Abstract
In Salmonella, the rod substructure of the flagellum is a periplasmic driveshaft that couples the torque generated by the basal body motor to the extracellular hook and filament. The rod subunits self-assemble, spanning the periplasmic space and stopping at the outer membrane when a mature length of ∼22 nm is reached. Assembly of the extracellular hook and filament follow rod completion. Hook initiation requires that a pore forms in the outer membrane and that the rod-capping protein, FlgJ, dislodges from the tip of the distal rod and is replaced with the hook-capping protein, FlgD. Approximately 26 FlgH subunits form the L-ring around the distal rod that creates the pore through which the growing flagellum will elongate from the cell body. The function of the L-ring in the mature flagellum is also thought to act as a bushing for the rotating rod. Work presented here demonstrates that, in addition to outer membrane pore formation, L-ring formation catalyzes the removal of the FlgJ rod cap. Rod cap removal allows the hook cap to assemble at the rod tip and results in the transition from rod completion in the periplasm to extracellular hook polymerization. By coupling the rod-to-hook switch to outer membrane penetration, FlgH ensures that hook and filament polymerization is initiated at the appropriate spatial and temporal point in flagellar biosynthesis.
INTRODUCTION
The bacterial flagellum exemplifies nanoscale engineering that microbes have been perfecting over billions of years (1). This supramolecular structure is composed of approximately 25 distinct protein subunits and self-assembles to final lengths of up to 20 μm, several times the length of the cell itself (2). The flagellum enables a bacterium to colonize surfaces, search for food, and avoid noxious substances in its environment (3). To orchestrate the ordered assembly of a flagellum, flagellated species of bacteria have evolved numerous regulatory mechanisms. Flagellar biosynthesis is regulated at the genetic level by mechanisms that couple gene expression to assembly (4, 5) and at the biomechanical level with many flagellar proteins possessing inherent self-assembly characteristics (6–8).
Synthesis of the flagellum begins with the assembly of a basal body composed of several substructures (9). First to assemble is the MS-ring within the cytoplasmic membrane, followed by the cytoplasm-facing C-ring, which serves as both the rotor for the flagellar motor and as an affinity site for the secretion and assembly of other flagellar structures (10). The flagellum-specific type three secretion (T3S) system assembles in the inner membrane within the MS- and C-rings. The flagellar T3S system secretes flagellar subunits from the cytoplasm to be assembled at the tip of the growing flagellum (11, 12). The rod assembles through the periplasmic space to the outer membrane and transmits the torque generated by the basal body to the extracellular hook and filament (13).
The rod initially assembles into a proximal rod structure that lies between the MS-ring and the cell wall and is composed of FliE and approximately 6 subunits each of FlgB, FlgC, and FlgF (14). It is thought that FliE acts as an adaptor that joins the axial rod to the planar MS-ring, with FlgB, FlgC, and FlgF assembling atop one another until the cell wall is reached (15, 16). The distal rod structure is about the same length as the proximal rod (∼11 nm) and is composed of approximately 26 FlgG subunits (14).
In order for rod assembly to proceed to the outer membrane, a hole must be made in the cell wall, a role fulfilled by FlgJ. FlgJ is a dual-domain protein composed of an N-terminal scaffolding domain required for polymerization of the distal rod and a C-terminal acetylmuramidase domain required for forming a hole in the cell wall (1). Once FlgJ has made a hole in the cell wall, about 26 subunits of the distal rod protein, FlgG, are secreted and assemble in two stacks underneath FlgJ, thereby spanning the remaining distance between the cell wall and the outer membrane (16, 17). What is not known is whether FlgJ assembles prior to proximal rod assembly or distal rod assembly. If FlgJ assembles prior to proximal rod assembly, then FlgJ would serve as a scaffold for 4 different proteins (FlgB, FlgC, FlgF, and FlgG). If the FlgJ cap assembles after proximal rod completion, then, similar to FlgD and FliD, which are scaffolds for FlgE (hook) and FliC (filament), respectively, FlgJ would serve as a scaffold for a single protein, FlgG.
After the distal rod has polymerized, a hole forms in the outer membrane for construction of the extracellular hook and filament to proceed from the cell surface. For this to happen, FlgA, FlgH, and FlgI are secreted via the Sec type II secretion system into the periplasm. FlgI forms the P-ring, whose assembly is facilitated by FlgA, and FlgH forms the L-ring (18–20). FlgI polymerizes around the distal rod in close proximity to the cell wall and directs assembly of the L-ring (17). Assembly of the L-ring forms a pore in the outer membrane required for hook polymerization to commence. In addition to forming a hole in the outer membrane, it has been suggested that the P- and L-rings function as bushings for the rod (21).
PL-ring completion and outer membrane pore formation is followed by hook polymerization. This requires that the FlgD rod scaffold replace the FlgJ scaffold in order for the hook (FlgE) to polymerize from the rod tip at the cell surface. Hook polymerization terminates by the action of a molecular ruler, FliK (22). FliK is secreted during hook polymerization, and when the hook reaches a minimal length of about 40 nm, the C terminus of FliK interacts with the FlhB component of the flagellar T3S system, resulting in a conformational change in FlhB (23, 24). FlhB specifies which substrates are secreted through the flagellar T3S system. Prior to FliK interaction, FlhB allows secretion of the rod-hook class of protein subunits. After hook completion and interaction with FliK, FlhB becomes specific for late, or filament class, substrate secretion. The hook-associated proteins (HAPs), FlgK, FlgL, and FliD, and the FlgM regulatory protein are secreted next. The assembly of the HAPs dislodges the hook scaffold FlgD. The filament then polymerizes beneath the FliD filament cap to lengths up to about 15 μm.
The enteric flagellum is composed of 11 protofilaments (9). Subunits that make up the axial structures, the rod hook filament, are added 5.5 subunits per turn of the helix. As a result, it takes two helical additions to add a full layer of subunits to the structure. Thus, the estimated 26 subunits of FlgG would include more than 4 helical additions. Based on mutations in the flgG gene that allow continuous polymerization of FlgG subunits, it was proposed that FlgG has an intrinsic stacking mechanism where subunits polymerize on each other and stop, which would result in addition of only 22 FlgG subunits (17). The mutations in flgG that allow continuous polymerization of FlgG subunits, called flgG*, produce filamentous rod structures whose lengths are controlled by the FliK molecular ruler (25).
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Detailed information about bacterial strains and plasmids used in this study are listed in Table 1. Cells were cultured in lysogeny broth (LB). The following antibiotic supplements were added as needed: chloramphenicol (12.5 μg ml−1), kanamycin (50 μg ml−1), tetracycline (15 μg ml−1), and anhydrotetracycline (ATc; 1 μg ml−1). Gene expression from the arabinose promoter was induced by addition of 0.2% l-arabinose. The generalized transducing phage of Salmonella enterica serovar Typhimurium P22 HT105/1 int-201 was used in all transductional crosses (26).
TABLE 1.
