Abstract
UDP-glucose pyrophosphorylase synthesizes UDP-glucose from UTP and glucose 1-phosphate and exists in almost all species. Most bacteria possess a GalU-type UDP-glucose pyrophosphorylase, whereas many cyanobacteria species do not. In certain cyanobacteria, UDP-glucose is used as a substrate for synthesis of exopolysaccharide cellulose in spite of the absence of GalU-type UDP-glucose pyrophosphorylase. Therefore, there should be an uncharacterized UDP-glucose pyrophosphorylase in cyanobacteria. Here, we show that all cyanobacteria possess a non-GalU-type bacterial UDP-glucose pyrophosphorylase, i.e., CugP, a novel family in the nucleotide triphosphate transferase superfamily. The expressed recombinant Synechocystis sp. strain PCC 6803 CugP had pyrophosphorylase activity that was highly specific for UTP and glucose 1-phosphate. The fact that the CugP gene cannot be deleted completely in Synechocystis sp. PCC 6803 suggests its central role as the substrate supplier for galactolipid synthesis. Galactolipids are major constituents of the photosynthetic thylakoid membrane and important for photosynthetic activity. Based on phylogenetic analysis, this CugP-type UDP-glucose pyrophosphorylase may have recently been horizontally transferred to certain noncyanobacteria.
INTRODUCTION
Glycosylation, which is catalyzed by various types of glycosyl transferases, is important for the biosynthesis of many biological molecules, e.g., polysaccharides, glycoproteins, and glycolipids. Nucleotide diphosphate (NDP)-sugars, which are the substrates for glycosylation, are mostly synthesized by nucleotidyl transferases that use a nucleotide triphosphate (NTP) and a sugar 1-phosphate (sugar-1P) as substrates. Various NDPs and sugars are combined into NDP-sugars in vivo. Of these, UDP-glucose (UDP-Glc) is used for the biosynthesis of cellulose (1) and glycogen (2), galactose metabolism (3), and addition of the glucose moiety in heteroglycans, glycoproteins (4–6), and glycolipids (7). Other NDP-sugars are utilized in the biosynthesis of other polysaccharides and in various glycosylation reactions.
UDP-Glc and the by-product inorganic pyrophosphate (PPi) are produced from UTP and Glc-1P by the UDP-Glc pyrophosphorylases (PPases) (EC 2.7.7.9) in the reversible reaction UTP + Glc-1P ↔ UDP-Glc + PPi. This enzyme is ubiquitous although its typical bacterial form, the GalU-type, is widely distributed throughout Bacteria including Proteobacteria (8), Firmicutes (9), and Actinobacteria (10, 11) and is distinct from its eukaryotic enzyme in amino acid sequence and three-dimensional structure (12–14). The best-known bacterial UDP-Glc PPase is GalU from Escherichia coli (15, 16). The crystal structures and the reaction mechanism of various GalU-type homologs have been elucidated (17–19). These bacterial GalU-type UDP-Glc PPases belong to a large superfamily of nucleotidyl transferases that also includes UDP-N-acetylglucosamine (GlcNAc) PPase, ADP-Glc PPase, CDP-Glc PPase, dTDP-Glc PPase, GDP-mannose (GDP-Man) PPase, and some yet uncharacterized enzymes. These enzymes probably diverged from a common ancestor in evolution (20).
There are many Glc-containing polysaccharides, such as glycogen and Glc-enriched exopolysaccharides, in cyanobacteria (21). Certain cyanobacteria also accumulate cellulose (β-1,4-glucan) (22, 23). Consistently, cyanobacterial genomes have been shown to encode many putative nucleotidyl transferases that might supply NDP-sugars (24, 25). Recently the GalU-type protein All3274 from the filamentous cyanobacterium Anabaena sp. strain PCC 7120 was biochemically revealed to be a UDP-Glc PPase (20). However, such a GalU-type enzyme is not present in all cyanobacteria. Specifically, although Thermosynechococcus is reported to possess a cellulose synthase to produce cellulose under conditions of light and low temperature (23), a GalU-type gene that could supply UDP-Glc to this cellulose has not been found in its genome (24). Given these observations, we hypothesized that an uncharacterized non-GalU-type UDP-Glc PPase may exist in such cyanobacteria. To test this hypothesis, we first focused on a common sequence feature that is shared only by GalU-type UDP-Glc PPases, UDP-GlcNAc PPases, and a putative enzyme family that is present in every species of cyanobacteria. Here, we report that the members of this previously uncharacterized family are cyanobacterial UDP-Glc PPases (thus denoted CugP).
MATERIALS AND METHODS
Sequence alignment and phylogram.
The sequences of the cyanobacterial proteins categorized as belonging to the bacterial nucleotidyl transferase superfamily (Pfam PF00483, NTP_transferase) (http://pfam.sanger.ac.uk) were obtained from NCBI (http://www.ncbi.nlm.nih.gov/) and CyanoBase (http://genome.microbedb.jp/cyanobase) databases. Sequence alignment was performed by the neighbor-joining method using Clustal X (26).
