Abstract
The phosphoenol phosphotransferase system (PTS) is a multicomponent signal transduction cascade that regulates diverse aspects of bacterial cellular physiology in response to the availability of high-energy sugars in the environment. Many PTS components are repressed at the transcriptional level when the substrates they transport are not available. In Escherichia coli, the transcription factor Mlc (for makes large colonies) represses transcription of the genes encoding enzyme I (EI), histidine protein (HPr), and the glucose-specific enzyme IIBC (EIIBCGlc) in defined media that lack PTS substrates. When glucose is present, the unphosphorylated form of EIIBCGlc sequesters Mlc to the cell membrane, preventing its interaction with DNA. Very little is known about Vibrio cholerae Mlc. We found that V. cholerae Mlc activates biofilm formation in LB broth but not in defined medium supplemented with either pyruvate or glucose. Therefore, we questioned whether V. cholerae Mlc functions differently than E. coli Mlc. Here we have shown that, like E. coli Mlc, V. cholerae Mlc represses transcription of PTS components in both defined medium and LB broth and that E. coli Mlc is able to rescue the biofilm defect of a V. cholerae Δmlc mutant. Furthermore, we provide evidence that Mlc indirectly activates transcription of the vps genes by repressing expression of EI. Because activation of the vps genes by Mlc occurs under only a subset of the conditions in which repression of PTS components is observed, we conclude that additional inputs present in LB broth are required for activation of vps gene transcription by Mlc.
INTRODUCTION
To thrive, a bacterium must sense and respond to carbohydrates in its environment. The phosphoenolpyruvate phosphotransferase system (PTS) is a central and far-reaching multicomponent signal transduction cascade that regulates many aspects of the bacterial response to carbohydrates, from chemotaxis and transport to storage and catabolism (1).
The PTS cascade is initiated by phosphotransfer from phosphoenolpyruvate (PEP) to enzyme I (EI) (Fig. 1). From here, phosphate is transferred sequentially to histidine protein (HPr) or one of its homologs and then to the multicomponent, sugar-specific enzymes II. There are more than 10 enzymes II encoded in the Vibrio cholerae genome, and the sugar specificities of only a few are known (2). While the enzymes IIA (EIIA) and IIB (EIIB) accept phosphate in turn, the enzyme IIC (EIIC) subunit, which spans the cytoplasmic membrane, transports PTS-dependent sugars into the cell, allowing them to be phosphorylated by EIIB. Thus, phosphate is channeled from PEP through PTS components to incoming PTS-dependent sugars.
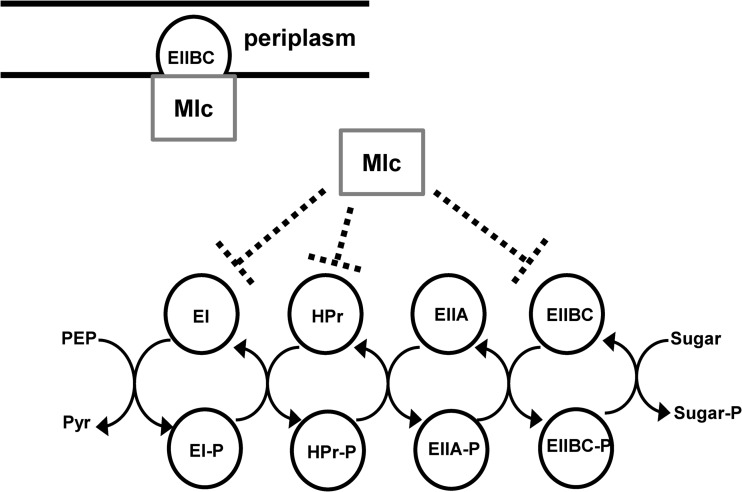
FIG 1.
Mlc interacts with the PTS. A schematic representation of the PTS, including the various ways in which E. coli Mlc is known to interact with PTS components.
The phosphorylation state of PTS components depends on the intracellular stores of PEP and the abundance of PTS-dependent sugars in the environment. For example, when PTS-dependent sugars are scarce and PEP is available as a phosphodonor, phosphate accumulates on PTS components. In contrast, when PTS sugars are abundant in the environment, sugar transport through the PTS removes phosphate from the general PTS components EI and HPr. These in turn receive phosphate from expressed sugar-specific PTS components that do not participate directly in transport of the available sugar, and these become dephosphorylated as well. Therefore, PTS components are ideal reporters of the overall nutritional status of the cell. In addition to facilitating the transport of high-energy carbohydrates, they coordinate the bacterial response to carbohydrate availability through direct interactions with other proteins (3, 4).
Transcription of many PTS components is repressed when they are not required for transport. For example, the Escherichia coli ROK (repressor Orf kinase) family transcription factor Mlc represses general as well as selected sugar-specific PTS components, such as the glucose-specific EIIBC (EIIBCGlc), when glucose is scarce (Fig. 1) (5–10). When glucose is abundant, Mlc is sequestered to the inner membrane through a direct interaction with the unphosphorylated form of the EIIB subunit of EIIBCGlc. This prevents its interaction with DNA (11–14).
Vibrio cholerae uses an exopolysaccharide and protein-based extracellular matrix to attach to surfaces. Synthesis of the Vibrio polysaccharide (VPS) biofilm exopolysaccharide is encoded by the vps genes (15). In addition, three proteins, RbmA, Bap1, and RbmC, are required for formation of contacts with other cells and with the surface (16). Synthesis of this extracellular matrix is tightly regulated. We recently determined that sugars transported by the PTS activate transcription of the vps genes and biofilm formation (17–20). Furthermore, even in the absence of PTS substrates, the network of Vibrio cholerae PTS components forms a complex web that regulates vps gene transcription and biofilm formation through multiple independent pathways (2, 20–23).