List of strains used in this study
| Strain | Genotype |
|---|---|
| TH9614 | flgG5664(G53C) |
| TH10082 | flgG5677(G53R) |
| TH10354 | flgG6705(G65R) |
| TH12005 | flgG6755(S64P) |
| TH12334 | flgG7327(G53S) |
| TH20653 | flgG8192(G53D) |
| TH13929 | ΔflgG7661 |
| TH13636 | ΔaraBAD972::flgH+I+A+ |
| TH20800 | flgG5664(G53C) ΔaraBAD972::flgH+I+A+ |
| TH20801 | flgG5677(G53R) ΔaraBAD972::flgH+I+A+ |
| TH20802 | flgG6705(G65R) ΔaraBAD972::flgH+I+A+ |
| TH20803 | flgG6755(S64P) ΔaraBAD972::flgH+I+A+ |
| TH20807 | flgG7327(G53S) ΔaraBAD972::flgH+I+A+ |
| TH20809 | flgG8192(G53D) ΔaraBAD972::flgH+I+A+ |
| TH20838 | flgG5677(G53R) ΔaraBAD1008::flgE+ |
| TH20839 | flgG5677(G53R) ΔaraBAD925::tetRA |
| TH12850 | flgG6705(G65R) ΔaraBAD972::flgH+I+A+ ΔfliK6137::tetRA |
| TH20840 | ΔflgH7662 ΔflgM5794::FCF |
| TH20842 | ΔflgH7662 ΔflgM5794::FCF ΔaraBAD1008::flgE+ |
| TH20843 | ΔflgH7662 ΔaraBAD925::tetRA |
| TH20844 | ΔflgH7662 ΔaraBAD1008::flgE+ |
| TH17693 | flgJ8013::3×HA |
| TH17820 | flgJ8021::3×HA ΔflgHI958 ΔaraBAD941::flgH+I+ fljBenx vh2 |
| TH20753 | flgJ8021::3×HA ΔflgHI958 ΔaraBAD941::flgH+I+ PflhDC8089::tetR PtetA fljBenx vh2 |
| TH20757 | flgJ8021::3×HA ΔflgHI958 ΔflgD6543 ΔaraBAD941::flgH+I+ PflhDC8089::tetR PtetA fljBenx vh2 |
| TH20765 | flgJ8013::3×HA ΔflgH7662 ΔaraBAD1001::flgH+ PflhDC8089::tetR PtetA |
| TH20769 | flgJ8013::3×HA ΔflgH7662 ΔflgD6543 ΔaraBAD1001::flgH+ PflhDC8089::tetR PtetA |
Isolation and measurement of HBBs.
Hook basal body (HBB) isolation was carried out by the methods described previously (27), with minor modifications. Flagellar samples were not collected by CsCl gradient centrifugation but were pelleted at 60,000 × g for 1 h using a Beckman 50.2Ti rotor at 4°C. Hook lengths were measured using NIH ImageJ 1.42q software.
Electron microscopy.
Purified HBB samples were negatively stained with 2% uranyl acetate on copper-coated grids. Images were captured using a Hitachi H-7100 electron microscope at an acceleration voltage of 125 kV.
SDS-PAGE sample preparation and immunoblotting.
Overnight cultures were diluted 1:100 into defined media (E-salts plus 0.2% glucose) with LB added to a final concentration of 20%. LB was added in order to provide a small amount of carrier protein to aid in the precipitation of secreted proteins during sample preparation. Cultures were grown at 37°C with aeration. ATc, when used, was added to cultures to a final concentration of 0.7 μg ml−1. Cells were harvested via centrifugation at 3,550 × g for 10 min in an Eppendorf 5430 benchtop centrifuge. Supernatants were then filtered through 0.45-μm cellulose acetate filters to remove any cells remaining after centrifugation. Supernatant protein was precipitated by the addition of 100% (wt/vol) trichloroacetic acid (Sigma) to a final concentration of 20 to 25% and precipitated on ice overnight. Following overnight precipitation, samples were spun at 14,000 rpm for 30 min in a Sorvall RC5-B centrifuge followed by two washes with −20°C acetone. Precipitated protein was resuspended in 30 to 40 μl 2× SDS sample buffer with 5% β-mercaptoethanol (βME) (recipe per Bio-Rad). Cell pellets were simply boiled in 2× SDS sample buffer with 5% βME. For SDS-PAGE, 10-well, 1-mm, 11% acrylamide gels were used. For supernatant samples, ∼2,000 to 3,000 optical density (OD) equivalents were loaded in each lane of the gel. For cell pellets, ∼100 to 200 OD equivalents were loaded. Protein was transferred to a 0.2-μm nitrocellulose membrane (Optitran BA-S 83 reinforced NC; GE Healthcare) by semidry transfer (Trans-Blot SD semidry transfer cell; Bio-Rad). After blocking in Tris-buffered saline (TBS) with 5% nonfat powdered milk for 30 min, blots were incubated with anti-HA monoclonal antibody from mouse ascites fluid (Covance), anti-DnaK mouse monoclonal antibody (Abcam), and/or anti-FlgE/anti-FlgM rabbit-derived antibody in TBST (TBS with 0.02% Tween 20) with 5% nonfat milk at 4°C with gentle agitation overnight.
Blots were then washed 3× with TBST and exposed to goat anti-mouse and/or anti-rabbit 2° antibodies (Dylight 800 and Li-Cor IRDye 680RD, respectively) in TBST with 10% milk for 1 h with agitation. After being washed of 2° antibodies, blots were imaged using a LI-COR Biosciences Odyssey infrared imaging system.
RESULTS
PL-ring gene overexpression suppresses motility defect in G53 filamentous rod mutants of distal rod gene flgG.
The rod component of the HBB grows to 22 nm and stops (25). This places the rod tip perpendicular to the outer membrane. The P-ring polymerizes around the 2 helical stacks of FlgG distal rod subunits. This is followed by L-ring formation, outer membrane penetration, and hook polymerization from the cell surface (Fig. 1). Hook polymerization continues until the molecular tape measure, FliK, is secreted through a hook with a minimal length of 40 nm. FliK then catalyzes a conformational change in the FlhB component of the flagellar T3S system that results in the secretion specificity switch from rod-hook secretion substrates to late, or filament-type, secretion substrates. Filamentous rod mutants in FlgG (flgG*) do not stop polymerizing at the outer membrane but do stop polymerizing when FliK catalyzes the secretion specificity switch after the rod has reached 40 nm or longer.
FIG 1.