Cloning of sll1558 and single-amino acid substitution.
sll1558 from Synechocystis sp. strain PCC 6803, which encodes the hypothetical UDP-Glc PPase Sll1558 was PCR amplified using PrimeSTAR Max DNA Polymerase (TaKaRa Bio, Otsu, Japan) and then directly cloned into the expression vector pET-28a(+) (Merck, Darmstadt, Germany) using In-Fusion HD Cloning kit reagents (TaKaRa) to produce sll1558-pET28a. The primers were pET28a-1F (5′-GGA TCC GAA TTC GAG CTC CGT C-3′), pET28a-2R (5′-CAT ATG GCT GCC GCG CGG CAC-3′), sll1558-1F28a (5′-CGC GGC AGC CAT ATG AAA GCC ATG ATT TTG GCC-3′), and sll1558-2R28a (5′-CTC GAA TTC GGA TCC TTA TTC CGG CTG GAG AAG-3′). To generate a single amino acid substitution within the Sll1558 protein product, the plasmid was amplified by two sets of primers (sll1558-3FA8G, 5′-GCC GGT GGC AAG GGC ACT CGG GTC AGA CCA ATC-3′; sll1558-4RA8G, 5′-GCC CTT GCC ACC GGC CAA AAT CAT GGC TTT CAT-3′) and self-combined using the aforementioned cloning reagents. These proteins were His tagged from the pET28a vector. The resulting DNA constructs were verified by dideoxy sequencing. These proteins were N-terminally His tagged, derived from the pET28a vector.
Expression, purification, and SDS-PAGE analysis of recombinant proteins.
E. coli strain C41 (DE3) carrying sll1558-pET28a or sll1558(A8G)-pET28a, which carries the sll1558 gene encoding the amino acid change A to G at residue 8, was cultured in 1 liter of LB medium at 37°C. When each culture reached an optical density at 600 nm (OD600) of 0.4 to 0.8, 1 mM isopropyl β-d-1-thiogalactopyranoside was added (final concentration, 100 μM), and the cells were grown at 18°C for 16 h to achieve induction. The cells from each culture were then collected by centrifugation at 4,220 × g for 15 min, suspended in 20 mM HEPES (pH 7.5) containing 100 mM NaCl and 10% (wt/vol) glycerol, and homogenized three times with a French press at 1,500 kg/cm−2. The soluble proteins were collected by centrifugation at 50,000 × g for 30 min. His-tagged Sll1558 and Sll1558 with the A8G substitution [Sll1558(A8G)] were purified by Ni-affinity column chromatography (HisTrap HP; GE Healthcare, Little Chalfont, United Kingdom), with the eluent consisting of a gradient of 30 to 430 mM imidazole in the aforementioned buffer system. Proteins in each fraction were subjected to SDS-PAGE, followed by Coomassie brilliant blue R-250 staining. Low-molecular-mass calibration kit standards (GE Healthcare) served as the molecular mass markers. The fraction enriched in each targeted protein was dialyzed against the aforementioned buffer to remove the imidazole. Protein concentration was assayed using the Bradford method (Bio-Rad, Hercules, CA) with bovine serum albumin as the standard.
Nucleotidyl transferase activity assay.
Nucleotidyl transferase activities were assayed at 37°C in the direction of NDP-sugar formation from NTP and sugar-1P. The reaction rate was determined as the change in the PPi concentration with time and measured using an EnzChek Pyrophosphate Assay kit (Molecular Probes, Life Technologies, Carlsbad, CA). The basic reaction medium contained 50 mM Tris-HCl (pH 7.5), 8 mM MgCl2, 200 μM 2-amino-6-mercapto-7-methylpurine ribonucleoside, 1 U of purine nucleoside phosphorylase, 30 mU of inorganic pyrophosphatase, and substrates. To reduce contaminating inorganic phosphate found in the substrate preparations, each reaction solution was preincubated for 20 min at 22°C and for 10 min at 37°C prior to the addition of the protein. For measurements of substrate specificity, all substrates were included in the reaction mixtures at a 200 μM concentration, and the purified enzymes were included at 30 μg ml−1. For measurements of kinetics, purified enzymes were included in the reaction mixtures at 4.5 to 18.0 μg ml−1.
Structure prediction based on previous studies.
The available Protein Data Bank (PDB) data of GalU-type UDP-Glc PPase (PDB 2PA4) (18), CDP-Glc PPase (PDB 1TZF) (27), and dTDP-Glc PPase (PDB 1H5T) (28) were used to compare the structures and substrate-binding regions of Sll1558.
Gene disruption and mutant analysis.
The sll1558 gene was inactivated by replacement with the chloramphenicol resistance cassette in a pPCRscript-derived DNA by the In-Fusion method using primers sll1558-5F (5′-CCG GGG GAT CCG CCC GAT GCG GAG CTG TGG GAC-3′), sll1558-6R (5′-CCG GAC ATC AGC GCT CAA CAA TTT TCC TCA AAC-3′), sll1558-7F (5′-ACG GTT AGC AGG CCT TCA CAG AAG AGA ACT GGG-3′), and sll1558-8R (5′-CGG CCG CTC TAG CCC GCA GGG CCT TTT CCC GTA-3′). Mutants were generated by natural transformation of Synechocystis cells with this DNA and selected on BG11 plates containing 20 μg m1−1 chloramphenicol. The segregation was examined by PCR with primers sll1558-9F (5′-GTG TGA TTG AGT TTG AGG-3′) and sll1558-10R (5′-TTT CCC CCA GTT CTC TTC-3′).
