Abstract
We present here the functional characterization of a third complete type II secretion system (T2SS) found in newly sequenced Pseudomonas aeruginosa strain PA7. We call this system Txc (third Xcp homolog). This system is encoded by the RGP69 region of genome plasticity found uniquely in strain PA7. In addition to the 11 txc genes, RGP69 contains two additional genes encoding a possible T2SS substrate and a predicted unorthodox sensor protein, TtsS (type II secretion sensor). We also identified a gene encoding a two-component response regulator called TtsR (type II secretion regulator), which is located upstream of the ttsS gene and just outside RGP69. We show that TtsS and TtsR constitute a new and functional two-component system that controls the production and secretion of the RGP69-encoded T2SS substrate in a Txc-dependent manner. Finally, we demonstrate that this Txc-secreted substrate binds chitin, and we therefore name it CbpE (chitin-binding protein E).
INTRODUCTION
The Gram-negative bacterium Pseudomonas aeruginosa is a ubiquitous organism found in highly diverse environments. It is also an opportunistic human pathogen responsible for a variety of chronic and acute infections in compromised patients and those afflicted with the genetic disease cystic fibrosis (1). The adaptation of P. aeruginosa to such diverse environments in the host is mediated by the fine coordination of a substantial arsenal of extracellular virulence factors (2). These factors include secreted and injected exoproteins as well as several extracellular organelles, enabling the pathogen to survive and proliferate in extremely hostile environments. The deployment of P. aeruginosa virulence factors is controlled by multiple signaling, transducing, and regulatory pathways (3), including a large array of two-component regulatory systems, which continuously sense the environment and allow the bacteria to modify their behavior in response to specific signals (4).
P. aeruginosa has become a model organism for the study of protein secretion, since it contains, sometimes in several copies, five of the six secretory machines found in Gram-negative bacteria (5). Of these, the type II secretion system (T2SS) is devoted to the extracellular transport of those exoproteins that require intracellular folding (6–8). Type II secretion is a two-step process in which exoproteins are first exported through the cytoplasmic membrane into the periplasm by either the Sec or Tat translocon, accompanied by the cleavage of the N-terminal signal peptide (9, 10). Their subsequent transport across the outer membrane is achieved by a macromolecular complex embedded in the envelope, the secreton (11). In P. aeruginosa, at least 12 different proteins constitute the Xcp type II secreton, organized into three subcomplexes. The inner membrane platform (XcpPRSYZ) is required to energize the secretion process (12). The next subcomplex (the multimer of the secretin XcpQ) forms a pore in the outer membrane through which each substrate is transported (13) by the action of the third secreton subcomplex, the pseudopilus (14).
The P. aeruginosa Xcp T2SS directs the extracellular targeting of a broad spectrum of at least 12 different exoproteins, including toxins and degradative enzymes (5). This system is regulated by quorum sensing and plays an important role in the global virulence of this bacterium (2, 15). P. aeruginosa possesses a second complete T2SS, the Hxc system, which functions only under phosphate-limiting growth conditions and is dedicated to the specific secretion of low-molecular-weight alkaline phosphatases (16, 17). Phylogenetic and functional comparisons of the Xcp and Hxc systems revealed that they constitute two distinct subfamilies of T2SSs. In contrast to Xcp, the Hxc system was acquired by PAO1 and other Pseudomonas species through horizontal gene transfer and contains an atypical pseudopilus that is specifically adapted to its substrate (18, 19). The coexistence of two complete and independent T2SSs in the same organism has also been reported for other pathogenic or nonpathogenic Gram-negative bacteria (20–24).
Among the nine P. aeruginosa strains fully sequenced and annotated to date (http://v2.pseudomonas.com/index.jsp) (25), PA7 is considered a taxonomic outlier. Although PA7 is at the border of the species, its 16S rRNA gene clearly places it within the P. aeruginosa species (26). Strain PA7 is a multidrug-resistant isolate from Argentina (26, 27). Although it is a nonrespiratory clinical isolate, it lacks genes for some of the major virulence factors, including the whole type III secretion system and its secreted effectors (ExoU, ExoT, ExoY, and ExoS) as well as the Xcp T2SS-dependent toxic exoprotein exotoxin A (ToxA), which are the major virulence factor during acute P. aeruginosa infections. It is therefore likely that this strain uses alternative mechanisms to mediate its virulence.
We present here the complete characterization of Txc, a new T2SS found in a region of genome plasticity of P. aeruginosa strain PA7.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Oligonucleotides are listed in Table 2. PCR was performed by using High Fidelity polymerase (Roche). Genomic DNA was isolated and purified by using the Pure Link genomic DNA minikit (Invitrogen). P. aeruginosa and Escherichia coli were grown at 37°C in Luria broth (LB) medium. The E. coli CC118λpir strain (28) was used to propagate pKNG208, while E. coli strains TG1, BL21(DE3), BTH101, TOP10F′, and DH10B were used for other plasmids. Plasmids were prepared with the QIAPrep spin kit (Qiagen). When required for plasmid selection in E. coli, LB agar medium was supplemented with kanamycin (Km) at 25 μg ml−1; ampicillin (Ap) at 50 μg ml−1, chloramphenicol (Cm) at 30 μg ml−1, streptomycin (Sm) at 30 μg ml−1, or tetracycline (Tc) at 15 μg ml−1. Recombinant plasmids were introduced into P. aeruginosa strain PA7 by conjugation using pRK2013 mobilization properties, as described previously (29). The resulting PA7 transconjugants were selected on LB agar supplemented with tetracycline (Tc) at 200 μg ml−1 or PIA (Pseudomonas isolation agar) medium (Difco) supplemented with carbenicillin (Cb) at 750 μg ml−1. To activate the Hxc system, cells were grown at 30°C under phosphate-limiting conditions by using proteose peptone medium (Difco) containing 0.4% glucose. Saccharomyces cerevisiae strain CRY1-2 was grown at 30°C in yeast extract-peptone-dextrose (YPD) medium (Difco). Plasmids were maintained in yeast by using uracil-deficient medium (Clontech). For selection, cycloheximide was used at 2.5 μg ml−1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| TG1 | F′ [traD36 proAB+ lacIq lacZΔM15] Δ(lac-proAB) | Laboratory collection |
| CC118λpir | (ara-leu) araD lacX74 rpsE rpoB argE(Am) recA1 RfR(λpir) | 28 |
| DH10B | F− endA1 recA1 nupG rpsL ΔlacX74 ϕ80lacZΔM15 araD139Δ(ara leu)7697 mcrA Δ(mrr-hsdRMS-mcrBC) λ− | Invitrogen |
| TOP10F′ | Similar to DH10B, with the F plasmid containing lacIq and Tn10 | Invitrogen |
| BL21(DE3) | F− ompT gal lon λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | 62 |
| BTH101 | F′ cya araD galE galK hsdR2 mcrA1 mcrB1 relA1; Smr | 35 |
| P. aeruginosa | ||
| PA7 | Nonrespiratory clinical isolate | 26 |
| PA7 Δtxc | txcPQRSTUVWXYZ deletion mutant in PA7 | This work |
| PA7::lacZ | PA7 strain carrying the lacZ promoterless fusion inserted into the att site | This work |
| PA7::PcbpE-lacZ | PA7 strain carrying the PcbpE-lacZ fusion inserted into the att site | This work |
| PA7::Ptxc-lacZ | PA7 strain carrying the Ptxc-lacZ fusions inserted into the att site | This work |
| S. cerevisiae CRY1-2 | Cycloheximide resistant (cyh2R) and uracil prototrophic (ura3) | 33 |
| Plasmids | ||
| pCR2.1 | TA cloning; lacZ ColE1 f1 ori; Apr Kmr | Invitrogen |
| pRK2013 | Tra+ Mob+ ColE1; Kmr | 63 |
| pBBR1mcs4 | Cloning vector; Apr Cbr | 64 |
| pCbpE | pBBR1MCS-4 containing the cbpE-His6 DNA sequence; Apr Cbr | This work |
| pTtsSc | pBBR1MCS-4 containing the ttsSc-His6 DNA sequence; Apr Cbr | This work |
| pTtsRc | pBBR1MCS-4 containing the ttsRc-His6 DNA sequence; Apr Cbr | This work |
| pT7.5 | Cloning vector with the T7 promoter; Apr | 65 |
| pCbpD | pT7.5 containing the C-terminal cbpD-His10 DNA sequence; Apr | This work |
| pLLX13 | Yeast capture plasmid; URA3 CEN6 ARSH4 oriV incP tra oriT; Tcr | 33 |
| pLLX8 | Plasmid containing cyh2 and bla genes; cyh2R Apr Cbr | 33 |
| pCV1407-1420 | Plasmid containing TS1 and TS2 sequences and cyh2R and bla genes | This work |
| pRGP69 | pLLX13 containing the ttsS-txcZ DNA region | This work |
| pKNG101 | Suicide vector; oriR6K mobRK2 sacBR+; Smr | 66 |
| pHP45Ω-Tc | Plasmid containing the Tc resistance cassette; Apr Tcr | 67 |
| pKNG208 | Tcr suicide vector; oriR6K mobRK2 sacBR+; Tcr | This work |
| pKNG208Δtxc | pKNG208 containing the 500-bp upstream and the 500-bp downstream DNA regions of the txc cluster; Tcr | This work |
| MiniCTX-lacZ | Ω-FRT-attP-MCS ori int oriT; Tcr | 30 |
| pPtxc-lacZ | Mini-CTX containing the transcriptional fusion Ptxc-lacZ | This work |
| pPcbpE-lacZ | Mini-CTX containing the transcriptional fusion PcbpE-lacZ | This work |
| pUT18C | Expression vector encoding the T18 fragment of cyaA; Apr | 37 |
| pKT25 | Expression vector encoding the T25 fragment of cyaA; Cmr | 37 |
| pT18-TtsSc | pUT18C containing the TtsSc DNA sequence; Apr | This study |
| pT18-Pal | pUT18C containing the Pal DNA sequence; Apr | E. Bouveret |
| pT25-TtsR | pKT25 containing the TtsR DNA sequence; Cmr | This study |
| pT25-TolB | pKT25 containing the TolB DNA sequence; Cmr | E. Bouveret |
Smr, streptomycin resistance; cyh2R, cycloheximide resistance; Apr, ampicillin resistance; Kmr, kanamycin resistance; Cbr, carbenicillin resistance; Tcr, tetracycline resistance; Cmr, chloramphenicol resistance.