We observed that Mlc activated V. cholerae biofilm formation in LB broth but not in a defined medium supplemented either with glucose, which is transported by the PTS, or pyruvate, which is PTS independent. Because E. coli Mlc was reported to regulate transcription of PTS components in defined medium, we questioned whether Mlc might function differently in V. cholerae. Here we have shown that V. cholerae Mlc represses transcription of the EI, HPr, and EIIBCGlc genes in both defined medium and LB broth and furthermore that E. coli Mlc rescues the biofilm defect of a V. cholerae Δmlc mutant. EI and EIIBCGlc indirectly repress V. cholerae biofilm formation at the transcriptional level (21). We have shown that repression of EI gene transcription by Mlc is responsible for activation of vps gene transcription and biofilm formation, while EIIBCGlc likely functions upstream of Mlc. Because activation of vps gene transcription occurs under only a subset of the growth conditions in which repression of EI and EIIBCGlc gene transcription is observed, we hypothesize that a second regulatory circuit present only in LB broth is required for activation of the vps genes. We previously showed that EIIAGlc activates V. cholerae biofilm formation in LB broth (20, 21). Here we provide evidence that EIIAGlc may be a component of the second regulatory circuit required for full activation of V. cholerae biofilm formation in LB broth.
MATERIALS AND METHODS
Bacteria strains, plasmids, and media.
Bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown in Luria-Bertani broth (LB) or a previously described defined medium supplemented with 0.5% (wt/vol) of the appropriate carbohydrate (20). When required, the following antibiotics were added to the growth medium: streptomycin (100 μg/ml), ampicillin (50 or 100 μg/ml as appropriate), carbenicillin (100 μg/ml), kanamycin (30 μg/ml), and tetracycline (10 μg/ml). For rescue experiments, protein expression was induced by adding arabinose to yield a final concentration of 0.02%. pBAD plasmids used as controls in rescue experiments included either the lacZ gene or the EIIAGlc gene in reverse coding orientation following the pBAD promoter as noted in Table 1. These plasmids were used because the empty pBAD vector was found to decrease bacterial growth. Control vectors were used with arabinose supplementation. Neither the control vectors nor arabinose supplementation affected biofilm formation.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strains | Genotype and/or description | Reference or source |
|---|---|---|
| E. coli | ||
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpirR6K; Kmr | 24 |
| V. cholerae | ||
| PW357 | MO10 ΔlacZ::vpsL→ lacZ; Smr | 25 |
| PW988 | PW357 ΔEIIAGlc | 20 |
| PW961 | PW357 ΔEI | 20 |
| PW1573 | PW357 ΔEI ΔEIIAGlc | This study |
| PW993 | PW357 Δmlc | 21 |
| PW1557 | PW357 ΔEI Δmlc | This study |
| PW1558 | PW357 ΔEI ΔEIIBCGlc | This study |
| PW1559 | PW357 ΔEl ΔEIIBCGlc Δmlc | This study |
| PW996 | PW357 ΔEIIBCGlc Δmlc | 21 |
| Plasmids | ||
| pBAD-TOPO | Arabinose-inducible expression vector used to generate His6/V5 epitope-tagged proteins | Life Technologies |
| pBAD-TOPO-lacZ | pBAD-TOPO containing the lacZ gene; provided as a control | Life Technologies |
| pBAD-TOPO-EIIAGlc rev | pBAD-TOPO containing the EIIAGlc gene in reverse, noncoding orientation | 20 |
| pBAD-TOPO- MlcV. cholerae | pBAD-TOPO containing full-length V. cholerae mlc::His6/V5 | This study |
| pBAD-TOPO-MlcE. coli | pBAD-TOPO containing full-length E. coli mlc::His6/V5 | This study |
Generation of deletion mutants.
Gene deletions were engineered as described previously using the splice overlap extension (SOE) method and double homologous recombination (25, 26). These primers and all others used in this study are available upon request.
Construction of pBADTOPO-mlc for biofilm complementation.
Constructs used for rescue experiments were pBAD-TOPO-mlcV. cholerae and pBAD-TOPO-mlcE. coli. pBADTOPO-mlcV. cholerae was generated by PCR amplification of V. cholerae mlc, excluding the native stop codon, and cloning of this PCR product into the pBAD-TOPO vector (Invitrogen) according to the manufacturer's instructions. Construction of pBAD-TOPO-mlcE. coli was generated by PCR amplification of mlc from the E. coli strain K-12, excluding the native stop codon. This was also cloned into the pBAD-TOPO vector.
Biofilm assays.
Vibrio cholerae strains to be tested were cultured overnight in LB broth and then diluted to an optical density at 655 nm (OD655) of 0.05 in the appropriate medium. Three hundred microliters of this cell suspension was aliquoted into three borosilicate tubes and incubated at 27°C for 21 to 24 h. Planktonic cells were removed, and planktonic growth was assessed by measuring the OD655. Three hundred microliters of 0.1 M phosphate-buffered saline (PBS) solution was added to the remaining adherent biofilm pellicle along with 1-mm diameter glass beads (30% [vol/vol]) (BioSpec Products, Inc.). Tubes were vortexed to disperse biofilm-associated cells, and the OD655 of the resultant cell suspension was measured to quantify biofilm formation. Total growth was calculated by adding the planktonic and biofilm measurements. Three independent replicates were included in each experiment, and experiments were repeated at least three times. Error bars represent the standard deviations, and a Student t test was used to assess statistical significance. P values of less than 0.05 were taken to indicate a statistically significant difference.
qRT-PCR analyses.
