Abstract
Streptococcus mutans, a major etiological agent of human dental caries, lives primarily on the tooth surface in biofilms. Limited information is available concerning the extracellular DNA (eDNA) as a scaffolding matrix in S. mutans biofilms. This study demonstrates that S. mutans produces eDNA by multiple avenues, including lysis-independent membrane vesicles. Unlike eDNAs from cell lysis that were abundant and mainly concentrated around broken cells or cell debris with floating open ends, eDNAs produced via the lysis-independent pathway appeared scattered but in a structured network under scanning electron microscopy. Compared to eDNA production of planktonic cultures, eDNA production in 5- and 24-h biofilms was increased by >3- and >1.6-fold, respectively. The addition of DNase I to growth medium significantly reduced biofilm formation. In an in vitro adherence assay, added chromosomal DNA alone had a limited effect on S. mutans adherence to saliva-coated hydroxylapatite beads, but in conjunction with glucans synthesized using purified glucosyltransferase B, the adherence was significantly enhanced. Deletion of sortase A, the transpeptidase that covalently couples multiple surface-associated proteins to the cell wall peptidoglycan, significantly reduced eDNA in both planktonic and biofilm cultures. Sortase A deficiency did not have a significant effect on membrane vesicle production; however, the protein profile of the mutant membrane vesicles was significantly altered, including reduction of adhesin P1 and glucan-binding proteins B and C. Relative to the wild type, deficiency of protein secretion and membrane protein insertion machinery components, including Ffh, YidC1, and YidC2, also caused significant reductions in eDNA.
INTRODUCTION
Streptococcus mutans, a major etiological agent of human dental caries, lives primarily on the tooth surface in biofilms. The ability of S. mutans to bind to and colonize the tooth surface is the result of many cell surface-localized and secreted factors and is significantly influenced by the availability of dietary sucrose. The multifunction adhesin P1 (also called antigen I/II, PAc, or SpaP) mediates sucrose-independent bacterial adherence to the tooth surface via interactions with agglutinin and other glycoproteins in the salivary pellicles (1, 2). Recently, it has been shown that P1, as well as other extracellular proteins, can also form amyloid and that amyloid is present in human dental plaque and in S. mutans biofilms (3). Sortase A (SrtA) is a membrane-localized transpeptidase that cleaves and covalently anchors cell surface-associated proteins containing an LPXTG motif to the peptidoglycan (4). SrtA deficiency in S. mutans results in defects in localization and activity of P1 and other proteins, and it significantly impairs biofilm formation (3, 5–7). In addition, sucrose-dependent adhesion is also integral to the ability of S. mutans to colonize a surface and form biofilms. The organism produces at least three glucosyltransferases (Gtfs; GtfB, GtfC, and GtfD; also known as Gtf-I, Gtf-SI, and Gtf-S) that utilize sucrose to generate adhesive glucose polymers known as glucans or mutans (8–10). These extracellular polymers function as a scaffold to facilitate bacterial adhesion to the tooth surface, promote bacterial cell-cell interactions and community behavior, help to create acidic microenvironments, and maintain integrity and stability of S. mutans biofilms (10–16). All of the Gtfs exhibit some degree of glucan-binding activity. Several additional cell surface-localized, nonenzymatic proteins also bind glucans with high affinity. There are at least four such glucan-binding proteins produced by S. mutans, GbpABCD (12). GbpC contains an LPXTG SrtA anchor motif, and similar to a deficiency in SrtA, a deficiency of GbpC results in impaired biofilm formation (17).
In addition to proteins and polysaccharides, extracellular DNA (eDNA) has also been demonstrated to be a major component of the extracellular matrix of bacterial biofilms (18). In Pseudomonas aeruginosa and several other bacteria, inclusion of DNase I in growth medium or pretreatment of bacterial inocula with DNase I resulted in drastic reduction, even loss of biofilm formation, compared to controls that received the heat-inactivated enzyme (18–20). Treatment of mature biofilms with DNase I also caused a reduction of biovolume/biomass and loss of 3-dimensional structure and mechanical stability of the biofilms (18, 19, 21–24). In Neisseria meningitidis, frequently carried pathogenic strains termed “settlers” were shown to depend on eDNA for biofilm formation, whereas biofilm formation by clonal complexes with low prevalence, termed “spreaders,” was eDNA independent (19).
While programmed cell death and lysis is widely considered the major source of eDNA (25–29), there is evidence that eDNA can also be actively released/secreted independently of cell lysis (18, 30, 31). In Enterococcus faecalis, growth in biofilms triggered eDNA release, resulting in increases of eDNA by more than 3 logs compared to the planktonic counterparts (30). Cell lysis-independent eDNA release is also well documented in Streptococcus gordonii, and such a release was recently shown to be triggered by hydrogen peroxide (21, 22). However, the molecular mechanisms that underlie eDNA release/secretion and eDNA-cellular interactions remain unknown.