TABLE 2.
Oligonucleotides used in this study
Cloning procedures for PcbpE-lacZ and Ptxc-lacZ transcriptional fusions.
The DNA fragments containing the putative promoter regions of cbpE (629 bp) and txc (555 bp) were PCR amplified with oligonucleotide pairs OFC7-3/OFC7-4 and OFC7-1/OFC7-2, respectively. The resulting DNA fragments were cloned into the mini-CTX–lacZ vector at the SmaI site. The resulting plasmids were used to generate chromosomal PcbpE-lacZ and Ptxc-lacZ fusions in strain PA7, as previously described (30, 31). The negative-control strain (PA7::lacZ) was generated by using the promoterless mini-CTX–lacZ vector. β-Galactosidase assays were carried out by using the spectrophotometric method described previously by Miller (32) with ortho-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate. Enzyme activities are reported in Miller units, expressed as a function of cell density measured at 600 nm. Assays were performed in triplicate.
Cloning procedure for plasmids expressing cbpD, cbpE, ttsSc, and ttsRc.
The DNA corresponding to CbpD, CbpE, and the cytoplasmic domains of TtsS (TtsSc) and TtsR (TtsRc) were PCR amplified with oligonucleotide pairs OFC1-76/OFC1-96, OFC7-5/OFC7-11, OFC7-86/OFC7-87, and OFC7-88/OFC7-89, respectively, and cloned into pCR2.1 by using the TA cloning kit (Invitrogen). For cbpE, ttsSc, and ttsRc, the corresponding 3′ oligonucleotides possess 6 histidine codons (10 histidine codons for cbpD) before the stop codon to allow immune detection or purification of their products. A cbpD DNA fragment was subcloned into pT7.5 at EcoRI sites. cbpE, ttsSc, and ttsRc DNA fragments were subcloned into the pBBR1-mcs4 vector at SpeI/EcoRV sites for cbpE and XhoI/HindIII sites for ttsSc and ttsRc.
Construction of gene capture plasmid pRGP69.
The gene capture plasmid was assembled by recombinational cloning in S. cerevisiae as described previously (33, 34). We used the vector pLLX13 linearized with NheI, two 1-kb targeting sequences (targeting sequence 1 [TS1] and TS2) (Fig. 1), and one PCR fragment (2.9 kb) amplified from the pLLX8 vector. The pLLX8 fragment was generated with oligonucleotides X8up and X8dw, possessing one end homologous to TS1 and the other one homologous to TS2 (Table 2). This fragment was inserted between the targeting sequences and provided the counterselection markers (cyh2R) that would be replaced by the captured genomic DNA. The TS1 and TS2 DNA regions were PCR amplified with oligonucleotide pairs OFC7-125/OFC7-126 and OFC7-127/OFC7-128, respectively, using PA7 genomic DNA as the template. To create capture plasmid pCV1407-1420, 200 ng of targeting sequences, 600 ng of the pLLX8 PCR product, and 200 ng of linearized pLLX13 were combined with 200 μl of lithium acetate-treated competent S. cerevisiae strain CRY1-2. Transformants were selected on uracil-deficient medium. Yeast DNA was purified (Zymoprep) and transformed into electrocompetent E. coli DH10B.
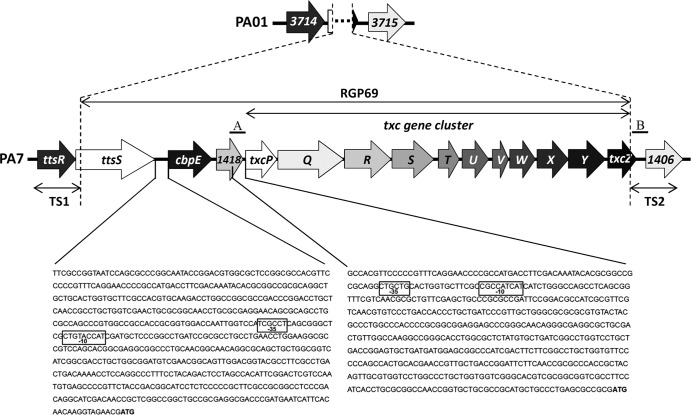
FIG 1.
Genetic organization of PA7 RGP69. The genomic island RGP69, uniquely present in P. aeruginosa strain PA7, possesses 14 predicted open reading frames encoding the 11 putative T2SS Txc components (TxcP to TxcZ) and 3 genes, a gene encoding a predicted chitin-binding protein (CbpE), a gene encoding an unorthodox sensor protein (TtsS), and PSPA7_1418, which is predicted to encode a protein belonging to the cytochrome b superfamily. RGP69 is externally flanked by the ttsR and PSPA7_1406 (1406) genes, which are homologous to the two contiguous genes PA3714 (3714) and PA3715 (3715), respectively, in strain PAO1. The two DNA sequences corresponding to the segments used to construct the PcbpE-lacZ and PtxcP-lacZ reporter fusions are also shown. The −10 and −35 boxes of the two predicted sigma 70 promoter regions of RGP69 are boxed in the corresponding DNA sequences. The ATG start codons of cbpE and txcP are indicated in boldface type. Also indicated are the two DNA regions TS1 and TS2, used to clone the RGP69 sequence in the gene capture vector pLLX13. A and B regions correspond to the DNA sequences (500 bp) cloned into pKNG208 to generate the suicide vector pKNG208Δtxc, which was used to generate the txc deletion mutant.
The DNA genomic fragment flanked by TS1 and TS2 DNA sequences was cloned by cotransforming 200 μl of competent S. cerevisiae CRY1-2 with 5 μg of PA7 sheared genomic DNA and 500 ng of PmeI-linearized pCV1407-1420. Transformation mixtures were plated onto uracil-deficient medium containing cycloheximide. The presence of captured P. aeruginosa sequences was checked by PCR on S. cerevisiae colonies. Yeast DNA was purified, and 100 ng was transformed into electrocompetent E. coli DH10B.
Construction of the PA7 Δtxc in-frame deletion mutant.
The suicide vector pKNG208 (Tc resistant), specifically adapted to multidrug-resistant strain PA7, was initially constructed to perform in-frame deletions in this strain. The 2.3-kb Tc resistance gene cassette was obtained from vector pHP45Ω-Tc by SmaI digestion followed by ligation into EcoRV of vector pCR2.1. The SpeI/NotI fragment was subcloned from this construct into pKNG101, resulting in pKNG208. To construct pKNG208Δtxc, the DNA regions (500 bp) corresponding to upstream and downstream segments of the txc cluster (Fig. 1) were PCR amplified by using genomic DNA and oligonucleotide pairs OFC7-129/OFC7-130 and OFC7-131/OFC7-132. The fragments were joined by overlapping PCR using primer pair OFC7-129/OFC7-132 and a mix of the two previously generated products as the templates. The PCR product was cloned into pCR2.1 by using the TA cloning kit (Invitrogen) and the 1-kb SpeI/NsiI fragment subcloned into pKNG208, yielding pKNG208Δtxc. This construct was introduced by conjugation into strain PA7 to generate the PA7 Δtxc strain.
Bacterial two-hybrid (BACTH) constructs and assays.
