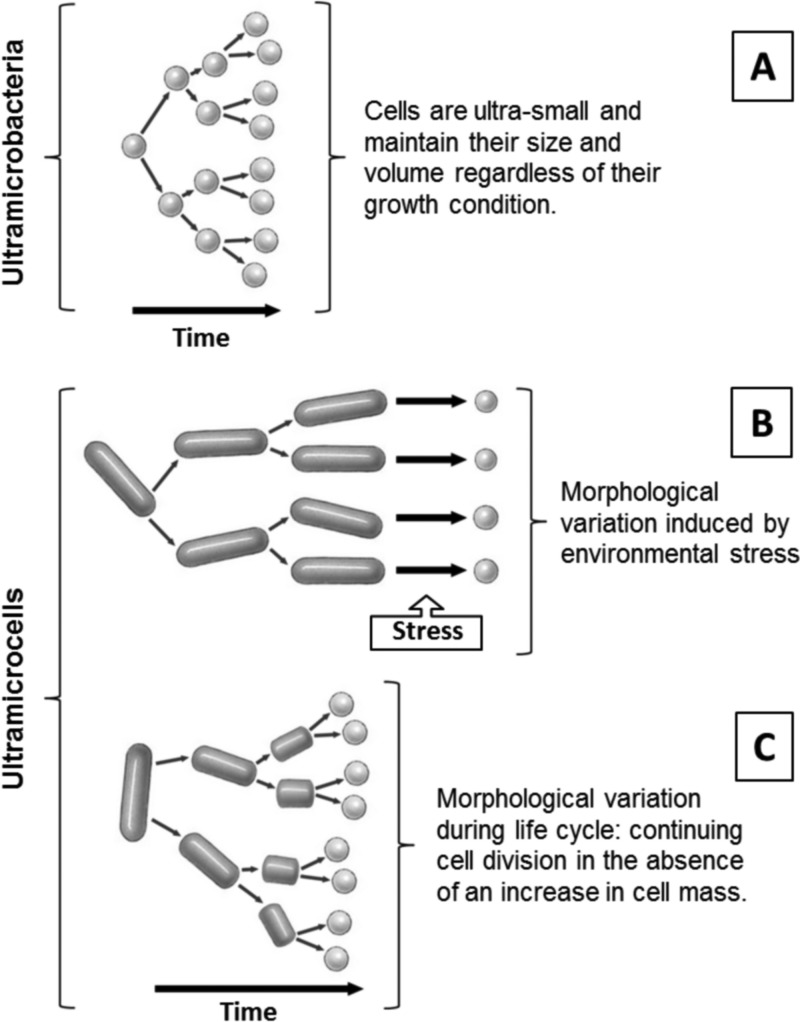
Different terminologies, such as ultramicrobacteria and ultramicrocells, are now used to classify ultrasmall microorganisms (reviewed in reference 23). The differences between ultramicrobacteria and ultramicrocells are structural and are related to specific life histories (Fig. 1). “Ultramicrobacteria” (24) is a term used to describe bacterial species with cell volumes of <0.1 μm3 which maintain similar cell size and volume regardless of their growth condition (Fig. 1A). The term “ultramicrocells” refers to cells reduced in size and volume as a consequence of unfavorable environmental factors or those with life cycle changes in morphology from rod to small cocci (Fig. 1B and C). Ultramicrocells are usually stressed, starved, dormant, or under osmotic pressure and increase in size and volume when cultivated (25, 26). Reduction of the cell size and morphological transition to a cyst-like resting cell have been previously described as strategies to maintain cell integrity and function in extremely cold and salty environments (27–29).
FIG 1.
Diagram demonstrating the distinctions between ultramicrobacteria and ultramicrocells. (A) Ultramicrobacteria; (B) ultramicrocells formed in response to unfavorable environmental conditions, such as hyperosmotic stress; (C) ultramicrocells formed as a consequence of aging; rod-to-sphere transition during late-log and stationary phases of the bacterial life cycle.
Here, microscopy imaging, elemental analyses, DNA-based molecular approaches, and culturing were employed to characterize the ∼0.2-μm cell-like particles previously observed by Murray et al. (2) in the Lake Vida brine. The results led to the conclusion that the abundant ∼0.2-μm cell-like particles are Bacteria-related ultramicrocells. The growth state and means for survival of Lake Vida ultramicrocells in the natural brine environment remain under investigation.
MATERIALS AND METHODS
Brine sampling and cell size fractionation.
Samples were collected in November 2010 from an ∼20.4-m hole drilled near the middle of Lake Vida, at 77°23′19.73′′S, 161°55′52.99′′E. To reduce risk of contamination, instruments used for entry and sampling approaches were cleaned with a dichloromethane-methanol mix (1:1, vol/vol) (30). After drilling, brine infiltrated the borehole until it reached a depth of ∼11 m below the ice surface (see Fig. S1 in the supplemental material). The brine was removed using a submersible pump (Proactive SS Monsoon) prior to sample collection to allow for sampling of fresh brine in the borehole. Samples were collected at a depth of 18.4 m using the submersible pump and polytetrafluoroethylene (PTFE) tubing that guided the brine to two separate PTFE lines (see Fig. S1). One line was used to collect the brine into 2- and 10-liter high-density polyethylene (HDPE) bottles under a nitrogen atmosphere for biomass collection. The second line guided the brine into an anaerobic glove bag (AtmosBag; Aldrich), where samples were collected for microscopy and geochemical analyses. Anoxic conditions were applied during all processes of sampling, filtering, and fixing the brine. Collected samples were stored at −10°C until processed.
All filtration was conducted under anoxic conditions at −10°C. For size fractionation, brine biomass of ≥0.2 μm was harvested on 0.22-μm-pore-size Sterivex filters (Millipore) and stored in a sucrose lysis buffer (31) for DNA extraction and on 0.22-μm-pore-size 142-mm membrane filters (Supor, Pall) and stored in RNAlater stabilization solution (Life Technologies) for total RNA extraction. The filtrate, composed of cells that passed through the 0.22-μm-pore-size filters, was collected in 1-liter glass bottles under anaerobic conditions and incubated for 30 days at −10°C. After incubation, the filtrate was passed again through 0.22-μm-pore-size Sterivex filters, and the cells that passed through the 0.22-μm-pore-size filters (the brine ultrasmall microbial assemblage [LVBrUMA]) were collected for DNA extraction and culturing. For DNA extraction, cells were collected on 0.1-μm-pore-size Supor-100 filters (Pall) and stored in a sucrose lysis buffer at −80°C. For culturing, the filtrate was preserved in 20% glycerol (vol/vol) and stored at −80°C.
Microscopy (confocal, scanning electron microscopy [SEM], and scanning transmission electron microscopy [STEM]) and energy dispersive X-ray spectroscopy (EDS) analysis.
Confocal microscopy (Olympus FluoView 1000 confocal microscope) was used to determine cell abundance in Lake Vida brine. Brine samples were fixed with 3.7% anoxic formalin (vol/vol) or 0.5% anoxic glutaraldehyde (vol/vol) under anoxic conditions during sampling. Fixed cells in the brine were stained with SYBR Gold (Invitrogen, Carlsbad, CA, USA) for 10 min and filtered onto 0.22-μm-pore-size black polycarbonate filters (Millipore) under a nitrogen atmosphere.
Electron microscopy was used to assess the size and morphology of the Lake Vida brine microbial cells. For SEM, brine aliquots (0.2 to 1 ml) were fixed with 0.5% anoxic glutaraldehyde (vol/vol final concentration) under anoxic conditions and filtered onto 0.22-μm-pore-size polycarbonate and 0.02-μm-pore-size Anodisc filters (Whatman). Cells were dehydrated by an ethanol series (30, 50, 70, 90, and 100%) under ambient conditions, dried in air, and coated with 1 to 3 nm of iridium or platinum to prevent charging during image acquisition. For STEM, two cell preparation protocols were developed. In one protocol, cells from a drop (20 μl) of anoxic fixed brine were transferred to a LuxFilmTM 2-mm open-area grid on a copper support and subsequently negative stained with phosphotungstic acid (PTA; H3PW12O40) at pH 0.4 for 30 s. In the second protocol, anoxic fixed brine was first treated with 40 mM EDTA (sample ratio by volume of 9:1), pH 8.0, for 10 min at room temperature, and then the suspension was transferred to a carbon type B 300-mesh copper grid, negative stained with uranyl acetate [UA; UO2(CH3COO)2·2H2O] at pH 4 to 5 for 5 min, and washed in deionized water. All buffers, solutions, and stains for EM analyses were filtered through 0.02-μm-pore-size filters immediately before use. EM observations were performed with a Carl Zeiss Ultra55 field emission instrument. SEM images were acquired with an Everhart-Thornley or annular secondary electron detector at working distances of 4 to 5 mm and 2.0 keV accelerating voltage. Beam energies of 20 to 30 keV were used to acquire STEM images.
EDS was performed using an Oxford Instruments INCA 350 system equipped with an XMax 80-mm2 silicon drift detector to determine the elemental composition of the brine cells supported on filters and grids. EDS signal counts were collected for 240 s (live time) with an accelerating voltage of 4 to 5 keV and an analytical working distance of 8.5 mm.