Recent studies in S. mutans have shown that eDNA can result from cell death induced by the competence-stimulating/inducing peptides CSP/XIP and significantly influences biofilm formation (20, 32, 33). Several genes of the autolytic pathways, such as lrgAB and cidAB for the bacterial phage holin- and anti-holin-like proteins, respectively, and lytST for a two-component signal transduction system that regulates murein hydrolyase and holin expression, are regulated in response to environmental conditions (28, 34, 35), suggestive of regulation of eDNA release from cell lysis in response to environmental conditions. In an in vitro study using atomic force microscopy (AFM), Das et al. showed that eDNA enhanced S. mutans' adherence to a substratum, with a more significant impact on a hydrophobic surface than on a hydrophilic one (36). Klein et al. (37) also demonstrated that eDNA may be important for the formation and stability of S. mutans biofilms formed in the presence of sucrose and starch. In our current study, high-resolution field emission-scanning electron microscopy (FE-SEM) and functional assays were used to characterize eDNA production and its role in S. mutans adherence and biofilm formation. Our results revealed that in S. mutans, multiple routes exist for eDNA release, including cell lysis, membrane vesicles, and pathways of protein secretion and membrane insertion. In addition, the results further demonstrated that as an integral component of the biofilm matrices, eDNA in S. mutans plays an important role in biofilm formation and maturation.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. mutans UA159, NG8, and their derivatives (Table 1) (38, 39) were maintained in brain heart infusion (BHI; Difco Laboratories). For glucan-binding assays, S. mutans UA159 was grown to mid-exponential phase in ultrafiltered (10-kDa-molecular-mass-cutoff membrane) tryptone-yeast extract broth (UFTYE; pH 7.0) containing 1% (wt/vol) glucose (14). Streptococcus anginosus KSB8, harboring gtfB encoding S. mutans glucosyltransferase B, was grown in Trypticase soy broth (Difco Laboratories) (15). Solid media were prepared similarly, but agar (Difco Laboratories) was added at a concentration of 1.5% (wt/vol). When needed, kanamycin (1 mg/ml), spectinomycin (1 mg/ml), and/or erythromycin (10 μg/ml) were added to the growth medium. Unless stated otherwise, all cultures were grown aerobically in a 37°C chamber containing 5% CO2 under static conditions. For eDNA quantitation during growth under defined conditions, bacteria were grown in the chemically defined medium FMC (40). For biofilm formation, FMC containing glucose (20 mM) (FMCG), sucrose (10 mM) (FMCS), or glucose (18 mM) plus sucrose (2 mM) (FMCGS) was employed. In some cases, semidefined biofilm medium (BM) supplemented with glucose (BMG), sucrose (BMS), or glucose plus sucrose (BMGS) as indicated above was also used (41–43).
TABLE 1.
Bacterial strains used in this study
| Strain | Major property(ies)a | Reference |
|---|---|---|
| S. mutans UA159 | Wild-type, serotype c | 38 |
| S. mutans NG8 | Wild-type, serotype c | 39 |
| S. mutans SAB102 | UA159 derivative, srtA, Kanr | 5 |
| S. mutans 630NP | UA159 derivative, atlA, Kanr | 67 |
| S. mutans PC339 | NG8 derivative, srtA, Ermr | 3 |
| S. mutans SJ271 | SAB102/pDL278::srtA, Kanr, Sper | 5 |
| S. mutans AH374 | NG8 derivative, yidC1, Ermr | 70 |
| S. mutans AH378 | NG8 derivative, yidC2, Kanr | 70 |
| S. mutans AH329 | NG8 derivative, ffh, Ermr | 70 |
| S. mutans AH312 | NG8 derivative, scRNA, Kanr | 70 |
| S. mutans AH307 | NG8 derivative, ftsY, Kanr | 70 |
| S. anginosus KSB8 | S. anginosus harboring gtfB of S. mutans | 15 |
Kanr, Sper, and Ermr indicate kanamycin, spectinomycin, and erythromycin resistance, respectively.
Biofilm formation and microscopic analysis.
Bacterial biofilms were grown in FMC or BM on 96-well plates or hydroxylapatite (HA) discs vertically deposited in 24-well plates (Corning, NY) (42, 44, 45). Biofilm mass on 96-well plates was measured spectrophotometrically following crystal violet staining (42). For analysis using FE-SEM, biofilm samples on HA discs were fixed in 2.5% glutaraldehyde (EM grade) in pH 7.2 phosphate buffer overnight at 4°C, rinsed in sodium phosphate-buffered saline (PBS; 20 mM potassium phosphate buffer, pH 7.2, 0.9% [wt/vol] sodium chloride), dehydrated with increasing concentrations of ethanol at 25%, 50%, 75% twice, 90% twice, and 100% twice, and then subjected to critical point drying in a carbon dioxide critical point drier (Electron Microscopy Sciences, PA). The dried specimens were mounted on an aluminum stub and coated with 10-nm-thick carbon using a Carbon Coater 208C (Cressington Scientific Instruments, Watford, United Kingdom) and analyzed using a Hitachi 480 FE-SEM (Hitachi Ltd., Tokyo, Japan) under an acceleration voltage of 3 kV. Analysis by confocal laser scanning microscopy (CLSM) was carried out similarly to previously described protocols (46, 47).
