Abstract
Borrelia species are unique in the bacterial world in possessing segmented genomes which sometimes contain over 20 genetic elements. Most elements are linear and contain covalently closed hairpin ends requiring a specialized process, telomere resolution, for their generation. Hairpin telomere resolution is mediated by the telomere resolvase, ResT. Although the process has been studied extensively in vitro, the essential nature of the resT gene has precluded biological studies to further probe the role of ResT. In this work, we have generated a B. burgdorferi strain that carries an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible resT gene controlled by a tightly regulated promoter. ResT is expressed in this strain at ∼14,000 monomers per cell, similar to the ∼15,000 monomers observed for the parental strain. We demonstrate ResT depletion with a half-life of 16 h upon IPTG washout. ResT depletion resulted in arrested growth 48 h after washout. Interestingly, not all spirochetes died after ResT washout, and at least 15% remained quiescent and could be resuscitated even at 2 weeks postwashout. Significant levels of DNA synthesis were not observed upon growth arrest, suggesting that ResT might interact directly or indirectly with factors controlling the initiation or elongation of DNA synthesis. Analysis of the linear plasmids lp17 and lp28-2 showed that the linear forms of these plasmids began to disappear and be replaced by higher-molecular-weight forms by 24 h post-IPTG washout. Treatment of DNA from the ResT-depleted strain with ResT in vitro revealed the presence of replicated telomeres expected in replication intermediates.
INTRODUCTION
Lyme disease, caused by the bacterium Borrelia burgdorferi and related species (1–4), is the most commonly reported vector-borne illness in North America and Europe (5), with a significant presence in other regions of the Northern Hemisphere (6, 7). A unique feature of B. burgdorferi is its segmented genome. The prototype B31 genome consists of a single linear chromosome of approximately 1 Mb, as well as a mixture of approximately 20 linear and circular plasmids (8, 9). To overcome the end replication problem, the ends of the linear replicons are terminated by covalently closed hairpin telomeres (10–12). Replication of the linear elements is believed to proceed bidirectionally (Fig. 1) from an internal origin of replication (13–15), resulting in a circular, head-to-head, tail-to-tail dimer intermediate. The dimer intermediate is processed by telomere resolution, a DNA breakage and reunion reaction that results in cleavage at the dimer junctions followed by ligation of complementary strands to generate the hairpin telomeres (16–19).
FIG 1.
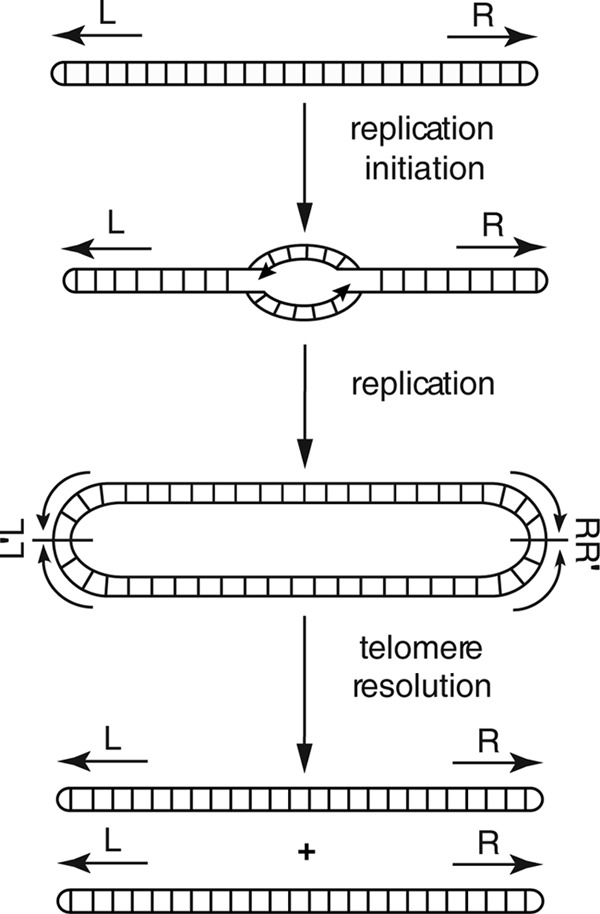
Replication pathway for linear replicons in Borrelia species. Arrows indicate hairpin telomeres at the left (L) and right (R) ends. Initiation occurs at the central origin (13), and complete replication results in the formation of a head-to-head (L′L)–tail-to-tail (R-R′) dimer. The lines bisecting the junctions denote axes of 180° rotational symmetry. Where in the cell telomere resolution occurs is unknown, as is whether telomere resolution is concerted at both ends or whether it occurs independently at each end. This figure is adapted from reference 20 with permission.
Telomere resolution is carried out by the telomere resolvase, ResT, encoded by the circular plasmid cp26 (20). ResT has been extensively characterized, and its mechanism is well defined (12, 21–32). The enzyme is mechanistically similar to type IB topoisomerases and tyrosine recombinases. It promotes telomere resolution through a two-step transesterification involving covalent linkage of tyrosine 335 to the DNA substrate at the cleavage site followed by a nucleolytic attack on the phosphotyrosine linkage by the free 5′ hydroxyl on the opposite DNA strand. Reversal of ResT activity has been proposed as the driving force for generating telomere exchanges between the linear Borrelia replicons and for mediating the continual rearrangements observed in the linear plasmids of this genus (18, 24, 33).
Due to its important function in the DNA replication process, resT is an essential gene in B. burgdorferi. Previous attempts to knock out resT have been unsuccessful unless resT has been provided in trans (30, 34, 35). Conditional disruption of essential genes has also not been possible in B. burgdorferi (36, 37). However, the recent development of inducible expression systems in B. burgdorferi has provided a powerful new approach in the arsenal of genetic tools available (38–40). In particular, the expression vector pJSB104 (39), in which the expressed gene is regulated by a double lac operator and the lac repressor, has allowed for tight regulation and the generation of conditional mutants in two essential genes: the dedA orthologue bb0250 (41) and the gene for the response regulator protein, rrp2 (42). Here we report the use of the pJBS104 expression system to generate a conditional mutation in the essential B. burgdorferi telomere resolvase resT. This has allowed us to study the effects of ResT depletion in vivo for the first time. Using this system, we have observed the effects of ResT depletion on the spirochetes, as well as the state of the linear DNA molecules.
MATERIALS AND METHODS
Plasmid construction.
Primers and strains used in this work are listed in Table S1 and Table S2 in the supplemental material, respectively. To facilitate the cloning of resT, the pJSB104 expression vector (39) was first modified by the addition of the multiple-cloning site (MCS) from pJLB12g (43). The MCS was amplified by PCR using primers B2155 and B2156 and inserted in the pJet 1.2 blunt cloning vector (Fermentas). The MCS was then recovered by digestion with HindIII and cloned into the HindIII site of pJSB104 to generate pJSB104MCS.
To construct the inducible resT plasmid, pNB12, the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter PpQE30 was linked to resT by overlap extension PCR. In the first PCR (PCR1), the PpQE30 promoter was amplified from pJSB104 using primers B2145 and B2149. In PCR2, the resT gene was amplified from B31-A (GCB908) genomic DNA using primers B2150 and B2169. In PCR3, the product from PCR1 was used as a primer along with B2169 to fuse the PpQE30 promoter to the resT gene amplified in PCR2. The resulting product was ligated into pJet 1.2 blunt. The insert was recovered by BamHI-XhoI digestion and cloned into BamHI-XhoI-digested pJSB104MCS to generate pNB12. The integrity of the final construct was verified by DNA sequencing.