Cell and unidentified particle dimensions were measured using ImageJ (http://imagej.nih.gov/ij). A total of 465 cells and particles from SEM and STEM micrographs were measured to obtain representative size distributions.
Grazing angle X-ray diffraction.
Grazing angle X-ray diffraction (gXRD) patterns were acquired with a Bruker D8 Advance diffractometer using Cu(Kα) radiation (λ = 1.5405 Å) and an NaI scintillation detector to identify the inorganic phase(s) of iron-containing precipitates that coated the ultrasmall cells. A 0.2-μm-pore-size polycarbonate filter with a tan-colored mat of captured cells and brine material in suspension was used for the measurements. The diffractometer was equipped with parallel beam optics (Goebel mirror) and thin film receiving slit accessories to enable measurement at an angle of 1°. Diffraction patterns were acquired between 10 and 80°, 2θ (coherent scattering angle), with a data spacing of 0.02° at 3 s per step.
Cultivation of heterotrophic bacteria from brine fractions.
Cultivation efforts were conducted to compare isolate recovery and identity in 0.22-μm-pore-size-filtered and unfiltered brine and followed the approach used in previous reports (2, 32). Glycerol stocks of 0.22-μm-pore-size-filtered and unfiltered brine were inoculated and incubated under aerobic growth conditions at three different temperatures, 2°C, 10°C, and 21°C, on five different solid media prepared with Bacto agar for all but medium iii, which was purchased as agar medium: (i) R2A (Difco), (ii) R2A-5% NaCl, (iii) marine agar (Difco)-5% NaCl, (iv) casein (Difco), and (v) malt extract (for fungi; Bacto). Plate counts were determined after 120 days for the 0.22-μm-pore-size-filtered brine and after 110 days for the unfiltered brine. A subset of CFU was selected based on morphological characteristics for isolation and identification based on 16S rRNA gene phylogeny.
Nucleic acid extractions and 16S rRNA gene and phylogenetic analyses.
Genomic DNA was extracted from both 0.22-μm-pore-size Sterivex filters and the 0.22-μm filtrate collected on 0.1-μm-pore-size filters, according to Massana et al. (31). The genomic DNA extracted was quantified using PicoGreen (Life Technologies) on a Spectromax Gemini (Molecular Devices).
Total RNA was extracted from cells collected on 0.22-μm-pore-size 142-mm filters using a modified Totally RNA protocol (Life Technologies) (2). The volume of the 0.22- to 0.1-μm filtrate was limited and thus was not available for RNA extraction. In brief, filters were cut up using RNase-free blades in petri dishes and added to 50-ml conical tubes, and then acid phenol-chloroform was used as an initial separation step along with an additional phenol-chloroform extraction, followed by two chloroform extractions. Linear polyacrylamide was added to the first isopropanol precipitation step to facilitate nucleic acid precipitation. Two DNase I treatments (Life Technologies) were performed to ensure no genomic DNA contamination, followed by purification using an RNeasy kit (Qiagen). RNA was quantified using RiboGreen (Life Technologies). cDNA primed with random hexamers was reverse transcribed (SuperScript III; Life Technologies). No RNA was extracted from the 0.1- to 0.22-μm-cell-size fraction.
Molecular profiles of 16S rRNA gene fragments were conducted by denaturing gradient gel electrophoresis (DGGE; D-Code System; Bio-Rad), where (i) complementary reverse-transcribed DNA (crDNA) from cells ≥0.2 μm, (ii) genomic DNA from cells ≥0.2 μm, and (iii) genomic DNA from the 0.1- to 0.22-μm-cell-size fraction were amplified with the primers targeting the V3 variable region of the 16S rRNA gene: BactGC358f and 517r (33) following previously used conditions (34), with a total of 28 cycles. DGGE also followed a standard protocol (34) using a linear gradient of 30% to 65% denaturants. Lake Vida brine cloned 16S rRNA genes from 2005 samples (2) were used as DGGE molecular markers to identify comigrating bands. GelCompar II version 6.5 (Applied Maths) was used in gel image analysis, DGGE banding patterns based on presence and absence were compared using Pearson's correlation coefficient (r), and cluster analyses were performed according to the unweighted-pair group method with average linkages (UPGMA) using the Statistica version 6.1 software package (StatSoft). The average linkage distance between the three samples is the arithmetic mean of distances between all band pairs in different samples.
A 16S rRNA gene clone library was constructed to phylogenetically identify components of the 0.1- to 0.22-μm-cell-size assemblage. Products of four PCRs (24 cycles each) of genomic DNA amplified with the primers Bact27F and Univ1391R (35) were gel purified and pooled for cloning using the TOPO TA cloning kit (Life Technologies), according to the manufacturer's recommendation. Transformants were screened using blue-white screening. Plasmids were extracted by the QIAprep Spin miniprep kit (Qiagen) and sequenced with BigDye V3.1 (Life Technologies) using vector primers T7 and T3. Additionally, PCR amplicons (primers Bact27F and Univ1301R) from bacterial culture genomic DNAs were directly sequenced. DNA fragments were sequenced on a Prism 3730 DNA Analyzer (Life Technologies).
Sequences were trimmed and assembled, edited (BioEdit) (36), and checked for chimeras using Bellerophon (http://greengenes.lbl.gov) (37). To identify nearest neighbors, sequences were compared to the GenBank database by BLASTn (38) to the nucleotide collection (nr). Sequence alignments were constructed with the nearest alignment space termination (NAST) algorithm (39), and then a phylogenetic tree was constructed by neighbor-joining analysis (Mega 5) (40) with 1,000 bootstrap replicates. Evolutionary distances were computed using the Jukes-Cantor model.
Comparative phylogenetic analyses were performed among clone libraries from LVBrUMA, the Lake Vida brine 2005 clone library (2), the microbial assemblage within Lake Vida ice cover (34), and the Blood Falls (a subglacial outflow from the Taylor Glacier) microbial assemblage (41) following the methods of Murray et al. (2). Briefly, data sets downloaded from GenBank were aligned (NAST; 1,068 aligned base positions for all sequences), and then distance matrices were calculated for each library. Nonredundant data sets were generated for each library at a distance of 0.01 using Mothur (42). UniFrac (43) was used to calculate statistical comparisons among the libraries to investigate their associations with and without abundance weight of the microbial assemblage components.
Nucleotide sequence accession numbers.
16S rRNA gene sequences generated in this study are listed under accession numbers JX136966 to JX136986 for LVBrUMA and under KF384117 to KF384126 for representatives of the cultivated isolates.
RESULTS
Microscopy and elemental analyses.
Confocal microscopy observations of brine cells stained with a nucleic acid-targeted fluorescent stain indicated nucleic acid content in the cell-like particles (2). Coccoid cells with a consistent diameter of ∼0.2 μm accounted for 6.13 ± 1.65 107 cells ml−1, while larger cells (0.2 to 1.5 μm in length) accounted for 1.47 ± 0.25 105 cells ml−1.
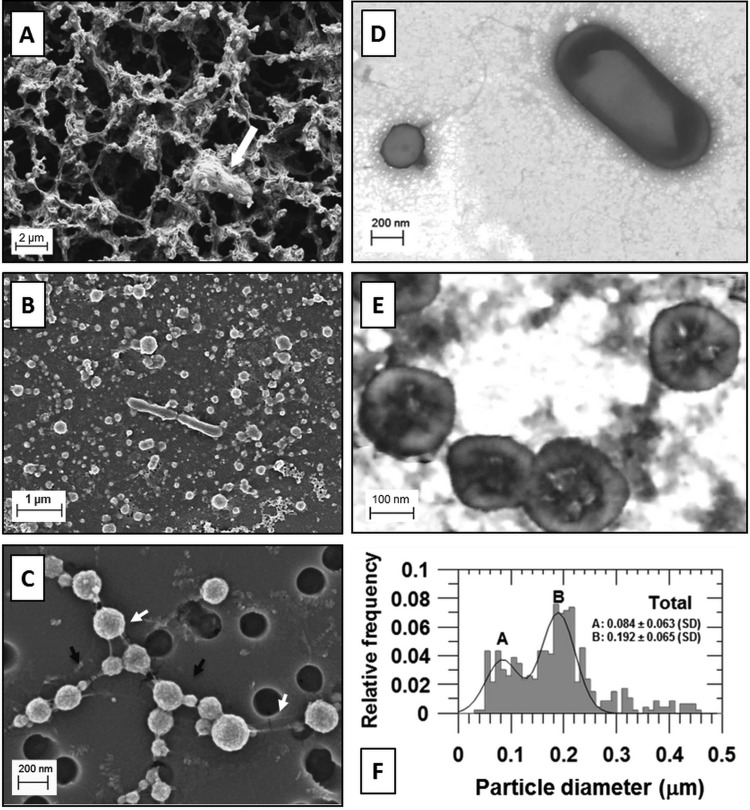
The two cell size populations were confirmed by EM (Fig. 2). SEM of brine fixed in glutaraldehyde (1 ml), filtered onto 0.2-μm-pore-size polycarbonate filters, and dried in air revealed a dense network of filamentous material colocated with cells and particles (Fig. 2A). The addition of an ethanol dehydration series after fixation and filtration resulted in disruption of the dense filamentous network, allowing visualization of a mat of cells and organic and inorganic material covering the filter surface (Fig. 2B). Coccoid cells of ∼0.2 μm in diameter, larger cells such as rod-shaped bacteria (∼0.4 μm by ∼1.5 μm), and abundant smaller particles (nanoparticles) ranging in size from approximately 0.020 to 0.140 μm in diameter were observed (Fig. 2B).