According to a procedure described previously by Battesti and Bouveret (35), two recombinant proteins were engineered for this assay, first between the T18 subunit of the adenylate cyclase and TtsSc (PCR amplified with oligonucleotides OFC7-117/OFC7-118) by using the XbaI and BamHI restriction sites of pUT18C and second between the T25 subunit of the adenylate cyclase and TtsR (PCR amplified with oligonucleotides OFC7-119/OFC7-120) by using the restriction sites XbaI and BamHI of pKT25. E. coli strain BTH101 was cotransformed with various combinations of T25 and T18 constructs, selected on plates of LB agar with Ap and Km, and incubated overnight at 30°C. Selected strains were resuspended in 3 ml of LB medium plus Ap and Km and incubated overnight at 30°C. Fifteen microliters of the bacterial cultures were spotted onto LB agar medium containing 0.5 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) at 40 μg/ml. The blue coloration appears after 24 h of incubation at 30°C. β-Galactosidase assays were carried out by using the spectrophotometric method described previously by Miller (32) with ONPG as a substrate. Enzyme activities are reported in Miller units, expressed as a function of cell density measured at 600 nm. Assays were performed in triplicate.
Preparation and analysis of P. aeruginosa protein fractions.
PA7 strains were grown at 37°C in LB medium. A volume of culture equivalent to 50 units at an optical density at 600 nm (OD600) was centrifuged for 10 min at 2,000 × g to separate cells from extracellular medium. For the intracellular soluble fraction, the cell pellet was washed with 15 ml of 10 mM Tris-HCl (pH 7.4) and centrifuged for 10 min at 2,000 × g at 4°C. The pellet was resuspended in 15 ml 10 mM Tris-HCl (pH 7.4)–1 mM phenylmethylsulfonyl fluoride (PMSF) and lysed by passage through a French press at 15,000 pressure units (lb/in2) three times. After removal of the unbroken cells by centrifugation for 5 min at 2,000 × g at 4°C, the cell lysate was centrifuged for 30 min at 120,000 × g at 4°C to pellet the membranes. The supernatant corresponding to the soluble cytoplasmic and periplasmic fractions was precipitated with 10% trichloroacetic acid (TCA) for 1 h at 4°C. Samples were centrifuged for 30 min at 15,000 × g at 4°C, and the pellet was washed with 90% acetone and resuspended in SDS-PAGE sample buffer at 0.2 OD600 units per μl. Aliquots of 10 μl were analyzed by SDS-PAGE (10.5% resolving gel), and the bands were visualized by immunoblotting with a specific primary antibody followed by chemiluminescence detection (Supersignal West Pico; Pierce). The primary antibodies used in this study are rabbit anti-CbpD (a gift from J. Tommassen) (at 1/5,000), rabbit anti-CbpE (peptide-based antibody [ThermoFischer] at 1/2,500), or rabbit anti-LasB (1/5,000). Two specific CbpE peptides (corresponding to amino acids 410 to 429 and 478 to 497 of the mature protein) were synthesized by ThermoFischer and used to generate anti-CbpE antibody with the aim of avoiding cross-reaction with the homolog CbpD. For extracellular protein fractions, culture supernatants were precipitated with 10% TCA for 1 h at 4°C. Samples were centrifuged for 30 min at 15,000 × g at 4°C, and the pellets were washed with 90% acetone and resuspended in SDS-PAGE sample buffer to 0.5 OD600 units per μl. Aliquots of 8 μl were analyzed by SDS-PAGE followed by immunoblotting.
Chitin-binding assay.
For the chitin-binding assay, 100 μl of prewashed chitin beads (New England BioLabs) in chitin-binding buffer (50 mM Tris-HCl [pH 8.0], 1 mM EDTA, 500 mM NaCl, 0.1% Tween 100) was incubated with a 200-μl solution containing the four purified proteins at 200 μg ml−1 each in chitin-binding buffer. The sample was incubated on a rotating wheel for 1 h 30 min at 4°C. The supernatant (unbound fraction) was separated from the beads (bound fraction) by gravity flow after 5 min at room temperature. The supernatant was collected, and the beads were washed twice with 200 μl of chitin-binding buffer. Fifteen microliters of each sample was separated by SDS-PAGE followed by Coomassie blue staining to determine the amount of proteins in each fraction. As a control, 60 μl of the solution containing the four purified proteins was diluted with 240 μl of chitin-binding buffer, and 15 μl was analyzed on an SDS-PAGE gel (total fraction).
Four purified proteins were used in this assay. Purified P. aeruginosa elastase (LasB) and exotoxin A (ToxA) were purchased from Nagase ChemteX Corp. (Kyoto, Japan) and List Biological Laboratories, Inc. (Campbell, CA), respectively. His6-tagged CbpE and His10-tagged CbpD were purified as follows: E. coli BL21(DE3) cells harboring plasmid pCbpE or pCbpD, both inducible with IPTG, were grown overnight on LB at 37°C. This preculture was used to inoculate 6 or 4 liters (for CbpE and CbpD, respectively) of Terrific Broth medium (Sigma-Aldrich) at an initial OD600 of 0.4. At an OD600 of 0.8, the temperature was decreased from 37°C to 17°C, IPTG was added at 100 μM, and the cells were grown for 24 h. Bacteria were pelleted by centrifugation, resuspended in lysis buffer (50 mM Tris [pH 8.0], 300 mM NaCl, 1 mM EDTA, 0.5 μg ml−1 lysozyme, 1 mM PMSF), submitted to three freeze-thawing cycles, and sonicated after the addition of DNase at 20 μg ml−1 and MgCl2 at 20 mM. Pellet and soluble fractions were separated by centrifugation for 30 min at 16,000 × g. Soluble fractions containing the His-tagged protein (CbpE or CbpD) were loaded onto a 5-ml HisTrap nickel column (Pharmacia) on an Äkta system (Amersham Biosciences); preequilibrated in a solution containing 50 mM Tris, 300 mM NaCl, and 10 mM imidazole (pH 8.0); and eluted by washing with a solution containing 50 mM Tris and 300 mM NaCl (pH 8.0) in the presence of 250 mM imidazole. His-tagged CbpE and CbpD in the eluted fractions were concentrated on a Centricon device with a cutoff of 3 kDa. CbpE was subjected to an extra purification step through a Superdex 200 gel filtration column equilibrated in a solution containing 50 mM Tris and 150 mM NaCl (pH 8.0) and further concentrated with a Centricon device with a cutoff of 3 kDa. Final concentrations of CbpE and CbpD were evaluated by using a BioSpec-nano instrument (Shimadzu-Biotech), at 5.1 and 7.8 mg ml−1, respectively.
RESULTS
Identification of a third complete T2SS in P. aeruginosa strain PA7.
Among the numerous regions of genomic plasticity (RGP) found in the recently sequenced genome of P. aeruginosa PA7, 18 (named RGP63 to RGP80) are unique to this strain (26). Analysis of one of these unique regions, RGP69, using the Pseudomonas Genome Database website (http://v2.pseudomonas.com/search.jsp) (25) revealed a cluster of 14 genes encoding a likely complete T2SS, including machinery components, secreted substrate, and a regulatory protein (Fig. 1). The 11 T2SS “core” genes txcP to txcZ (PSPA7_1417 to PSPA7_1407) present in RGP69 are arranged unidirectionally and may constitute a single operon. However, such a T2SS genetic organization, often recovered in T2SS gene clusters (8), has never been reported for P. aeruginosa, suggesting that this represents a new type of P. aeruginosa T2SS. Since this is the third T2SS to be identified in P. aeruginosa, we named it Txc, for third homolog to Xcp. RGP69 also contains cbpE (PSPA7_1419), a gene encoding a 497-amino-acid protein, a putative substrate for the Txc T2SS. Indeed, CbpE possesses a predicted N-terminal signal peptide used by Sec-dependent proteins to cross the inner membrane, a pathway also followed by T2SS-secreted proteins. According to genome annotation (Pseudomonas Genome Database), this protein belongs to the N-acetylglucosamine (GlcNAc)-binding family and has 28% amino acid identity to the chitin-binding protein CbpD (PSPA7_4667), a P. aeruginosa exoprotein known to be secreted by the Xcp T2SS (36). We thus called this putative Txc T2SS substrate CbpE for chitin-binding protein E.
Another gene in RGP69, ttsS (PSPA7_1420), encodes a predicted unorthodox sensor protein that we called TtsS for type two secretion sensor. Unorthodox sensor proteins are complex histidine kinases in which the three phosphotransfer domains (transmitter, receiver, and histidine phosphotransferase [Hpt]) are combined in a single protein. Just upstream of the ttsS gene is a gene, ttsR (PSPA7_1421), encoding a predicted response regulator that we called TtsR for type two secretion regulator. Such a location suggests that TtsR may work in concert with TtsS to regulate the downstream T2SS cluster and its putative substrate. The remaining gene on RGP69 is PSPA7_1418; it is located between cbpE and the first gene (txcP) of the T2SS cluster and is annotated as a predicted inner membrane protein with a putative cytochrome b superfamily signature.
Using in silico tools, we further explored the regulation of the RGP69 region. Two putative σ70 promoters are predicted in the RGP69 sequence by the bacterial promoter prediction program BProm (SoftBerry). The −35 and −10 boxes of the two promoters are located upstream of the cbpE gene and the first txc gene, txcP (Fig. 1). This promoter prediction supports our proposed operon structure of the txc genes.
The RGP69-encoded sensor protein TtsS forms a two-component system with the upstream-encoded response regulator TtsR.