Chlorophyll (Chl) content was determined after extraction with 100% methanol as described previously (29). Total photosynthetic activity of cells was measured as oxygen evolution supported by 0.2 mM Na2CO3 in BG11 medium using a Clark-type electrode (Oxygraph, Hansatech, United Kingdom) at a chlorophyll concentration of 2.5 μg ml−1 (30).
RESULTS AND DISCUSSION
Sequence features of nucleotidyl transferases.
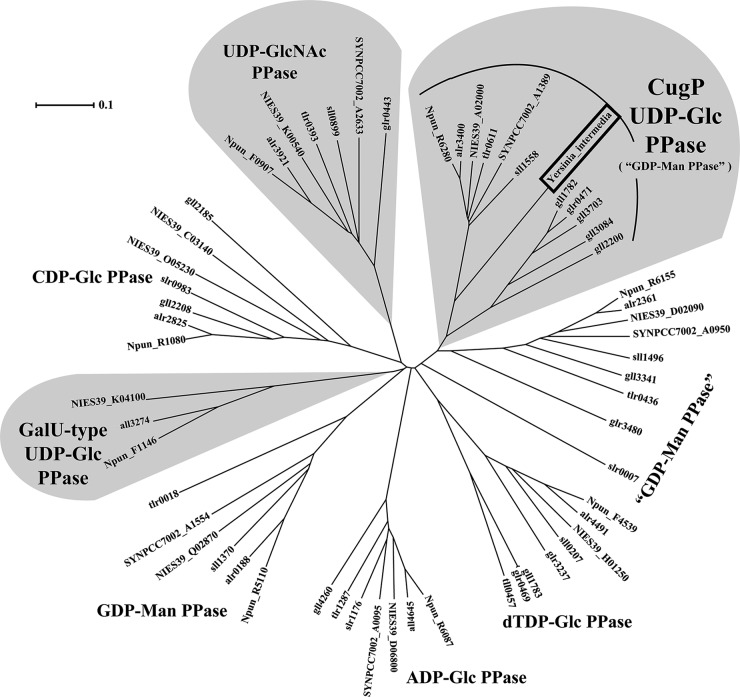
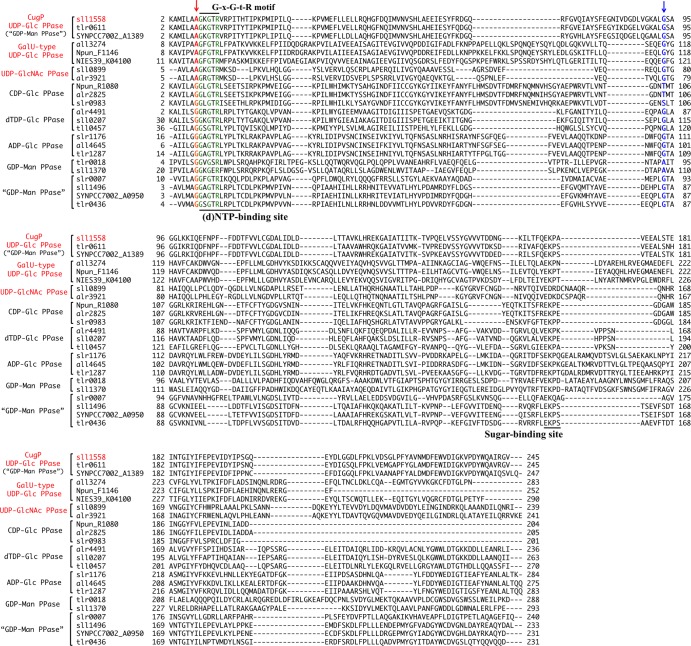
The phylogeny of the nucleotidyl transferase superfamily (protein family [Pfam], NTP_transferase) in cyanobacterial species was analyzed based on alignment of full-length amino acid sequences of the transferases (Fig. 1). It is of note that GalU-type UDP-Glc PPase is found in only a few species (Anabaena sp. PCC 7120, Arthrospira platensis NIES-39, and Nostoc punctiforme ATCC 29133). On the other hand, many other enzymes such as dTDP-Glc PPase, CDP-Glc PPase, ADP-Glc PPase, UDP-GlcNAc PPase, and as yet uncharacterized nucleotidyl transferases are found in most species of cyanobacteria. We searched for common residues in the GalU-type UDP-Glc PPase and UDP-GlcNAc PPase sequences. As reported previously, the G-X-G-T-R motif is highly conserved in all sequences as a part of the (d)NTP-binding site. We found that one residue just prior to the motif is characteristically conserved: Ala is present in GalU-type UDP-Glc PPase, UDP-GlcNAc PPase, and one uncharacterized family (provisionally annotated GDP-Man PPase), whereas Gly is present in all of the others (Fig. 2). Since this residue was the only candidate that might be responsible for substrate discrimination, we expressed and characterized the activities of Sll1558, the putative Synechocystis sp. PCC 6803 GDP-Man PPase, and its Ala-8-Gly mutant Sll1558(A8G), which we chose because that cyanobacterium is a well-studied model for cyanobacteria in general.