For measurements of transcription in mid-log phase, cells were harvested at an OD655 of 0.5 and resuspended in 1 ml of TRIzol (Invitrogen). For measurements of transcription in stationary-phase cultures, a mid-log culture of V. cholerae was used to prepare a suspension with an OD655 of 0.05 in 90-mm petri dishes. This was incubated for 19 h at 28°C. All cells were then removed to 50-ml conical tubes, pelleted, resuspended in 2.5 ml of TRIzol, and diluted 1:1 with fresh TRIzol. Total RNA was prepared from 1 ml of the TRIzol suspension as directed by the manufacturer. cDNA was synthesized using the SuperScript III kit (Invitrogen) as directed. Transcripts were quantified by quantitative reverse transcription-PCR (qRT-PCR) using the iTaq SYBR green kit (Bio-Rad) along with the Step One Plus real-time PCR system (Applied Biosystems). Transcript levels were normalized to clpX. Fold expression was determined using the ΔΔCT method. Three experimental replicates were performed in each trial, and multiple trials were performed. A Student t test was used to calculate statistical significance. P values of less than 0.05 were taken to be statistically significant.
RESULTS AND DISCUSSION
V. cholerae Mlc promotes biofilm formation in LB broth only.
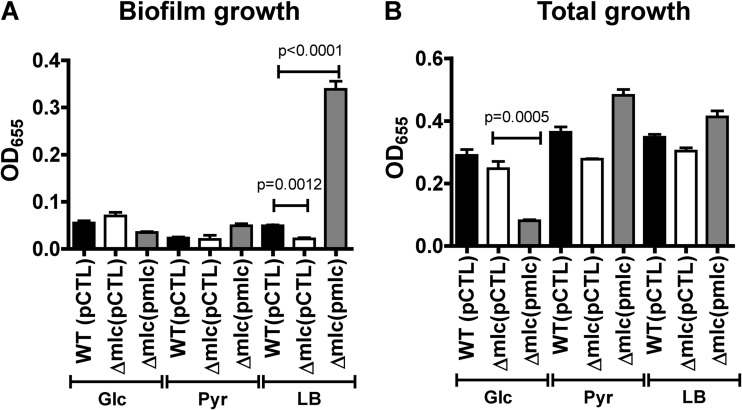
We previously showed that Mlc activates biofilm formation in LB broth, which has relatively small amounts of PTS sugars (17, 21). To study its role more precisely, we repeated these experiments, including chemically defined medium containing glucose, a PTS substrate, or pyruvate, a PTS-independent substrate. As shown in Fig. 2A, deletion of mlc decreased biofilm formation in LB broth but not in defined medium supplemented with either glucose or pyruvate. Overexpression of Mlc from an arabinose-inducible promoter rescued the biofilm defect of a Δmlc mutant but had no effect on biofilm formation in defined media (Fig. 2A). Overexpression of Mlc did decrease total cell growth in defined medium containing glucose, consistent with repression of PTS components by Mlc and hence a reduction in glucose transport under these conditions (Fig. 2B).
FIG 2.
Mlc activates V. cholerae biofilm formation in LB broth only: biofilm-associated growth (A) or total growth (B) was measured for wild-type V. cholerae (WT) or a Δmlc mutant carrying either the pBAD-lacZ control vector (pCTL) or the same plasmid expressing Mlc (pmlc). Static biofilms were formed in LB broth and defined medium supplemented with glucose (Glc) or pyruvate (Pyr).
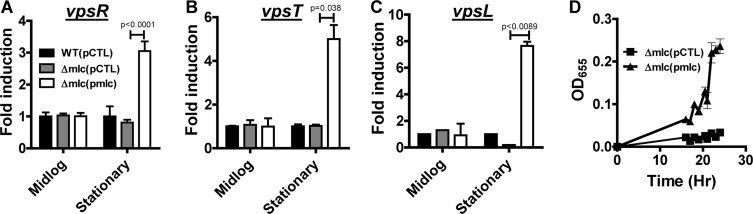
A change in V. cholerae biofilm formation is often correlated with a similar change in transcription of the vpsA-vpsQ genes, which encode the synthesis of the VPS biofilm exopolysaccharide, as well as the two master regulators of these genes, vpsR and vpsT (15, 27, 28). To determine if Mlc regulated transcription of these genes, we measured mRNA levels by qRT-PCR for wild-type V. cholerae, a Δmlc mutant, or a Δmlc mutant rescued with wild-type mlc in cells harvested from an LB culture in mid-log phase. Surprisingly, there was no evidence of regulation of the vps genes by Mlc in exponentially growing cells (Fig. 3A to C). We then performed a similar experiment using cells harvested from stationary-phase cultures. In this case, transcriptional activation of vps genes by Mlc was observed. This suggested to us that Mlc might have its largest effect on biofilm formation in later stages of growth. To test this, we performed a time course comparing biofilm formation by a Δmlc mutant and that by a Δmlc mutant overexpressing wild-type mlc. As shown in Fig. 3D, although Mlc expression was induced at the initiation of the experiment, biofilm formation did not increase until stationary phase was reached. Deletion or overexpression of Mlc had no effect on total growth (data not shown).
FIG 3.
Mlc activates vps gene transcription and biofilm formation in later stages of growth in LB broth. qRT-PCR analysis of transcription of vpsR (A), vpsT (B), or vpsL (C) in wild-type V. cholerae carrying the pBAD-lacZ control or a Δmlc mutant carrying the pBAD-lacZ control vector (pCTL) or the same plasmid expressing Mlc (pmlc). Cells were harvested from LB broth in either the mid-log or stationary phase of growth. Values are normalized to wild-type measurements, which were set to 1. (D) Quantification of biofilm formation over time by a Δmlc mutant carrying either a pBAD-EIIAGlc rev control vector (pCTL) or a vector encoding Mlc.