According to the Pseudomonas Genome Database website, the RGP69 gene encoding the sensor TtsS is predicted to be organized in an operon with the gene encoding the response regulator TtsR, located directly upstream of but outside RGP69 (Fig. 1). Whereas RGP69 is present only in strain PA7 of P. aeruginosa, TtsR is highly conserved in all sequenced strains of P. aeruginosa, where it is therefore an orphan regulator. In order to verify that TtsS and TtsR form a two-component system, we tested their interaction using the Escherichia coli bacterial two-hybrid (BACTH) method developed previously by Karimova and collaborators (37). To this end, we cloned the DNA fragments corresponding to the cytoplasmic domain of TtsS (TtsSc) and full-length TtsR (Fig. 2a) in pUT18C and pKT25, creating plasmids pT18-TtsSc and pT25-TtsR, respectively. We showed that TtsS interacts with the cognate response regulator TtsR (Fig. 2b and c). Such direct interactions between a regulator and a sensor protein usually mean that they work in concert as transcriptional regulators (38, 39). We therefore proposed that TtsR and TtsS may form a new and functional two-component system.
FIG 2.
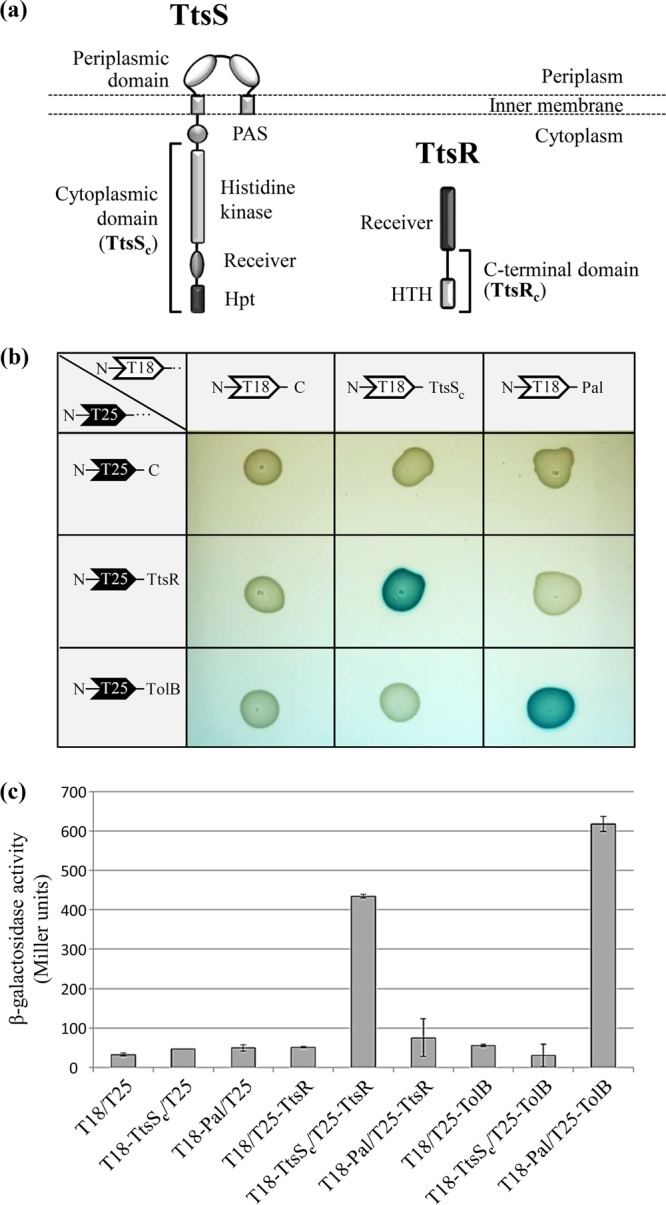
TtsS and TtsR interact together to form a two-component system. (a) Modular structures of the unorthodox sensor TtsS and the response regulator TtsR. The cytoplasmic domain of TtsS (TtsSc) and the C-terminal domain of TtsR (TtsRc) used for transcriptional and secretion experiments are indicated by brackets. All the other characteristic domains of the unorthodox sensor and NarL-type response regulator are indicated (Hpt for histidine phosphotransferase and HTH for helix-turn-helix). (b) Binary interactions between TtsSc and full-length TtsR determined by using the BACTH assay. TtsSc and TtsR were fused to the C termini of Bordetella pertussis adenylate cyclase fragments T18 and T25, leading to plasmids pT18-TtsSc and pT25-TtsR. Various T18 and T25 plasmid combinations were cotransformed into E. coli cya strain BTH101 and plated onto LB agar–IPTG–X-gal medium. Functional complementation between the T18 and T25 fragments, which occurs only upon interactions of the hybrid proteins, triggers the expression of the cya gene and yields blue colonies. The E. coli TolB and Pal interacting proteins were used as a positive control. (c) Interactions were quantified by β-galactosidase assays in biological triplicates for all tested interactions.
The txc gene cluster and the cbpE gene are positively regulated by the TtsR/TtsS two-component system.
In order to study the regulation of the txc and cbpE genes, we created reporter constructs containing their putative promoter sequences. The 629- and 555-bp regions upstream of cbpE and txcP, respectively (Fig. 1), were used to construct the PcbpE-lacZ and PtxcP-lacZ chromosomal transcriptional fusions. When these fusions were introduced into PA7, only background β-galactosidase activity was detected in strains grown under a variety of conditions, including LB, low temperature, minimal medium, calcium and phosphate starvation, MgCl2 supplementation, and rich/poor medium supplemented or not with colloidal chitin or GlcNAc (data not shown).
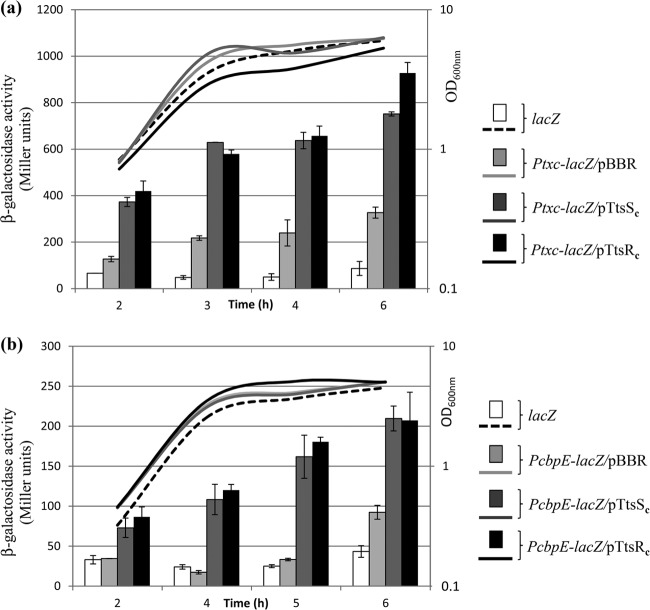
Because of the low level of activity of the PcbpE-lacZ and Ptxc-lacZ transcriptional fusions under all tested laboratory conditions, it is likely that the expression of the RGP69 genes is strictly regulated by the environment. We also reasoned that the upstream-encoded two-component system TtsR/TtsS could be involved in the regulation of the cbpE and txc genes. In two-component systems, the overproduction of the full-length or the cytoplasmic domain of a sensor protein can artificially mimic its activation. This can lead to the transcriptional activation of the target promoters, presumably through an increase in the sensor baseline kinase activity on cognate response regulators (40–42). A similar artificial activation of two-component system target genes was also observed with overproduction of the response regulator (38). We cloned the DNA regions encoding the C-terminal cytoplasmic domain of TtsS (TtsSc) and the C-terminal domain of TtsR (TtsRc) (Fig. 2a) into the constitutive expression vector pBBR1-MCS4, creating plasmids pTtsSc and pTtsRc, respectively. In order to monitor the effects of TtsS and TtsR on the two promoters identified in RGP69, TtsSc and TtsRc were overproduced in the PA7::PcbpE-lacZ and PA7::PtxcP-lacZ reporter strains. In the four strains and throughout the growth phases, we observed a >2-times increase in β-galactosidase activity as a result of TtsSc and TtsRc overproduction (Fig. 3). The TtsS sensor and the TtsR response regulator therefore positively regulate the expression of the downstream cbpE and txc T2SS genes. The observation that TtsS and TtsR regulate the same targets fully validates their association in a functional two-component system. Moreover, the coregulation of the cbpE and txc genes by TtsS/TtsR further supports a functional link between the two corresponding proteins, such as the secretion of CbpE by the Txc T2SS.
FIG 3.
The TtsS/TtsR two-component system positively regulates cbpE and txc gene expressions. Shown are β-galactosidase activities of Ptxc-lacZ (a) and PcbpE-lacZ (b) transcriptional fusions (in Miller units) in PA7::Ptxc-lacZ (Ptxc-lacZ) and PA7::PcbpE-lacZ (PcbpE-lacZ) strains overproducing TtsSc or TtsRc or not (pBBR). β-Galactosidase activities were also measured in the strain bearing the promoterless lacZ fusion, PA7::lacZ (lacZ), as a negative control. The growth curves of the strains are also shown (OD600). The experiment was reproduced three times. Samples were duplicated for each experiment, and the corresponding standard deviations were calculated.