FIG 1.
Phylogram and sequence alignment of cyanobacterial nucleotidyl transferases. The phylogram of the nucleotidyl transferases from Synechocystis sp. PCC 6803, Anabaena sp. PCC 7120, Arthrospira platensis NIES-39, Synechococcus sp. PCC 7002, Thermosynechococcus elongatus BP-1, Nostoc punctiforme ATCC 29133, and Gloeobacter violaceus PCC 7421 and the proteobacterium Yersinia intermedia (boxed) nucleotidyl transferase sequence. Families of UDP-Glc PPase and UDP-GlcNAc PPase are highlighted with a gray background.
FIG 2.
Sequence alignment of the NTP transferase domain of the nucleotidyl transferases shown in Fig. 1. The highly conserved residues of the G-X-G-T-R motif are indicated in green. The residues that correspond to position 8 in Sll1558 are highlighted in red (Ala) and orange (Gly). The residues that are aligned with Gly-93 in Sll1558 are in blue. Gly-93 in Sll1558 corresponds to Gly-117 in the GalU-type UDP-Glc PPase from Corynebacterium glutamicum, as shown in Fig. S2 in the supplemental material.
Purified Sll1558 has a specific UDP-Glc PPase activity.
We expressed Sll1558 as a His-tagged protein, which we purified by nickel-affinity chromatography (see Fig. S1 in the supplemental material). According to our SDS-PAGE study, the molecular mass of Sll1558 is that predicted by the translated sequence of sll1558 (45 kDa). The enzyme activity of this product was measured through the liberation of a by-product PPi using a combination of various NTPs (UTP, dTTP, CTP, ATP, and GTP) and sugar-1Ps (Glc-1P, GlcNAc-1P, Gal-1P, and Man-1P). We found that Sll1558 showed activity only in the presence of the combination of UTP and Glc-1P. Notably, it showed practically no activity toward any other substrates or combinations, including Man-1P, even though Sll1558 and related homologs had provisionally been considered to be GDP-Man PPases, as annotated in CyanoBase. dTTP does not substitute for UTP in reactions with Sll1558, a finding that contrasts with the ability of dTTP to act as a substrate of the GalU-type UDP-Glc PPase from Xanthomonas campestris (8) and Sphingomonas elodea ATCC 31461 (31).
Activity and substrate affinity comparison between Sll1558 and Sll1558(A8G).
To further examine our prediction about nucleoside discrimination at the Ala residue, we evaluated the possible role of the Ala-8 residue of Sll1558 by site-directed replacement of this residue with Gly. After purification, the substrate specificity and enzyme kinetics of Sll1558(A8G) were measured as described above for Sll1558. Sll1558(A8G) also exhibited the PPase activity only for a combination of UTP and Glc-1P, as with wild-type Sll1558.
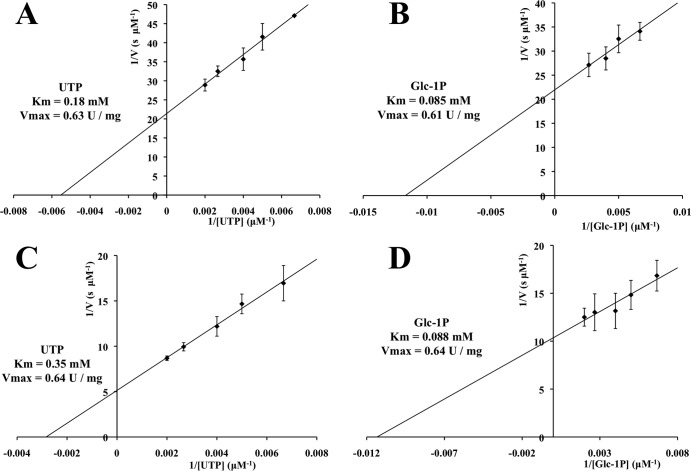
According to Lineweaver-Burk plots (Fig. 3), the Km for UTP and Km for Glc-1P for Sll1558(A8G) were 0.35 mM and 0.088 mM, and for Sll1558 they were 0.18 mM and 0.085 mM, respectively (Table 1). Thus, the Km for UTP was found to be approximately 2-fold higher than that of the wild type, and the Km for Glc-1P was comparable to the wild-type level. This finding suggests that the Ala-8 residue plays a slight role in the UTP binding. Notably, the Vmax and kcat values for Sll1558(A8G) were comparable to those of the wild-type enzyme (Table 1). The Km values for Sll1558 were similar to those of GalU-type enzymes from bacteria other than cyanobacteria, whereas the Vmax and kcat were about 60- to 200-fold lower than those of these counterpart enzymes (8–10). On the other hand, the kinetics parameters of Sll1558 were found to be similar to those of All3274, the GalU-type UDP-Glc PPase from Anabaena sp. PCC 7120 (20).