FIG 2.
The two cell size populations and nanoparticles of Lake Vida brine. (A) SEM of fixed brine prepared on a 0.2-μm-pore-size polycarbonate filter, air-dried (not dehydrated). A microbial cell trapped on the filamentous network is indicated by the white arrow. (B) SEM of fixed and dehydrated brine prepared on a 0.2-μm-pore-size polycarbonate filter showing the brine cells surrounded by a dense layer of organic and inorganic material covering the filter (volume of brine filtered, 1 ml). (C) SEM of fixed and dehydrated Lake Vida ultrasmall microbial cells prepared on a 0.2-μm-pore-size polycarbonate filter (volume of brine filtered, 0.5 ml). Nanoparticles attached to the cell surface (indicated by black arrows) and uncharacterized filaments connecting the cells (indicated by white arrows). (D) STEM of fixed and dehydrated brine cells treated with EDTA and stained with UA, pH 4 to 5. A cell of ∼0.2 μm in diameter is shown on the left of the image, and a rod-shaped bacterial cell is shown on the right. (E) STEM of ultrasmall brine cells stained with PTA, pH 0.4. Scale bars are shown in each micrograph. (F) Size distribution of cells and unidentified nanoparticles in Lake Vida brine.
Filtration of smaller brine volumes (≤500 μl), followed by ethanol dehydration steps, washed away more of the residue, facilitating the observation of the ultrasmall cells (Fig. 2C and 3A). The 0.2-μm pores of the polycarbonate filter provided a convenient particle size reference and indicated a size range of 0.03- to 0.25-μm particles (Fig. 2C). The ultrasmall cells were spherical, with a granular appearance or coating. In addition, fiber-like filaments connecting cells and particles were observed, likely a remnant of the filamentous material shown in Fig. 2A.
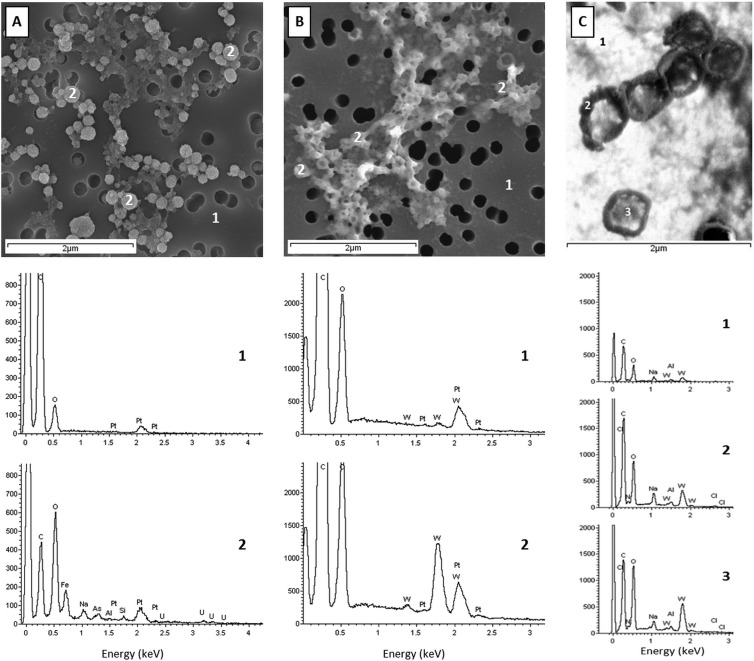
FIG 3.
EDS analyses of the brine ultrasmall cells. (A) EDS analyses of cells stained with uranyl acetate [UA; UO2(CH3COO)2·2H2O], pH 4 to 5; (B) EDS analyses of cells stained with UA followed by phosphotungstic acid stain (PTA; H3PW12O40), pH 0.4. Cells were fixed, stained, deposited on a polycarbonate filter, and coated with platinum. Numbers 1 and 2 mark the spots where EDS analyses were measured: 1, the background; 2, the cells. (C) EDS analyses during STEM observations of cells stained with PTA, pH 0.4, and deposited on a LuxFilm grid. Numbers 1, 2, and 3 mark the spots where EDS analyses were measured: 1, background; 2, electron-dense edge of the ∼0.2-μm cell; and 3, less electron dense in the middle of the cell. The weak nitrogen X-ray emission line is swamped by intense C and O signals, the latter due to the Fe,O coating. Elements identified by EDS: C, carbon; N, nitrogen; O, oxygen; Fe, iron; Na, sodium; As, arsenic; Pt, platinum; W, tungsten; Al, aluminum; Si, silicon; Cl, chlorine; and U, uranium.
When filtered cells were treated with a 0.4 pH phosphotungstic acid stain, the granular coating was removed and cells had a flattened appearance but were still round and exhibited a depressed center (Fig. 3B). Treatment of the brine with EDTA also removed the external coating that surrounded the cells (Fig. 2C), similar to the PTA stain (Fig. 3B). Particles substantially <0.2 μm in size were no longer distinctly observed. The very small size and less rigid structures may have allowed them to pass through the filter. Clearly, preparation of the brine for EM imaging was a critical step that revealed various features according to treatment.
STEM micrographs of negative-stained ultrasmall cells showed the effects of different preparative steps that removed the granular coating (Fig. 2D and E and 3C). Ultrasmall and rod-shaped cells did not appear to be collapsed (Fig. 2D) when treated with EDTA followed by uranyl acetate stain at pH 4 to 5, as did those treated with PTA (Fig. 2E and 3C). STEM observations showed an electron-dense layer surrounding a less electron-dense region, characteristic of negative-stained microbial cells in which the membrane takes up stain to provide contrast, permitting identification of cellular components. In addition to the individual cocci, diplococcic morphology was observed by visualization of cell envelope connections (Fig. 2E).
EDS measurements were performed to investigate the chemical nature of the apparent granular coating of 0.2-μm cells prepared under aerobic conditions and to provide additional evidence that these particles are indeed cells by colocation of negative stain with membranes. Figure 3A shows an SEM image of cells prepared as described for Fig. 2C, but with the addition of UA at pH 4 to 5. EDS spectra were acquired at cell locations and on the polycarbonate filter. The position labeled “1” in Fig. 3A has carbon and oxygen signals ascribed to the polycarbonate filter and a clear peak at 2.05 keV due to the Mα atomic emission line of Pt. Positions labeled “2” in Fig. 3A, located at cells, show a distinct iron peak at 0.71 keV. In addition, the oxygen-to-carbon peak intensity ratios are reversed compared to those of position 1, suggesting significantly more oxygen than carbon at cells. Significantly, weak but definitive signals due to the M-shell family of uranium lines were observed with a maximum at 3.172 keV (Mα) when the electron beam was focused on the cells but not when the polycarbonate filter was irradiated. This observation is highly suggestive of UA-stained 0.2-μm cells (44). To support this assertion, 10 EDS measurements, each of both ∼0.2- and ∼0.10-μm cells, were performed. Eighty percent of the 0.2-μm cells showed a definitive uranium signal, while only 30% of the ∼0.1-μm fraction had a spectrum similar to that shown in Fig. 3A. We hypothesize that there is a distinction in size between actual cells and nanoparticles, which were both coated with a robust iron and oxygen (Fe,O)-rich coat and had a granular appearance. Cells tended to be larger and took up the UA stain, while noncells (lacking the UA stain) were smaller.
Brine treated with EDTA or PTA at low pH removed the Fe,O-rich coating, which resulted in nonspherical cell shapes and collapsed cells on the filter (Fig. 3B). EDS spectra acquired from the filter (Fig. 3B, position 1) showed robust C, O, and Pt signals and a weak tungsten peak (W, Mα = 1.78 keV). However, spectra obtained from cell locations (Fig. 3B, position 2) showed a more intense signal due to the tungsten M-shell family of atomic lines. EDS spectra acquired from STEM specimens prepared with PTA also had a strong tungsten signal at cell positions and a weaker one on the carbon-coated TEM grid (Fig. 3C). Strong C and O peaks were accompanied by a weak N signal in the spectra of cells, while the background yielded weaker C and O peaks and no N signal (Fig. 3C, spectrum 1). The higher resolution of this image shows that both cell edges, membranes and centers, took up the PTA stain, yielding a W signal well above background levels.
EDS measurements showed that the granular coat and the spherical shape of the ultrasmall cells and nanoparticles were associated in the presence of iron (Fig. 3A). The Fe,O coat indiscriminately covers virtually all material in the brine under certain conditions (Fig. 2B and C and 3A). Grazing angle X-ray diffraction experiments were performed on a thick layer of fixed, filtered, and dehydrated brine to determine if the Fe,O material was a crystalline solid. The gXRD pattern was characteristic of an amorphous material; (bio)minerals such as ferrihydrite, goethite, or lepidocrocite, common in precipitated aqueous ferric solutions, were not observed (not shown).