V. cholerae Mlc represses transcription of PTS components in cells cultured in defined media and LB broth.
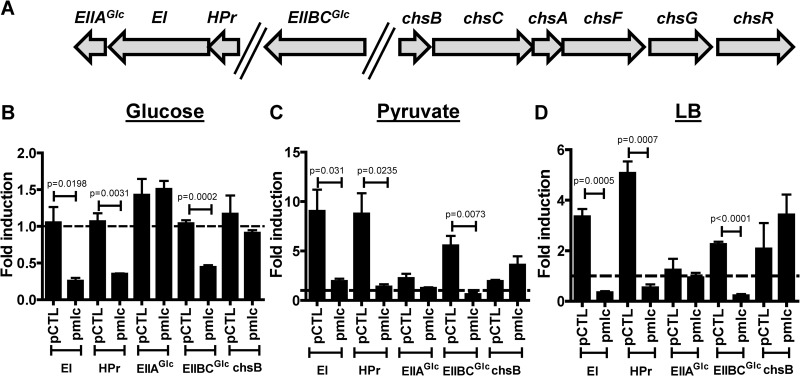
Because multiple studies of E. coli Mlc have demonstrated repression of PTS components in cells cultured in defined medium and because it was recently suggested that V. cholerae Mlc activates transcription of chsB, which encodes a chitobiose-specific EII component (29), we questioned whether Mlc might have a fundamentally different function in V. cholerae than in E. coli (6, 9, 30). To investigate this, we used qRT-PCR to measure transcription of the EI, HPr, EIIAGlc, and EIIBCGlc genes and chsB in cells cultured in LB broth or in defined medium supplemented with glucose or pyruvate. Measurements were made in wild-type cells and a Δmlc mutant carrying either a control plasmid or a plasmid encoding Mlc and normalized to the wild-type value. Therefore, values greater than 1 indicate that transcription is increased relative to the wild-type measurement, whereas values less than 1 indicate decreased transcription. As shown in Fig. 4A, the HPr, EI, and EIIAGlc genes form a putative operon, while the EIIBCGlc gene and chsB are in different chromosomal locations. In defined medium supplemented with glucose, deletion of mlc had very little effect on gene transcription (Fig. 4B). In defined medium supplemented with pyruvate or in LB broth, deletion of mlc greatly increased transcription of the EI, HPr, and EIIBCGlc genes but had no effect on the EIIAGlc gene or chsB (Fig. 4C and D). This suggests that Mlc represses transcription of the EI, HPr, and EIIBCGlc genes but not chsB or the EIIAGlc gene when V. cholerae is cultured in LB broth or defined medium supplemented with pyruvate, both of which contain a low level of or no PTS substrates.
FIG 4.
In the presence of non-PTS substrates, native levels of V. cholerae Mlc repress transcription of the genes encoding selected PTS components; when overexpressed, Mlc represses transcription under all conditions. (A) Operon structure corresponding to the transcripts measured. (B) qRT-PCR analysis comparing transcription of the genes encoding the specified PTS components in a Δmlc mutant carrying either the control vector pBAD-lacZ (pCTL) or a similar plasmid encoding Mlc. Strains were cultured in minimal medium supplemented with glucose (B), minimal medium supplemented with pyruvate (C), or LB broth (D). Transcript levels were normalized to measurements for wild-type V. cholerae carrying the same control vector and cultured in the same medium. The broken line indicates the wild-type transcription level, which was set to 1. Transcription was measured in cells harvested in the mid-log phase of growth.
To determine whether Mlc was inactive in the presence of glucose due to a change in the cofactor concentration or due to inadequate amounts of Mlc available for binding to DNA, we overexpressed Mlc from a plasmid using an inducible promoter. In all the media tested, overexpression of Mlc in a Δmlc mutant background repressed transcription of the EI, HPr, and EIIBCGlc genes (Fig. 4B to D). These data suggest that when Mlc is available, it represses gene transcription in the presence of both a PTS-dependent substrate, such as glucose, and a PTS-independent substrate, such as pyruvate. From these observations, we conclude that transcriptional regulation of PTS components by Mlc is similar in V. cholerae and E. coli. Specifically, Mlc represses transcription of the general PTS components in LB broth and defined medium supplemented with pyruvate, both of which contain a low level of or no PTS substrates. Repression of PTS components by Mlc in defined medium supplemented with the PTS substrate glucose is dependent on Mlc availability. We hypothesize that EIIBCGlc, which is predicted to be predominantly in its unphosphorylated form when V. cholerae is cultured in medium containing glucose, sequesters V. cholerae Mlc to the inner membrane, preventing it from binding to DNA. Thus, regulation by Mlc is observed only when excess Mlc is provided by overexpression. Furthermore, similar to what is observed in E. coli, transcription of EIIAGlc is independent of Mlc. Although the E. coli HPr, EI, and EIIAGlc genes are also in an operon in E. coli, an additional promoter just upstream of the EIIAGlc gene is responsible for its different pattern of transcription (31). This is consistent with the key role of EIIAGlc in directing the cell to utilize non-PTS carbohydrates, which demands its presence even when PTS substrates are scarce.
V. cholerae and E. coli EIIBCGlc and Mlc are highly similar.
Our data suggest that V. cholerae and E. coli Mlc similarly regulate gene transcription. To explore this further, we compared the amino acid sequences of V. cholerae and E. coli Mlc. These proteins are 46% identical and 69% similar (Fig. 5A). Furthermore, the amino acids that form the helix-turn-helix motif and that participate in Zn2+ binding are conserved. The action of ROK domain regulators, such as NagC, is often modulated by a carbohydrate cofactor (32). However, there is no evidence that E. coli Mlc binds to or coregulates with a carbohydrate. Some have attributed this to the conservation of only 4 out of the 5 sugar binding residues of ROK domain proteins (33). Interestingly, all 5 sugar-binding residues are conserved in V. cholerae Mlc.
FIG 5.
V. cholerae and E. coli Mlc and EIIBGlc are highly similar. (A) An alignment of the V. cholerae (Vc) and E. coli (Ec) Mlc proteins, which are 46% identical and 69% similar. Zn2+-binding residues, which are shown in blue, are conserved. Residues predicted to be involved in sugar binding, which are shown in green, are conserved in V. cholerae Mlc. In E. coli Mlc, residue 194 is not conserved. The helix-turn-helix motifs of each protein are shown in red. (B) An alignment of the cytoplasmic portions of V. cholerae (Vc) and E. coli (Ec) EIIBCGlc, which are 74% identical and 84% similar. In both panels A and B, the residues boxed in pink are known to be required for the interaction between EIIBCGlc and Mlc.