The resT knockout plasmid, pNB13, was also built through overlap extension PCR. First, 500 bp flanking resT on either side in cp26 was amplified using primers B2209 and B2210 (3′ flank) and B2213 and B2214 (5′ flank). A kanamycin resistance cassette was then amplified using primers B2211 and B2212, and similar to the construction of pNB12, the kanamycin cassette was then joined to the 3′ flanking region; subsequently, this fragment was fused to the 5′ flanking region. The final PCR product was cloned in pJet 1.2 blunt.
Construction of a conditional resT knockouts in B. burgdorferi.
B. burgdorferi was grown in BSK II medium (44) prepared in-house and supplemented with 5% rabbit serum (Cedar Lane). B. burgdorferi transformations were performed as described previously (45, 46). PCR was performed on genomic DNA to screen for the presence of the insert. Construction of a conditional resT knockout (GCB2127) was done as shown in Fig. S2A in the supplemental material. The parent strain, B31-A (GCB908), was first transformed with the resT-inducible plasmid pNB12, followed by disruption of the endogenous resT gene by transformation with the knockout plasmid pNB13. For pNB12 transformants, primers B2180 and B2181 were used for screening. Potential resT knockout clones were further analyzed using primers B2251 and B2252, which anneal in the homologous recombination target; the native resT gene is larger than the kanamycin cassette, which is inserted into this site and gives a larger band than that observed in the case of the insertion. Recombinants were confirmed by DNA sequencing.
A second approach (see Fig. S2B in the supplemental material) to constructing a strain with conditional expression of resT (GCB2103) was to transform strain GCB51 (30), which contains a disrupted resT gene and a complementing plasmid carrying the B. hermsii resT gene (pBSV2BhresT) with pNB12 (Smr). The plasmid with the B. hermsii resT gene was then displaced by growth in increasing concentrations of streptomycin as described in the supplemental material.
Growth of conditional resT knockouts.
Conditional mutants were grown in BSK II medium supplemented with 1 mM IPTG and 100 μg/ml of streptomycin at 35°C until reaching a density of approximately 1 × 108 spirochetes per ml. The spirochetes were collected by centrifugation at 6,000 × g at 4°C, resuspended in 5 ml of sterile phosphate-buffered saline (PBS), and then centrifuged again. The spirochetes were resuspended in sterile PBS and used to seed fresh cultures at 1 × 106 spirochetes per ml with or without IPTG. Spirochetes were diluted 1:1 with fresh BSK II medium at 48 h to ensure that cells remained in log phase during the course of growth. For growth curves, spirochetes were enumerated at 24-h intervals by counting in a Petroff-Hausser counting chamber.
Western blotting.
B. burgdorferi spirochetes were grown as described above for the conditional resT knockout. The spirochetes were washed and resuspended in BSK II medium once and this culture used to start each time point sample at 1 × 106 spirochetes per ml without IPTG. At 8, 16, 24, 32, and 48 h, aliquots containing at least 6 × 107 spirochetes were harvested by centrifugation at 6,000 × g for 15 min at 4°C. To prepare whole-cell lysates, B. burgdorferi cells were lysed using SDS-PAGE loading buffer (125 mM Tris-HCl [pH 6.8], 4% SDS, 20% glycerol, 1.4 M 2-mercaptoethanol, and 0.2% bromophenol blue) at 95°C for 4 min. A volume of whole-cell lysate generated from 2.0 × 107 spirochetes was loaded in each well of 7% SDS-PAGE gels for separation with a 5% stacking gel. Samples were blotted onto a nitrocellulose membrane (Amersham Hydrobond-ECL). The membrane was blocked in 5% nonfat dry milk in TBS-T (20 mM Tris-HCl, 154 mM NaCl, 0.1% Tween 20 [pH 7.4]). ResT bands were detected using ResT antiserum (SACRI Antibody Services, University of Calgary) at a dilution of 1/10,000 as a primary antibody and horseradish peroxidase-conjugated anti-rabbit antibody (Cedar Lane) at a dilution of 1/5,000 as the secondary antibody prepared in TBS-T containing 3% nonfat dry milk. 3,3′,5,5′-Tetramethylbenzidine (TMB) liquid substrate (Sigma; T0565) was used for colorimetric detection according to the manufacturer's instructions.
Spirochete length and DNA content measurements.
The parent B. burgdorferi strain, B31-A (GCB908), and the conditionally expressing resT strain (GCB2127) were grown to late log phase, and IPTG washout was performed as described above. Cultures were sampled every 24 h, and each sample was stained with 4′,6-diamidino-2-phenylindole (DAPI) to a final concentration of 100 μM for length measurements (Invitrogen) or with Hoechst 33258 (for DNA content measurements) in a total of 10 μl. The stained spirochetes were finally mixed with 1 μl of Fluoromount (Sigma-Aldrich) in preparation for microscopy. Slides were imaged on a Leica DMIRE2 fluorescence microscope with a Hamamatsu ORCA-ER camera. An exposure time of 500 ms was used for capture of all images. Fluorescence intensity was measured using ImageJ software (NIH). Individual spirochetes were selected, and the background was subtracted from the raw integrated fluorescence to give the corrected total fluorescence. Length was measured by tracing the length of the spirochete in ImageJ with comparison to a calibrated scale bar generated by the microscope.
Field inversion gels and Southern blotting.
Total genomic B. burgdorferi was analyzed on field inversion gels and blotted as previously described (16). Probes were generated to bp 12980 to 13727 in lp17 using primers B2275 and B2276 through PCR. Similarly, a probe was generated to bp 18725 to 19374 of lp28-2 through PCR. Following ResT treatment, additional probes were needed for the left end of lp17 and the right end of lp28-2. These were generated by PCR using primers B2317 and B2318 for lp17 and B2331 and B2332 for lp28-2. Probes were purified using a QIAquick PCR purification kit (Qiagen) before radioactive labeling with the Random Primers DNA Labeling System (Invitrogen). Labeling was performed as per the manufacturer's instructions, using [α-32P]dCTP. The radioactive signals from the bands were detected by exposure using phosphor screens (PerkinElmer) for exposures between 1 h and up to overnight. The screens were imaged using a Packard Cyclone imager (Packard) and OptiQuant imaging software.
Multiplex PCR for analysis of plasmid content.
Multiplex PCR was performed for analysis of plasmid content as described by Bunikis et al. (47). Briefly, genomic B. burgdorferi DNA was purified as described above, and 100 ng of total genomic DNA was used as the template for detection of linear and circular plasmids. PCRs were run on a 3% MetaPhor agarose gel and stained with GelRed (Cedar Lane).
ResT treatment of B. burgdorferi DNA.
Total B. burgdorferi genomic DNA was purified from spirochetes grown under noninducing conditions after 72 h. The DNA was digested with 10 U of either EcoRI for detection of the left-end lp17 telomere or BamHI for detection of the right-end lp28-2 telomere. Digestions of 1 μg of B. burgdorferi genomic DNA were carried out at 37°C for 2 h in the appropriate reaction buffer as recommended by the manufacturer (NEB) in a final volume of 20 μl. The buffers were adjusted to 100 mM NaCl, 10 mM EDTA, 100 μg/ml of bovine serum albumin (BSA), and 5 mM spermidine prior to incubation with 200 ng (150 nM) of purified His-tagged recombinant ResT (a gift from Kerri Kobryn, University of Saskatchewan) in a final volume of 25 μl. The reaction was allowed to continue for 2 h at 30°C, and then the reaction mixture was immediately loaded on a 1% agarose gel. Specific DNA fragments were detected through Southern blotting hybridization for the telomeres as described above.