The majority (96%) of the brine particles (cells and unidentified particles) observed by electron microscopy were ≤0.5 μm (Fig. 2F). Half of the particles (52%) ranged from 0.150 to 0.250 μm, and of these, 0.192 ± 0.065-μm coccoid cells (Gaussian curve B) comprised 56 to 60% of the total particles measured. Unidentified nanoparticles, 0.084 ± 0.063 μm (Gaussian labeled A, Fig. 2F), comprised 31 to 36% of the total particles measured. Differences in size distributions were identified when comparing particles observed by SEM compared to STEM and when comparing the particles with the presence and absence of amorphous Fe,O in the preparation (see Fig. S2 in the supplemental material). In the SEM observations, cells and nanoparticles appeared with similar abundance. The nanoparticles, however, were not detected in the STEM observations and were rare in preparations in the absence of the amorphous Fe,O coat, as discussed previously.
Heterotrophic bacterium cultivation.
Aerobic cultivation efforts with the ≤0.2-μm brine fraction at three different temperatures and with five different media yielded a total of 27 CFU isolated at either 10 or 20°C on the R2A-5% NaCl plates after 120 days. The 2°C incubation temperature and other four media tested did not yield growth after 120 days. Fifteen CFU were isolated and classified as being related to three Actinobacteria: Microbacterium lacus strain R-43968 (13 isolates; 98.8% identity across the 16S rRNA gene), Microbacterium testaceum strain S15-M13 (1 isolate; 99.8% identity across the 16S rRNA gene), and Kocuria marina strain KMM 3905 (1 isolate; 99.8% identity across the 16S rRNA gene) (Table 1). Observations by epifluorescence microscopy in cultures from LV10fR510-15 (Microbacterium lacus related), LV10fR510-15 (M. testaceum related), and LV10fR520-8 (Kocuria marina related) indicated rod morphology for all three strains (1.8 μm in length, 0.3 μm in diameter). In addition, after 10 days, small coccoid cells (average diameter of 0.235 μm) were observed to cooccur with the larger cells in cultures of LV10fR510-15 and LV10fR510-15. K. marina did not present any change in morphology. Marinobacter- and Psychrobacter-affiliated bacteria, commonly isolated in the unfiltered brine (2, 32) (see below), were not isolated from the ≤0.2-μm brine fraction. Isolation temperatures are indicated in Table 1.
TABLE 1.
Summary of the heterotrophic bacterial strains isolated from Lake Vida brine collected in 2010a
| Lake Vida brine isolate(s) (GenBank ID) | No. of isolates | Medium (temp [°C]) | Closest cultivated relative (GenBank ID) | % identity (nt) | Closest-relative notes (reference) |
|---|---|---|---|---|---|
| LV10fR510-15 (KF384117) | 13 | R2A-5% NaCl (10 and 20) | Microbacterium lacus strain R-43968 (FR691402) | 98.8 (1,185/1,199) | Psychrophilic irregular rod-shaped actinobacterium isolated from Lundström Lake, Shackleton Range, Antarctica (56) |
| LV10fR510-14 (KF384118) | 1 | R2A-5% NaCl (10) | Microbacterium testaceum strain S15-M13 (AM234161) | 99.8 (1,197/1,199) | Actinobacterium isolated from radioactive waste depository Tomsk-7, Siberia, Russia (77) |
| LV10fR520-8 (KF384119) | 1 | R2A-5% NaCl (20) | Kocuria marina strain KMM 3905 (AY211385) | 99.8 (1,203/1,205) | Halotolerant denitrifier coccoid actinobacterium isolated from marine sediment from East Siberian Sea (78) |
| LV10R510-5 (KF384120); LV10R510-8 (KF384121) | 20 | R2A-5% NaCl and marine agar-5% NaCl (10 and 20) | Marinobacter sp. strain LV10S (EU908281) | 99.5 (1,211/1,217) | Psychrotolerant, halophilic rod-shaped gammaproteobacterium isolated from Lake Vida brine collected in 2005 (32) |
| LV10R510-11A (KF384122); LV10MA510-1 (KF384123) | 8 | R2A-5% NaCl and marine agar-5% NaCl (10 and 20) | Marinobacter sp. strain ELB17 (AY518678) | 99.2 (1,221/1,230) | Psychrophilic denitrifier rod-shaped gammaproteobacterium isolated from east lobe of Lake Bonney, Antarctica, at a 17-m depth (60) |
| LV10R510-2 (KF384124) | 1 | R2A-5% NaCl (10) | Marinobacter psychrophilus strain R-36953 (FR691434) | 98.6 (1,214/1,230) | Psychrophilic rod-shaped gammaproteobacterium isolated from Forlidas Pond, Pensacola Mountains, Antarctica (56) |
| LV10R520-6 (KF384125) | 3 | R2A-5% NaCl and casein agar (10 and 20) | Psychrobacter sp. strain LV414 (EU908288) | 99.5 (1,209/1,214) | Psychrophilic, halophilic coccoid gammaproteobacterium isolated from Lake Vida brine collected in 2005 (32) |
| LV10R520-3 (KF384126) | 1 | R2A-5% NaCl (20) | Arthrobacter halodurans strain JSM 078085 (EU583729) | 97.2 (1,170/1,203) | Halotolerant denitrifier actinobacterium with rod-coccus growth cycle isolated from South China Sea seawater (61) |
16S rRNA gene sequences of the isolates were compared to GenBank, and the nearest related cultivated bacterium is listed along with GenBank accession number. Isolate identifier: brine with (LV10f) and without (LV10) size fractionation. GenBank accession numbers for Lake Vida type isolates are listed. Type isolates were selected to represent the group of isolates. Nomenclature as follow: sample/medium/temperature-isolate number. Examples: LV10fR520-8 (LV10f/R5/20-8) is Lake Vida brine collected in 2010 and fractionated, isolate cultivated on R2A-5% NaCl, at 20°C, isolate number 8; LV10MA510-1 (LV10/MA5/10-1) is Lake Vida brine collected in 2010, isolate cultivated on marine agar-5% NaCl, at 10°C, isolate number 1. Isolate 16S rRNA gene sequence GenBank accession numbers are in parentheses.
Cultivation efforts with the unfiltered brine yielded a total of 335 CFU after 110 days from which the 33 selected for isolation were phylogenetically affiliated with Marinobacter sp. strain LV10S (20 isolates; 99.5% identity), Marinobacter sp. strain ELB17 (8 isolates; 99.2% identity), Marinobacter psychrophilus strain R-36953 (1 isolate, 98.6% identity), Psychrobacter sp. strain LV414 (3 isolates at 10 and 20°C, 99.5% identity), and Arthrobacter halodurans strain JSM 078085 (1 isolate; 97.2% identity) (Table 1; see also Fig. 5). 16S rRNA gene sequence from the Marinobacter-related isolate LV10R510-8 was 100% identical to LVBrUMA clone sequences. Fungi were not detected in either brine fraction.
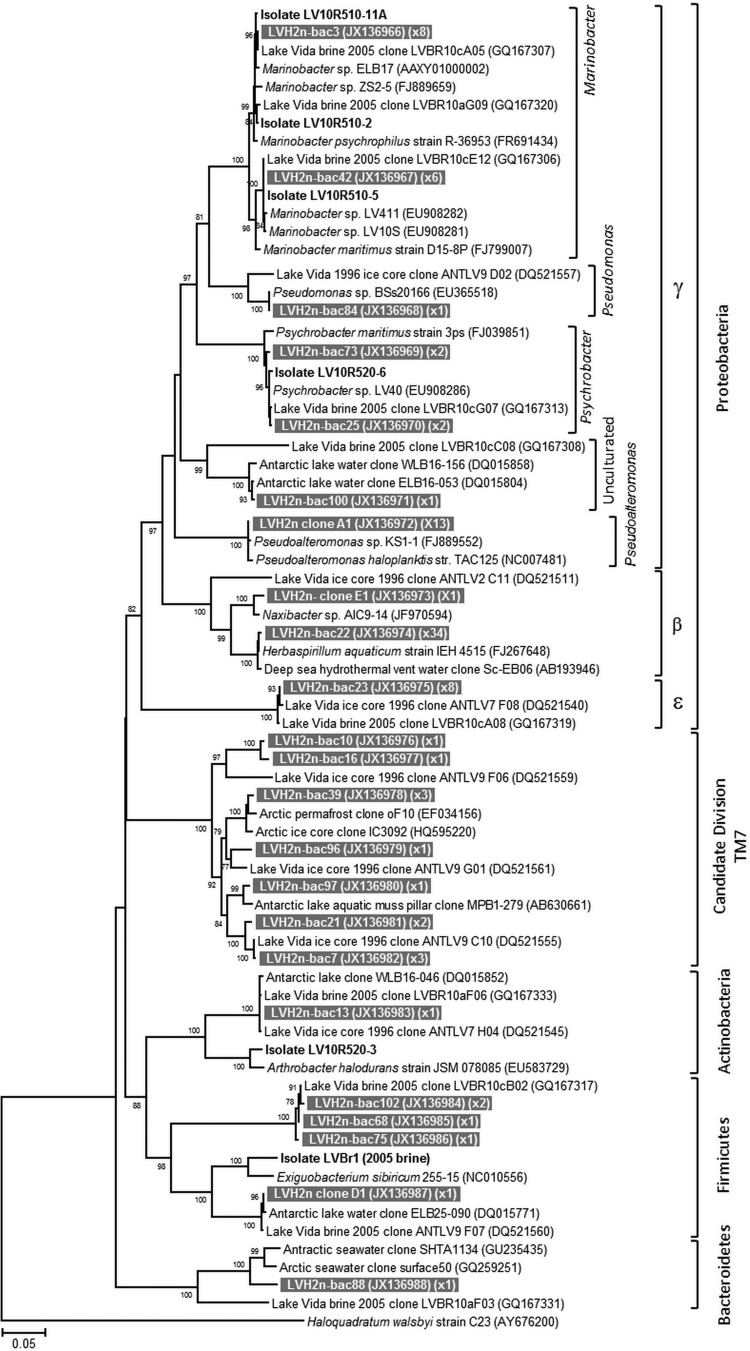
FIG 5.