When expressed at native levels, we did not observe transcriptional regulation by Mlc in defined medium supplemented with the PTS substrate glucose (Fig. 4B). We hypothesized that, similar to E. coli Mlc, V. cholerae Mlc might interact with the cytoplasmic portion of EIIBCGlc, resulting in sequestration to the membrane. As shown in Fig. 5B, the cytoplasmic domain of V. cholerae EIIBCGlc is 74% identical and 84% similar to that of E. coli. Cys421, Arg424, Val449, Ala451, and Gln456 of EIIBCGlc, as well as Phe136 of Mlc, are essential for the interaction of E. coli EIIB with Mlc (34, 35). As shown in Fig. 5, with the exception of Ala451, these residues are conserved in V. cholerae EIIBCGlc. Because E. coli and V. cholerae EIIBCGlc and Mlc are highly similar, it seems likely that these two proteins also interact in V. cholerae.
E. coli mlc can rescue a V. cholerae Δmlc mutant.
Because of the similarity of E. coli and V. cholerae Mlc, we questioned whether E. coli Mlc expressed from a plasmid could rescue the biofilm defect of a V. cholerae Δmlc mutant. As shown in Fig. 6, both E. coli and V. cholerae Mlc rescued the biofilm formation defect of a V. cholerae Δmlc mutant. This demonstrates that the role played by V. cholerae Mlc in activation of biofilm formation can also be fulfilled by E. coli Mlc.
FIG 6.
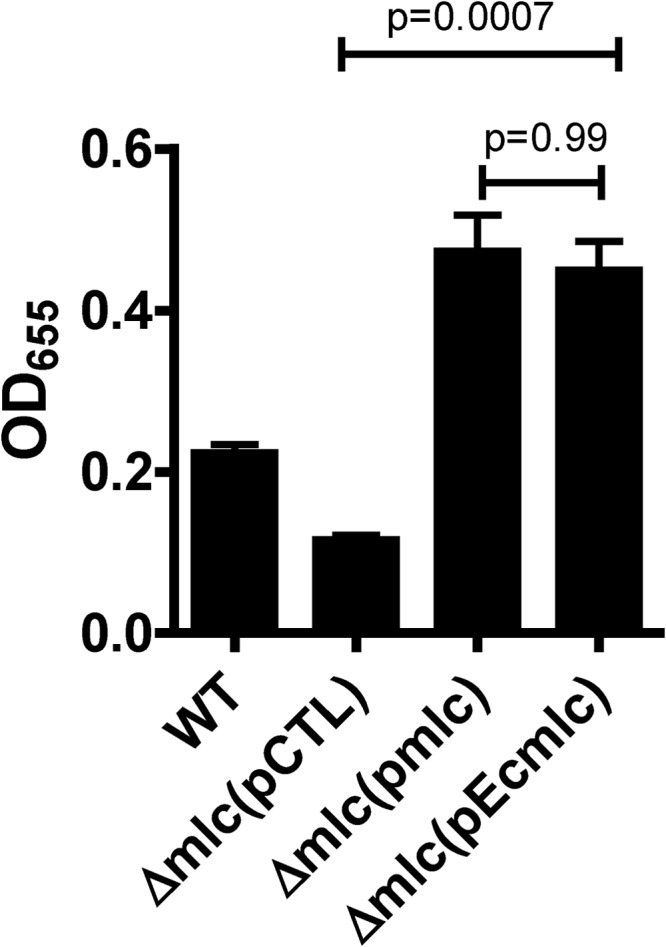
E. coli Mlc rescues the biofilm formation defect of a V. cholerae Δmlc mutant. Quantification of biofilm formation by wild-type V. cholerae (WT) and a V. cholerae Δmlc mutant carrying the control vector pBAD-lacZ (pCTL), pBAD-TOPO-MlcV. cholerae (pmlc), or pBAD-TOPO-MlcE. coli (pEcmlc). All cultures were supplemented with arabinose.
Mlc activates biofilm formation by repression of EI.
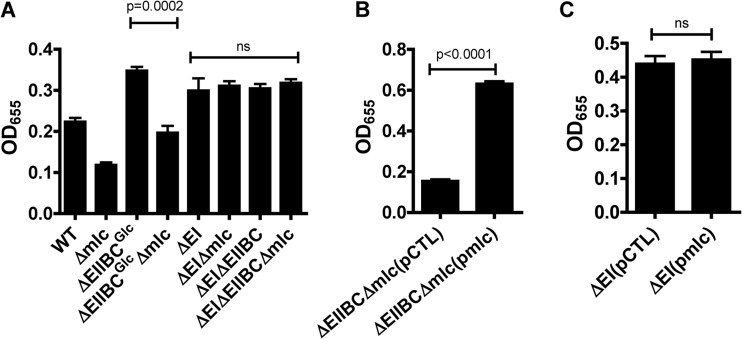
Transcription of the genes encoding EI and EIIBCGlc is repressed by Mlc. We previously showed that EI and EIIBCGlc repress vps gene transcription and biofilm formation by cells cultured in LB broth (21). We hypothesized that one or both of these proteins might be downstream of Mlc in a regulatory pathway. As shown in Fig. 7A, deletion of EIIBCGlc increased biofilm formation. Mutation of Mlc in this background, however, decreased biofilm formation to wild-type levels. This result is consistent with a model in which the primary function of EIIBCGlc in regulation of biofilm formation is to sequester Mlc to the membrane. Furthermore, this suggests that Mlc must act on another protein to activate biofilm formation. In agreement with this model, overexpression of Mlc in a Δmlc ΔEIIBCGlc mutant background increased biofilm formation (Fig. 7B). This demonstrates that activation of biofilm formation by Mlc is not dependent on EIIBCGlc.