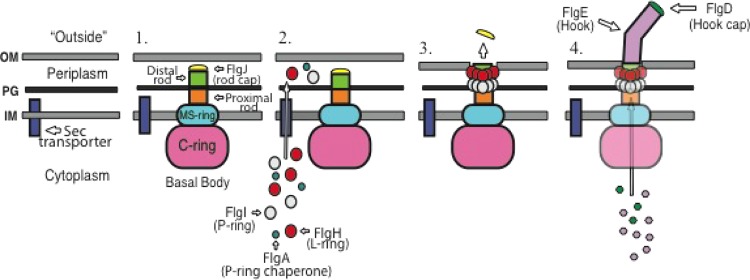
Model for the PL-ring-mediated switch from rod to hook polymerization during flagellar morphogenesis. The assembly of the flagellum is a hierarchical process. (Step 1) The first structures to assemble are the MS-ring in the inner membrane (IM) and the cytoplasmic C-ring. The inner membrane components of the flagellar T3S system are believed to assemble within the MS-ring, while the cytoplasmic components of the flagellar T3S system associate with the C-ring. The first secreted subunits to assemble are FliE, FlgB, FlgC, and FlgF, which form the proximal rod. FlgJ, the rod scaffold, associates with the rod at some point during this process in order to form a hole in the cell wall. FlgG polymerizes to form the distal rod on top of the proximal rod and underneath FlgJ. Distal rod assembly ceases once the rod has reached the outer membrane (OM), positioning the tip of the rod perpendicular to the outer membrane. PG, peptidoglycan. (Step 2) The subunits required for PL-ring formation, FlgA, FlgH, and FlgI, are secreted into the periplasm through the Sec secretion system. Once in the periplasm, FlgA assists FlgI in the assembly of the P-ring around the distal rod. FlgH forms the L-ring around the distal rod in close proximity to the P-ring. (Step 3) Formation of the L-ring around the distal rod by FlgH forms a pore in the outer membrane and also causes the rod scaffold, FlgJ, to dissociate from the tip of the distal rod into the extracellular environment. (Step 4) Once the rod cap has been dislodged from the rod, FlgD is able to form the hook scaffold and direct assembly of the hook by FlgE.
Rod completion and PL-ring formation are uncoupled processes. The proteins required for PL-ring formation, FlgA, FlgH, and FlgI, are secreted into the periplasm through the Sec type II secretion system. FlgA facilitates polymerization of the FlgI P-ring structural subunits around the distal rod. Formation of the P-ring is followed by assembly of the L-ring by FlgH directly adjacent to the P-ring.
We wondered if overexpression of the PL-ring components suppresses the flgG* motility defect. It was thought that by increasing the pool of PL-ring components available in the periplasm, the PL-ring might be able to form while the rod was still perpendicular to the outer membrane and allow the flagellar structure to penetrate the outer membrane and grow beyond the cell surface. All 25 flgG* mutants isolated to date were tested for the ability of PL-ring gene overexpression to suppress the flgG* motility defect. Only one allele, the G53R substitution, showed a high degree of suppression when the flgA, flgH, and flgI genes were overexpressed (Fig. 2). Three other single-amino-acid substitution mutants, G53D, G53S, and S64P, also showed suppression at a low but reproducible level (Fig. 2). We presumed that these FlgG alleles polymerized slowly enough to allow PL-rings to form at short rod lengths, thereby allowing motility.
FIG 2.
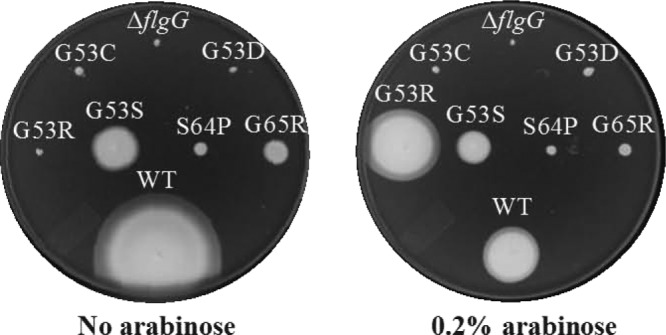
Overexpression of flgA, flgH, and flgI suppresses the motility defect of the flgG* G53R filamentous rod mutant. The genes required for PL-ring formation (flgA, flgH, and flgI) were cloned into the arabinose-inducible ΔaraBAD locus in every filamentous rod mutant isolated to date. The chromosomal copies of flgA, flgH, and flgI were left intact in the flg operon. The motility defect of the G53R mutant was suppressed to nearly WT levels when inoculated in soft agar with 0.2% arabinose added. WT, DaraBAD::flgAHI.
We then tested if PL-ring genes could form on an flgG* polyrod mutant if we did not demand suppression of the motility defect using an fliK null mutant background. In the absence of FliK, FlgG polymerization continues, resulting in polyrod structures that measured up to a micron in length (17). When the flgG* G65R mutant was grown under the PL-ring overexpression conditions, the strain remained defective in motility. The polyrod structures of the flgG* (G65R) fliK double mutant grown under PL-ring gene overexpression were isolated and examined by electron microscopy to see if PL-rings could form on polyrods (Fig. 3). In this background, the filamentous rod was accompanied by many P-rings along the length of the distal rod, and some single L-rings had assembled around the rod in very close proximity to the most distal P-ring. When this occurred, rod formation ceased and hook polymerization commenced at the PL-ring junction. This suggested that FlgI will form P-rings around FlgG indefinitely so long as there is a sufficient length of rod to form a P-ring around. However, formation of the L-ring only occurs at the tip of the rod in very close proximity to the P-ring. Therefore, when the rod of an flgG* mutant continues to polymerize after the addition of a P-ring, a gap between the P-ring and the rod tip is formed before FlgH can associate with both the rod tip and the P-ring, thereby preventing formation of the L-ring by FlgH.
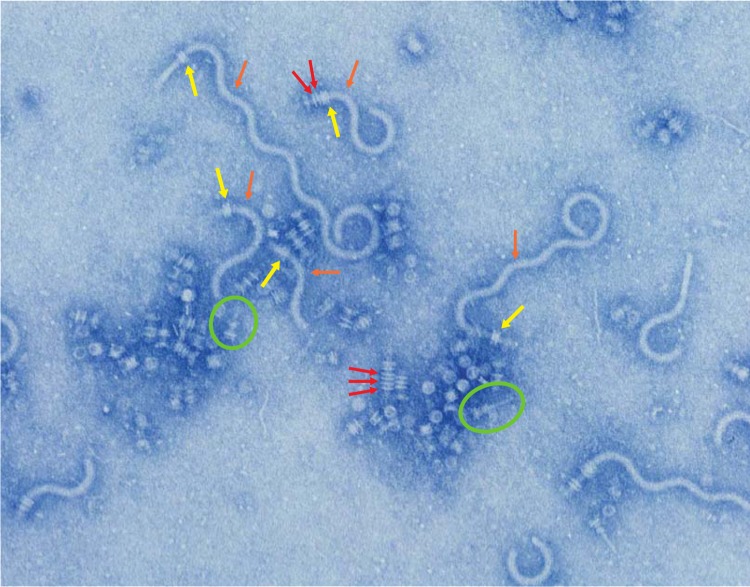
FIG 3.
PL-ring assembly triggers a switch from rod polymerization to hook polymerization in the flgG*(G65R) ΔfliK ΔaraBAD::flgH+I+A+ background. The flgG* G65R mutant produces distal rods that fail to stop polymerizing at the WT length of ∼12 nm, forming aberrantly long rod structures termed filamentous rods. When the intact genes required for PL-ring assembly (flgH+, flgI+, and flgA+) are overexpressed in this background, rods of a variety of lengths are produced. Many P-rings (red arrows) are able to form along the length of the distal rod in this background. When a PL-ring (yellow arrows) forms, the switch from rod to hook polymerization occurs. Deletion of the hook length gene fliK results in hooks (orange arrows) that are hundreds of nanometers long, known as polyhooks. Two injectisome structures are circled in green. The injectisome shares many common structural features with the flagellum and possesses its own T3S system needed for the construction of the injectisome as well as to translocate effector proteins into host cells in order to establish infection.