Measurement of eDNA and extraction of chromosomal DNA.
eDNA in planktonic cultures, and in biofilms in some cases, was measured by spectrofluorometry (30) and spectrophotometry (48, 49). Brief sonication (10 s, twice at 10% energy level, with 2 min on ice between treatments) was used to dechain streptococcal cells or disperse biofilms using a Branson digital sonifier (model S-250D; Branson Ultrasonics Co, CT) (47). Aliquots of sonicated cultures were used to enumerate bacteria by plating in triplicate on BHI agar plates. The remaining cells were spun down by centrifugation at 3,250 × g at 4°C for 15 min and removed using 0.22-μm syringe filters. For fluorometry, cell-free supernatants were treated with Sytox green (Invitrogen), a cell-impermanent dye that stains extracellular nucleic acids with green fluorescence (30). In addition, an AccuBlue high-sensitivity double-stranded DNA (dsDNA) quantitation kit (Biotium, Hayward, CA), which reliably detects dsDNA ranging from 0.2 to 100 ng, was also used by following the manufacturer's instructions. All samples were excited at 485 nm with emission at 535 nm measured using a Synergy HT multiple-mode microplate reader (BioTek). For spectrophotometry, cell-free supernatants were mixed with 2 volumes of absolute ethanol and a 1/10 volume of sodium acetate (3 M, pH 5.2) containing 1 mM EDTA (final concentration) (48, 49). Following precipitation at −80°C overnight, eDNA was collected by centrifugation at 21,130 × g at 4°C for 20 min, washed once with ice-cold 70% ethanol, air dried, and then dissolved in sterile deionized water. The DNA concentration was then analyzed using a NanoDrop 2000 spectrophotometer (Thermo Scientific). The eDNA data were further normalized to colony-forming units (CFU). For chromosomal DNA extraction, S. mutans UA159 was grown in UFTYE (pH 7.0) plus 1% glucose to mid-exponential phase. The cell culture was centrifuged at 5,500 × g, 4°C, for 10 min, and the pellets were used to isolate chromosomal DNA with the MasterPure DNA purification kit (Epicenter Technologies, Madison, WI) as recommended by the manufacturer.
DNase, RNase, proteinase K, dextranase, and formic acid treatment.
Biofilms grown for 24 h on HA discs were treated with proteinase K (Sigma) at 2.5 mg/ml at 37°C for 1 h, DNase I (Promega) at 2 U/μl at 37°C for 1 h, RNase H (Invitrogen) at 2 U/μl at 37°C for 1 h, dextranase (also glucanase, Sigma) at 0.1 mg/ml, pH 6.5, at 4°C overnight, or 88% formic acid at room temperature for 1 h (50). Negative controls received buffer only (for proteinase K and dextranase) or heat-inactivated enzyme (for DNase I and RNase H). To evaluate the impact of eDNA on biofilm formation, DNase I (100 μg/ml; Sigma) was included in the biofilm culture medium for the entire 24-h incubation period or for the initial 5 h only. To remove the DNase, the plates/discs were washed with basal medium, and biofilms were allowed to continue to grow in fresh medium. Controls received heat-inactivated enzyme. To assess the role of the eDNA matrix on biofilm stability, 24-h biofilms grown in FMCGS on 24-well plates were treated with DNase I (100 μg/ml) for 30 min and then washed in PBS containing 0.2% SDS for 20 min or sonicated using a Branson digital sonifier at 10% power level for 5 and 10 s, respectively. The loosened cells were discarded, and the remaining biofilms were stained using crystal violet and quantitated as described above (42).
Adherence assays.
The impact of eDNA on S. mutans' adherence was examined by evaluating its binding to experimental salivary pellicle (sHA) or to glucans in the pellicle (gsHA) formed on the HA surface (9). Mid-exponential-phase S. mutans cells grown in UFTYE-glucose were washed three times in ice-cold adsorption buffer (50 mM KCl, 1.0 mM KPO4, 1.0 mM CaCl2, and 0.1 mM MgCl2, pH 6.5). The resulting cell suspension was briefly sonicated to dechain the cells and adjusted to an optical density at 600 nm (OD600) of ∼1.0 ± 0.05, with cell numbers counted on a Petroff-Hausser cell-counting chamber (Hausser Scientific Partnership, Horsham, PA). GtfB was prepared from culture supernatants of S. anginosus KSB8 and purified to near homogeneity by using HA column chromatography as described by Wunder et al. (51). GtfB activity was measured by the incorporation of [14C]glucose from labeled sucrose (NEN Research Products, MA) into glucans (9).