Neighbor-joining phylogenetic tree based on near-complete 16S rRNA gene sequences from LVBrUMA and distance-affiliated sequences. The alignment utilized fragment sizes of 1,458 nucleotide positions. The bootstrap values of 1,000 repetitions with support values above 75 are represented at nodes. The scale bar denotes the divergence percentage between sequences. LVBrUMA environmental clones are represented by gray boxes with white text. GenBank accession numbers for the nucleotide sequences are in the first set of parentheses, following the sequence name. The number of clones related to the same sequence in the clone library (0.01 distance) is in the second set of parentheses. Lake Vida brine heterotrophic strains isolated and characterized in this study are in bold (exception: LV05br1 was isolated from brine collected in 2005); the nomenclature is the same as described in the Table 1 footnote.
Molecular analyses.
The microbial assemblages of the >0.2-μm brine fraction and LVBrUMA were initially compared by PCR-DGGE profiling of the 16S rRNA gene (Fig. 4). Cluster analysis showed that DNA and cDNA profiles from the >0.2-μm brine fraction had a linkage distance of 0.8 (Fig. 4, lanes 1 and 2, respectively) and clustered separately from the LVBrUMA DNA profile (lane 3), with a linkage distance of 1.2. The LVBrUMA 16S rRNA profile shared only six phylotypes out of 30 with the unfiltered brine microbial assemblage cDNA banding profile (Fig. 4A to F). Based on comigration, DGGE bands B, D, E, and F were related to Psychrobacter sp., two uncultured epsilonproteobacteria sequences, and Marinobacter sp.
FIG 4.

DGGE profile and cluster analysis of PCR-amplified 16S rRNA gene fragments and 16S cRNA from Lake Vida microbial assemblages. Samples were loaded as follows: lane 1, amplified DNA from the unfiltered brine; lane 2, amplified cDNA from the unfiltered brine; lane 3, amplified DNA from LVBrUMA. L, ladder, Lake Vida 16S rRNA gene clones phylogenetically related to uncultured Verrucomicrobia bacterium (GQ167324) (1), Psychrobacter sp. (GQ167313) (2), uncultured epsilonproteobacterium (GQ167319) (3), uncultured flavobacterium (GQ167312) (4), Marinobacter sp. (GQ167320) (5), Sphaerochaeta sp. (GQ167322) (6). The dendrogram, calculated based on the presence and absence of the amplified 16S rRNA gene partial fragments, shows the linkage distance between the samples. Letters A to F indicate the six phylotypes detected in LVBrUMA DNA and the unfiltered brine cDNA.
Analysis of 98 16S rRNA gene sequences from the LVBrUMA clone library showed that the 0.1- to 0.2-μm-size fractionated cells were affiliated with five bacterial phyla: Proteobacteria (Betaproteobacteria, Gammaproteobacteria, Epsilonproteobacteria), Actinobacteria, Firmicutes, Bacteroidetes, and candidate division TM7 (Fig. 5). The majority of sequences, 80%, were affiliated with the phylum Proteobacteria. Comparison between the 2005 16S rRNA gene clone library data of the brine microbial assemblage collected on a 0.2-μm-pore-size filter (LVBr) (2) and the 2010 ultrasmall microbial assemblage, LVBrUMA, revealed that the phylogenetic composition of the two cell size fractions had some overlap, though a number of LVBrUMA-specific sequences were detected. Gammaproteobacteria was the main class detected in both size fractions (38% of both assemblages). As shown in Fig. 5, sequences closely affiliated with Psychrobacter (98.6 to 99.1% identity), Marinobacter (98.6 to 99% identity), and the strain Pseudoalteromonas haloplanktis TAC125 (99.3% identity) prevailed in the Gammaproteobacteria class of the LVBrUMA fraction. The second most abundant group detected in the LVBrUMA fraction (35% of sequences) was affiliated with Betaproteobacteria, where sequences were closely affiliated with Herbaspirillum aquaticus strain IEH4515 (99.7% identity), which dominated the sequences detected. Betaproteobacteria-affiliated sequences were not detected in LVBr. Additionally, Deltaproteobacteria, Spirochaeta, and Lentisphaera sequences (comprising 1%, 7%, and 16% of LVBr, respectively) were not detected in LVBrUMA. Candidate division TM7-affiliated LVBrUMA sequences were more closely related to those recovered in Lake Vida ice at 15.9 m (Fig. 5; 94% identity) (34) than to those in the LVBr clone library (84% identity) (2).
The compositions of LVBrUMA and LVBr 16S rRNA gene clone libraries for sequences binned at a distance of 0.03 were 78% similar. Comparison of LVBrUMA with other environmental 16S rRNA gene clone libraries showed that LVBrUMA formed an external cluster with LVBr and the 15.9-m-deep portion of Lake Vida ice cover (see Fig. S3A in the supplemental material) when phylogenetic distance and the distribution of sequences were taken into account. Phylogenetic distance and sequence presence/absence analyses showed LVBrUMA clustering with the 15.9-m-deep ice core assemblage of the lake (see Fig. S3B).
DISCUSSION
Electron microscopy and elemental analyses.
The detection of membrane boundaries and stain signals above background by electron microscopy and EDS indicates that Lake Vida brine ∼0.2-μm cell-like particles are cellular entities. The observation of morphological variability, such as diplococci, cells connected by membrane-septa, presence of a capsular structure, and formation of aggregates, provides additional support to the conclusion that they are cells.
EDS measurements and grazing angle X-ray diffraction showed that the cells are surrounded by an amorphous iron and oxygen (Fe,O) layer. Aqueous Fe2+ intrinsic to Lake Vida brine may have oxidized to Fe3+ in air during sample preparation (after anoxic fixation), which then forms insoluble hydrous ferric oxide at near neutral pH, forming precipitates on cell and particle surfaces (45). To overcome this side effect and observe cells free of iron precipitates, PTA (pH 0.4) and EDTA were used to keep the iron in solution. Alternatively, the cells could be covered by an amorphous Fe,O layer in the natural brine environment. For example, Lake Vida brine contains a network of uncharacterized fiber-like filaments connecting cells and particles (Fig. 2A and C) (2), and we detected microorganisms (e.g., Pseudoalteromonas antarctica NF3) (46, 47) that are recognized EPS producers. Microbial cells enriched with exopolysaccharides (which carry a net negative electric charge and have the capacity to bind Fe2+ ions) are known to serve as nucleation sites for Fe(III) encrustation formed from oxidation reactions (48, 49). The production of EPS matrices by cells in cold and freezing environments is a strategy used for cryoprotection (50).
The nature of the smaller nanoparticles of 0.084 ± 0.063 μm, observed by SEM, remains under investigation. As the nanoparticles were more frequently observed in the presence of amorphous Fe,O, they could be a consequence from the nucleation of amorphous Fe,O on cellular debris, viral particles, other organic matter, or inorganic particles present in the brine. Evidence, including nanoparticle size, morphology, and attachment to the cell surfaces, motivates the hypothesis that some of the nanoparticles might be extracellular membrane vesicles (MV; see Fig. S4 in the supplemental material) (51). Vesicle production has been reported as a temperature stress response used by mesophiles to remove undesirable soluble and insoluble components (e.g., misfolded proteins) from the bacterial envelope (52). Protein misfolding in cold and freezing environments is a crucial obstacle that cells need to overcome for survival. Cold-adapted bacteria from the genera Marinobacter, Psychrobacter, and Pseudoalteromonas have been described as producing large numbers of vesicles (20 to 100 nm) attached to cells and extracellular polymeric substances (EPS) in cultures grown at temperatures from 4°C to 15°C (46, 53). Alternatively, the nanoparticles could also be representative of inclusion bodies (aggregated proteins associated with cell aging) (54, 55) or viral particles which could serve as nucleation sites precipitating amorphous Fe,O material. The role of viral particles is currently unknown in Lake Vida, though recognizable tailed bacteriophage particles were not observed by electron microscopy.
Composition of the assemblage of Lake Vida ultrasmall cells.