We tested the hypothesis that the abrupt switch from rod assembly to hook assembly in the G65R fliK null background upon formation of the PL-ring was a result of PL-ring-dependent removal of FlgJ. As discussed above, FlgJ performs two roles in the construction of the flagellum: penetration of the cell wall and acting as a scaffold for FlgG (distal rod) subunit polymerization. The FlgD hook cap remains associated with a completed hook until it is dislodged by FlgK polymerization. In a similar manner, we wondered if FlgJ remains associated with the tip of the rod following rod completion, awaiting the addition of L-ring subunits to dislodge it. In doing so, FlgJ could act as a cap to prevent assembly of the hook scaffold, FlgD, prior to outer membrane penetration.
Loss of FlgM or FlgE overexpression partially restores the motility defect of an flgH deletion allele.
In the original characterization of flgG* alleles, about 25% of the polyrod structures that form in the absence of both FliK and the PL-rings had attached polyhooks (17). This suggested that PL-rings were not absolutely required for the removal of FlgJ and the transition from rod to hook polymerization. This is consistent with an earlier study showing that overexpression of FlgE (hook) from a high-copy-number plasmid vector partially suppresses the nonmotile phenotype of a mutant lacking L-rings (28). This suggests that the interaction of FlgJ at the rod tip is weak enough that continuous passage of FlgE subunits eventually dislodges FlgJ, but L-ring formation is required for efficient removal of FlgJ. In the case of an L-ring-defective strain (ΔflgH), we expect there will be 6 to 8 basal body structures per cell. If flgE overexpression dislodged FlgJ from only a single structure, the suppression of the motility might be hindered by the accumulation of FlgM, since it would be secreted by a single basal structure and may still accumulate to an extent that σ28-dependent transcription of late flagellar genes, including the flagellin gene fliC (or fljB) and chemosensory genes, is limited by excess cellular FlgM. We tested whether loss of the rod cap in the absence of the L-ring occurred at a high enough frequency with normal rod structures (flgG+) such that removal of FlgM alone could suppress the motility defect in a ΔflgH strain.
When a ΔflgH mutant is inoculated in soft agar and allowed to incubate for a day or two, some motility is observed for a fraction of the cells, giving the colony a speckled phenotype with small satellite colonies forming around the original, nonmotile cells (Fig. 4A). This indicates that in the absence of L-rings, some cells occasionally are able to form at least one functional flagellum. When this same protocol was followed for a strain deleted for both flgH and flgM, the colony still has a speckled phenotype, but the motility diameter of the colony was about twice that of the ΔflgH mutant (Fig. 4A). This indicated that the full σ28-dependent transcription in a ΔflgH background increased the likelihood of L-ring-independent flagellar formation. When flgE was overexpressed from the arabinose locus in the ΔflgH ΔflgM double mutant, the diameter of the colony on soft agar was slightly larger still (Fig. 4A). This demonstrates that some transition from rod to hook polymerization and outer membrane penetration by the flagellar structure will occur in the absence of the L-ring; however, the L-ring is required for this process to occur with close to 100% efficiency.
FIG 4.
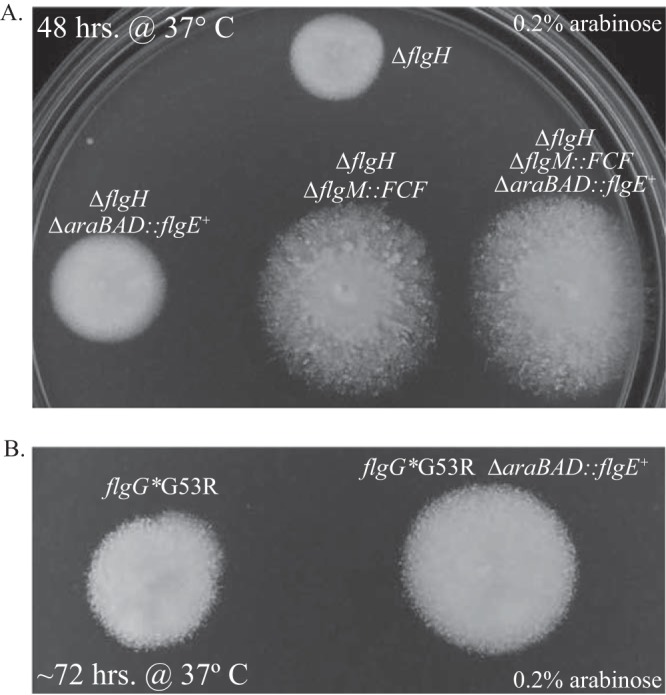
Nonmotile phenotype of mutants defective in PL-ring assembly can be partially suppressed by knocking out the anti-sigma factor flgM and/or overexpressing the hook protein, FlgE. Mutants that are unable to form PL-rings are largely nonmotile. The speckled phenotype of these mutants on soft agar indicates that the switch from rod to hook polymerization can occur without the PL-ring but at only a fraction of WT levels. (A) The motility defect of a mutant lacking L-rings (ΔflgH) is partially suppressed by overexpressing FlgE and/or knocking out flgM. (B) The phenotype of the flgG* G53R mutant in soft agar is similar to that of the ΔflgH mutant. By overexpressing the subunits required for PL-ring assembly, motility was restored to a large degree (Fig. 3), suggesting that this mutant is defective in PL-ring assembly. Similar to a ΔflgH mutant, the motility defect of the G53R mutant can be partially suppressed by overexpressing the hook protein, FlgE.
As discussed in the previous section, we managed to isolate a single, nonmotile flgG* mutant (G53R) whose motility defect was suppressed by overexpression of PL-ring genes. We reasoned that the nonmotile phenotype of this mutant was due to a poor affinity of FlgH or FlgI for FlgGG53R that resulted in a ring-less rod. If this were the case, it was expected that the motility defect would be partially suppressed by overexpressing flgE, as is the case in a ΔflgH background. When flgE was overexpressed in the flgGG53R background, the diameter of the colonies inoculated in motility plates was about twice that observed for the flgGG53R strain with normal flgE expression (Fig. 4B).
FlgJ accumulates in the culture supernatant.