To study the impact of eDNA on S. mutans adherence to an apatitic surface, HA beads (Bio-Rad) were incubated with clarified and filter sterilized whole saliva to form an experimental salivary pellicle, with and without inclusion of S. mutans chromosomal DNA (at 0.2 and 2 μg, respectively). For bacterial adherence to gsHA, the sHA beads were exposed to saturating amounts of GtfB and incubated with sucrose (100 mM) and eDNA (0.2 and 2 μg) at 37°C for 4 h to allow glucan formation on the surface (9). The radiolabeled bacterial cells were obtained from UFTYE containing 10 μCi/ml [3H]thymidine (Perkin-Elmer Life and Analytical Sciences, Boston, MA). Bacterial cells (9 × 109 S. mutans cells/ml) were incubated with sHA or gsHA (formed with or without eDNA). After 60 min, the beads were washed using adsorption buffer to remove unbound cells, and the number of adherent cells was determined by liquid scintillation counting (9).
Preparation of MVs.
Membrane vesicles (MVs) were prepared by following the protocol of Rivera et al. (52, 53). Briefly, two sets of 100-ml S. mutans cultures were grown in FMC medium with glucose, one to early exponential phase (OD600 of ∼0.3) and the other overnight. Following centrifugation at 6,000 × g at 4°C for 10 min, the cell-free culture supernatants were filtered first through 0.45-μm syringe filters (Pall Life Science) followed by filtration through 0.22-μm filters (Millipore) to remove residual cells. MVs in the cell-free supernatants were collected and washed once with sterile PBS after centrifugation at 100,000 × g at 4°C for 1 h. After washing, the pellets were resuspended in 200 μl PBS. The MV yield was quantitated by measuring protein concentration using a micro-bicinchoninic (BCA) assay (Pierce) and the protein profile by SDS-PAGE and Western blot analysis as described previously (43, 54). For eDNA estimation in MVs, the vesicles were first hydrolyzed using GES reagent (5 M guanidinium thiocyanate, 100 mM EDTA, and 0.5% [vol/vol] Sarkosyl) (55) prior to fluorometric assay as described above. For morphological analysis, aliquots of MV preparations were stained negatively using 1% uranyl acetate and analyzed using transmission EM (TEM) and cryo-EM. In addition, aliquots were streaked onto BHI agar to verify the absence of viable cells in the preparations.
Statistical methods.
Quantitative data were analyzed using the Student t test. A P value of 0.05 or less is considered statistically significant.
RESULTS
S. mutans produces DNase I-sensitive extracellular nanofibers.
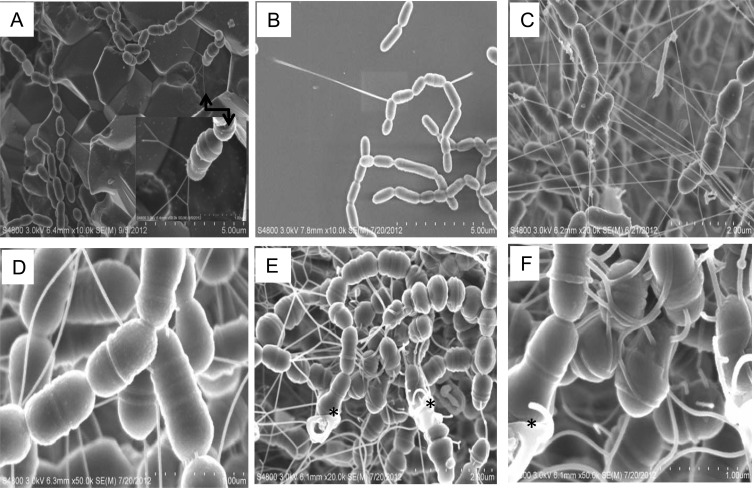
During growth in biofilms, S. mutans was shown to produce a fibrous substance of ∼15 nm in diameter and various lengths, as shown under FE-SEM (Fig. 1). The nanofibers in mature, older biofilms were plentiful and often concentrated around broken cells or cell debris with floating open ends (Fig. 1C to F). In contrast, the nanofibers in early biofilms appeared as a more scattered, but still structured, network with ends of the fibers connected to cells and to the substratum (Fig. 1A and B). To investigate the chemical nature of the nanofibers, biofilms grown on HA discs were treated with DNase I, RNase H, proteinase K, formic acid, and dextranase (also glucanase). The nanofibrous materials completely disappeared following DNase I treatment and were significantly reduced in abundance by dextranase and formic acid (see Fig. S1 in the supplemental material). In contrast, little difference was observed when samples were treated with RNase H or proteinase K (see Fig. S1), suggesting that the nanofibers were primarily comprised of extracellular DNA.
FIG 1.
FE-SEM analysis of the eDNA nanofibers. S. mutans UA159 was grown in BM-glucose on hydroxylapatite (A and C to F) and silicon (B) discs for 4 (A and B) and 24 (C to F) h. Panels A to D show a network of eDNA nanofibers connecting cells to substratum (A and B) and cells to cells (C and D); panels E and F show eDNA from cell lysis that was concentrated around broken cells (indicated by asterisks) and often floating with open ends. The inset in panel A shows a close-up of the area indicated by an arrow. Images in panels A and B were taken at 10,000×, C and E at 20,000×, and D and F at 50,000× magnification.
Destruction of eDNA significantly reduces biofilm formation.