CbpE is secreted in PA7 by the Txc T2SS.
As described above, the overproduction of TtsSc or TtsRc triggers the expression of the cbpE and txc genes, which are poorly expressed under conditions of laboratory culture. We therefore used strains grown under inducing conditions (i.e., overproducing TtsSc or TtsRc) to monitor the production and localization of CbpE. The secretion of CbpE in the extracellular milieu was monitored by Western blotting experiments using the peptide-based CbpE antibody. The analysis of the extracellular fraction showed that CbpE is secreted by PA7 only when TtsSc or TtsRc is overproduced (Fig. 4a and b, lanes 2). This observation is in agreement with the increased transcriptional activity of the cbpE promoter observed under these conditions (Fig. 3). Two bands are specifically detected by the CbpE antibody. The lower band (labeled with an asterisk in Fig. 4) may correspond to a CbpE degradation product. The extracellular instability of CbpE, often observed in our experiments (Fig. 4), could be the consequence of its degradation by the secreted extracellular proteases, since a similar instability was also reported for its homolog CbpD (36).
FIG 4.
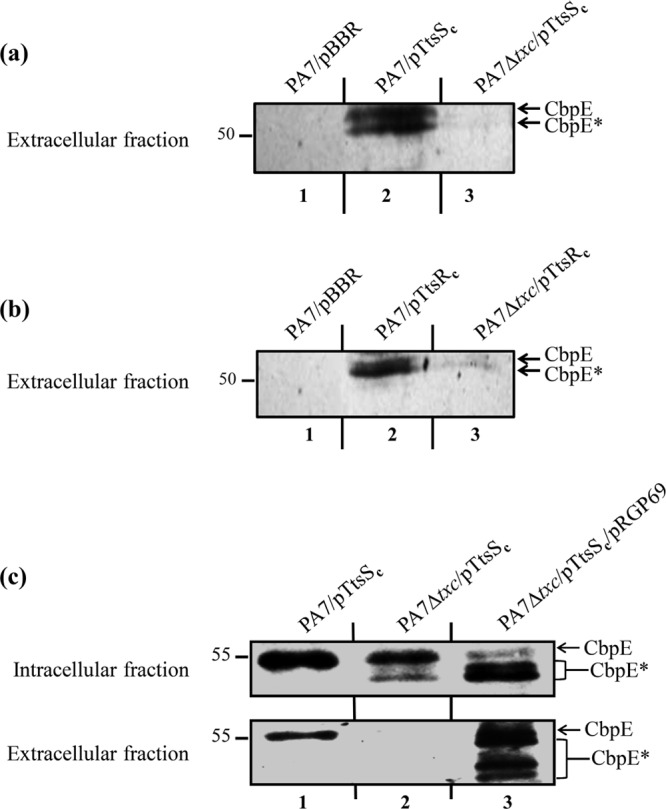
Txc-dependent secretion of CbpE by strain PA7 overproducing TtsSc or TtsRc. (a and b) Immunoblots of extracellular protein fractions from wild-type PA7 (PA7) or Txc-deficient PA7 (PA7 Δtxc) carrying the empty vector (pBBR), pTtsSc (a), or pTtsRc (b), probed with anti-CbpE antibody. (c) Immunoblots of intracellular (top) and extracellular (bottom) fractions from wild-type PA7 (PA7) and the Txc-deficient PA7 strain (PA7 Δtxc) carrying pTtsSc alone or pTtsSc and pRGP69, probed with anti-CbpE antibody. CbpE and degradation products of CbpE are indicated by asterisks. Molecular mass markers (in kDa) are indicated on the left.
To address the role of Txc in the secretion of CbpE, we analyzed its secretion in a Txc-deficient strain. A PA7 strain with a nonpolar chromosomal deletion of the 11 txc genes was engineered by double-crossover events using the pKNG208 suicide vector. Wild-type (WT) and Txc-deficient PA7 strains carrying pTtsSc or pTtsRc were grown under inducing conditions, and intracellular soluble and extracellular protein fractions were analyzed by Western blotting with the anti-CbpE antibody (Fig. 4). CbpE is specifically absent in the extracellular fraction of the Txc-deficient strain overproducing TtsSc or TtsRc (Fig. 4a and b, lanes 3, and c, lane 2), while it is present intracellularly only in a mutant strain overproducing TtsSc (Fig. 4c, lane 2). The secretion defect of the PA7 Δtxc strain can be rescued by introducing plasmid pRGP69 carrying the 15.4-kb RGP69 genomic region, cloned by using the yeast gene capture method. This result demonstrates that CbpE secretion in PA7 is mediated by the Txc T2SS. This secretion strongly implies the assembly of a functional Txc T2SS, in agreement with the higher level of transcriptional activity of the txc promoter observed under conditions of TtsSc or TtsRc overproduction (Fig. 3).
CbpE is a chitin-binding protein.
GlcNAc is the monomer unit of chitin found in the exoskeleton of arthropods and the fungal cell wall. It is also implicated in bacterial development, adherence, and signal transduction but can also be used as a carbon source (43–48). The PA7 CbpE protein is annotated as a GlcNAc-binding protein and has 40% identity at the amino acid level with the GlcNAc-binding protein archetype, GbpA of Vibrio cholerae (GI:15601566) (43). The alignment of the two proteins moreover reveals that CbpE presents the same modular organization as GbpA (Fig. 5a). Similar to GbpA, and according to Wong and collaborators (49), CbpE possesses a predicted domain for chitin and mucin binding (D1) in its N terminus. This D1 domain belongs to the pfam03067 family, which was found to be associated with a wide variety of chitin-binding proteins (50). The predicted D2 and D3 domains of CbpE may confer to this protein the capacity to bind the bacterial cell surface. Finally, a second chitin-binding domain (D4) is predicted in the C terminus of CbpE. CbpE also shares 28% identity and the D1 and D4 chitin-binding domains with the well-characterized P. aeruginosa chitin-binding protein D (CbpD), which is secreted by the Xcp T2SS (Fig. 5a) (36).
FIG 5.
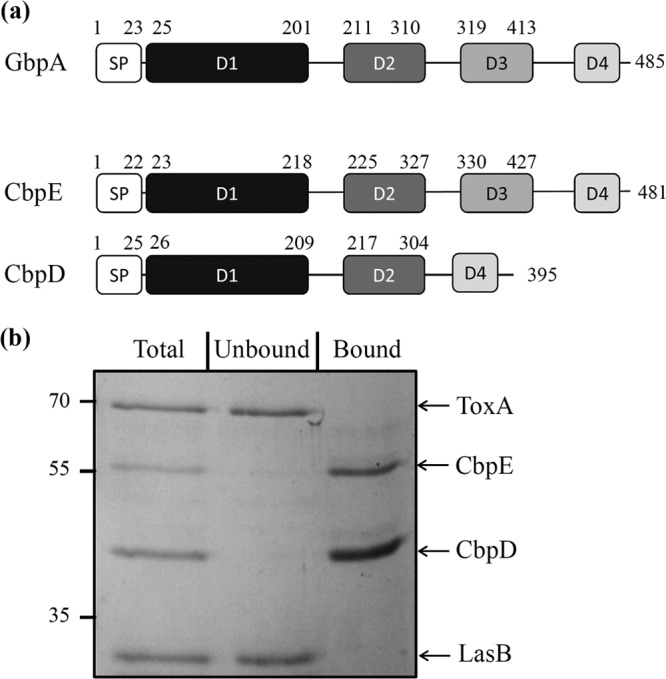
CbpE is a chitin-binding protein. (a) Schematic representation of the presence of the functional domains of GbpA (GI:15601566) in CbpE (PSPA7_1419) and CbpD (PSPA7_4667). The domains represented are the signal peptide (SP), the GbpA D1 chitin- and mucin-binding domain, the GbpA D2 and D3 bacterial surface-binding domains, and the GbpA D4 chitin-binding domain. Signal peptide domains were predicted by the SignalP 4.1 server (http://www.cbs.dtu.dk/services/SignalP/). When present, the D1, D2, and D3 domains were predicted by homology modeling of CbpE and CbpD based on the structure of GbpA (PDB accession number 2XWX), using Modeler software (68). The D4 domain in CbpE and CbpD was defined based on a sequence alignment of the C-terminal sequences of CbpE, CbpD, and GbpA by using the Multalin Web server (http://multalin.toulouse.inra.fr/multalin/multalin.html). (b) To test the chitin-binding affinity of CbpE, a mixture of affinity-purified CbpE and CbpD and purified LasB and ToxA exoproteins (Total) was incubated with chitin beads according to the protocol described in Materials and Methods. Incubation with chitin beads resulted in the precipitation of the exoproteins interacting with chitin (Bound), while the exoproteins without affinity for chitin remained in the soluble fraction (Unbound). Samples were analyzed by SDS-PAGE followed by Coomassie blue staining. Molecular mass markers (in kDa) are indicated on the left.