The results described above demonstrate that PL-ring formation on the tip of the distal rod results in a transition from rod to hook polymerization. One mechanism that would account for this observation is that L-ring formation dislodges the rod scaffold, which is made up of FlgJ protein subunits. Removal of the FlgJ rod scaffold might be required to allow the hook scaffold, which is composed of FlgD subunits, to form on the rod tip and the transition to hook polymerization. We tested the hypothesis that PL-ring formation was coupled to release of FlgJ from the rod. Previous work suggested that FlgJ dissociates or dislodges from the rod and, under overexpression conditions, is accumulated in the periplasmic space (29). The filament cap protein, FliD, forms a pentamer that associates with the filament tip and acts as a scaffold for filament polymerization (7). Wild-type (WT) cells possess about six flagella; therefore, they only require that ∼30 FliD subunits be translocated from the cytoplasm to the filament tips. FlgJ has also been postulated to form a cap composed of only a few subunits (30). If this were the case, one would expect that the amount of FlgJ dislodged into the supernatant would be small. As such, a sensitive method to assay for the presence of extracellular FlgJ was needed. To that end, the influenza virus-derived hemagglutinin (HA) tag was inserted into the chromosomal flgJ gene in the linker region between the N- and C-terminal domains believed to be unstructured (31). The resulting FlgJ-3×HA chimera (TH17693) exhibited wild-type motility and allowed for sensitive and unambiguous identification of FlgJ, expressed from its native, chromosomal locus, by standard Western blot analysis.
An overnight culture of TH17693 was diluted 1:100 into 100 ml of liquid media and grown to an OD at 600 nm (OD600) of 0.1, at which point 15 ml of culture was placed on ice. This was repeated at each 0.1 OD600 unit up to an OD600 of 1.1. Cells were removed by centrifugation and filtration, and the secreted protein was concentrated and subjected to Western blot analysis probing for HA. Contrary to previous findings where FlgJ produced by a plasmid overexpression system accumulated in the periplasm, we observed that chromosomally expressed FlgJ was present in the secreted extracellular fraction. The presence of extracellular FlgJ was detected at an OD of ∼0.6 (Fig. 5). Previous investigations may have failed to detect FlgJ in the supernatant due to insufficient culture volumes. Even with ≥15-ml cultures, detection of FlgJ-3×HA was difficult.
FIG 5.

FlgJ is secreted extracellularly. Fifteen ml of culture from TH17693 (flgJ8013::HA) was collected at each OD600 unit from 0.4 to 1.1. Cells were removed, and the supernatant protein was concentrated and analyzed by SDS-PAGE using anti-HA antibody. Anti-DnaK antibody was added as a cell lysis control.
FlgH is responsible for the accumulation of FlgJ in the supernatant.
FlgH forms a pore in the outer membrane; therefore, it was assumed to be required for FlgJ (or any other secreted flagellar protein) to accumulate in the supernatant. We hypothesized that FlgH was responsible for the accumulation of extracellular FlgJ by destabilizing or dislodging FlgJ from the rod. To test this model, a series of strains was constructed with either the L-ring structural gene flgH or both flgH and the P-ring structural gene flgI deleted from their chromosomal location in the flg operon and placed under the control of the arabinose-inducible araBAD promoter (ParaBAD). The flagellar master regulon, flhDC, was also placed under the control of the tetA promoter (PtetA), which is induced by addition of either tetracycline (Tc) or its nonantibiotic analog, anhydrotetracycline (ATc), to the growth medium. This inducible promoter allows for the controlled time of induction of the flagellar regulon and subsequent flagellum assembly. This was accomplished by replacing the flhDC promoter region with PtetA and an adjacent tetR repressor gene, resulting in the ΔPflhDC8089::tetR PtetA allele. Thus, flagellar gene expression and flagellum assembly could be synchronized with the addition of ATc.
Overnight cultures were diluted into liquid media and grown to early- to mid-log phase (OD600 of ∼0.3). At this point, ATc was added to induce flhDC expression and subsequent flagellar basal body-rod construction. At 10 min post-flhDC induction, either arabinose or saline was added to the cultures. Ten-ml aliquots of each culture were taken every 10 min for 1 h following arabinose induction. mRNA translation was arrested with the addition of spectinomycin, and the cells were placed on ice. As before, cells were removed by centrifugation followed by filtration, and the supernatant samples were prepared for Western blot analysis. It was found that FlgJ-HA was detected in the supernatant ∼30 to 40 min after flhDC induction with the subsequent addition of arabinose and continued to accumulate throughout the remainder of the experiment (Fig. 6B). Cultures that received ATc without the subsequent addition of arabinose expressed FlgJ, but this was only found in the cellular fraction. Without arabinose induction, we were unable to detect any FlgJ in the supernatant, even at 70 min post-ATc induction. The intensity of the FlgJ-HA band in the cellular fraction remained stable over the course of the experiment, in contrast to the supernatant bands, which increase from one time point to the next. Furthermore, the HA signal detected in the cellular fractions appeared not as a single band, as in the supernatant samples, but as two bands ∼5 kDa apart and of about the same intensity (Fig. 6A). This indicates that either cytoplasmic or periplasmic FlgJ was subject to degradation.
FIG 6.
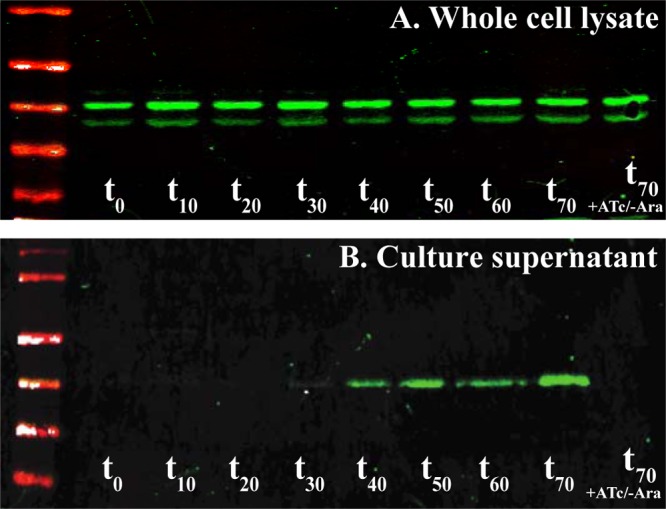
Secretion of FlgJ into the supernatant is dependent on PL-ring formation. Liquid culture of TH20753 (flgJ8021::3×HA ΔflgHI958 ΔaraBAD941::flgH+I+ PflhDC8089::tetR PtetA fljBenx vh2) was grown to an OD600 of ∼0.3, at which point anhydrotetracycline (ATc) was added to induce flhDC expression and basal body assembly. After 10 min of incubation with ATc, arabinose (Ara) was added to the culture to induce expression of flgH+. Samples were collected at 10-min intervals. Translation was arrested by the addition of spectinomycin, and the cells were placed on ice. Pellet and supernatant samples from each time point were subjected to Western blot analysis with probing for FlgJ-HA with anti-HA antibody. Time point zero (t0) represents samples taken after 10 min of incubation with ATc immediately prior to addition of Ara to the culture. (A) Levels of FlgJ-HA in the whole-cell lysates remained constant for the duration of the experiment. The upper band is full-length FlgJ-HA, and the lower band is presumed to be partially degraded FlgJ-HA. (B) In the supernatant samples, FlgJ-3×HA began to accumulate at ∼30 min after the addition of arabinose and continued to accumulate throughout the remainder of the experiment. In the supernatant, FlgJ-HA is present in the full-length form only. The supernatant of the sample that received ATc and saline (t70 + ATc/−Ara) had no detectable FlgJ-HA.