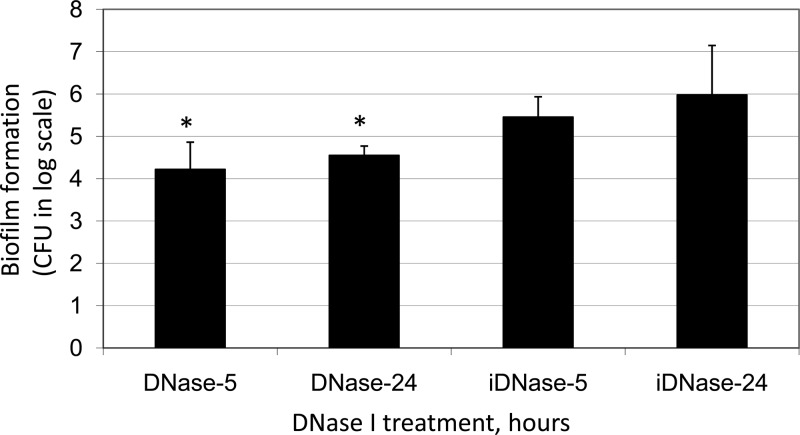
To examine the effects of eDNA on biofilm formation, S. mutans biofilms were grown in FMC medium that included DNase I. It was first carried out using 96-well culture plates. Relative to controls that received the heat-inactivated enzyme, biofilm formation by S. mutans grown in the presence of DNase I was reduced significantly, as estimated by spectrophotometry following crystal violet staining (data not shown). This effect was further analyzed using HA discs, a commonly used in vitro tooth model (44, 56), which were vertically deposited in 24-well plates with S. mutans grown in FMC containing glucose (18 mM) and sucrose (2 mM). In comparison, biofilm formation during growth in the presence of DNase I was reduced by more than 1 log in CFU (P < 0.001) compared to the controls that received the heat-inactivated enzyme (Fig. 2).
FIG 2.
Effect of DNase I on biofilm formation. S. mutans biofilms were grown in FMC with glucose (18 mM) and sucrose (2 mM) as the carbohydrate sources on hydroxylapatite discs vertically deposited in 24-well plates with inclusion of DNase I (DNase) for 5 and 24 h, respectively. Controls received the heat-inactivated enzyme (iDNase). Data presented here represent the averages (± standard deviations) from more than three independent sets of experiments, with an asterisk indicating statistical difference at P < 0.001 compared to the controls.
eDNA enhances glucan-mediated adherence by S. mutans.
Following growth in biofilm medium containing sucrose, the eDNA nanofibers visualized by SEM appeared to be highly interwoven within the extracellular glucans (see Fig. S2 in the supplemental material). This suggests an interaction that is also consistent with the observation that eDNA nanofibers were reduced significantly following dextranase treatment (see Fig. S1) and a role for eDNA in S. mutans biofilm formation as evidenced by the inhibitory effect of treatment with DNase I under these growth conditions (Fig. 2). To assess the impact of eDNA on S. mutans adherence, chromosomal DNA was included in adherence assays, and the ability of the bacterium to bind to saliva-coated hydroxyapatite (sHA) or sHA with surface-formed glucans (gsHA) was compared to controls prepared without eDNA. The S. mutans-derived Gtfs are known to be incorporated into the salivary pellicle in an enzymatically active form, rapidly producing glucans in situ when sucrose is available (9). In turn, S. mutans binds avidly to glucans synthesized by Gtfs on sHA, which is a key step for the sucrose-mediated biofilm initiation and accumulation on tooth surfaces (16). As shown in Fig. 3, glucans formed on sHA (gsHA) significantly increased S. mutans adherence to the apatite surface compared to sHA without glucans (sHA only), which is in agreement with previous studies (9, 10, 16, 51). More importantly, the addition of eDNA significantly increased bacterial adherence to gsHA but not to sHA.
FIG 3.
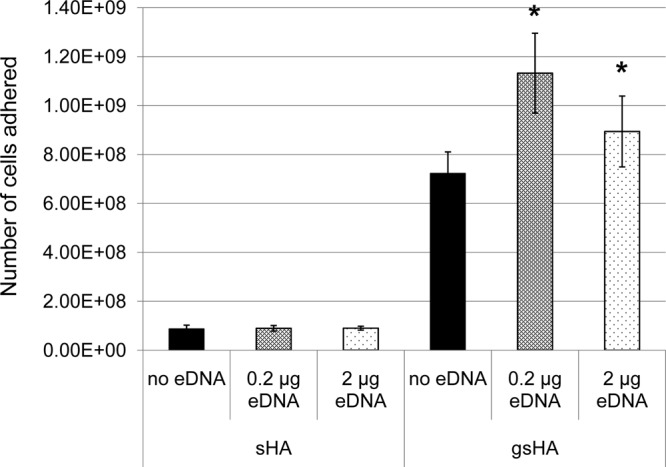
S. mutans adhesion to apatitic surfaces in presence and absence of eDNA. sHA, saliva-coated hydroxyapatite; gsHA, glucan formed on saliva-coated hydroxyapatite. An asterisk indicates that there is a statistical difference for the samples with eDNA compared to controls without eDNA (P < 0.05).
eDNA deficiency affects mechanical stability of S. mutans biofilms.