RESULTS
Construction of a conditional resT mutant.
An inducible resT plasmid construct was generated by insertion of the wild-type (wt) resT gene from B. burgdorferi B31-A into a derivative of expression vector pJSB104 (39) to generate pNB12 (see Fig. S1 in the supplemental material) as described in Materials and Methods. The resulting construct has the resT gene under the control of the tightly regulated lac-inducible PpQE30 promoter, which carries two lac operators to maximize repression (one between −35 and −10 and the second overlapping the transcriptional start site). The construct also carries a constitutively expressed lacI gene under the control of the B. burgdorferi flaB promoter and displays >800-fold repression of the target gene in B. burgdorferi (39).
Construction of a strain in which the native resT gene was inactivated was accomplished using two different approaches, as shown in Fig. S2 in the supplemental material. First, the high-passage-number B. burgdorferi strain B31-A was transformed with pNB12 (see Fig. S2A). This strain was then transformed with the knockout plasmid pNB13, which deleted the entire resT locus through allelic exchange, replacing it with a kanamycin resistance cassette. Transformants were recovered in as little as 50 nM IPTG; however, as expected, no transformants were recovered when IPTG was absent. The recovered clones were analyzed for the presence of the kanamycin cassette, as well as for the complete deletion of the native resT gene, by PCR and confirmed through DNA sequencing. The final construct was GCB2127 (see Table S1 in the supplemental material).
Additionally, a conditional mutant was independently recovered through transformation of the previously reported strain GCB51 (30) with the conditionally expressing plasmid pNB12 (see Fig. S2B in the supplemental material). GCB51 carries an inactivated resT gene on cp26 but is viable because it carries the orthologous resT gene from Borrelia hermsii on a shuttle vector (pBSV2BhresT). All transformants were found to contain both the newly introduced pNB12 (Smr) and pBSV2BhresT (Kmr). Displacement of pBSV2BhresT (final construct, GCB2103) was achieved by increasing the concentration of streptomycin in the media until clones were recovered that were resistant to streptomycin but sensitive to kanamycin (see the supplemental material).
Characterization of the conditional resT mutant.
To assess the effect of IPTG washout on ResT levels in the cell, Western blotting was performed using polyclonal antiserum to purified ResT (Fig. 2A). The conditional mutant carrying the full resT deletion (GCB2127) was grown to late log phase with IPTG, then harvested by centrifugation, and resuspended in fresh BSK II medium without IPTG. Spirochetes were sampled at various times and whole-cell lysates were assessed for ResT content by Western blotting. Under inducing conditions (see Fig. S3 in the supplemental material), ResT was produced at approximately the same level in the conditional mutant (14,000 ± 2,000 monomers per spirochete) as in the wild-type parental strain, B31-A (15,000 ± 1,000 monomers per spirochete). However, after IPTG washout (Fig. 2A), ResT levels gradually decreased at 8, 16, and 24 h, and ResT was barely visible at 32 h and undetectable at 48 h postwashout. Analysis of the data revealed a half-life of 16 h for ResT after IPTG washout (Fig. 2B).
FIG 2.
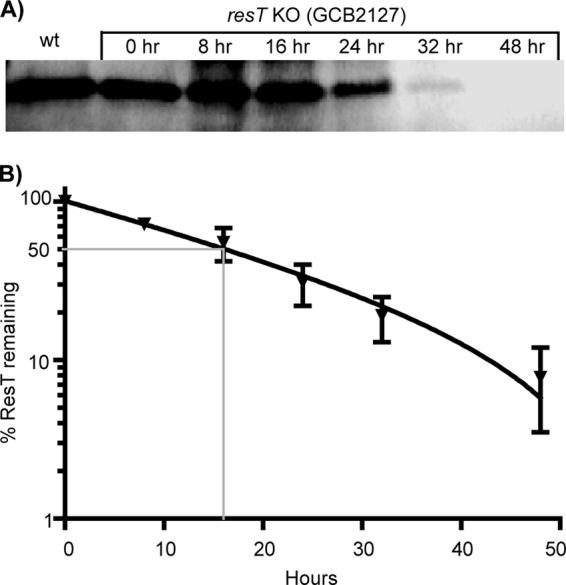
ResT half-life in B. burgdorferi. (A) To measure the half-life of ResT protein in the conditional expression strain (GCB2127), a time course analysis was performed using Western blotting following IPTG washout. B. burgdorferi was grown for 48 h in the presence of 1 mM IPTG until it reached late exponential phase. Following IPTG washout, the culture was diluted to 1.0 × 106 spirochetes/ml (time zero) and incubation was continued for 48 additional hours, with aliquots removed at the indicated times. For Western blotting, 2.0 × 107 spirochetes were loaded per well as described in Materials and Methods. ResT from the wild-type parent, B31-A, is also shown. Band intensities were quantified and analyzed using AlphaImager software (Alpha Innotech). (B) The half-life of ResT was calculated by plotting the percentage of ResT remaining versus time. Data in the graph represent two separate experiments. The calculated ResT half-life was 16 h.
To assess the effects of ResT depletion, B. burgdorferi was grown in BSK II medium under inducing conditions (plus IPTG) to late log phase prior to collection of the bacteria through centrifugation, which were then resuspended at 1 × 106 spirochetes/ml in fresh BSK II medium with or without IPTG. The bacteria were assessed for growth through daily counting using dark-field microscopy. After 48 h, the bacteria were diluted with an equal volume of fresh medium with or without IPTG to avoid entrance into stationary phase. For the first 48 h, the mutant (plus or minus IPTG) grew at the same rate as the wild-type, B31-A (Fig. 3A). However, 48 h after IPTG washout, the conditional resT mutant was unable to continue growing and remained at the same cell density at 72 h, decreasing slightly by 96 h. Growth was not affected in strain B31-A or in the induced culture. Growth arrest was also observed in the conditional mutant carrying the partial resT deletion (GCB2103 [data not shown]). The strain carrying the complete resT deletion (GCB2127) was used for the remainder of experiments in the work presented here.
FIG 3.