FIG 3.
Lineweaver-Burk plots of the enzyme activity of Sll1558 (A and B) and A8G-Sll1558 (C and D). As a fixed substrate, 1 mM Glc-1P was included in the experiments for panels A and C, or 2 mM UTP was included in the experiments for panels B and D. Protein concentrations: 4.5 μg ml–−1 (A and B), 18.0 μg ml−1 (C), and 9.0 μg ml−1 (D). The error bars indicate standard deviations (for A, B, and C, n = 3; for D, n = 6).
TABLE 1.
Kinetic parameters for Sll1558 and Sll1558(A8G)
| Protein and substrate | Vmax (U mg−1) | Km (mM) | kcat (min−1) | kcat/Km (min−1 mM−1) |
|---|---|---|---|---|
| Sll1558 | ||||
| UTP | 0.63 | 0.18 | 28 | 160 |
| Glc-1P | 0.61 | 0.085 | 27 | 320 |
| Sll1558(A8G) | ||||
| UTP | 0.64 | 0.35 | 29 | 83 |
| Glc-1P | 0.64 | 0.088 | 29 | 330 |
Distribution of the GalU-type and CugP-type enzymes, two types of UDP-Glc PPase.
Herein, we designated Sll1558 and its homologs as CugP-type UDP-Glc PPase after cyanobacterial UDP-Glc PPase. Because all cyanobacterial genomes examined to date encode a CugP-type UDP-Glc PPase, this enzyme appears to be present in all cyanobacteria (Fig. 1), whereas the GalU-type UDP-Glc PPase is present in only certain species (20). As shown in Fig. 1, a phylogram of all related enzymes in the cyanobacterial nucleotidyl transferase superfamily confirmed such a distribution in cyanobacteria. It must be mentioned also that the cyanobacterial species that possess the GalU-type also possess the CugP-type UDP-Glc PPase.
According to the structure of the phylogram, the GalU-type and CugP-type enzyme sequences diverged early on from each other or from other related enzymes during the evolution of the nucleotidyl transferase superfamily. It is thus possible that CugP-type UDP-Glc PPases evolved independently within this superfamily. When a BLAST search was performed for the whole NCBI database, the GalU-type PPases were found to be widely distributed throughout the bacterial kingdom, whereas the CugP-type could be found in only very limited species other than cyanobacteria. These limited species included certain, but not all, of the Beta- and Gammaproteobacteria. The sequence similarity among the cyanobacterial and proteobacterial CugP-type enzymes is much greater (E value, 1e−122 to 1e−147) than that of their GalU-type counterparts (E value, 1e−28 to 1e−32). Notably, the proteobacterial sequences have clearly diverged from the inside of the cyanobacterial CugP cluster (e.g., Yersinia intermedia, Methylotenera mobilis, and Vibrio ichthyoenteri) (Fig. 1), suggesting that the proteobacterial CugP-type enzymes have been acquired by horizontal gene transfer from cyanobacteria rather recently. Conversely, although the GalU-type enzymes were, in general, widely distributed in Bacteria, they were found in only some filamentous cyanobacteria and may, therefore, have also been acquired through another horizontal transfer from bacteria (e.g., Poribacteria and Anaerolinea thermophila) to the limited species of cyanobacteria or may have been lost in many cyanobacterial lineages. We speculate that the GalU-type enzyme may supply UDP-Glc to certain glycosyltransferases specific for cell differentiation in those filamentous cyanobacteria.
Structural implications of the motif for substrate specificity.
According to the crystal structure and sequence similarities, the overall structure of the whole catalytic domain seems to be conserved between GalU-type UDP-Glc PPase, CDP-Glc PPase, dTDP-Glc PPase, and other subfamilies of the nucleotidyl transferases. Because CugP-type UDP-Glc PPase is also included in this superfamily, we discuss structural implications of the motif for substrate specificities based on the known structures. These enzymes bind (d)NTP and sugar-1P sequentially in the substrate-binding pocket, which can be divided into a (d)NTP-binding site and a sugar-binding site (18, 31). In the former site, three highly conserved residues directly interact with the pyrimidine/purine base of (d)NTP by hydrogen bonds (Fig. 4; see also Fig. S2 in the supplemental material). However, these hydrogen bonds are mostly derived from the main-chain amide of an Ala or Gly residue, and, therefore, the specificity determination for (d)NTP has not clearly been elucidated. For example, the amide NH of Gly-117 appears to discriminate uracil of UTP in UDP-Glc PPase from cytosine of CTP (see Fig. S2) but is also conserved in other nucleotidyl transferases. The Ala residue that is highlighted in this study as a possible marker for the UTP-utilizing enzymes directly interacts by hydrogen bonding with the C-2 carbonyl of uracil, but very similar hydrogen bonding is found between the Gly-8 and C-2 carbonyl of cytosine in CDP-Glc PPase (see Fig. S2). Thus, the precise mechanism to determine the specificity for UTP still remains elusive. However, it must be mentioned that the spatial arrangement of the Gly residue (Gly-21/Gly-9/Gly-9 in GalU/CugP/CDP-Glc PPase) next to the mutated Ala position is markedly different, resulting in distinct hydrogen bonding to cytosine in CDP-Glc PPase or to ribose in GalU-type UDP-Glc PPase (Fig. 4; see also Fig. S2 in the supplemental material). This may be the reason why the Km of Sll1558(A8G) for UTP increased slightly. Thus, it is still possible that the highlighted Ala residue, which is exclusively conserved in the UTP-utilizing transferases, is somehow involved in discrimination of the uracil of UTP as a substrate. To test this hypothesis, it will be necessary to study the substrate discrimination of the UTP-utilizing transferases by a thorough mutagenesis survey.