To test the effect of eDNA on the mechanical stability of biofilms, 24-h biofilms were grown in FMC containing glucose and sucrose on 24-well plates and then treated with DNase I followed by SDS washes and sonication for 5 or 10 s. Biofilms that were treated with DNase I were more easily disrupted and washed away, particularly after brief sonication, than were control biofilms that received the heat-inactivated enzyme (see Fig. S3 in the supplemental material). These results confirm that S. mutans eDNA acts as a functional component of the biofilm matrix, influencing mechanical stability, consistent with findings for several other bacterial species (18, 19, 57).
Growth in biofilms increases eDNA production.
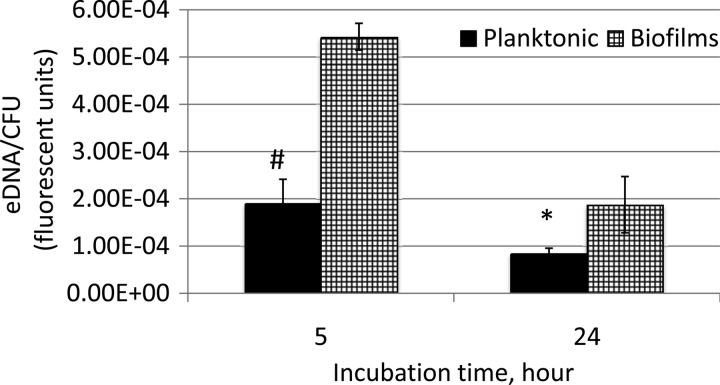
To assess whether growth in biofilms influences eDNA production, S. mutans cultures were grown either in planktonic culture or in sessile form on HA discs that were vertically oriented in 24-well plates. Growth in biofilms yielded a significantly larger amount of eDNA per CFU (Fig. 4). This was particularly evident in the early biofilm, with an increase of approximately 3-fold compared to the planktonic counterpart. Due to nutrient limitation and toxic metabolite accumulation, overnight cultures are known to have significantly more dead, broken cells. While the 24-h cultures demonstrated more total eDNA than the earlier 5-h cultures, the younger cultures contained significantly more eDNA when normalized for CFU, especially during growth in biofilms. An effort was also made to examine the effects of carbohydrate source and availability, aeration, and dATP on eDNA production. However, no significant differences were observed between the groups under the conditions studied, except growth under anaerobic conditions (see Fig. S4 in the supplemental material). Relative to aerobic conditions with 5% CO2, growth under anaerobic conditions led to a significant reduction of eDNA (P < 0.05) (see Fig. S4).
FIG 4.
eDNA production by S. mutans. S. mutans UA159 was grown planktonically in biofilm medium plus glucose (20 mM) in 5-ml Falcon tubes (solid bars) or in biofilms on hydroxylapatite discs vertically deposited in 24-well plates (hatched bars) for 5 and 24 h. Data presented here represent the averages (± standard deviations) from more than 3 independent experiments, with statistic differences at P < 0.01 (#) and P < 0.05 (*) compared to the planktonic counterparts.
Iron limitation influences growth and causes alterations in cell morphology.
Iron is a micronutrient known to be required for DNA synthesis and many other metabolic processes (58). To examine the effect of iron limitation on S. mutans eDNA production and biofilm formation, S. mutans was grown in modified BMG (mBMG) medium in which ferric chloride was omitted. Relative to complete BMG, omission of ferric chloride in mBMG had little impact on the bacterial growth rate (data not shown), but it did significantly reduce the final optical densities (OD600) of the cultures. In addition, cultures grown in mBMG ceased growth 3.5 h earlier than cells growing in complete BMG. Similar results were also observed during growth in mBMGS and mBGS (data not shown). Measurable eDNA in 5-h cultures grown in mBMG was slightly reduced compared to levels of cultures grown in regular BMG, but the differences were not statistically significant (P > 0.05) (data not shown). Similar results were observed with mFMC, which contained no added iron (see Fig. S4 in the supplemental material). Unlike cells grown in regular BM in which nanofibers were apparent when viewed by FE-SEM, no eDNA nanofibers were visualized in 24-h biofilms grown in mBMG (see Fig. S5). In addition, the cells grown under these conditions appeared to be elongated and rod-like (see Fig. S5). As a semidefined medium, BM contains 2 g liter−1 of Casamino Acids (Bacto 223050; BD), which is an acid hydrolysate of casein with low but unspecified concentrations of iron. S. mutans requires only nanomoles of iron to grow (59). It appears that endogenous iron derived from the Casamino Acids was sufficient to meet S. mutans' initial requirement for growth but ultimately became depleted as the cultures continued to grow, affecting bacterial growth and eDNA production.