Size fractionation has been demonstrated to be valuable in accessing different components of microbial assemblages in which new and not readily cultivable bacteria have been enriched (5, 16) or molecular approaches used (15, 21) to access more diversity than otherwise apparent. The cultivation efforts in this study were limited to aerobic strategies, similar to what has been successful in previous efforts (2, 32); however, this approach potentially has not recovered the extent of diverse cultivable strains inhabiting the brine. The previous cultivation efforts recovered a limited diversity of facultative aerobes (Marinobacter sp. and Psychrobacter sp.) using both aerobic and anaerobic enrichment culture attempts. Cultivation efforts recovered different bacteria from the two cell size fractions and add to the extent of diversity recovered from Lake Vida brine. In all cases, we isolated organisms that were closely related (>97% sequence identity) to organisms from cold habitats but did not have identical 16S rRNA gene sequences to previously isolated strains. Micrococcaceae (strain LV10fR510-15), which represented the majority of the 0.2-μm-brine-fraction isolates, were closely related (98.8% identity) to Microbacterium lacus strain R-43968 isolated from a microbial mat at Lundström Lake, West Antarctica (56). Relatives of other 0.1- to 0.2-μm-brine-fraction strains (strain LV10fR510-14 and LV10fR520-8) were derived from northern high-latitude ecosystems in or near Siberia (Table 1).
The unfiltered brine cultivars (32 Gram-negative and one Gram-positive bacteria) yielded strains dominated by Marinobacter sp., similar to what has been previously reported for Lake Vida brine (2, 32), in which Psychrobacter sp. and Marinobacter sp. have repeatedly been cultivated. These genera are well known for inhabiting cold and salty environments (57–59). Two new strain-level Marinobacter sp. cultivars closely related to strains isolated from other Antarctic lake systems (56, 60) (Table 1) were isolated in this study, though no Marinobacter isolates were detected in the ≤0.2-μm brine fraction. Three Psychrobacter isolates (closely related to LV414) (32) and one Gram-positive Arthrobacter-related isolate (strain LV10R520-3) were also identified among the unfiltered brine cultures. The Arthrobacter strain was the first identified in this ecosystem and was phylogenetically affiliated with a halotolerant actinobacterium cultivated from seawater (61) and more distantly related to Arthrobacter sp. inhabiting Antarctic dry valley soils (62). In the anoxic brine habitat, the three Actinobacteria spp. and the Arthrobacter sp. likely grow using a fermentative metabolism, while those organisms related to Marinobacter sp. and Psychrobacter sp. could potentially use nitrate as a terminal electron acceptor.
Similar to the results of the cultivation efforts, the molecular profiling and sequencing results of the size-fractionated brine samples indicated overlap in assemblage composition as shown by the phylogenetic tree clusters, though pointed to distinct assemblage components between the cell size fractions. Size fractionation would not necessarily eliminate DNA-containing compromised cells from passing the 0.22-μm membrane. Those 16S rRNA gene sequences that distinguished the LVBrUMA and the >0.2-μm brine fraction resembled well-known bacterial groups (Herbaspirillum, Pseudoalteromonas) and candidate division TM7; none of these LVBrUMA-specific sequences were related to organisms brought into culture in this study or were detected in previous Lake Vida brine or ice 16S rRNA gene profiling surveys (2, 34). The prevalence of Betaproteobacteria-related Herbaspirillum sp. in the LVBrUMA (36% of the library) was unusual, given that we had not detected this group previously and that this genus is mostly known to inhabit soil environments where a number of the members of the genus are aerobic mesophiles capable of nitrogen fixation (63). The enrichment of other ultrasmall Betaproteobacteria clades by size fractionation has been reported in studies from Greenland ice, deep groundwater, and deep-sea hydrothermal vents (5, 15).
Sequences affiliated with Pseudoalteromonas haloplanktis strain TAC125 were also abundant (∼10%) in the LVBrUMA clone library. The genus Pseudoalteromonas is ubiquitous in marine environments (64), and P. haloplanktis, a psychrophilic moderate halophilic bacterium isolated from Antarctic seawater (65), has the capacity to grow at low temperatures and is well adapted to high salinities, with optimal growth between 1.5% and 3.5% of NaCl. Strain TAC125 has been shown to cope with membrane fluidity challenges at low temperatures by expressing lipid desaturases and has high expression of genes involved in the syntheses of biofilm (65). Together, these findings suggest that both Herbaspirillum and Pseudoalteromonas genera harbor strains with the potential for ultrasmall cell morphology, which, to our knowledge, is the first report of this cellular state for both genera.
Nature of the ultrasmall cells.
None of the bacterial groups detected in the LVBrUMA have been reported as ultramicrobacteria (small cells that maintain their size and volume regardless of the growth condition). The morphology observed in the brine ultrasmall microbial cells is comparable to that of cells with pleomorphic life cycles (emerging to ultrasmall cells in late life cycle stage), cyst-like resting cells, and cells exposed to stressful osmotic conditions (66, 67). Morphological transition is a genetically controlled phenomenon prevalent in bacterial life cycles and correlated to aging (68–70). For example, Actinobacteria, including Arthrobacter (66, 71) and Microbacterium (72), can be pleomorphic, meaning they can shift to ultrasmall cells during their life cycle. Likewise, unfavorable environmental conditions have been attributed to small cell size in Pseudomonas, Micrococcus, and Arthrobacter, which have been described with reduced cellular activity and size and a thick cellular envelope (73, 74). Cyst-like resting cells from Arthrobacter globiformis isolated from Siberian permafrost were characterized as dwarf forms of 0.2 μm in diameter, exhibiting a thick cell wall with an external capsular layer and low-electron-dense cytoplasm (66), similar to what we observed in this study with the Lake Vida ultrasmall microbial cells. Further, hyperosmotic stress can result in cell shrinkage as a consequence of water loss (Fig. 1B) (75). Bacteria with thick cell walls (Gram-positive bacteria), such as Actinobacteria and Firmicutes, typically do not change morphology in response to hyperosmotic stress. However, cells with a thin cell wall (Gram-negative bacteria), such as Proteobacteria, can shrink and become distorted and smaller, often acquiring a spherical shape. The characterization of ∼0.2-μm cells presented in this report is consistent with descriptions of stressed bacteria in extreme environments and provides strong evidence that LVBrUMA are ultramicrocells.
Lake Vida brine ultrasmall microbial cells are in accordance with the lowest theoretical size for a free-living prokaryotic cell, which is estimated to be a sphere of 250 to 300 nm in diameter (76). However, their means of survival in the natural brine environment remain under investigation. Studies of extreme environments such as Lake Vida brine and its ultrasmall microbial community provide additional impetus to understand the existence of life in freezing brines (potential exohabitats, such as in icy worlds) and to help determine the minimum size of a biological cell expected to be found in these subzero and hypersaline environments.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by NSF ANT-0739681 to A.E.M. and C.H.F. and by NSF ANT-0739698 to P.T.D; E.K. was supported by Fulbright/CAPES-BRAZIL grant 2163-08-8. A.S.I. and the SEM facility at SFSU were supported by NSF grants 0821619 and 0949176.
Thanks are extended to Clive Hayzelden, SFSU, for assistance on the SEM and STEM and Christiano Garnett Marques Brum, Arecibo Observatory, for guidance in data analyses. K. C. King, Desert Research Institute, and Tara N. Thomas, University of Nevada, are graciously appreciated for reviewing the manuscript. Special thanks to members of the 2010 Lake Vida Expedition team, Hilary Dugan, Bernd Wagner, Fabien Kenig, Brian Glazer, Seth Young, Peter Glenday, and Jay Kyne, and to Raytheon Polar Services for logistic support.
No potential conflicts of interests are declared.
Footnotes
Published ahead of print 11 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00276-14.