FIG 4.
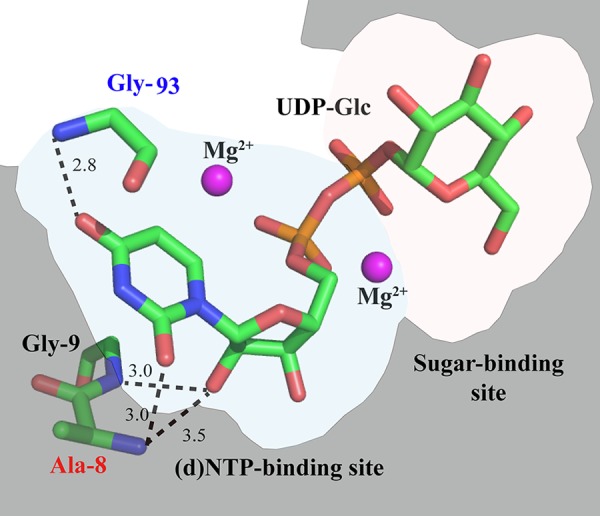
A predicted model of substrate binding in Sll1558 (CugP-type UDP-Glc PPase). This model is based on the crystal structure of GalU-type UDP-Glc PPase from Corynebacterium glutamicum in complex with magnesium and UDP-Glc (2PA4) (18). Only a few key residues and substrates are shown (green, carbon; blue, nitrogen; red, oxygen; and orange, phosphorus). The broken lines denote hypothetical hydrogen bonds.
Role of CugP-type UDP-Glc PPase in cyanobacteria.
To better understand the role of cyanobacterial CugP-type UDP-Glc PPases, we attempted to disrupt sll1558 in the Synechocystis sp. PCC 6803 genome. However, we could not produce such a mutant because cyanobacteria typically contain multiple copies of their chromosomes, and, consequently, the mutated Synechocystis sp. PCC 6803 did not cleanly segregate, suggesting that the CugP-type UDP-Glc is essential for survival of Synechocystis (see Fig. S3 in the supplemental material). In this context, it must be mentioned that a galactolipid, monogalactosyldiacylglycerol (MGDG), which is essential in Synechocystis, is supplied from UDP-Glc via monoglucosyldiacylglycerol (32). The galactolipids MGDG and digalactosyldiacylglycerol are major constituents of the photosynthetic thylakoid membrane in cyanobacteria and in plant chloroplasts. They bind to the membrane-spanning photosynthetic complexes and thereby maintain them in a functional state (33). Hence, the role of UDP-Glc PPases in providing UDP-Glc must be essential for growth and survival in cyanobacteria.
As a preliminary survey, we compared this nonsegregated mutant with the wild type. The chlorophyll (Chl) content was lower in the mutant than in the wild type (mutant, 3.47 ± 0.51 μg Chl/108 cells; wild type, 4.22 ± 0.04 μg Chl/108 cells; n = 3). The whole photosynthetic activity from water to CO2 was higher in the mutant than in the wild type (mutant, 263 ± 40 μmol O2/mg Chl/h; wild type, 138 ± 7 μmol O2/mg Chl/h; n = 4). These facts may reflect modified membrane biogenesis in the partially disrupted mutant (see Fig. S3 in the supplemental material). To further characterize the phenotype, it is essential to achieve the complete segregation of the mutant.
Because galactolipids are specific to photosynthetic membranes, the unique CugP-type enzyme, found mainly in cyanobacteria, may be the result of an evolutionary event in oxygenic photosynthesis. Certain exopolysaccharides secreted by cyanobacteria, including Synechocystis, often contain glucose as the major constituent (21). However, it is unknown if they are essential for survival. Cellulose is selectively deposited in a thermophilic cyanobacterium, Thermosynechococcus, under conditions of low temperature and light (23). So, a CugP-type Tlr0611 appears to be the sole enzyme to provide UDP-Glc for biosynthesis of cellulose and might be induced under such conditions in this cyanobacterium. Regional distribution of GalU-type PPases in certain cyanobacteria may account for variation in the requirement of exopolysaccharides. Further functional studies of the CugP-type and GalU-type enzymes are needed to understand the role of UDP-Glc in cyanobacteria.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Grants-in-Aid for Scientific Research from MEXT and PRESTO from JST (to R.N.) and by Grants-in-Aid for Scientific Research and the GCOE program “From the Earth to Earths” from MEXT and CREST from JST (to M.I.).