To test whether CbpE is a true chitin-binding protein, we performed a chitin-binding assay on purified proteins. To this end, CbpE and its homolog CbpD were recombinantly produced as C-terminally His-tagged proteins in E. coli and purified by affinity chromatography. In addition, and as negative controls, we used purified elastase (LasB) and exotoxin A (ToxA), two exoproteins secreted by the P. aeruginosa Xcp T2SS and not predicted to bind chitin. The chitin-binding assay was performed by incubating chitin beads with a mixture of the four purified proteins. As presented in Fig. 5b, the recombinant CbpE protein was recovered like CbpD in the chitin-bound fraction, while LasB and ToxA were found, as expected, in the unbound fraction. This result shows that the Txc-secreted protein CbpE, like the Xcp-secreted protein CbpD, is a chitin-binding protein, which is in agreement with the prediction of 2 chitin-binding domains in these proteins (Fig. 5a).
Coexistence of three functional T2SSs in the same P. aeruginosa strain.
The xcp and hxc T2SS gene clusters are present on the PA7 genome (Fig. 6a). We tested the functionality of the Xcp T2SS in strain PA7 by SDS-PAGE and immunoblotting of extracellular fractions. We found that two Xcp substrates, CbpD and LasB, are secreted into the extracellular medium by WT and Txc-deficient PA7 strains. This demonstrates the functionality and the specificity of the Xcp T2SS in PA7 (Fig. 6b). Similarly, the Hxc T2SS substrate LapC is recovered in the extracellular fraction of WT and txc-deficient PA7 strains grown under conditions of phosphate starvation, conditions which are required for hxc induction (Fig. 6b). Our data therefore indicate that the Xcp and Hxc T2SSs are functional and independent of the Txc T2SS in PA7. Hence, with the addition to the Txc T2SS described in the present study, P. aeruginosa PA7 has three independent and functional T2SSs at its disposal.
FIG 6.
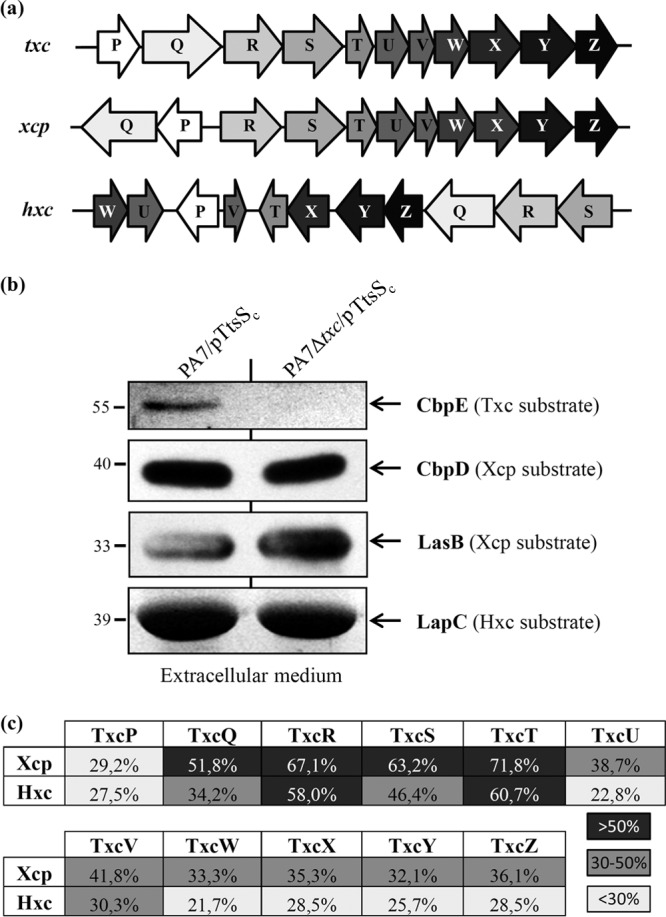
Coexistence of three functional T2SSs in strain PA7. (a) Genetic organization of the three T2SS gene clusters in PA7. (b) Comparative analysis of PA7 T2SS secretomes. The extracellular protein fractions of PA7 strains were analyzed by SDS-PAGE followed by Coomassie blue staining for LapC secretion by the Hxc T2SS (LapC identity was confirmed by mass spectrometry), immunodetection with anti-CbpE for CbpE secretion by the Txc T2SS, and immunodetection with anti-CbpD and anti-LasB for CbpD and LasB secretion by the Xcp T2SS. Molecular mass markers (in kDa) are indicated on the left. (c) Sequence relatedness between Txc components and their counterparts in the Xcp and Hxc T2SSs. The percent identities over the complete protein sequences are indicated. Low (<30%), medium (30 to 50%), and high (>50%) identities are shown.
DISCUSSION
We report here the complete functional characterization of a new T2SS found in P. aeruginosa strain PA7. Our data revealed that this Txc T2SS, its unorthodox sensor TtsS, and the secreted protein CbpE are all encoded by a region of genome plasticity unique to this strain. While the Txc T2SS is limited to PA7 among P. aeruginosa strains, similar txc-cbpE clusters have been recovered from other members of the Pseudomonas genus, such as P. putida strain ND6 (51) and P. fluorescens strain F113 (52). However, and in contrast to what was mentioned previously by Redondo-Nieto et al. (52), the txc cluster of P. fluorescens strain F113 is not a “second xcp cluster” but a new T2SS cluster, as we demonstrate in this study. In addition, similar genetic and/or physical associations between a chitin-binding protein and a T2SS can also be found in other Gram-negative bacteria, such as Yersinia enterocolitica, V. cholerae, Legionella pneumophila, and Escherichia coli K-12 (22, 53–55). The T2SS therefore appears to be well suited for the transport of GlcNAc-binding proteins that need to be released into the medium to provide adherence properties or generate carbon sources in the case of chitinases. Interestingly, GlcNAc-binding proteins that enter host organisms use the alternative T3SS, which allows direct injection into the host (56, 57).
Our data revealed that the Txc T2SS is regulated by a two-component system constituted of the unorthodox sensor TtsS and the response regulator TtsR. Unorthodox sensor proteins are complex histidine kinases in which the three phosphotransfer domains (transmitter, receiver, and Hpt) are combined in a single protein and are thought to participate in complex phosphorelay networks (4, 58). Five unorthodox sensor proteins have been described so far for P. aeruginosa PAO1 (4). The gene encoding TtsS, present only in PA7, therefore constitutes the sixth unorthodox sensor described for P. aeruginosa.
The discovery of a new T2SS in P. aeruginosa strain PA7 brings the number of complete and functional T2SSs coexisting in P. aeruginosa strain PA7 to three: the Xcp, Hxc, and Txc systems. In silico analysis of the xcp, hxc, and txc gene clusters revealed three different genetic organizations (Fig. 6a), suggesting that the txc T2SS gene cluster is not a simple duplication of one system or the other. While xcp and hxc gene clusters are organized into several divergent operons, the txc genes are all in the same orientation and may constitute one single operon. The identity levels between each component of the three T2SS systems reveal a globally higher degree of relatedness between Txc and Xcp than between Txc and Hxc (Fig. 6c). This observation is supported by a previous phylogenetic analysis involving 145 T2SS major pseudopilin genes (18). In this phylogenetic tree, the txc major pseudopilin gene txcT is located in the same monophylogenetic group as xcpT, while hxcT is nested within a large cluster of betaproteobacteria, indicating its likely acquisition by horizontal gene transfer. In support of this conclusion, the following several lines of evidence suggest that, in contrast to the hxc cluster, RGP69 was not acquired by horizontal gene transfer: (i) scars of the RGP69 extremities can be identified in other P. aeruginosa strains, such as PAO1 (Fig. 1), suggesting that this region was originally present in the common P. aeruginosa ancestor and was lost by excision in most contemporary strains; (ii) the similar GC% contents between RGP69 and the rest of the PA7 genome suggest that RGP69 was not acquired by horizontal gene transfer; and (iii) in T2SSs, pseudopilins must be matured by a prepilin peptidase to be functional (59, 60). Because no prepilin peptidases are encoded by RGP69, the Txc pseudopilins probably use the general prepilin peptidase XcpA/PilD (PSPA7_5164), which is also used by the Xcp and Hxc pseudopilins (16, 59, 61). This dependency on a distant gene suggests a loss of the txc cluster in other P. aeruginosa strains instead of an acquisition in PA7.