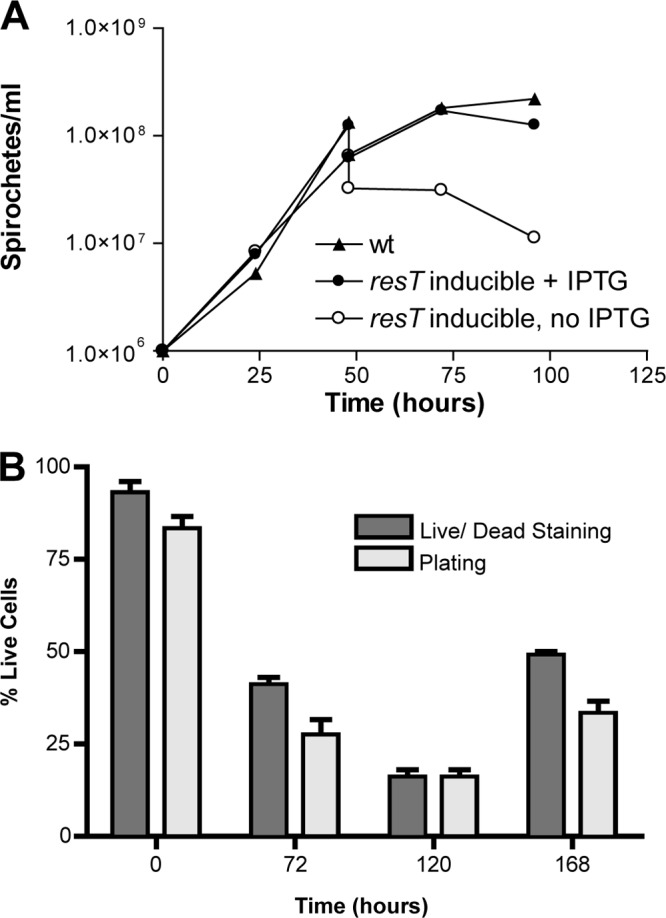
(A) Growth curves of the resT knockout mutant GCB2127. B. burgdorferi cultures were grown in BSK II medium with and without IPTG at 35°C, and spirochetes were counted every 24 h. After 48 h, the cultures were diluted with an equal volume of medium to avoid entry into stationary phase and were counted before and after dilution at the 48-h time point. Growth of the wild type, B31-A (GCB908), was compared to growth of the resT-inducible strain (GCB2127). (B) Spirochetes from cultures at the indicated times following IPTG washout were stained with the LIVE/DEAD BacLight staining kit (Invitrogen) to determine if cells were alive or dead. Spirochetes were analyzed by fluorescence microscopy for staining with Syto-9 (live) or propidium iodide (dead). Ten fields of view were analyzed for each time point. Living spirochetes were also enumerated by dilution in duplicate of the cultures and plating in BSK II medium plus IPTG in 96-well microtiter plates. The percent living cells for counting was based upon the total number of spirochetes counted by dark-field microscopy. At each time point, the mean percentage of living cells ± the standard error is plotted for both the LIVE/DEAD staining and plating methods.
Cultures were assessed for the viability of spirochetes through LIVE/DEAD staining and by plating in liquid medium (Fig. 3B), which have been reported to give similar results (48). At 72 h postwashout, a decrease in viable cells to between 27 and 41% (plating and LIVE/DEAD staining, respectively) was observed. At 120 h, 16% viability was observed by both methods, and at 168 h, 33 and 49% living spirochetes were observed by LIVE/DEAD staining and plating, respectively. The increase in living spirochetes by LIVE/DEAD staining at 168 h resulted from a lysis of the dead cells, rendering them uncountable and thereby changing the relative proportions of living and dead cells. Living spirochetes could be rescued at 1 or 2 weeks postwashout by the addition fresh BSK II medium containing IPTG. The absence of ResT, therefore, appears to be lethal to most spirochetes, while some remain quiescent and can be resuscitated by providing ResT at 7 or 14 days.
To assess the possibility of a cell division phenotype resulting from a depletion of ResT, spirochete length was measured as described in Materials and Methods. The conditional resT mutant was grown under noninducing conditions as described for Fig. 3 and then imaged with the DNA stain DAPI and examined by fluorescence microscopy at each time point (Fig. 4). ResT depletion did not result in any increase in spirochete length relative to that of the wild-type strain; all fell within the 10- to 30-μm range, as expected for Borrelia burgdorferi (49), and no filamentous forms were observed. Therefore, a specific defect in cell division, expected to result in longer spirochetes or filamentous forms, does not appear to occur upon ResT depletion.
FIG 4.
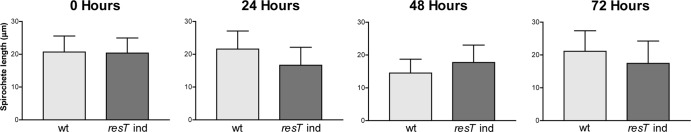
Determination of the average spirochete length of the wild-type (GCB908) and resT-inducible (GCB2127) strains. The length of individual DAPI-stained spirochetes was determined at the indicated times after IPTG washout using ImageJ (NIH) as described in Materials and Methods. The average length ± the standard error is plotted above for each time point for 100 measured spirochetes.
Finally, in view of the fact that the spirochetes ceased to double after 48 h of ResT depletion, it was of interest to determine if they continued to replicate their DNA. To analyze DNA levels, wild-type controls or spirochetes at various times after IPTG washout were stained with Hoechst 33258, a DNA-specific fluorescent dye previously used to quantify bacterial DNA content (50). Fluorescence intensity of individual spirochetes was determined by analysis of fluorescence micrographs at several time points (Fig. 5). To ensure that an increase in DNA content would be observed under our experimental conditions, spirochetes were stained and examined with increasing exposure times to ensure that exposure was in the linear range. Additionally, spirochetes were stained at five times the typical cell density to demonstrate that the Hoechst dye was in excess and that increased DNA content would be detectable under the conditions used. Indeed, this was the case, as a 5-fold increase in cell density did not result in a decrease of fluorescent signal of individual spirochetes (data not shown). Examining the resT mutant strain using these conditions revealed that DNA synthesis did not continue in ResT-depleted spirochetes after the cessation of cell division. In fact, the total fluorescence of the spirochetes decreased slightly relative to that of the wild type.
FIG 5.
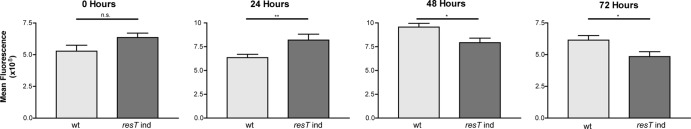
Determination of the relative DNA content of wild-type (GCB908) versus resT-inducible spirochetes (GCB2127) by quantification of fluorescence intensity of Hoechst 33258-stained spirochetes in fluorescence micrographs. Images of fluorescent spirochetes were analyzed for total fluorescence intensity by collecting the integrated fluorescence density using ImageJ (NIH) and correcting for the fluorescence background. The mean fluorescence intensity ± the standard error is plotted. Statistical significance was determined by a two-tailed unpaired t test and is indicated above the graphs (*, P < 0.05; **, P < 0.01; n.s., not significant).
The effect of ResT depletion on the state of linear plasmids.
To assess the effect of ResT depletion upon the state of the linear DNA molecules, DNA was purified from the mutant strain GCB2127 at 72 h after IPTG washout and run on a 0.65% agarose field inversion gel (Fig. 6). In the absence of ResT, some plasmid bands were obviously missing from the uninduced mutant (lane 1) compared to the induced mutant and to the wild-type strain (lanes 2, 3, and 4, respectively). Especially evident were the well-separated lp17 and bands in the 28-kb range which were present in the wild type as well as induced mutant but absent in the ResT-depleted strain (white arrowheads). The presence of multiple plasmids of both linear and circular forms made both identification and observance of new forms (replicative intermediates) difficult. We therefore used Southern blotting hybridization to analyze the state of lp17 and lp28-2 (two plasmids that can unambiguously be probed for) after IPTG washout (Fig. 7). After 24 h, a band in the higher-molecular-weight region appeared for both lp17 (Fig. 7A) and lp28-2 (Fig. 7B), which is roughly double the size of each of the plasmids and likely represents a dimeric replication intermediate (Fig. 1). By 72 h post-IPTG washout, both lp17 and lp28-2 disappeared. The linear plasmid disappearance at 72 and 96 h suggested either plasmid loss or conversion to heterogeneous forms (e.g., a population of partially replicated molecules) which resulted in a smear rather than discrete bands during the gel run. To determine whether plasmids were being lost upon ResT depletion, a multiplex PCR assay (47) was performed to determine plasmid content. It was observed that the ResT-depleted mutant strain maintained the full plasmid complement as seen in the wild-type control and that none of the linear plasmids were lost (data not shown).