Footnotes
Published ahead of print 11 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01591-14.
REFERENCES
- 1.Romling U. 2002. Molecular biology of cellulose production in bacteria. Res. Microbiol. 153:205–212. 10.1016/S0923-2508(02)01316-5 [DOI] [PubMed] [Google Scholar]
- 2.Alonso MD, Lomako J, Lomako WM, Whelan WJ. 1995. A new look at the biogenesis of glycogen. FASEB. J. 9:1126–1137 [DOI] [PubMed] [Google Scholar]
- 3.Holden HM, Rayment I, Thoden JB. 2003. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J. Biol. Chem. 278:43885–43888. 10.1074/jbc.R300025200 [DOI] [PubMed] [Google Scholar]
- 4.Roth J. 1995. Compartmentation of glycoprotein biosynthesis, p 237–312 In Montreuil J, Vliegenthart JFG, Shachter H. (ed), Glycoproteins. Elsevier Science, Amsterdam, The Netherlands [Google Scholar]
- 5.Verbert A. 1995. From Glc3MangGlcNAc2-protein to Man5GlcNAc2-protein: transfer en bloc, p 145–152 In Montreuil J, Vliegenthart JFG, Shachter H. (ed), Glycoproteins. Elsevier Science, Amsterdam, The Netherlands [Google Scholar]
- 6.Silbert JE, Sugumaran G. 1995. Intracellular membranes in the synthesis, transport, and metabolism of proteoglycans. Biochim. Biophys. Acta 1241:371–384. 10.1016/0304-4157(95)00011-9 [DOI] [PubMed] [Google Scholar]
- 7.Sandhoff K, van Echten G, Schroder M, Schnabel D, Suzuki K. 1992. Metabolism of glycolipids: the role of glycolipid-binding proteins in the function and pathobiochemistry of lysosomes. Biochem. Soc. Trans. 20:695–699 [DOI] [PubMed] [Google Scholar]
- 8.Bosco M, Machtey M, Iglesias A, Aleanzi M. 2009. UDPglucose pyrophosphorylase from Xanthomonas spp. Characterization of the enzyme kinetics, structure and inactivation related to oligomeric dissociation. Biochimie 91:204–213. 10.1016/j.biochi.2008.09.001 [DOI] [PubMed] [Google Scholar]
- 9.Ma Z, Fan H-j, Lu C-p. 2011. Molecular cloning and analysis of the UDP-Glucose Pyrophosphorylase in Streptococcus equi subsp. zooepidemicus. Mol. Biol. Rep. 38:2751–2760. 10.1007/s11033-010-0420-8 [DOI] [PubMed] [Google Scholar]
- 10.Lai X, Wu J, Chen S, Zhang X, Wang H. 2008. Expression, purification, and characterization of a functionally active Mycobacterium tuberculosis UDP-glucose pyrophosphorylase. Protein Expr. Purif. 61:50–56. 10.1016/j.pep.2008.05.015 [DOI] [PubMed] [Google Scholar]
- 11.Asencion Diez MD, Peiru S, Demonte AM, Gramajo H, Iglesias AA. 2012. Characterization of recombinant UDP-and ADP-glucose pyrophosphorylases and glycogen synthase to elucidate glucose-1-phosphate partitioning into oligo- and polysaccharides in Streptomyces coelicolor. J. Bacteriol. 194:1485–1493. 10.1128/JB.06377-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mollerach M, Lopez R, Garcia E. 1998. Characterization of the galU gene of Streptococcus pneumoniae encoding a uridine diphosphoglucose pyrophosphorylase: a gene essential for capsular polysaccharide biosynthesis. J. Exp. Med. 188:2047–2056. 10.1084/jem.188.11.2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flores-Diaz M, Alape-Giron A, Persson B, Pollesello P, Moos M, von Eichel-Streiber C, Thelestam M, Florin I. 1997. Cellular UDP-glucose deficiency caused by a single point mutation in the UDP-glucose pyrophosphorylase gene. J. Biol. Chem. 272:23784–23791. 10.1074/jbc.272.38.23784 [DOI] [PubMed] [Google Scholar]
- 14.Mollerach M, Garcia E. 2000. The galU gene of Streptococcus pneumoniae that codes for a UDP-glucose pyrophosphorylase is highly polymorphic and suitable for molecular typing and phylogenetic studies. Gene 260:77–86. 10.1016/S0378-1119(00)00468-6 [DOI] [PubMed] [Google Scholar]
- 15.Weissborn AC, Liu Q, Rumley MK, Kennedy EP. 1994. UTP: alpha-d-glucose-1-phosphate uridylyltransferase of Escherichia coli: isolation and DNA sequence of the galU gene and purification of the enzyme. J. Bacteriol. 176:2611–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hossain SA, Tanizawa K, Kazuta Y, Fukui T. 1994. Overproduction and characterization of recombinant UDP-glucose pyrophosphorylase from Escherichia coli K-12. J. Biochem. 115:965–972 [DOI] [PubMed] [Google Scholar]