During the last decade, increasing numbers of Gram-negative bacteria have been shown to express multiple T2SSs. These bacteria generally possess one constitutively expressed machine, while additional systems are generally subject to specific regulation. This is the case for Y. enterocolitica (22), Xanthomonas campestris pv. vesicatoria (21), enterotoxigenic E. coli (20), Dickeya dadantii (24), Stenotrophomonas maltophilia (23), and P. aeruginosa PAO1 (16). This observation was also verified for P. aeruginosa PA7, which possesses one constitutive T2SS, the Xcp system, and two additional T2SSs, the Hxc and Txc systems, which are subject to specific regulation. The reason why T2SS multiplicity has been favored over the extension of the general T2SS to additional substrates remains unclear. In a previous study, we presented evidence that the Hxc T2SS of P. aeruginosa was present to fulfill specific functions that are incompatible with the general Xcp T2SS (18). In this case, we propose that the Txc T2SS provides specific properties required for the proper secretion of CbpE, which, for unknown reasons, cannot be secreted by the general Xcp system. It is possible that the remaining nonaffiliated gene of RGP69, PSPA7_1418, located between cbpE and the first gene (txcP) of the T2SS cluster (Fig. 1), plays a key role in Txc T2SS specificity. In support of this assumption, homologs of PSPA7_1418 have been recovered in other T2SSs involved in chitin-binding secretion. This is the case for the two other Pseudomonas Txc systems, found in P. fluorescens (52) and P. putida (51), but also for the Yts1 T2SS of Y. enterocolitica (22). In conclusion, we propose that, when possible, compatible T2SS-secreted exoproteins are exported by using the same general machine. This is illustrated by the P. aeruginosa Xcp T2SS, which secretes >12 different exoproteins. However, when T2SS substrates have specific, incompatible constraints, a dedicated machine is developed by the bacteria. The identification of a third nonconstitutive T2SS, the Txc system, in P. aeruginosa suggests that the CbpE protein secreted by this system may be incompatible with the two existing T2SSs. Comparison of this protein with the Xcp-compatible substrate CbpD (Fig. 5a) revealed sufficient differences justifying such specificities. An understanding of the nature of subtle differences that target specific substrates to their cognate secretion machineries will certainly provide key information on this still mysterious protein secretion mechanism.
ACKNOWLEDGMENTS
F.C. is funded by a Ph.D. grant from the French government.
We are grateful to Steve Lory, Christophe Bordi, Sophie Bleves, and Bérengère Ize for fruitful discussions and Ben Field for English corrections. Plasmids pT18-Pal and pT25-TolB were kindly provided by Emmanuelle Bouveret.
Footnotes
Published ahead of print 18 April 2014
REFERENCES
- 1.Driscoll JA, Brody SL, Kollef MH. 2007. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67:351–368. 10.2165/00003495-200767030-00003 [DOI] [PubMed] [Google Scholar]
- 2.Kipnis E, Sawa T, Wiener-Kronish J. 2006. Targeting mechanisms of Pseudomonas aeruginosa pathogenesis. Med. Mal. Infect. 36:78–91. 10.1016/j.medmal.2005.10.007 [DOI] [PubMed] [Google Scholar]
- 3.Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. 2012. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 76:46–65. 10.1128/MMBR.05007-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigue A, Quentin Y, Lazdunski A, Mejean V, Foglino M. 2000. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol. 8:498–504. 10.1016/S0966-842X(00)01833-3 [DOI] [PubMed] [Google Scholar]
- 5.Bleves S, Viarre V, Salacha R, Michel GP, Filloux A, Voulhoux R. 2010. Protein secretion systems in Pseudomonas aeruginosa: a wealth of pathogenic weapons. Int. J. Med. Microbiol. 300:534–543. 10.1016/j.ijmm.2010.08.005 [DOI] [PubMed] [Google Scholar]
- 6.Hirst TR, Holmgren J. 1987. Conformation of protein secreted across bacterial outer membranes: a study of enterotoxin translocation from Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 84:7418–7422. 10.1073/pnas.84.21.7418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardie KR, Schulze A, Parker MW, Buckley JT. 1995. Vibrio spp. secrete proaerolysin as a folded dimer without the need for disulphide bond formation. Mol. Microbiol. 17:1035–1044. 10.1111/j.1365-2958.1995.mmi_17061035.x [DOI] [PubMed] [Google Scholar]
- 8.Douzi B, Filloux A, Voulhoux R. 2012. On the path to uncover the bacterial type II secretion system. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367:1059–1072. 10.1098/rstb.2011.0204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pugsley AP, Possot O. 1993. The general secretory pathway of Klebsiella oxytoca: no evidence for relocalization or assembly of pilin-like PulG protein into a multiprotein complex. Mol. Microbiol. 10:665–674. 10.1111/j.1365-2958.1993.tb00938.x [DOI] [PubMed] [Google Scholar]
- 10.Voulhoux R, Ball G, Ize B, Vasil ML, Lazdunski A, Wu LF, Filloux A. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 20:6735–6741. 10.1093/emboj/20.23.6735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauvonnet N, Pugsley AP. 1998. The requirement for DsbA in pullulanase secretion is independent of disulphide bond formation in the enzyme. Mol. Microbiol. 27:661–667. 10.1046/j.1365-2958.1998.00722.x [DOI] [PubMed] [Google Scholar]
- 12.Chen YL, Hu NT. 2013. Function-related positioning of the type II secretion ATPase of Xanthomonas campestris pv. campestris. PLoS One 8:e59123. 10.1371/journal.pone.0059123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durand E, Bernadac A, Ball G, Lazdunski A, Sturgis JN, Filloux A. 2003. Type II protein secretion in Pseudomonas aeruginosa: the pseudopilus is a multifibrillar and adhesive structure. J. Bacteriol. 185:2749–2758. 10.1128/JB.185.9.2749-2758.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douzi B, Ball G, Cambillau C, Tegoni M, Voulhoux R. 2011. Deciphering the Xcp Pseudomonas aeruginosa type II secretion machinery through multiple interactions with substrates. J. Biol. Chem. 286:40792–40801. 10.1074/jbc.M111.294843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chapon-Herve V, Akrim M, Latifi A, Williams P, Lazdunski A, Bally M. 1997. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol. Microbiol. 24:1169–1178. 10.1046/j.1365-2958.1997.4271794.x [DOI] [PubMed] [Google Scholar]
- 16.Ball G, Durand E, Lazdunski A, Filloux A. 2002. A novel type II secretion system in Pseudomonas aeruginosa. Mol. Microbiol. 43:475–485. 10.1046/j.1365-2958.2002.02759.x [DOI] [PubMed] [Google Scholar]
- 17.Ball G, Viarre V, Garvis S, Voulhoux R, Filloux A. 2012. Type II-dependent secretion of a Pseudomonas aeruginosa DING protein. Res. Microbiol. 163:457–469. 10.1016/j.resmic.2012.07.007 [DOI] [PubMed] [Google Scholar]
- 18.Durand E, Alphonse S, Brochier-Armanet C, Ball G, Douzi B, Filloux A, Bernard C, Voulhoux R. 2011. The assembly mode of the pseudopilus: a hallmark to distinguish a novel secretion system subtype. J. Biol. Chem. 286:24407–24416. 10.1074/jbc.M111.234278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaborina O, Holbrook C, Chen Y, Long J, Zaborin A, Morozova I, Fernandez H, Wang Y, Turner JR, Alverdy JC. 2008. Structure-function aspects of PstS in multi-drug-resistant Pseudomonas aeruginosa. PLoS Pathog. 4:e43. 10.1371/journal.ppat.0040043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strozen TG, Li G, Howard SP. 2012. YghG (GspSbeta) is a novel pilot protein required for localization of the GspSbeta type II secretion system secretin of enterotoxigenic Escherichia coli. Infect. Immun. 80:2608–2622. 10.1128/IAI.06394-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szczesny R, Jordan M, Schramm C, Schulz S, Cogez V, Bonas U, Buttner D. 2010. Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv vesicatoria. New Phytol. 187:983–1002. 10.1111/j.1469-8137.2010.03312.x [DOI] [PubMed] [Google Scholar]
- 22.Shutinoski B, Schmidt MA, Heusipp G. 2010. Transcriptional regulation of the Yts1 type II secretion system of Yersinia enterocolitica and identification of secretion substrates. Mol. Microbiol. 75:676–691. 10.1111/j.1365-2958.2009.06998.x [DOI] [PubMed] [Google Scholar]
- 23.Karaba SM, White RC, Cianciotto NP. 2013. Stenotrophomonas maltophilia encodes a type II protein secretion system that promotes detrimental effects on lung epithelial cells. Infect. Immun. 81:3210–3219. 10.1128/IAI.00546-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrandez Y, Condemine G. 2008. Novel mechanism of outer membrane targeting of proteins in Gram-negative bacteria. Mol. Microbiol. 69:1349–1357. 10.1111/j.1365-2958.2008.06366.x [DOI] [PubMed] [Google Scholar]
- 25.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39:D596–D600. 10.1093/nar/gkq869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy PH, Tetu SG, Larouche A, Elbourne L, Tremblay S, Ren Q, Dodson R, Harkins D, Shay R, Watkins K, Mahamoud Y, Paulsen IT. 2010. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS One 5:e8842. 10.1371/journal.pone.0008842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita Y, Tomida J, Kawamura Y. 2012. Primary mechanisms mediating aminoglycoside resistance in the multidrug-resistant Pseudomonas aeruginosa clinical isolate PA7. Microbiology 158:1071–1083. 10.1099/mic.0.054320-0 [DOI] [PubMed] [Google Scholar]