REFERENCES
- 1.Doran PT, Fritsen CH, McKay CP, Priscu JC, Adams EE. 2003. Formation and character of an ancient 19-m ice cover and underlying trapped brine in an “ice-sealed” east Antarctic lake. Proc. Natl. Acad. Sci. U. S. A. 100:26–31. 10.1073/pnas.222680999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray AE, Kenig F, Fritsen CH, McKay CP, Cawley KM, Edwards R, Kuhn E, McKnight DM, Ostrom NE, Peng V, Ponce A, Priscu JC, Samarkin V, Townsend AT, Wagh P, Young SA, Yung PT, Doran PT. 2012. Microbial life at −13°C in the brine of an ice-sealed Antarctic lake. Proc. Natl. Acad. Sci. U. S. A. 109:20626–20631. 10.1073/pnas.1208607109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karl DM, Bird DF, Bjorkman K, Houlihan T, Shackelford R, Tupas L. 1999. Microorganisms in the accreted ice of Lake Vostok, Antarctica. Science 286:2144–2147. 10.1126/science.286.5447.2144 [DOI] [PubMed] [Google Scholar]
- 4.Priscu JC, Adams EE, Lyons WB, Voytek MA, Mogk DW, Brown RL, McKay CP, Takacs CD, Welch KA, Wolf CF, Kirshtein JD, Avci R. 1999. Geomicrobiology of subglacial ice above Lake Vostok, Antarctica. Science 286:2141–2144. 10.1126/science.286.5447.2141 [DOI] [PubMed] [Google Scholar]
- 5.Miteva VI, Brenchley JE. 2005. Detection and isolation of ultrasmall microorganisms from a 120,000-year-old Greenland glacier ice core. Appl. Environ. Microbiol. 71:7806–7818. 10.1128/AEM.71.12.7806-7818.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haller CM, Rölleke S, Vybiral D, Witte A, Velimirov B. 2000. Investigation of 0.2 μm filterable bacteria from the Western Mediterranean Sea using a molecular approach: dominance of potential starvation forms. FEMS Microbiol. Ecol. 31:153–161. 10.1111/j.1574-6941.2000.tb00680.x [DOI] [PubMed] [Google Scholar]
- 7.Rappé MS, Connon Sa Vergin KL, Giovannoni SJ. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630–633. 10.1038/nature00917 [DOI] [PubMed] [Google Scholar]
- 8.Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, Bibbs L, Eads J, Richardson TH, Noordewier M, Rappé MS, Short JM, Carrington JC, Mathur EJ. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242–1245. 10.1126/science.1114057 [DOI] [PubMed] [Google Scholar]
- 9.Dmitriev VV, Suzina NE, Rusakova TG, Gilichinskii DA, Duda VI. 2001. Ultrastructural characteristics of natural forms of microorganisms isolated from permafrost grounds of eastern Siberia by the method of low-temperature fractionation. Dokl. Bio. Sci. 378:304–306. 10.1023/A:1019203800276 [DOI] [PubMed] [Google Scholar]
- 10.Dmitriev VV, Suzina NE, Barinova ES, Duda VI, Boronin AM. 2004. An electron microscopic study of the ultrastructure of microbial cells in extreme biotopes in situ. Mikrobiologiia 73:832–840. 10.1007/s11021-005-0014-y [DOI] [PubMed] [Google Scholar]
- 11.Dmitriev VV, Suzina NE, Rusakova TG, Petrov PY, Oleinikov RR, Esikova TZ, Kholodenko VP, Duda VI, Boronin AM. 2011. Electron microscopic detection and in situ characterization of bacterial nanoforms in extreme biotopes. Mikrobiologiia 77:46–54. 10.1134/S0026261708010062 [DOI] [PubMed] [Google Scholar]
- 12.Bae HC, Cota-Robles EH, Casida LE. 1972. Microflora of soil as viewed by transmission electron microscopy. Appl. Microbiol. 23:637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iizuka T, Yamanaka S, Nishiyama T, Hiraishi A. 1998. Isolation and phylogenetic analysis of aerobic copiotrophic ultramicrobacteria from urban soil. J. Gen. Appl. Microbiol. 44:75–84. 10.2323/jgam.44.75 [DOI] [PubMed] [Google Scholar]
- 14.Duda VI, Suzina NE, Esikova TZ, Akimov VN, Oleinikov RR, Polivtseva VN, Abashina TN, Shorokhova AP, Boronin AM. 2009. A cytological characterization of the parasitic action of ultramicrobacteria NF1 and NF3 of the genus Kaistia on chemoorganotrophic and phototrophic bacteria. FEMS Microbiol. Ecol. 69:180–193. 10.1111/j.1574-6941.2009.00696.x [DOI] [PubMed] [Google Scholar]
- 15.Miyoshi T, Iwatsuki T, Naganuma T. 2005. Phylogenetic characterization of 16S rRNA gene clones from deep-groundwater microorganisms that pass through 0.2-micrometer-pore-size filters. Appl. Environ. Microbiol. 71:1084–1088. 10.1128/AEM.71.2.1084-1088.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn MW, Stadler P, Wu QL, Pöckl M. 2004. The filtration-acclimatization method for isolation of an important fraction of the not readily cultivable bacteria. J. Microbiol. Methods 57:379–390. 10.1016/j.mimet.2004.02.004 [DOI] [PubMed] [Google Scholar]
- 17.Comolli LR, Baker BJ, Downing KH, Siegerist CE, Banfield JF. 2009. Three-dimensional analysis of the structure and ecology of a novel, ultra-small archaeon. ISME J. 3:159–167. 10.1038/ismej.2008.99 [DOI] [PubMed] [Google Scholar]
- 18.Baker BJ, Comolli LR, Dick GJ, Hauser LJ, Hyatt D, Dill BD, Land ML, Verberkmoes NC, Hettich RL, Banfield JF, Mountain I. 2010. Enigmatic, ultrasmall, uncultivated Archaea. Proc. Natl. Acad. Sci. U. S. A. 107:8806–8811. 10.1073/pnas.0914470107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO. 2002. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature 417:63–67. 10.1038/417063a [DOI] [PubMed] [Google Scholar]
- 20.Naganuma T, Miyoshi T, Kimura H. 2007. Phylotype diversity of deep-sea hydrothermal vent prokaryotes trapped by 0.2- and 0.1-micron-pore-size filters. Extremophiles 11:637–646. 10.1007/s00792-007-0070-5 [DOI] [PubMed] [Google Scholar]
- 21.Nakai R, Abe T, Takeyama H, Naganuma T. 2011. Metagenomic analysis of 0.2-μm-passable microorganisms in deep-sea hydrothermal fluid. Mar. Biotechnol. 13:900–908. 10.1007/s10126-010-9351-6 [DOI] [PubMed] [Google Scholar]
- 22.Geissinger O, Herlemann DPR, Mörschel E, Maier UG, Brune A. 2009. The ultramicrobacterium “Elusimicrobium minutum” gen. nov., sp. nov., the first cultivated representative of the termite group 1 phylum. Appl. Environ. Microbiol. 75:2831–2840. 10.1128/AEM.02697-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cavicchioli R, Ostrowski M. 2003. Ultramicrobacteria, p 1–8 In Battista J. (ed), Encyclopedia of life sciences. John Wiley & Sons, West Sussex, United Kingdom [Google Scholar]
- 24.Torrella F, Morita RY. 1981. Microcultural study of bacterial size changes and microcolony and ultramicrocolony formation by heterotrophic bacteria in seawater. Appl. Environ. Microbiol. 41:518–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hahn MW, Lünsdorf H, Wu Q, Höfle MG, Boenigk J, Stadler P, Lu H, Schauer M, Ho MG. 2003. Isolation of novel ultramicrobacteria classified as Actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442–1451. 10.1128/AEM.69.3.1442-1451.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutz BA, Kieft TL. 2004. Phylogenetic characterization of dwarf archaea and bacteria from a semiarid soil. Soil Biol. Biochem. 36:825–833. 10.1016/j.soilbio.2004.01.012 [DOI] [Google Scholar]
- 27.Litchfield CD. 1998. Survival strategies for microorganisms in hypersaline environments and their relevance to life on early Mars. Meteorit. Planet. Sci. 33:813–819. 10.1111/j.1945-5100.1998.tb01688.x [DOI] [PubMed] [Google Scholar]
- 28.Gilichinsky DA. 2001. Permafrost model of extraterrestrial habitat, p 271–295 In Horneck G, Baumstark-Khan C. (ed), Astrobiology. The quest for the conditions of life. Springer-Verlag, Berlin, Germany [Google Scholar]
- 29.Ponder MA, Thomashow MF, Tiedje JM. 2008. Metabolic activity of Siberian permafrost isolates, Psychrobacter arcticus and Exiguobacterium sibiricum, at low water activities. Extremophiles 12:481–490. 10.1007/s00792-008-0151-0 [DOI] [PubMed] [Google Scholar]
- 30.Doran PT, Fritsen CH, Murray AE, Kenig F, McKay CP, Kyne JD. 2008. Entry approach into pristine ice-sealed lakes—Lake Vida, East Antarctica, a model ecosystem. Limnol. Oceanogr. Methods 6:542–547. 10.4319/lom.2008.6.542 [DOI] [Google Scholar]
- 31.Massana R, Murray AE, Preston CM, DeLong EF. 1997. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl. Environ. Microbiol. 63:50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mondino LJ, Asao M, Madigan MT. 2009. Cold-active halophilic bacteria from the ice-sealed Lake Vida, Antarctica. Arch. Microbiol. 191:785–790. 10.1007/s00203-009-0503-x [DOI] [PubMed] [Google Scholar]
- 33.Murray AE, Hollibaugh JT, Orrego C. 1996. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl. Environ. Microbiol. 62:2676–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosier AC, Murray AE, Fritsen CH. 2007. Microbiota within the perennial ice cover of Lake Vida, Antarctica. FEMS Microbiol. Ecol. 59:274–288. 10.1111/j.1574-6941.2006.00220.x [DOI] [PubMed] [Google Scholar]
- 35.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. Wiley, New York, NY [Google Scholar]
- 36.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 37.Huber T, Faulkner G, Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319. 10.1093/bioinformatics/bth226 [DOI] [PubMed] [Google Scholar]