FIG 6.
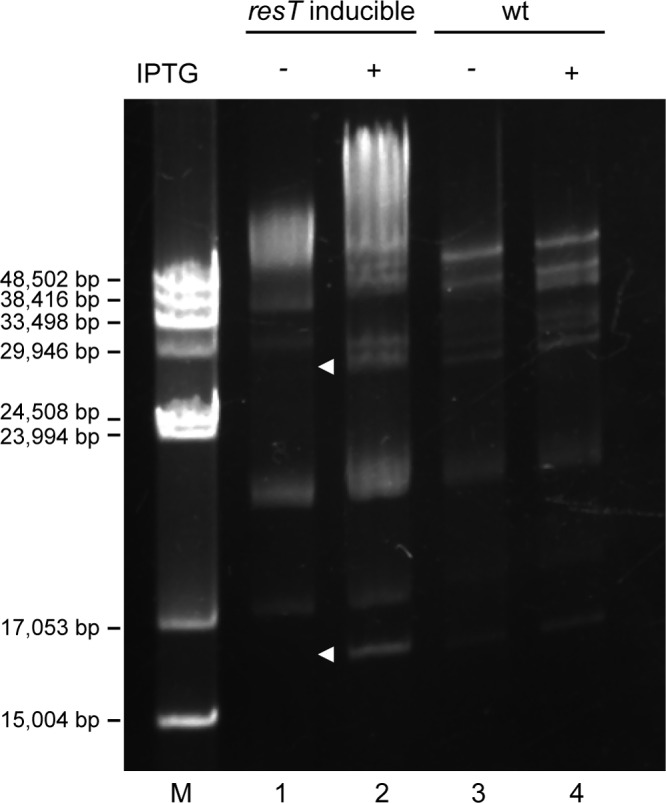
Field inversion gel of isolated B. burgdorferi genomic DNA. The resT-inducible strain GCB2127 (lanes 1 and 2) was grown for 48 h with or without IPTG, and the genomic content was compared to that of the wild type, B31-A (GCB908) (lanes 3 and 4). The gel was stained with ethidium bromide. M, lambda monocut size marker (NEB).
FIG 7.
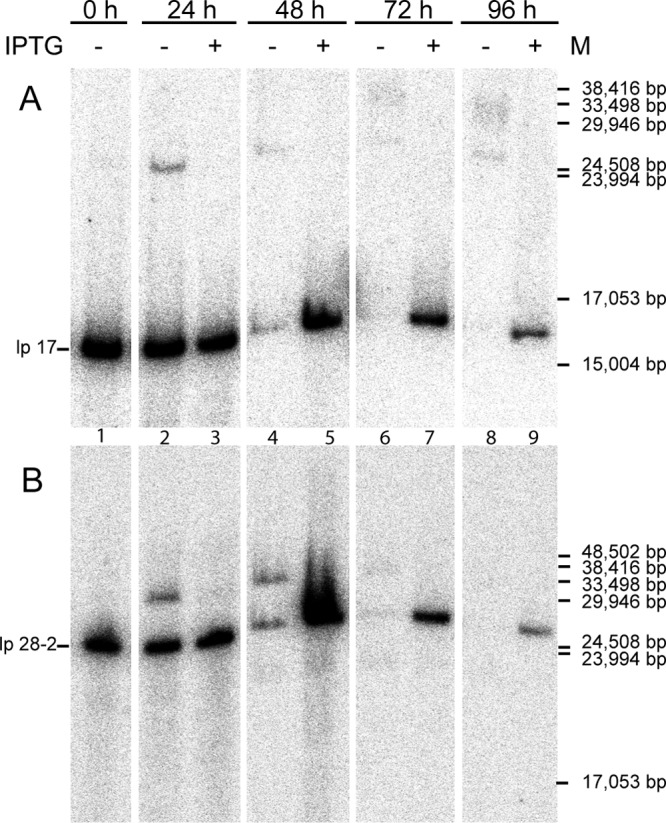
Southern blot of genomic DNA from a resT-inducible strain (GCB2127). Spirochetes were grown either with or without IPTG, and the culture was sampled at 0, 24, 48, 72, and 96 h. DNA was extracted and run on a 0.65% agarose field inversion gel and transferred to a nylon membrane for Southern blotting. Probes were generated against the left end of lp17 (A) as well as the right end of lp28-2 (B) as described in Materials and Methods, using PCR primers B2275 and B2276 for lp17 and B2295 and B2296 for lp28-2.
To determine if the linear plasmids had been converted into the expected replication intermediates (Fig. 1) in the ResT-depleted strains, purified ResT was added to genomic DNA, with the expectation that replicative intermediates would be resolved to regenerate linear plasmids. A Southern blot was performed for lp17; however, no linear plasmid was generated by treatment with ResT in vitro (data not shown). ResT reactions were performed either alone or after treatment of the DNA with topoisomerase I, as ResT does not function on negatively supercoiled DNA (20, 22).
To simplify detection of replicated telomeres (L′L and RR′ [Fig. 1]), smaller DNA fragments were generated by digesting the DNA with restriction enzymes which cut near the telomeres (Fig. 8A). These fragments were then treated with ResT to determine if a replicated telomere was present in the DNA. Southern blots from such an experiment for lp17 and lp28-2 are shown in Fig. 8B and C, respectively. Cleavage of DNA from the wild-type control with EcoRI generated the expected 5,684-bp fragment for the left end of lp17 (Fig. 8B, lane 2). Additional treatment with ResT (lane 3) did not result in any changes. In contrast, treatment of the ResT-depleted mutant resulted in two bands: a 5,684-bp fragment and a second fragment of about twice the size (lane 5), expected for the left end when present as a replicated telomere fragment. Treatment with ResT after EcoRI resulted in the disappearance of the larger left-end fragment (lane 6) and intensification of the lower fragment, as expected for telomere resolution of the left-end replicated telomere.
FIG 8.