S. mutans produces membrane vesicles that contain eDNA.
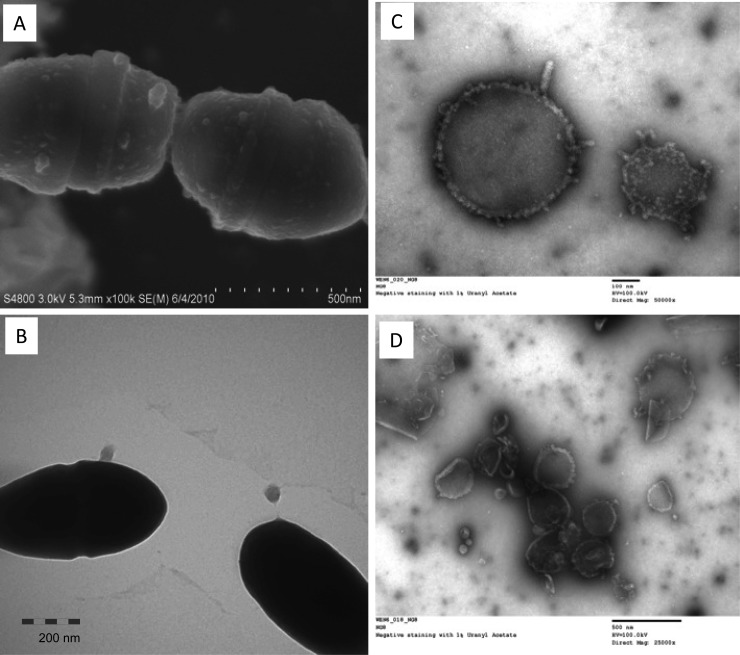
In Gram-negative bacteria, MVs have long been considered a source of eDNA (18, 30, 60). Recently, the Gram-positive organisms Staphylococcus aureus and Bacillus anthracis have also been reported to produce membrane-derived vesicles (61, 62) which play a role in delivery of bacterial effector molecules to the host cells (52, 63). In our initial experiments, we tested whether S. mutans produces MVs. Such vesicles were identified by electron microscopy following ultracentrifugation of cell-free supernatants from overnight cultures (Fig. 5). Electron microscopy of S. mutans also demonstrated the presence of small blebs associated with the cells (Fig. 5A), some of which appeared to be pinching off (Fig. 5B). Under TEM, when negatively stained with uranyl acetate, cell-free supernatants were shown to possess vesicular structures typical of membrane vesicles (Fig. 5C and D). When measured using a micro-BCA assay, the early-exponential-phase cultures had 2.87-fold more MVs than the overnight cultures (P < 0.05).
FIG 5.
EM analysis of S. mutans membrane vesicles. S. mutans UA159 (A) and NG8 (B to D) were grown in BHI broth (B to D) and in biofilm medium on hydroxylapatite discs (A) overnight. FE-SEM (A) and TEM (B) analysis show small blebs on the cell surfaces. Panels C and D show EM images of vesicular structures in cell-free supernatants of NG8 following negative staining with 1% uranyl acetate. Similar vesicles were also seen with UA159 (not shown). Images were taken at magnifications of 50,000× (A), 30,000× (B), 50,000× (C), and 25,000× (D).
To assess whether MVs can serve as a vehicle for eDNA release in S. mutans, MVs derived from early-exponential-phase and overnight cultures were hydrolyzed and tested for the presence of eDNA. Results showed that MVs of both early-exponential-phase and overnight cultures contained eDNA. In comparison, MVs from the early-exponential-phase cultures contained 2.82-fold more eDNA than those prepared from the overnight cultures (P < 0.05).
SrtA deficiency affects membrane vesicle formation.
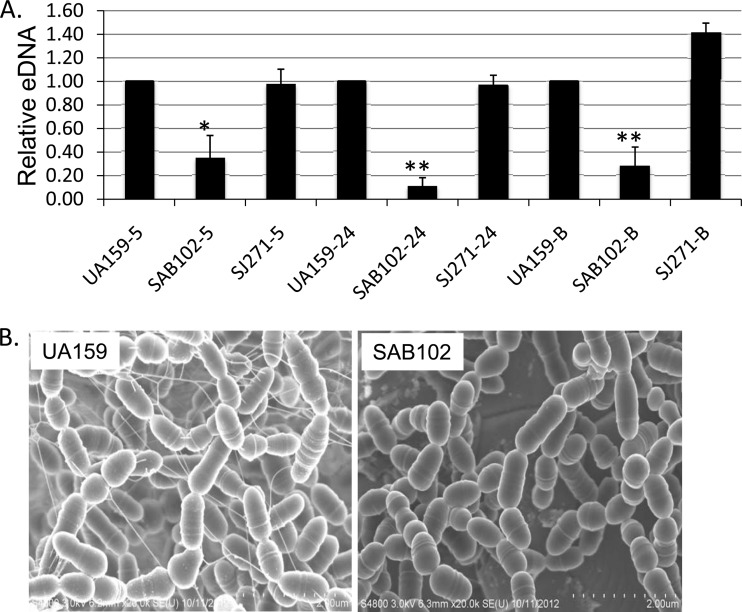
Bacterial adherence and biofilm formation by S. mutans is known to be severely impaired in the absence of SrtA (3, 5–7), an enzyme known for anchoring cell surface proteins bearing an LPXTG motif to the cell wall peptidoglycan in this bacterium (6, 64, 65). To assess whether S. mutans eDNA release is influenced by SrtA, an srtA-deficient mutant, SAB102 (5), was analyzed by both spectrofluorometry and FE-SEM. Compared to levels for its parental strain, UA159, significant reductions in the amount of eDNA in cell-free supernatants were measured from SAB102 in early-exponential-phase cultures and especially in overnight cultures and biofilms (Fig. 6A). When viewed by FE-SEM (Fig. 6B), the SrtA-deficient mutant had notably lower levels of biofilm formation than its parent strain, consistent with previous findings (3, 5–7). Relative to the UA159 parental strain, which displayed substantial eDNA, no eDNA nanofibers were visualized in the biofilms of the SrtA-deficient mutant (Fig. 6B). Similar results were also observed with S. mutans NG8 (data not shown), another commonly used laboratory strain (Table 1).