FIG 7.
Evidence that Mlc activates biofilm formation through repression of EI. Quantification of biofilms formed by wild-type V. cholerae and Δmlc, ΔEI, and ΔEI Δmlc mutants (A), a ΔEIIBCGlc Δmlc mutant carrying either a control plasmid (pCTL) or a plasmid encoding Mlc (pmlc) (B), or wild-type V. cholerae and a ΔEI mutant carrying either a control plasmid (pCTL) or a plasmid encoding Mlc (pmlc) (C). P values are shown above. ns, not significant.
As shown in Fig. 7A and C, deletion or overexpression of mlc had no effect on biofilm formation by a ΔEI mutant. Because we have demonstrated that Mlc represses transcription of the EI gene, EI represses biofilm formation, and Mlc has no effect on biofilm formation in the absence of EI, we hypothesize that Mlc activates V. cholerae biofilm formation indirectly by repressing expression of EI.
Evidence that EIIAGlc is a component of a second regulatory pathway required in LB broth for derepression of biofilm formation in the absence of EI.
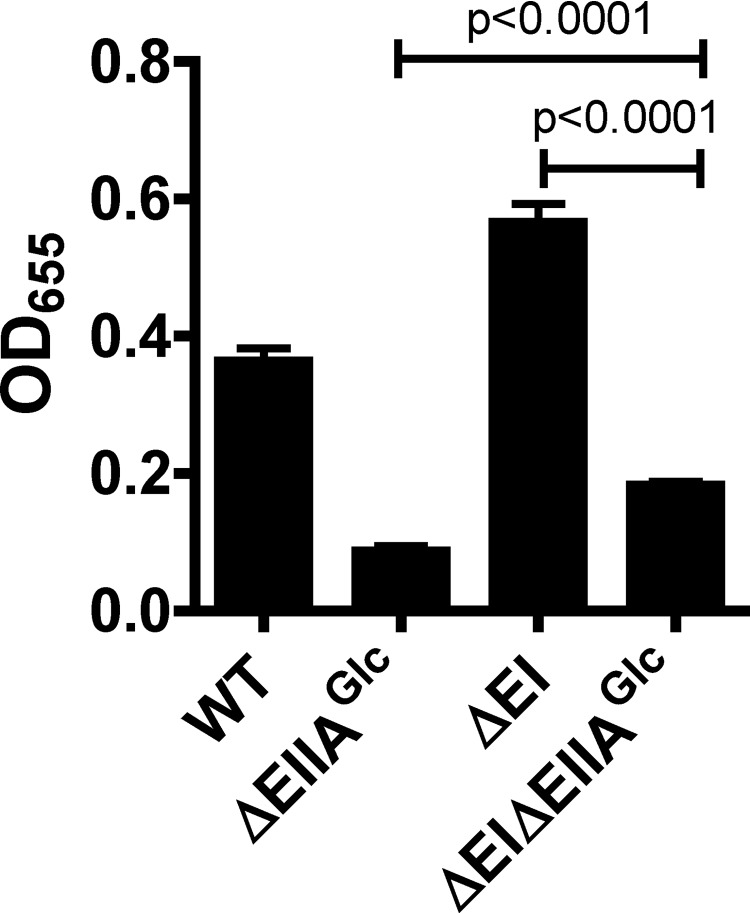
Although Mlc repressed EI when V. cholerae was cultured in either LB broth or minimal medium supplemented with pyruvate (Fig. 4C and D), biofilm formation was activated by Mlc in LB broth only. Therefore, we hypothesized that repression of EI gene transcription by Mlc was necessary but not sufficient for activation of vps gene transcription and V. cholerae biofilm formation. We previously reported that EIIAGlc activates biofilm formation in LB broth and that overexpression of Mlc only partially rescues biofilm formation in a Δmlc ΔEIIAGlc mutant background (20, 21). We suggested that expression of EIIAGlc might be regulated by Mlc (21). However, our current findings show that this is not the case (Fig. 4). Another possibility is that EIIAGlc is a component of a second regulatory pathway required for activation of biofilm formation by Mlc. Because Mlc activates biofilm formation through repression of EI, we evaluated the effect of a ΔEIIAGlc gene deletion on biofilm formation by a ΔEI mutant. As shown in Fig. 8, deletion of the EIIAGlc gene in a ΔEI mutant background greatly reduced biofilm formation. However, repression of biofilm formation by EI was significant even in the ΔEIIAGlc mutant background. These findings support a model in which the EI and EIIAGlc proteins function in independent regulatory pathways.
FIG 8.
Derepression of biofilm formation in the absence of EI requires EIIAGlc. Quantification of biofilm formation by ΔEI, ΔEIIAGlc, and ΔEI ΔEIIAGlc mutants in LB broth.
Our previous results show that EIIAGlc does not activate biofilm formation in minimal medium supplemented with pyruvate (20). The role of EIIAGlc in a particular growth condition is governed by its interactions with other proteins (22). We hypothesize that activation of biofilm formation by EIIAGlc in LB broth reflects its interaction with an as yet unexplored partner.
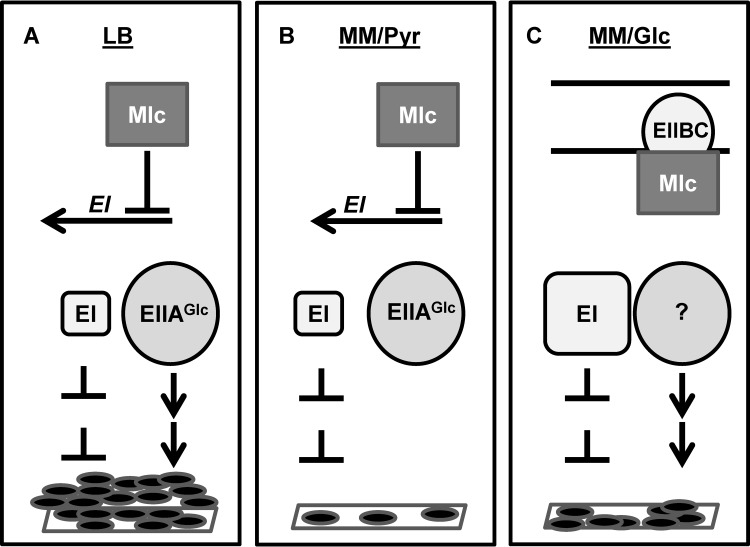
Model for regulation of biofilm formation by Mlc.