If FlgH pore formation dislodged the rod cap, FlgJ should appear in the supernatant before or at the same time as other secreted flagellar proteins. To determine if this was the case, the same protocol as that used before was followed but with assaying for FlgE (hook) and FlgM in addition to FlgJ-HA by Western blotting. FlgE polymerizes on top of FlgG and is the first extracellular component of flagellum assembly. FlgM is an anti-sigma factor that binds σ28 in the cytoplasm, thereby preventing expression of flagellum class 3 promoters of genes required after HBB completion, such as filament and chemosensory-associated genes. Once hook synthesis has completed, the export apparatus of the basal body undergoes a secretion specificity switch from rod-hook substrate specificity to late-type substrate specificity including FlgM. If it is true that FlgH displaces FlgJ during L-ring formation and outer membrane penetration, FlgJ, FlgE, and FlgM would also be expected to accumulate in the supernatant around the same time following induction with ATc and arabinose. Moreover, neither FlgE nor FlgM should be found in the secreted fraction prior to FlgJ. After performing another time course experiment assaying for all three proteins, it was found that they are all detected in the supernatant at the same point in time, about 30 to 40 min after ATc and arabinose induction. In the absence of arabinose in the culture, no amount of FlgJ-HA, FlgE, or FlgM was detected (Fig. 7A).
FIG 7.
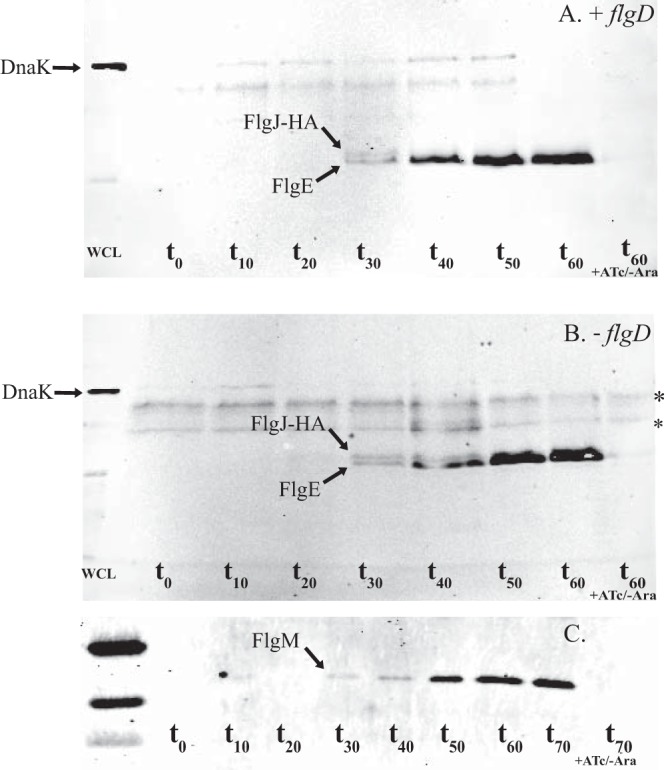
Outer membrane penetration and the secretion of flagellar subunits is dependent on FlgH and independent of the hook-capping protein, FlgD. The protocol described in the legend to Fig. 6 was followed, this time probing for FlgJ-HA, FlgE, and DnaK (as a cell lysis control). (A) FlgE and FlgJ-HA appear in the supernatant at the same time point (∼30 min after addition of arabinose). (B) Western blot of TH20757 (flgJ8021::3×HA ΔflgHI958 ΔflgD6543 ΔaraBAD941::flgH+I+ PflhDC8089::tetR PtetA fljBenx vh2) probing for FlgE, FlgJ-HA, and DnaK. Knocking out flgD does not affect the secretion of FlgJ-HA or FlgE into the culture supernatant. (C) FlgM appeared in the supernatant at the same time as FlgE and FlgJ-HA following addition of arabinose to TH20753 (flgJ8021::3×HA ΔflgHI958 ΔaraBAD941::flgH+I+ PflhDC8089::tetR PtetA fljBenx vh2). WCL, whole-cell lysate. t0, sample taken ∼10 min post-ATc induction, immediately before addition of arabinose. t60 + Atc/−Ara, control samples induced with anhydrotetracycline but not arabinose. *, nonspecific band.
The preceding lines of evidence demonstrate that FlgJ accumulates in the supernatant, and that the presence of FlgJ in the spent growth medium is dependent on L-ring formation. However, the possibility that FlgH forms a pore in the outer membrane but that FlgJ is dislodged from the rod by a different mechanism could not be ruled out (i.e., pore formation and the switch from rod to hook are simultaneous but mechanically independent events). If this were the case, the likely agent responsible for dislodging FlgJ was expected to be FlgD, the hook-capping protein, which assembles immediately after L-ring formation. FlgD forms a cap on top of the distal rod and acts as a scaffold for hook polymerization by FlgE. In the absence of FlgD, FlgE is secreted but unable to polymerize. Therefore, FlgD must be the subunit that polymerizes once FlgJ is dislodged in order for construction of the hook and filament to proceed. To determine if FlgD, and not FlgH, was responsible for dislodging FlgJ, the strain used in the preceding experiments (TH20753) was deleted for flgD. If FlgD was responsible for dislodging FlgJ, one would expect that FlgJ (and FlgE and FlgM) would be absent from culture supernatant after performing the protocol described above. When this experiment was performed, the resulting Western blot was almost indistinguishable from the previous blots. The absence of FlgD did not affect detection of FlgJ and FlgE in the supernatant, and the absence of FlgD did not affect the point in time following ATc and arabinose induction at which FlgJ and FlgE were detected (Fig. 7B).
FlgJ remains stably associated with the rod in the absence of FlgH.
It was still possible that the FlgJ detected in culture supernatants in the preceding experiments was just cytoplasmic FlgJ secreted into the supernatant following outer membrane penetration by FlgH. In other words, the results did not prove that FlgJ forms a structure that caps the distal rod and awaits displacement by the L-ring. The possibility that the distal rod had no cap and premature FlgD/FlgE assembly was prevented via a different mechanism seemed implausible, but it was a possibility that could be ruled out.
To determine if FlgJ formed a stably associated rod cap, basal bodies before and after formation of the L-ring were purified. Two 500-ml cultures of strain TH20769 (ΔflgH ΔaraBAD::flgHWT PflhDC8089::tetR PtetA flgJ-HA ΔflgD) were grown to an OD600 of ∼0.5 and induced with ATc. After ∼10 min, one culture was induced with arabinose and the other with saline. The cultures were incubated an additional 25 min, arrested with spectinomycin, and allowed to incubate for five more minutes before being placed on ice. Cells from both cultures were pelleted and gently lysed. Basal bodies from both cultures were concentrated via ultracentrifugation, denatured with SDS sample loading buffer, and analyzed by Western blotting.