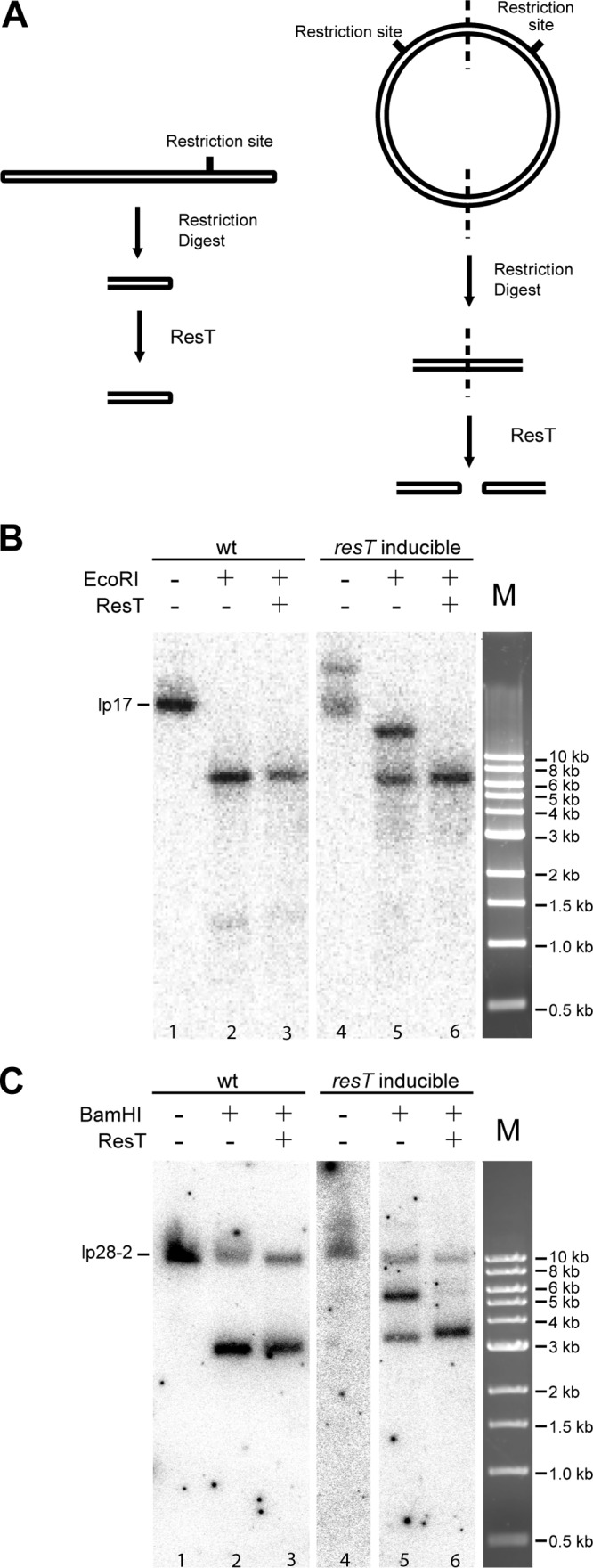
Treatment of DNA from the resT-inducible strain (GCB2127) and the wild-type parent (GCB908) with the telomere resolvase ResT. (A) Schematic of the experimental design. To simplify detection of telomere ends, DNA was digested with a restriction enzyme to liberate either a free telomere or the replicated telomere junction. ResT activity on the replicated junction generates free ends, which were detected by Southern blotting hybridization. (B and C) Southern blots of DNA that was isolated 72 h after IPTG washout and digested with EcoRI or BamHI prior to being treated with ResT. DNA was run on a 1% agarose gel and transferred to a nylon membrane for Southern blotting hybridization with probes for the lp17 (B) and lp28-2 (C) left ends.
Similar results were obtained with lp28-2. For this plasmid, BamHI was used to liberate the left-end 3,274-bp fragment (Fig. 8C, lane 2). BamHI cleavage was not complete and could not be driven to completion with more enzyme (data not shown). No change resulted from ResT treatment of the DNA from the wt strain (lane 3). However, as for lp17, treatment of the ResT-depleted mutant resulted in two bands: the 3,274-bp fragment and a second fragment of twice that size (lane 5), expected for the left end when present as a replicated telomere fragment. Treatment with ResT after BamHI resulted in the disappearance of the replicated telomere fragment (lane 6) and intensification of the lower fragment, as expected for resolution of the left-end replicated telomere.
DISCUSSION
A previous attempts to disrupt the essential resT gene resulted only in the recovery of merodiploids where the bacteria maintained both the disrupted gene and a functional copy of resT (34). Similarly, disruption of resT in B. burgdorferi was achieved only when the ortholog to resT from the relapsing-fever spirochete B. hermsii was provided in trans prior to gene disruption (30). In this work, we have generated a strain with conditional expression of ResT in the pJSB104 system carried on a B. burgdorferi shuttle vector. Protein expression is driven from a phage T5 promoter controlled by two lac operators (39).
Characterization of the conditional resT mutant.
Levels of ResT expression in the conditional mutant grown in the presence of 1 mM IPTG (GCB2127) were estimated from a Western blot to be 14,000 monomers per cell, similar to ResT levels (15,000 monomers per cell) observed in the wild-type parent strain, B31-A (GCB908). It is of interest that ResT was expressed to the same level from the lac inducible PpQE30 promoter on the multicopy shuttle vector as from its native promoter at the endogenous location on the low-copy-number cp26. The relative strength of the two promoters is not known. It may be fortuitous that the expression levels in the two situations were almost identical, or there may be some additional regulatory mechanisms in place for ResT expression (e.g., transcriptional or translational autoregulation) that keep ResT expression at a relatively constant level.
At 24 h post-IPTG washout, when ∼30% of the ResT was still present (Fig. 2), replication intermediates were observed (Fig. 7). In contrast to the appearance of replication intermediates at 24 h, cellular growth was at wild-type levels through 48 h, when ResT was present at only ∼10% of the wild-type level. At this time point, only a small amount of lp17 and lp28-2 migrated as linear monomers. The appearance of the growth arrest phenotype therefore coincided with the shift in gel migration of most of the linear plasmids. Whether the physical state of the linear replicons or the low levels of ResT are responsible for growth arrest is not currently known.
At 48 h, all spirochetes were still alive, as determined by LIVE/DEAD staining, but a reduction of live spirochetes was observed at 72 h and thereafter by LIVE/DEAD staining and plating. The spirochetes that did not die remained nondividing for extended periods. Interestingly, living but nondividing spirochetes could be rescued by inoculation into fresh media containing IPTG at 1 or 2 weeks post-IPTG washout. The presence of ResT is, therefore, required for active growth. However, a significant fraction (∼15 to 25%) of ResT-depeleted spirochetes can survive for lengthy periods and be revived by renewed ResT synthesis. The reason for lethality versus survival in the spirochete population is not currently understood.
Before DNA segregation and cell division can occur, sister DNA molecules must be completely unlinked from one another so that each sister can be partitioned into a new daughter cell (51). It was expected that ResT-depleted B. burgdorferi would be unable to complete chromosomal dimer resolution and hence cell division. Such spirochetes would be expected to become long and filamentous, similar to a bb0250 (inner membrane protein) knockout that was unable to complete cell division (41). Surprisingly, ResT-depleted spirochetes did not display a filamentous phenotype, suggesting that there may not be an intimate spatial and temporal coupling between cell division and telomere resolution in B. burgdorferi.
We also investigated whether ResT-depleted spirochetes would continue to accrue DNA once they had stopped dividing. As seen by Hoechst 33258 staining of the spirochetes (Fig. 5), the DNA content did not increase and in fact slightly decreased relative to that of the wild-type control at 48 and 72 h after IPTG washout. The shutdown in DNA synthesis upon ResT depletion suggests that ResT may interact directly with components of the replication machinery involved with the initiation and/or elongation steps of DNA replication. Further studies will be required to elucidate such interactions.
State of the linear DNA in ResT-depleted B. burgdorferi.
Although it has been previously shown that ResT can bind and resolve replicated telomere substrates in vivo (15, 16) and in vitro (17, 18, 20), a resT mutant and its effect upon the segmented genome have not been previously reported. By depleting ResT in our conditional mutant, we were able to promote the accumulation of replication intermediates. A complete disappearance of the linear monomer forms of both lp17 and lp28-2 was observed by 72 h post-IPTG washout (Fig. 7). Moreover, as early as 24 h after washout, putative replication intermediates roughly double the size of the monomeric plasmids were observed. At early time points (24 and 48 h), only a single, more slowly migrating dimer-sized plasmid band was observed for each plasmid. These results are in agreement with current thoughts on replication of the linear Borrelia replicons (Fig. 1). At the later time points (72 and 96 h), additional bands were observed at higher molecular weights for lp17, while the bands disappeared altogether for lp28-2. The additional higher bands likely represent more complex unresolved forms of the linear plasmids, perhaps the result of branched structures resulting from further partial replication and/or stalled replication forks. The disappearance of the hybridization signal likely results from the heterogeneity of the larger complex products and possibly from decreased efficiency of transfer to the membrane of high-molecular-weight DNA.