- 28.Herrero M, de Lorenzo V, Timmis KN. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sana T, Laubier A, Bleves S. 2014. Gene transfer: conjugation. In Filloux A, Ramos J-L. (ed), Pseudomonas methods and protocols, in press Springer, New York, NY: [DOI] [PubMed] [Google Scholar]
- 30.Hoang TT, Kutchma AJ, Becher A, Schweizer HP. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59–72. 10.1006/plas.1999.1441 [DOI] [PubMed] [Google Scholar]
- 31.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- 32.Miller JH. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33.Wolfgang MC, Kulasekara BR, Liang X, Boyd D, Wu K, Yang Q, Miyada CG, Lory S. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:8484–8489. 10.1073/pnas.0832438100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ball G, Filloux A, Voulhoux R. 2014. A method to capture large DNA fragments from genomic DNA. In Filloux A, Ramos J-L. (ed), Pseudomonas methods and protocols, in press Springer, New York, NY: [DOI] [PubMed] [Google Scholar]
- 35.Battesti A, Bouveret E. 2012. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58:325–334. 10.1016/j.ymeth.2012.07.018 [DOI] [PubMed] [Google Scholar]
- 36.Folders J, Tommassen J, van Loon LC, Bitter W. 2000. Identification of a chitin-binding protein secreted by Pseudomonas aeruginosa. J. Bacteriol. 182:1257–1263. 10.1128/JB.182.5.1257-1263.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karimova G, Ullmann A, Ladant D. 2001. Protein-protein interaction between Bacillus stearothermophilus tyrosyl-tRNA synthetase subdomains revealed by a bacterial two-hybrid system. J. Mol. Microbiol. Biotechnol. 3:73–82 http://www.horizonpress.com/backlist/jmmb/v/v3/v3n1/07.pdf [PubMed] [Google Scholar]
- 38.Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55:368–380. 10.1111/j.1365-2958.2004.04402.x [DOI] [PubMed] [Google Scholar]
- 39.Sivaneson M, Mikkelsen H, Ventre I, Bordi C, Filloux A. 2011. Two-component regulatory systems in Pseudomonas aeruginosa: an intricate network mediating fimbrial and efflux pump gene expression. Mol. Microbiol. 79:1353–1366. 10.1111/j.1365-2958.2010.07527.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. U. S. A. 103:171–176. 10.1073/pnas.0507407103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uhl MA, Miller JF. 1994. Autophosphorylation and phosphotransfer in the Bordetella pertussis BvgAS signal transduction cascade. Proc. Natl. Acad. Sci. U. S. A. 91:1163–1167. 10.1073/pnas.91.3.1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aiba H, Nakasai F, Mizushima S, Mizuno T. 1989. Evidence for the physiological importance of the phosphotransfer between the two regulatory components, EnvZ and OmpR, in osmoregulation in Escherichia coli. J. Biol. Chem. 264:14090–14094 [PubMed] [Google Scholar]
- 43.Folders J, Algra J, Roelofs MS, van Loon LC, Tommassen J, Bitter W. 2001. Characterization of Pseudomonas aeruginosa chitinase, a gradually secreted protein. J. Bacteriol. 183:7044–7052. 10.1128/JB.183.24.7044-7052.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhowmick R, Ghosal A, Das B, Koley H, Saha DR, Ganguly S, Nandy RK, Bhadra RK, Chatterjee NS. 2008. Intestinal adherence of Vibrio cholerae involves a coordinated interaction between colonization factor GbpA and mucin. Infect. Immun. 76:4968–4977. 10.1128/IAI.01615-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi KH, Seo JY, Park KM, Park CS, Cha J. 2009. Characterization of glycosyl hydrolase family 3 beta-N-acetylglucosaminidases from Thermotoga maritima and Thermotoga neapolitana. J. Biosci. Bioeng. 108:455–459. 10.1016/j.jbiosc.2009.06.003 [DOI] [PubMed] [Google Scholar]
- 46.Izano EA, Amarante MA, Kher WB, Kaplan JB. 2008. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74:470–476. 10.1128/AEM.02073-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirn TJ, Jude BA, Taylor RK. 2005. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438:863–866. 10.1038/nature04249 [DOI] [PubMed] [Google Scholar]
- 48.Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. 2004. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U. S. A. 101:2524–2529. 10.1073/pnas.0308707101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong E, Vaaje-Kolstad G, Ghosh A, Hurtado-Guerrero R, Konarev PV, Ibrahim AF, Svergun DI, Eijsink VG, Chatterjee NS, van Aalten DM. 2012. The Vibrio cholerae colonization factor GbpA possesses a modular structure that governs binding to different host surfaces. PLoS Pathog. 8:e1002373. 10.1371/journal.ppat.1002373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. 2013. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41:D348–D352. 10.1093/nar/gks1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Tils D, Bladel I, Schmidt MA, Heusipp G. 2012. Type II secretion in Yersinia—a secretion system for pathogenicity and environmental fitness. Front. Cell. Infect. Microbiol. 2:160. 10.3389/fcimb.2012.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Redondo-Nieto M, Barret M, Morrissey J, Germaine K, Martinez-Granero F, Barahona E, Navazo A, Sanchez-Contreras M, Moynihan JA, Muriel C, Dowling D, O'Gara F, Martin M, Rivilla R. 2013. Genome sequence reveals that Pseudomonas fluorescens F113 possesses a large and diverse array of systems for rhizosphere function and host interaction. BMC Genomics 14:54. 10.1186/1471-2164-14-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DebRoy S, Dao J, Soderberg M, Rossier O, Cianciotto NP. 2006. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc. Natl. Acad. Sci. U. S. A. 103:19146–19151. 10.1073/pnas.0608279103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Connell TD, Metzger DJ, Lynch J, Folster JP. 1998. Endochitinase is transported to the extracellular milieu by the eps-encoded general secretory pathway of Vibrio cholerae. J. Bacteriol. 180:5591–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Francetic O, Belin D, Badaut C, Pugsley AP. 2000. Expression of the endogenous type II secretion pathway in Escherichia coli leads to chitinase secretion. EMBO J. 19:6697–6703. 10.1093/emboj/19.24.6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li S, Zhang L, Yao Q, Li L, Dong N, Rong J, Gao W, Ding X, Sun L, Chen X, Chen S, Shao F. 2013. Pathogen blocks host death receptor signalling by arginine GlcNAcylation of death domains. Nature 501:242–246. 10.1038/nature12436 [DOI] [PubMed] [Google Scholar]
- 57.Pearson JS, Giogha C, Ong SY, Kennedy CL, Kelly M, Robinson KS, Lung TW, Mansell A, Riedmaier P, Oates CV, Zaid A, Muhlen S, Crepin VF, Marches O, Ang CS, Williamson NA, O'Reilly LA, Bankovacki A, Nachbur U, Infusini G, Webb AI, Silke J, Strasser A, Frankel G, Hartland EL. 2013. A type III effector antagonizes death receptor signalling during bacterial gut infection. Nature 501:247–251. 10.1038/nature12524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gooderham WJ, Hancock RE. 2009. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 33:279–294. 10.1111/j.1574-6976.2008.00135.x [DOI] [PubMed] [Google Scholar]
- 59.Bally M, Filloux A, Akrim M, Ball G, Lazdunski A, Tommassen J. 1992. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol. Microbiol. 6:1121–1131. 10.1111/j.1365-2958.1992.tb01550.x [DOI] [PubMed] [Google Scholar]
- 60.Pugsley AP. 1993. Processing and methylation of PuIG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol. Microbiol. 9:295–308. 10.1111/j.1365-2958.1993.tb01691.x [DOI] [PubMed] [Google Scholar]
- 61.Bleves S, Voulhoux R, Michel G, Lazdunski A, Tommassen J, Filloux A. 1998. The secretion apparatus of Pseudomonas aeruginosa: identification of a fifth pseudopilin, XcpX (GspK family). Mol. Microbiol. 27:31–40. 10.1046/j.1365-2958.1998.00653.x [DOI] [PubMed] [Google Scholar]
- 62.Studier FW, Moffatt BA. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113–130. 10.1016/0022-2836(86)90385-2 [DOI] [PubMed] [Google Scholar]
- 63.Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652. 10.1073/pnas.76.4.1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. 10.1016/0378-1119(95)00584-1 [DOI] [PubMed] [Google Scholar]
- 65.Tabor S, Richardson CC. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. U. S. A. 82:1074–1078. 10.1073/pnas.82.4.1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaniga K, Delor I, Cornelis GR. 1991. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene 109:137–141. 10.1016/0378-1119(91)90599-7 [DOI] [PubMed] [Google Scholar]
- 67.Prentki P, Krisch HM. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303–313. 10.1016/0378-1119(84)90059-3 [DOI] [PubMed] [Google Scholar]
- 68.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. 2006. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics Chapter 5:Unit 5.6. 10.1002/0471250953.bi0506s15 [DOI] [PMC free article] [PubMed] [Google Scholar]