Samples induced with only ATc (+flhDC) had a prominent FlgJ band, whereas those induced with both ATc and arabinose (+flhDC, +flgH) had only a very faint FlgJ band (Fig. 8). The presence of FlgJ following the lengthy basal body purification indicates that in the absence of the L-ring, FlgJ forms a stable rod-associated structure.
FIG 8.
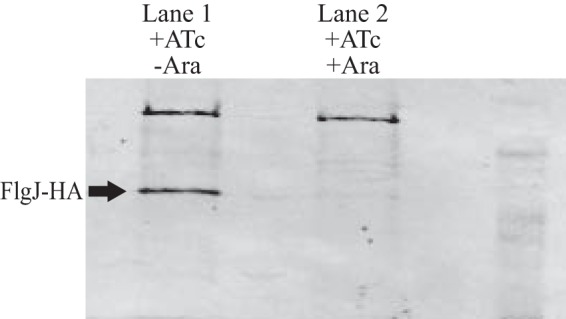
Rod scaffold protein FlgJ remains stably associated with the basal body until it is dislodged by FlgH. Two 500-ml aliquots of TH20769 (flgJ8013::3×HA ΔflgH7662 ΔflgD6543 ΔaraBAD1001::flgH+ PflhDC8089::tetR PtetA) were grown to mid-log phase and induced to construct basal bodies by adding anhydrotetracycline (ATc). After 10 min, one culture received arabinose (Ara) to induce expression of the L-ring protein, FlgH. The other culture received saline. Following an additional 25 min of incubation, mRNA translation was blocked in both cultures by the addition of spectinomycin. Basal bodies from both cultures were purified and subjected to SDS-PAGE. Western blot analysis probing for FlgJ-HA revealed that without L-ring formation, FlgJ-HA remained associated with the basal body (lane 1). The culture, which was induced to express FlgH, had only a trace amount of FlgJ-HA associated with purified basal bodies (lane 2).
DISCUSSION
The flagellar basal body of S. Typhimurium is composed of 22 different structural proteins followed by the hook, filament, and 3 hook-associated proteins (HAPs). The rod cap, hook cap, and hook length control protein FliK are transiently associated during HBB assembly. The periplasmic chaperone FlgA is required for assembly of the P-ring, and secretion chaperones FlgN (for FlgK and FlgL), FliT (for FliD), and FliS (for FliC/FljB) are required for efficient secretion of late secretion substrates. The FliK, FlgM, and σ28 proteins are required for the transition to the assembly of HAPs and filament and expression of chemosensory genes. Finally, motor force generators MotA and MotB are required as stators to drive flagellum rotation using the energy of the proton motive force. Thus, 38 different proteins are known to be required for the assembly of a functional flagellum. A remarkable feature of this structure is the requirement that subunits self-assemble onto the growing structure (32).
The process of self-assembly requires that some proteins are transiently associated with the structure at specific stages of the assembly process. The first structural subunit that is transiently associated is the rod scaffold protein FlgJ. FlgJ is thought to assemble prior to proximal rod formation, because severely truncated flgJ mutants produced MS-rings without rods (10). After rod completion, the FlgJ cap on the distal rod must be removed before the hook scaffold, FlgD, can assemble to allow hook subunits to polymerize onto the completed distal rod. The rod grows to a length of ∼22 nm, which places the terminal end of the rod at the outer membrane. Mutants in the distal rod gene flgG, called flgG* alleles, were described that resulted in rod lengths of ∼50 nm. Filaments that grow on these extended rod structures grow in the periplasm rather than outside the cell (17). The PL-ring structure was required for the transition from rod assembly to filament assembly for the formation of periplasmic flagella, but the hook was not required (25). Deletion of the hook gene did not prevent the growth of periplasmic flagella in the flgG* mutant strains. This led to a model in which the wild-type distal rod subunit FlgG polymerizes on top of another assembled FlgG subunit only once (17). An interaction between stacked FlgG subunits prevents further polymerization of FlgG subunits and would terminate rod growth at ∼22 nm. The flgG* alleles are defective in this FlgG stacking interaction that prevents further FlgG polymerization. Termination of rod growth at ∼22 nm would place the rod tip at the outer membrane. The PL-ring assembles around the distal rod and forms a pore in the outer membrane so that polymerization of the hook is external from the cell surface.
This checkpoint in flagellum assembly that couples (i) rod growth termination with (ii) PL-ring outer membrane pore assembly and (iii) the initiation of hook assembly predicts that a component of this checkpoint is associated with removal of the FlgJ rod scaffold to be replaced by the FlgD hook scaffold. The N-terminal 152 amino acids of FlgJ are required for rod polymerization, while the remaining C-terminal 164 amino acids include the muramidase domain required for rod penetration of the cell wall (30). FlgJ also has a heptad repeat (HR) domain of hydrophobic residues near its C terminus. Deletion of the FlgJ HR domain produced basal structures with P-rings but lacking the L-ring. This suggested that FlgJ does not dissociate from completed rods, but its presence at the rod tip is required for L-ring formation. Furthermore, FlgD, which is present in the secreted fraction in wild-type strains, is absent from the secreted fraction for flgJ mutants defective in L-ring assembly. This further supports the model in which the L-ring forms a pore for flagellar growth outside the cell.
The two remaining mechanisms for FlgJ scaffold removal we considered were that L-ring assembly displaced FlgJ or that the FlgD hook scaffold could dislodge FlgJ and assemble in its place. Because FlgD is continuously secreted into the periplasm prior to rod and PL-ring completion, the most plausible mechanism was that L-ring formation resulted in FlgJ removal from the rod tip.
The flgG* mutant strains missing hook-length control protein FliK have a polyrod phenotype, where rod polymerization is uncontrolled and rods as long as 1 μm have been observed (17). When the PL-ring formation genes flgA, flgH, and flgI were overexpressed in an flgG* fliK mutant background, we observed that PL-rings formed on polyrod structures and that their formation resulted in the transition from polyrod to polyhook formation. This supported the hypothesis that L-ring formation was responsible for FlgJ removal from the rod tip. Thus, we set up experiments to explore the possibility of FlgJ removal by L-ring formation.
It was previously reported that FlgJ was not secreted from the cell and was found only in the periplasmic fraction (29). We were concerned, because this result was obtained in strains that highly overexpressed FlgJ from a plasmid expression system. We suspected that, like the filament cap FliD, there were only 5 subunits of FlgJ per basal body, making it difficult to detect such a small amount of protein that might be released into the spent growth medium upon L-ring formation. We constructed an HA-tagged version of the chromosomal flgJ gene, placing the HA between the N-terminal scaffold domain and C-terminal muramidase domain of FlgJ. The resulting FlgJ-HA construct had wild-type motility, indicating that FlgJ-HA could form a perfectly functional rod cap and the HA tag would facilitate detection using anti-HA antibodies. Using this chromosomal FlgJ-HA construct, we could detect FlgJ in the secreted cell fraction. This suggested that L-ring pore formation in the outer membrane released FlgJ into the culture supernatant and not into the periplasm. By placing the L-ring structural gene (flgH) expression under the control of an arabinose-inducible promoter, we further showed that extracellular FlgJ required flgH expression. Furthermore, the presence of hook (FlgE) and FlgM in the secreted fraction does not occur before the appearance of secreted FlgJ. Finally, FlgJ was stably associated with basal structures lacking the L-ring, further supporting a model in which L-ring formation is coupled to the removal of FlgJ from the rod tip.