At 72 h, when no linear monomer forms of free lp17 or lp28-2 could be observed (Fig. 7), the expected replicated telomeres that could be resolved by ResT in vitro were observed (Fig. 8B and C, lanes 5 and 6), providing the first in vivo observation of these replication intermediates, which are ordinarily too short-lived to be detected. In addition, free telomere ends were liberated from lp17 and lp28-2 after digestion with restriction endonucleases (Fig. 8). Some high-molecular-weight plasmid forms, therefore, appear to contain unreplicated telomeres, indicating either that DNA replication in these molecules did not progress to completion or that these telomeres had been resolved with the residual ResT remaining in the cell. Telomere resolution can occur independently at the two ends of replicating N15 prophage DNA (52), and such a scenario could exist in B. burgdorferi to generate higher-molecular-weight replicative intermediates with only one resolved telomere. Nonetheless, the situation in B. burgdorferi appears to be more complex, with a population of partially replicated molecules accumulating.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kerri Kobryn for providing purified His-tagged ResT protein used for antibody production and in vitro experiments and for helpful discussions and comments on the manuscript. We also thank John Blevins for providing the inducible expression plasmid pJSB104 and Genevieve Chaconas for technical support.
This work was supported by grant MOP 53086 from the Canadian Institutes of Health Research (http://www.cihr-irsc.gc.ca/e/193.html). G.C. holds a Canada Research Chair in the Molecular Biology of Lyme Borreliosis (http://www.chairs-chaires.gc.ca/home-accueil-eng.aspx) and a Scientist Award from Alberta Innovates Health Solutions (http://www.ahfmr.ab.ca/).
Footnotes
Published ahead of print 18 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01435-13.
REFERENCES
- 1.Steere AC, Coburn J, Glickstein L. 2004. The emergence of Lyme disease. J. Clin. Invest. 113:1093–1101. 10.1172/JCI200421681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanek G, Wormser GP, Gray J, Strle F. 2012. Lyme borreliosis. Lancet 379:461–473. 10.1016/S0140-6736(11)60103-7 [DOI] [PubMed] [Google Scholar]
- 3.Radolf JD, Caimano MJ, Stevenson B, Hu LT. 2012. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat. Rev. 10:87–99. 10.1038/nrmicro2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samuels DS, Radolf JD. 2010. Borrelia: molecular biology, host interaction and pathogenesis. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 5.Dennis DT, Hayes EB. 2002. Epidemiology of Lyme borreliosis, p 251–280 In Gray JS, Kahl O, Lane RS, Stanek G. (ed), Lyme borreliosis: biology, epidemiology and control. CAB International, Wallingford, United Kingdom [Google Scholar]
- 6.Korenberg EI, Gorelova NB, Kovalevskii YV. 2002. Ecology of Borrelia burgdorferi sensu lato in Russia, p 175–200 In Gray JS, Kahl O, Lane RS, Stanek G. (ed), Lyme borreliosis: biology, epidemiology and control. CAB International, Wallingford, United Kingdom [Google Scholar]
- 7.Miyamoto K, Masuzawa T. 2002. Ecology of Borrelia burgdorferi sensu lato in Japan and East Asia, p 201–222 In Gray JS, Kahl O, Lane RS, Stanek G. (ed), Lyme borreliosis: biology, epidemiology and control. CAB International, Wallingford, United Kingdom [Google Scholar]
- 8.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586. 10.1038/37551 [DOI] [PubMed] [Google Scholar]
- 9.Casjens S, Palmer N, van Vugt R, Huang WH, Stevenson B, Rosa P, Lathigra R, Sutton G, Peterson J, Dodson RJ, Haft D, Hickey E, Gwinn M, White O, Fraser CM. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490–516. 10.1046/j.1365-2958.2000.01698.x [DOI] [PubMed] [Google Scholar]
- 10.Barbour AG, Garon CF. 1987. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science 237:409–411. 10.1126/science.3603026 [DOI] [PubMed] [Google Scholar]
- 11.Hinnebusch J, Barbour AG. 1991. Linear plasmids of Borrelia burgdorferi have a telomeric structure and sequence similar to those of a eukaryotic virus. J. Bacteriol. 173:7233–7239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tourand Y, Deneke J, Moriarty TJ, Chaconas G. 2009. Characterization and in vitro reaction properties of 19 unique hairpin telomeres from the linear plasmids of the Lyme disease spirochete. J. Biol. Chem. 284:7264–7272. 10.1074/jbc.M808918200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picardeau M, Lobry JR, Hinnebusch BJ. 1999. Physical mapping of an origin of bidirectional replication at the centre of the Borrelia burgdorferi linear chromosome. Mol. Microbiol. 32:437–445. 10.1046/j.1365-2958.1999.01368.x [DOI] [PubMed] [Google Scholar]
- 14.Picardeau M, Lobry JR, Hinnebusch BJ. 2000. Analyzing DNA strand compositional asymmetry to identify candidate replication origins of Borrelia burgdorferi linear and circular plasmids. Genome Res. 10:1594–1604. 10.1101/gr.124000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beaurepaire C, Chaconas G. 2005. Mapping of essential replication functions of the linear plasmid lp17 of B. burgdorferi by targeted deletion walking. Mol. Microbiol. 57:132–142. 10.1111/j.1365-2958.2005.04688.x [DOI] [PubMed] [Google Scholar]
- 16.Chaconas G, Stewart PE, Tilly K, Bono JL, Rosa P. 2001. Telomere resolution in the Lyme disease spirochete. EMBO J. 20:3229–3237. 10.1093/emboj/20.12.3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaconas G. 2005. Hairpin telomeres and genome plasticity in Borrelia: all mixed up in the end. Mol. Microbiol. 58:625–635. 10.1111/j.1365-2958.2005.04872.x [DOI] [PubMed] [Google Scholar]
- 18.Chaconas G, Kobryn K. 2010. Structure, function, and evolution of linear replicons in Borrelia. Annu. Rev. Microbiol. 64:185–202. 10.1146/annurev.micro.112408.134037 [DOI] [PubMed] [Google Scholar]
- 19.Kobryn K. 2007. The linear hairpin replicons of Borrelia burgdorferi, p 117–140 In Meinhardt F, Klassen R. (ed), Microbial linear plasmids. Springer, Berlin, Germany [Google Scholar]
- 20.Kobryn K, Chaconas G. 2002. ResT, a telomere resolvase encoded by the Lyme disease spirochete. Mol. Cell 9:195–201. 10.1016/S1097-2765(01)00433-6 [DOI] [PubMed] [Google Scholar]
- 21.Bankhead T, Chaconas G. 2004. Mixing active site components: a recipe for the unique enzymatic activity of a telomere resolvase. Proc. Natl. Acad. Sci. U. S. A. 101:13768–13773. 10.1073/pnas.0405762101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bankhead T, Kobryn K, Chaconas G. 2006. Unexpected twist: harnessing the energy in positive supercoils to control telomere resolution. Mol. Microbiol. 62:895–905. 10.1111/j.1365-2958.2006.05423.x [DOI] [PubMed] [Google Scholar]
- 23.Deneke J, Burgin AB, Wilson SL, Chaconas G. 2004. Catalytic residues of the telomere resolvase ResT: a pattern similar to, but distinct from tyrosine recombinases and type IB topoisomerases. J. Biol. Chem. 279:53699–53706. 10.1074/jbc.M409001200 [DOI] [PubMed] [Google Scholar]