Based on our observations, we present the following model for activation of V. cholerae biofilm formation by the transcription factor Mlc (Fig. 9). In media that do not contain PTS substrates, such as LB broth and minimal medium supplemented with pyruvate, Mlc decreases but does not eliminate transcription of PTS components. Thus, there is residual repression of biofilm formation by EI, and deletion of EI increases biofilm formation in these media (2, 21). In LB broth, a second, independent regulatory pathway that includes EIIAGlc activates biofilm formation, resulting in formation of a biofilm (Fig. 9A). In this scenario, deletion of either mlc or the EIIAGlc gene decreases biofilm formation. EIIAGlc does not activate biofilm formation in minimal medium supplemented with pyruvate. Therefore, biofilm formation by wild-type V. cholerae is completely repressed by the small amount of EI that is made (Fig. 9B). Further repression of biofilm formation by deletion of Mlc and the concomitant derepression of EI is not possible. In minimal medium supplemented with the PTS substrate glucose, Mlc is sequestered to the membrane by the unphosphorylated form of EIIBCGlc (Fig. 9C). Because Mlc is inactive, EI is maximally expressed, and deletion of mlc does not further alter biofilm formation. Surface accumulation is observed under these growth conditions. Therefore, a second, biofilm-activating pathway may be operative.
FIG 9.
A model for regulation of V. cholerae biofilm formation by the transcription factor Mlc. (A) LB broth (LB): biofilm formation reflects decreased expression of the biofilm repressor EI due to repression by Mlc and activation of biofilm formation by a signal transduction pathway involving EIIAGlc. (B) Minimal medium supplemented with pyruvate (MM/Pyr): Mlc partially represses EI transcription. However, the small amount of EI in the cell maximally represses biofilm formation because the biofilm-activating pathway involving EIIAGlc is not present. (C) Minimal medium supplemented with glucose (MM/Glc): Mlc is inactive due to sequestration to the membrane by the unphosphorylated form of EIIBCGlc. This prevents repression of EI transcription by Mlc. In spite of repression by EI, a biofilm does form. This may indicate the presence of a second regulatory pathway that activates biofilm formation.
ACKNOWLEDGMENTS
This work was supported by NIH grant AI 50032 to P.I.W. D.R.S. was supported by NIH training grant 5T32HD055148-05.
Footnotes
Published ahead of print 25 April 2014
REFERENCES
- 1.Lengeler JW, Jahreis K. 2009. Bacterial PEP-dependent carbohydrate: phosphotransferase systems couple sensing and global control mechanisms. Contrib. Microbiol. 16:65–87. 10.1159/000219373 [DOI] [PubMed] [Google Scholar]
- 2.Houot L, Chang S, Absalon C, Watnick PI. 2010. Vibrio cholerae phosphoenolpyruvate phosphotransferase system control of carbohydrate transport, biofilm formation, and colonization of the germfree mouse intestine. Infect. Immun. 78:1482–1494. 10.1128/IAI.01356-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70:939–1031. 10.1128/MMBR.00024-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saier MH., Jr 1989. Protein phosphorylation and allosteric control of inducer exclusion and catabolite repression by the bacterial phosphoenolpyruvate:sugar phosphotransferase system. Microbiol. Rev. 53:109–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim SY, Nam TW, Shin D, Koo BM, Seok YJ, Ryu S. 1999. Purification of Mlc and analysis of its effects on the pts expression in Escherichia coli. J. Biol. Chem. 274:25398–25402. 10.1074/jbc.274.36.25398 [DOI] [PubMed] [Google Scholar]
- 6.Plumbridge J. 1998. Control of the expression of the manXYZ operon in Escherichia coli: Mlc is a negative regulator of the mannose PTS. Mol. Microbiol. 27:369–380. 10.1046/j.1365-2958.1998.00685.x [DOI] [PubMed] [Google Scholar]
- 7.Plumbridge J. 1998. Expression of ptsG, the gene for the major glucose PTS transporter in Escherichia coli, is repressed by Mlc and induced by growth on glucose. Mol. Microbiol. 29:1053–1063. 10.1046/j.1365-2958.1998.00991.x [DOI] [PubMed] [Google Scholar]
- 8.Plumbridge J. 1999. Expression of the phosphotransferase system both mediates and is mediated by Mlc regulation in Escherichia coli. Mol. Microbiol. 33:260–273. 10.1046/j.1365-2958.1999.01462.x [DOI] [PubMed] [Google Scholar]
- 9.Tanaka Y, Kimata K, Inada T, Tagami H, Aiba H. 1999. Negative regulation of the pts operon by Mlc: mechanism underlying glucose induction in Escherichia coli. Genes Cells 4:391–399. 10.1046/j.1365-2443.1999.00268.x [DOI] [PubMed] [Google Scholar]
- 10.Hosono K, Kakuda H, Ichihara S. 1995. Decreasing accumulation of acetate in a rich medium by Escherichia coli on introduction of genes on a multicopy plasmid. Biosci. Biotechnol. Biochem. 59:256–261. 10.1271/bbb.59.256 [DOI] [PubMed] [Google Scholar]
- 11.Joyet P, Bouraoui H, Ake FM, Derkaoui M, Zebre AC, Cao TN, Ventroux M, Nessler S, Noirot-Gros MF, Deutscher J, Milohanic E. 2013. Transcription regulators controlled by interaction with enzyme IIB components of the phosphoenolpyruvate:sugar phosphotransferase system. Biochim. Biophys. Acta 1834:1415–1424. 10.1016/j.bbapap.2013.01.004 [DOI] [PubMed] [Google Scholar]