- 38.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 39.Desantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:394–399. 10.1093/nar/gkl244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikucki JA, Priscu JC. 2007. Bacterial diversity associated with Blood Falls, a subglacial outflow from the Taylor Glacier, Antarctica. Appl. Environ. Microbiol. 73:4029–4039. 10.1128/AEM.01396-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. 2011. UniFrac: an effective distance metric for microbial community comparison. ISME J. 5:169–172. 10.1038/ismej.2010.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayat MA. 1993. Stains and cytochemical methods. Plenum Press, New York, NY [Google Scholar]
- 45.Cotton FA, Wilkinson G, Murillo CA, Bochmann M. 1972. Advanced inorganic chemistry, 6th ed. John Wiley & Sons Ltd., New York, NY [Google Scholar]
- 46.Nevot M, Deroncele V, López-Iglesias C, Bozal N, Guinea J, Mercade E. 2006. Ultrastructural analysis of the extracellular matter secreted by the psychrotolerant bacterium Pseudoalteromonas antarctica NF3. Microb. Ecol. 51:501–507. 10.1007/s00248-006-9065-5 [DOI] [PubMed] [Google Scholar]
- 47.Hinsa-Leasure SM, Koid C, Tiedje JM, Schultzhaus JN. 2013. Biofilm formation by Psychrobacter arcticus and the role of a large adhesin in attachment to surfaces. Appl. Environ. Microbiol. 79:3967–3973. 10.1128/AEM.00867-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frankel RB. 2003. Biologically induced mineralization by bacteria. Rev. Mineral. Geochem. 54:95–114. 10.2113/0540095 [DOI] [Google Scholar]
- 49.Miot J, Benzerara K, Morin G, Kappler A, Bernard S, Obst M, Férard C, Skouri-Panet F, Guigner J-M, Posth N, Galvez M, Brown GE, Guyot F. 2009. Iron biomineralization by anaerobic neutrophilic iron-oxidizing bacteria. Geochim. Cosmochim. Acta 73:696–711. 10.1016/j.gca.2008.10.033 [DOI] [Google Scholar]
- 50.Krembs C, Deming JW. 2008. The role of exopolymers in microbial adaptation to sea ice, p 247–264 In Margesin R, Schinner F, Marx JC, Gerday G. (ed), Psychrophiles: from biodiversity to biotechnology. Springer Verlag, Berlin, Germany [Google Scholar]
- 51.Kulp A, Kuehn MJ. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64:163–184. 10.1146/annurev.micro.091208.073413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McBroom AJ, Kuehn MJ. 2007. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63:545–558. 10.1111/j.1365-2958.2006.05522.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frias A, Manresa A, de Oliveira E, López-Iglesias C, Mercade E. 2010. Membrane vesicles: a common feature in the extracellular matter of cold-adapted Antarctic bacteria. Microb. Ecol. 59:476–486. 10.1007/s00248-009-9622-9 [DOI] [PubMed] [Google Scholar]
- 54.Nyström T. 2007. A bacterial kind of aging. PLoS Genet. 3:2355–2357. 10.1371/journal.pgen.0030224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. 2008. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl. Acad. Sci. U. S. A. 105:3076–3081. 10.1073/pnas.0708931105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peeters K, Hodgson DA, Convey P, Willems A. 2011. Culturable diversity of heterotrophic bacteria in Forlidas Pond (Pensacola Mountains) and Lundström Lake (Shackleton Range), Antarctica. Microb. Ecol. 62:399–413. 10.1007/s00248-011-9842-7 [DOI] [PubMed] [Google Scholar]
- 57.Gilichinsky D, Rivkina E, Bakermans C, Shcherbakova V, Petrovskaya L, Ozerskaya S, Ivanushkina N, Kochkina G, Laurinavichuis K, Pecheritsina S, Fattakhova R, Tiedje JM. 2005. Biodiversity of cryopegs in permafrost. FEMS Microbiol. Ecol. 53:117–128. 10.1016/j.femsec.2005.02.003 [DOI] [PubMed] [Google Scholar]
- 58.Zhang D-C, Li H-R, Xin Y-H, Chi Z-M, Zhou P-J, Yu Y. 2008. Marinobacter psychrophilus sp. nov., a psychrophilic bacterium isolated from the Arctic. Int. J. Syst. Evol. Microbiol. 58:1463–1466. 10.1099/ijs.0.65690-0 [DOI] [PubMed] [Google Scholar]
- 59.Ayala-del-Río HL, Chain PS, Grzymski JJ, Ponder Ma Ivanova N, Bergholz PW, Di Bartolo G, Hauser L, Land M, Bakermans C, Rodrigues D, Klappenbach J, Zarka D, Larimer F, Richardson P, Murray A, Thomashow M, Tiedje JM. 2010. The genome sequence of Psychrobacter arcticus 273-4, a psychroactive Siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Appl. Environ. Microbiol. 76:2304–2312. 10.1128/AEM.02101-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ward BB, Priscu JC. 1997. Detection and characterization of denitrifying bacteria from a permanently ice-covered Antarctic Lake. Hydrobiologia 347:57–68. 10.1023/A:1003087532137 [DOI] [Google Scholar]
- 61.Chen Y-G, Tang S-K, Zhang Y-Q, Li Z-Y, Yi L-B, Wang Y-X, Li W-J, Cui X-L. 2009. Arthrobacter halodurans sp. nov., a new halotolerant bacterium isolated from sea water. Antonie Van Leeuwenhoek 96:63–70. 10.1007/s10482-009-9336-5 [DOI] [PubMed] [Google Scholar]
- 62.Cary SC, McDonald IR, Barrett JE, Cowan DA. 2010. On the rocks: the microbiology of Antarctic Dry Valley soils. Nat. Rev. Microbiol. 8:129–138. 10.1038/nrmicro2281 [DOI] [PubMed] [Google Scholar]
- 63.Baldani JI, Baldani VLD, Dobereiner J. 2005. Genus III. Herbaspirillum, p 629–636 In Garrity GM. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2 Springer, New York, NY [Google Scholar]
- 64.Lovejoy C, Bowman JP, Hallegraeff GM. 1998. Algicidal effects of a novel marine Pseudoalteromonas isolate (class Proteobacteria, gamma subdivision) on harmful algal bloom species of the genera Chattonella, Gymnodinium, and Heterosigma. Appl. Environ. Microbiol. 64:2806–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Médigue C, Krin E, Pascal G, Barbe V, Bernsel A, Bertin PN, Cheung F, Cruveiller S, D'Amico S, Duilio A, Fang G, Feller G, Ho C, Mangenot S, Marino G, Nilsson J, Parrilli E, Rocha EPC, Rouy Z, Sekowska A, Tutino ML, Vallenet D, von Heijne G, Danchin A. 2005. Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 15:1325–1335. 10.1101/gr.4126905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Soina VS, Mulyukin AL, Demkina EV, Vorobyova EA, El-Registan GI. 2004. The structure of resting bacterial populations in soil and subsoil permafrost. Astrobiology 4:345–358. 10.1089/ast.2004.4.345 [DOI] [PubMed] [Google Scholar]
- 67.Joshi HM, Toleti RS. 2009. Nutrition induced pleomorphism and budding mode of reproduction in Deinococcus radiodurans. BMC Res. Notes 2:123–123. 10.1186/1756-0500-2-123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lange R, Hengge-Aronis R. 1991. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor S. J. Bacteriol. 173:4474–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Young KD. 2006. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 70:660–703. 10.1128/MMBR.00001-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kageyama A, Morisaki K, Omura S, Takahashi Y. 2008. Arthrobacter oryzae sp. nov. and Arthrobacter humicola sp. nov. Int. J. Syst. Evol. Microbiol. 58:53–56. 10.1099/ijs.0.64875-0 [DOI] [PubMed] [Google Scholar]
- 71.Demkina EV, Soina VS, El Registan GI, Zviagintsev DG. 2000. Reproductive resting forms of Arthrobacter globiformis. Mikrobiologiia 69:377–382. 10.1007/BF02756739 [DOI] [PubMed] [Google Scholar]
- 72.Takeuchi M, Hatano K. 1998. Union of the genera Microbacterium Orla-Jensen and Aureobacterium Collins et al. in a redefined genus Microbacterium. Int. J. Syst. Bacteriol. 3:739–747 [DOI] [PubMed] [Google Scholar]
- 73.Suzina NE, Mulyukin AL, Dmitriev VV, Nikolaev YA, Shorokhova AP, Bobkova YS, Barinova ES, Plakunov VK, El-Registan GI, Duda VI. 2006. The structural bases of long-term anabiosis in non-spore-forming bacteria. Adv. Space Res. 38:1209–1219. 10.1016/j.asr.2005.09.020 [DOI] [Google Scholar]
- 74.Mulyukin AL, Suzina NE, Duda VI, El'-Registan GI. 2008. Structural and physiological diversity among cystlike resting cells of bacteria of the genus Pseudomonas. Mikrobiologiia 77:512–523. 10.1134/S0026261708040127 [DOI] [PubMed] [Google Scholar]
- 75.Pianetti A, Battistelli M, Citterio B, Parlani C, Falcieri E, Bruscolini F. 2009. Morphological changes of Aeromonas hydrophila in response to osmotic stress. Micron 40:426–433. 10.1016/j.micron.2009.01.006 [DOI] [PubMed] [Google Scholar]
- 76.Knoll A, Osborn MJ, Baross J, Berg HC, Pace NR, Sogin M. 1999. Size limits of very small microorganisms: proceedings of a workshop. National Research Council Space Studies Board. National Academy Press, Washington, DC [Google Scholar]
- 77.Nedelkova M, Merroun ML, Rossberg A, Hennig C, Selenska-Pobell S. 2007. Microbacterium isolates from the vicinity of a radioactive waste depository and their interactions with uranium. FEMS Microbiol. Ecol. 59:694–705. 10.1111/j.1574-6941.2006.00261.x [DOI] [PubMed] [Google Scholar]
- 78.Kim SB, Nedashkovskaya OI, Mikhailov VV, Han SK, Kim KO, Rhee MS, Bae KS. 2004. Kocuria marina sp. nov., a novel actinobacterium isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 54:1617–1620. 10.1099/ijs.0.02742-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.