ACKNOWLEDGMENTS
We thank Shin-Ichi Aizawa for performing the electron microscopy presented in this work.
This work was supported by grant GM056141 from the National Institutes of Health to K.T.H.
We have no conflicts of interest to declare.
Footnotes
Published ahead of print 18 April 2014
REFERENCES
- 1.Erhardt M, Namba K, Hughes KT. 2010. Bacterial nanomachines: the flagellum and type III injectisome. Cold Spring Harb. Perspect. Biol. 2:a000299. 10.1101/cshperspect.a000299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6:455–465. 10.1038/nrmicro1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 8:634–644. 10.1038/nrmicro2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aldridge P, Hughes KT. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160–165. 10.1016/S1369-5274(02)00302-8 [DOI] [PubMed] [Google Scholar]
- 5.Chilcott GS, Hughes KT. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694–708. 10.1128/MMBR.64.4.694-708.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minamino T, Macnab RM. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yonekura K, Maki S, Morgan DG, DeRosier DJ, Vonderviszt F, Imada K, Namba K. 2000. The bacterial flagellar cap as the rotary promoter of flagellin self-assembly. Science 290:2148–2152. 10.1126/science.290.5499.2148 [DOI] [PubMed] [Google Scholar]
- 8.Yonekura K, Maki-Yonekura S, Namba K. 2002. Growth mechanism of the bacterial flagellar filament. Res. Microbiol. 153:191–197. 10.1016/S0923-2508(02)01308-6 [DOI] [PubMed] [Google Scholar]
- 9.Macnab RM. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77–100. 10.1146/annurev.micro.57.030502.090832 [DOI] [PubMed] [Google Scholar]
- 10.Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S. 1992. Morphological pathway of flagellar assembly in Salmonella typhimurium. J. Mol. Biol. 226:433–446. 10.1016/0022-2836(92)90958-M [DOI] [PubMed] [Google Scholar]
- 11.Emerson SU, Tokuyasu K, Simon MI. 1970. Bacterial flagella: polarity of elongation. Science 169:190–192. 10.1126/science.169.3941.190 [DOI] [PubMed] [Google Scholar]
- 12.Iino T. 1969. Polarity of flagellar growth in Salmonella. J. Gen. Microbiol. 56:227–239. 10.1099/00221287-56-2-227 [DOI] [PubMed] [Google Scholar]
- 13.Berg HC. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19–54. 10.1146/annurev.biochem.72.121801.161737 [DOI] [PubMed] [Google Scholar]
- 14.Jones CJ, Macnab RM, Okino H, Aizawa S. 1990. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J. Mol. Biol. 212:377–387. 10.1016/0022-2836(90)90132-6 [DOI] [PubMed] [Google Scholar]
- 15.Homma M, Kutsukake K, Hasebe M, Iino T, Macnab RM. 1990. FlgB, FlgC, FlgF and FlgG. A family of structurally related proteins in the flagellar basal body of Salmonella typhimurium. J. Mol. Biol. 211:465–477 [DOI] [PubMed] [Google Scholar]
- 16.Jones CJ, Macnab RM. 1990. Flagellar assembly in Salmonella typhimurium: analysis with temperature-sensitive mutants. J. Bacteriol. 172:1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chevance FF, Takahashi N, Karlinsey JE, Gnerer J, Hirano T, Samudrala R, Aizawa S, Hughes KT. 2007. The mechanism of outer membrane penetration by the eubacterial flagellum and implications for spirochete evolution. Genes Dev. 21:2326–2335. 10.1101/gad.1571607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Homma M, Komeda Y, Iino T, Macnab RM. 1987. The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J. Bacteriol. 169:1493–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones CJ, Homma M, Macnab RM. 1989. L-, P-, and M-ring proteins of the flagellar basal body of Salmonella typhimurium: gene sequences and deduced protein sequences. J. Bacteriol. 171:3890–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nambu T, Kutsukake K. 2000. The Salmonella FlgA protein, a putativeve periplasmic chaperone essential for flagellar P ring formation. Microbiology 146(Part 5):1171–1178 [DOI] [PubMed] [Google Scholar]
- 21.DeRosier DJ. 1998. The turn of the screw: the bacterial flagellar motor. Cell 93:17–20. 10.1016/S0092-8674(00)81141-1 [DOI] [PubMed] [Google Scholar]
- 22.Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT. 2011. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J. 30:2948–2961. 10.1038/emboj.2011.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minamino T, Ferris HU, Moriya N, Kihara M, Namba K. 2006. Two parts of the T3S4 domain of the hook-length control protein FliK are essential for the substrate specificity switching of the flagellar type III export apparatus. J. Mol. Biol. 362:1148–1158. 10.1016/j.jmb.2006.08.004 [DOI] [PubMed] [Google Scholar]
- 24.Minamino T, Moriya N, Hirano T, Hughes KT, Namba K. 2009. Interaction of FliK with the bacterial flagellar hook is required for efficient export specificity switching. Mol. Microbiol. 74:239–251. 10.1111/j.1365-2958.2009.06871.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi N, Mizuno S, Hirano T, Chevance FF, Hughes KT, Aizawa S. 2009. Autonomous and FliK-dependent length control of the flagellar rod in Salmonella enterica. J. Bacteriol. 191:6469–6472. 10.1128/JB.00509-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis RW, Botstein D, Roth JR. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 27.Aizawa SI, Dean GE, Jones CJ, Macnab RM, Yamaguchi S. 1985. Purification and characterization of the flagellar hook-basal body complex of Salmonella typhimurium. J. Bacteriol. 161:836–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohnishi K, Homma M, Kutsukake K, Iino T. 1987. Formation of flagella lacking outer rings by flaM, flaU, and flaY mutants of Escherichia coli. J. Bacteriol. 169:1485–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nambu T, Minamino T, Macnab RM, Kutsukake K. 1999. Peptidoglycan-hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium. J. Bacteriol. 181:1555–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirano T, Minamino T, Macnab RM. 2001. The role in flagellar rod assembly of the N-terminal domain of Salmonella FlgJ, a flagellum-specific muramidase. J. Mol. Biol. 312:359–369. 10.1006/jmbi.2001.4963 [DOI] [PubMed] [Google Scholar]
- 31.Nambu T, Inagaki Y, Kutsukake K. 2006. Plasticity of the domain structure in FlgJ, a bacterial protein involved in flagellar rod formation. Genes Genet. Syst. 81:381–389. 10.1266/ggs.81.381 [DOI] [PubMed] [Google Scholar]
- 32.Minamino T, Namba K. 2004. Self-assembly and type III protein export of the bacterial flagellum. J. Mol. Microbiol. Biotechnol. 7:5–17. 10.1159/000077865 [DOI] [PubMed] [Google Scholar]