- 12.Lee SJ, Boos W, Bouche JP, Plumbridge J. 2000. Signal transduction between a membrane-bound transporter, PtsG, and a soluble transcription factor, Mlc, of Escherichia coli. EMBO J. 19:5353–5361. 10.1093/emboj/19.20.5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nam TW, Cho SH, Shin D, Kim JH, Jeong JY, Lee JH, Roe JH, Peterkofsky A, Kang SO, Ryu S, Seok YJ. 2001. The Escherichia coli glucose transporter enzyme IICB(Glc) recruits the global repressor Mlc. EMBO J. 20:491–498. 10.1093/emboj/20.3.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka Y, Kimata K, Aiba H. 2000. A novel regulatory role of glucose transporter of Escherichia coli: membrane sequestration of a global repressor Mlc. EMBO J. 19:5344–5352. 10.1093/emboj/19.20.5344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yildiz FH, Schoolnik GK. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. U. S. A. 96:4028–4033. 10.1073/pnas.96.7.4028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Absalon C, Van Dellen K, Watnick PI. 2011. A communal bacterial adhesin anchors biofilm and bystander cells to surfaces. PLoS Pathog. 7:e1002210. 10.1371/journal.ppat.1002210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kierek K, Watnick PI. 2003. Environmental determinants of Vibrio cholerae biofilm development. Appl. Environ. Microbiol. 69:5079–5088. 10.1128/AEM.69.9.5079-5088.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moorthy S, Watnick PI. 2004. Genetic evidence that the Vibrio cholerae monolayer is a distinct stage in biofilm development. Mol. Microbiol. 52:573–587. 10.1111/j.1365-2958.2004.04000.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moorthy S, Watnick PI. 2005. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol. Microbiol. 57:1623–1635. 10.1111/j.1365-2958.2005.04797.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houot L, Watnick PI. 2008. A novel role for enzyme I of the Vibrio cholerae phosphoenolpyruvate phosphotransferase system in regulation of growth in a biofilm. J. Bacteriol. 190:311–320. 10.1128/JB.01410-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houot L, Chang S, Pickering BS, Absalon C, Watnick PI. 2010. The phosphoenolpyruvate phosphotransferase system regulates Vibrio cholerae biofilm formation through multiple independent pathways. J. Bacteriol. 192:3055–3067. 10.1128/JB.00213-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pickering BS, Smith DR, Watnick PI. 2012. Glucose-specific enzyme IIA has unique binding partners in the Vibrio cholerae biofilm. mBio 3(6):e00228–00212. 10.1128/mBio.00228-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ymele-Leki P, Houot L, Watnick PI. 2013. Mannitol and the mannitol-specific enzyme IIB subunit activate Vibrio cholerae biofilm formation. Appl. Environ. Microbiol. 79:4675–4683. 10.1128/AEM.01184-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haugo AJ, Watnick PI. 2002. Vibrio cholerae CytR is a repressor of biofilm development. Mol. Microbiol. 45:471–483. 10.1046/j.1365-2958.2002.03023.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horton RM, Cai ZL, Ho SN, Pease LR. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528–535 [PubMed] [Google Scholar]
- 27.Yildiz FH, Dolganov NA, Schoolnik GK. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716–1726. 10.1128/JB.183.5.1716-1726.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casper-Lindley C, Yildiz FH. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 186:1574–1578. 10.1128/JB.186.5.1574-1578.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg T, Schild S, Reidl J. 2007. Regulation of the chitobiose-phosphotransferase system in Vibrio cholerae. Arch. Microbiol. 187:433–439. 10.1007/s00203-006-0207-4 [DOI] [PubMed] [Google Scholar]
- 30.Plumbridge J. 2000. A mutation which affects both the specificity of PtsG sugar transport and the regulation of ptsG expression by Mlc in Escherichia coli. Microbiology 146:2655–2663 [DOI] [PubMed] [Google Scholar]
- 31.De Reuse H, Danchin A. 1988. The ptsH, ptsI, and crr genes of the Escherichia coli phosphoenolpyruvate-dependent phosphotransferase system: a complex operon with several modes of transcription. J. Bacteriol. 170:3827–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pennetier C, Dominguez-Ramirez L, Plumbridge J. 2008. Different regions of Mlc and NagC, homologous transcriptional repressors controlling expression of the glucose and N-acetylglucosamine phosphotransferase systems in Escherichia coli, are required for inducer signal recognition. Mol. Microbiol. 67:364–377. 10.1111/j.1365-2958.2007.06041.x [DOI] [PubMed] [Google Scholar]
- 33.Schiefner A, Gerber K, Seitz S, Welte W, Diederichs K, Boos W. 2005. The crystal structure of Mlc, a global regulator of sugar metabolism in Escherichia coli. J. Biol. Chem. 280:29073–29079. 10.1074/jbc.M504215200 [DOI] [PubMed] [Google Scholar]
- 34.Nam TW, Jung HI, An YJ, Park YH, Lee SH, Seok YJ, Cha SS. 2008. Analyses of Mlc-IIBGlc interaction and a plausible molecular mechanism of Mlc inactivation by membrane sequestration. Proc. Natl. Acad. Sci. U. S. A. 105:3751–3756. 10.1073/pnas.0709295105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seitz S, Lee SJ, Pennetier C, Boos W, Plumbridge J. 2003. Analysis of the interaction between the global regulator Mlc and EIIBGlc of the glucose-specific phosphotransferase system in Escherichia coli. J. Biol. Chem. 278:10744–10751. 10.1074/jbc.M212066200 [DOI] [PubMed] [Google Scholar]