- 17.Thoden JB, Holden HM. 2007. The molecular architecture of glucose-1-phosphate uridylyltransferase. Protein Sci. 16:432–440. 10.1110/ps.062626007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thoden JB, Holden HM. 2007. Active site geometry of glucose-1-phosphate uridylyltransferase. Protein Sci. 16:1379–1388. 10.1110/ps.072864707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H, Choi J, Kim T, Lokanath NK, Ha SC, Suh SW, Hwang HY, Kim KK. 2010. Structural basis for the reaction mechanism of UDP-glucose pyrophosphorylase. Mol. Cells 29:397–405. 10.1007/s10059-010-0047-6 [DOI] [PubMed] [Google Scholar]
- 20.Kawano Y, Sekine M, Ihara M. 2013. Identification and characterization of UDP-glucose pyrophosphorylase in cyanobacteria Anabaena sp. PCC 7120. J. Biosci. Bioeng. 117:531–538. 10.1016/j.jbiosc.2013.10.015 [DOI] [PubMed] [Google Scholar]
- 21.De Philippis R, Vincenzini M. 1998. Exocellular polysaccharides from cyanobacteria and their possible applications. FEMS Microbiol. Rev. 22:151–175. 10.1111/j.1574-6976.1998.tb00365.x [DOI] [Google Scholar]
- 22.Nobles DR, Romanovicz DK, Brown RM. 2001. Cellulose in cyanobacteria. Origin of vascular plant cellulose synthase? Plant Physiol. 127:529–542. 10.1104/pp.010557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawano Y, Saotome T, Ochiai Y, Katayama M, Narikawa R, Ikeuchi M. 2011. Cellulose accumulation and a cellulose synthase gene are responsible for cell aggregation in the cyanobacterium Thermosynechococcus vulcanus RKN. Plant Cell Physiol. 52:957–966. 10.1093/pcp/pcr047 [DOI] [PubMed] [Google Scholar]
- 24.Nakamura Y, Kaneko T, Sato S, Ikeuchi M, Katoh H, Sasamoto S, Watanabe A, Iriguchi M, Kawashima K, Kimura T, Kishida Y, Kiyokawa C, Kohara M, Matsumoto M, Matsuno A, Nakazaki N, Shimpo S, Sugimoto M, Takeuchi C, Yamada M, Tabata S. 2002. Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res. 9:135–148. 10.1093/dnares/9.4.135 [DOI] [PubMed] [Google Scholar]
- 25.Kaneko T, Nakamura Y, Wolk CP, Kuritz T, Sasamoto S, Watanabe A, Iriguchi M, Ishikawa A, Kawashima K, Kimura T. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205–213. 10.1093/dnares/8.5.205 [DOI] [PubMed] [Google Scholar]
- 26.Larkin M, Blackshields G, Brown N, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 27.Koropatkin NM, Holden HM. 2004. Molecular structure of alpha-d-glucose-1-phosphate cytidylyltransferase from Salmonella typhi. J. Biol. Chem. 279:44023–44029. 10.1074/jbc.M407755200 [DOI] [PubMed] [Google Scholar]
- 28.Zuccotti S, Zanardi D, Rosano C, Sturla L, Tonetti M, Bolognesi M. 2001. Kinetic and crystallographic analyses support a sequential-ordered bi bi catalytic mechanism for Escherichia coli glucose-1-phosphate thymidylyltransferase. J. Mol. Biol. 313:831–843. 10.1006/jmbi.2001.5073 [DOI] [PubMed] [Google Scholar]
- 29.Midorikawa T, Matsumoto K, Narikawa R, Ikeuchi M. 2009. An Rrf2-type transcriptional regulator is required for expression of psaAB genes in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 151:882–892. 10.1104/pp.109.141390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hihara Y, Sonoike K, Ikeuchi M. 1998. A novel gene, pmgA, specifically regulates photosystem stoichiometry in the cyanobacterium Synechocystis species PCC 6803 in response to high light. Plant Physiol. 117:1205–1216. 10.1104/pp.117.4.1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silva E, Marques AR, Fialho AM, Granja AT, Sá-Correia I. 2005. Proteins encoded by Sphingomonas elodea ATCC 31461 rmlA and ugpG genes, involved in gellan gum biosynthesis, exhibit both dTDP-and UDP-glucose pyrophosphorylase activities. Appl. Environ. Microbiol. 71:4703–4712. 10.1128/AEM.71.8.4703-4712.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Awai K, Kakimoto T, Awai C, Kaneko T, Nakamura Y, Takamiya K-i, Wada H, Ohta H. 2006. Comparative genomic analysis revealed a gene for monoglucosyldiacylglycerol synthase, an enzyme for photosynthetic membrane lipid synthesis in cyanobacteria. Plant Physiol. 141:1120–1127. 10.1104/pp.106.082859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakurai I, Mizusawa N, Wada H, Sato N. 2007. Digalactosyldiacylglycerol is required for stabilization of the oxygen-evolving complex in photosystem II. Plant Physiol. 145:1361–1370. 10.1104/pp.107.106781 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.