- 24.Kobryn K, Chaconas G. 2005. Fusion of hairpin telomeres by the B. burgdorferi telomere resolvase ResT: implications for shaping a genome in flux. Mol. Cell 17:783–791. 10.1016/j.molcel.2005.02.025 [DOI] [PubMed] [Google Scholar]
- 25.Kobryn K, Burgin AB, Chaconas G. 2005. Uncoupling the chemical steps of telomere resolution by ResT. J. Biol. Chem. 280:26788–26795. 10.1074/jbc.M504530200 [DOI] [PubMed] [Google Scholar]
- 26.Kobryn K, Briffotaux J, Karpov V. 2009. Holliday junction formation by the Borrelia burgdorferi telomere resolvase, ResT: implications for the origin of genome linearity. Mol. Microbiol. 71:1117–1130. 10.1111/j.1365-2958.2008.06584.x [DOI] [PubMed] [Google Scholar]
- 27.Briffotaux J, Kobryn K. 2010. Preventing broken borrelia telomeres: ResT couples dual hairpin telomere formation to product release. J. Biol. Chem. 285:41010–41018. 10.1074/jbc.M110.150060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tourand Y, Kobryn K, Chaconas G. 2003. Sequence-specific recognition but position-dependent cleavage of two distinct telomeres by the Borrelia burgdorferi telomere resolvase, ResT. Mol. Microbiol. 48:901–911. 10.1046/j.1365-2958.2003.03485.x [DOI] [PubMed] [Google Scholar]
- 29.Tourand Y, Lee L, Chaconas G. 2007. Telomere resolution by Borrelia burgdorferi ResT through the collaborative efforts of tethered DNA binding domains. Mol. Microbiol. 64:580–590. 10.1111/j.1365-2958.2007.05691.x [DOI] [PubMed] [Google Scholar]
- 30.Tourand Y, Bankhead T, Wilson SL, Putteet-Driver AD, Barbour AG, Byram R, Rosa PA, Chaconas G. 2006. Differential telomere processing by Borrelia telomere resolvases in vitro but not in vivo. J. Bacteriol. 188:7378–7386. 10.1128/JB.00760-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lefas G, Chaconas G. 2009. High-throughput screening identifies three inhibitor classes of the telomere resolvase from the Lyme disease spirochete. Antimicrob. Agents Chemother. 53:4441–4449. 10.1128/AAC.00529-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriarty TJ, Chaconas G. 2009. Identification of the determinant conferring permissive substrate usage in the telomere resolvase, ResT. J. Biol. Chem. 284:23293–23301. 10.1074/jbc.M109.023549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaconas G. 2010. Replication of the B. burgdorferi genome and scrambling of the linear replicons through reverse telomere resolution, p 49–60 In Samuels DS, Radolf JD. (ed), Borrelia: molecular biology, host interaction and pathogenesis. Horizon Scientific Press, Norwich, United Kingdom [Google Scholar]
- 34.Byram R, Stewart PE, Rosa P. 2004. The essential nature of the ubiquitous 26-kilobase circular replicon of Borrelia burgdorferi. J. Bacteriol. 186:3561–3569. 10.1128/JB.186.11.3561-3569.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jewett MW, Byram R, Bestor A, Tilly K, Lawrence K, Burtnick MN, Gherardini F, Rosa PA. 2007. Genetic basis for retention of a critical virulence plasmid of Borrelia burgdorferi. Mol. Microbiol. 66:975–990. 10.1111/j.1365-2958.2007.05969.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brisson D, Drecktrah D, Eggers CH, Samuels DS. 2012. Genetics of Borrelia burgdorferi. Annu. Rev. Genet. 46:515–536. 10.1146/annurev-genet-011112-112140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosa PA, Cabello F, Samuels DS. 2010. Genetic manipulation of B. burgdorferi, p 189–219 In Samuels DS, Radolf JD. (ed), Borrelia: molecular biology, host interaction and pathogenesis. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 38.Gilbert MA, Morton EA, Bundle SF, Samuels DS. 2007. Artificial regulation of ospC expression in Borrelia burgdorferi. Mol. Microbiol. 63:1259–1273. 10.1111/j.1365-2958.2007.05593.x [DOI] [PubMed] [Google Scholar]
- 39.Blevins JS, Revel AT, Smith AH, Bachlani GN, Norgard MV. 2007. Adaptation of a luciferase gene reporter and lac expression system to Borrelia burgdorferi. Appl. Environ. Microbiol. 73:1501–1513. 10.1128/AEM.02454-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whetstine CR, Slusser JG, Zuckert WR. 2009. Development of a single-plasmid-based regulatable gene expression system for Borrelia burgdorferi. Appl. Environ. Microbiol. 75:6553–6558. 10.1128/AEM.02825-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang FT, Xu Q, Sikdar R, Xiao Y, Cox JS, Doerrler WT. 2010. BB0250 of Borrelia burgdorferi is a conserved and essential inner membrane protein required for cell division. J. Bacteriol. 192:6105–6115. 10.1128/JB.00571-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Groshong AM, Gibbons NE, Yang XF, Blevins JS. 2012. Rrp2, a prokaryotic enhancer-like binding protein, is essential for viability of Borrelia burgdorferi. J. Bacteriol. 194:3336–3342. 10.1128/JB.00253-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bono JL, Elias AF, Kupko JJ, III, Stevenson B, Tilly K, Rosa P. 2000. Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 182:2445–2452. 10.1128/JB.182.9.2445-2452.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barbour AG. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521–525 [PMC free article] [PubMed] [Google Scholar]
- 45.Dresser AR, Hardy P-O, Chaconas G. 2009. Investigation of the role of DNA replication, recombination and repair genes in antigenic switching at the vlsE locus in Borrelia burgdorferi: an essential role for the RuvAB branch migrase. PLoS Pathog. 5:e1000680. 10.1371/journal.ppat.1000680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hardy P-O, Chaconas G. 2013. The nucleotide excision repair system of Borrelia burgdorferi is the sole pathway involved in repair of DNA damage by UV light. J. Bacteriol. 195:2220–2231. 10.1128/JB.00043-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bunikis I, Kutschan-Bunikis S, Bonde M, Bergström S. 2011. Multiplex PCR as a tool for validating plasmid content of Borrelia burgdorferi. J. Microbiol. Methods 86:243–247. 10.1016/j.mimet.2011.05.004 [DOI] [PubMed] [Google Scholar]
- 48.Elias AF, Bono JL, Carroll JA, Stewart P, Tilly K, Rosa P. 2000. Altered stationary-phase response in a Borrelia burgdorferi rpoS mutant. J. Bacteriol. 182:2909–2918. 10.1128/JB.182.10.2909-2918.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317–1319. 10.1126/science.7043737 [DOI] [PubMed] [Google Scholar]
- 50.Tobiason DM, Seifert HS. 2006. The obligate human pathogen, Neisseria gonorrhoeae, is polyploid. PLoS Biol. 4:e185. 10.1371/journal.pbio.0040185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reyes-Lamothe R, Nicolas E, Sherratt DJ. 2012. Chromosome replication and segregation in bacteria. Annu. Rev. Genet. 46:121–143. 10.1146/annurev-genet-110711-155421 [DOI] [PubMed] [Google Scholar]
- 52.Ravin NV. 2003. Mechanisms of replication and telomere resolution of the linear plasmid prophage N15. FEMS Microbiol. Lett. 221:1–6. 10.1016/S0378-1097(03)00125-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