FIG 6.
Sortase A deficiency on eDNA production. (A) For quantitative analysis, S. mutans UA159, the sortase A-deficient mutant SAB102, and its complement strain, SJ271, which carries a wild-type copy of srtA in a multicopy shuttle vector, pDL271, were grown in the chemically defined medium FMC with glucose (20 mM) for 5-h cultures (−5) or in FMC with glucose (18 mM) plus sucrose (2 mM) for 24-h (−24) and for biofilm (−B) cultures. Biofilms were grown on HA discs that were vertically deposited in wells of 24-well plates. The eDNA in the cell-free supernatant was further normalized and expressed relative to levels for wild-type UA159. Data presented represent averages (± standard deviations) from more than 3 independent experiments, with * and ** indicating statistical difference at P < 0.01 and P < 0.001, respectively, compared to UA159 under the same conditions. Similar results were also obtained during growth in BM. (B) FE-SEM analysis of wild-type S. mutans (UA159) and the SrtA-deficient mutant (SAB102) biofilms on HA discs during growth in BM-glucose. Compared to the wild type, the SrtA-deficient mutant SAB102 consistently displayed few or no eDNA nanofibers in biofilms grown for 24 h. Images were taken at a magnification of 20,000×.
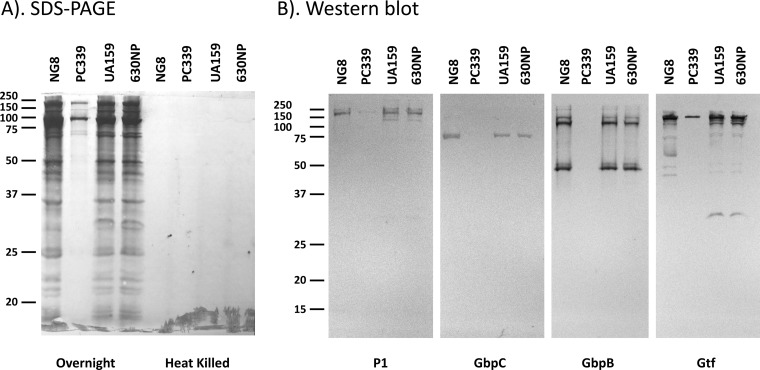
Efforts were also made to determine whether SrtA affects MV formation. No significant differences were identified in total protein or eDNA levels between MV preparations of the wild type and the SrtA-deficient mutant (data not shown). When viewed under TEM following negative staining with 1% uranyl acetate, no significant differences in quantity and morphology of the MVs between the SrtA mutant and the wild type were identified (data not shown). Interestingly, SDS-PAGE analysis revealed a significant reduction in the overall protein content of the purified MVs of the SrtA-deficient mutant compared to the wild type (Fig. 7A). SDS-PAGE separates proteins by molecular weight and small proteins and peptides easily run off the gel, while BCA is a quantitative assay that measures proteins of all sizes, including peptides. One possible explanation for the discrepancy between the SDS-PAGE data and the BCA results is that low-molecular-weight proteins/peptides cannot be detected by SDS-PAGE but were measured by BCA. A difference in MV protein content was not observed by SDS-PAGE when a mutant devoid of AtlA, an autolysin known to affect cell wall turnover and cell surface properties (66, 67), was evaluated (Fig. 7A and B). As a control, a set of cells was heat killed by incubating at 60°C for 45 min, and samples were prepared as described for live cells. No MVs were produced by the heat-killed cells (Fig. 7), suggesting that this is an active process in S. mutans and not the result of lysis of dead cells. Western blotting was used to further analyze specific proteins, and the anchoring of some proteins to the cell surface is known to be associated with SrtA (7, 68). The SrtA-deficient mutant consistently displayed less P1, glucan-binding proteins B and C, and glucosyltransferases than the wild type (Fig. 7B). No differences in amounts of the selected proteins were observed when the AtlA-deficient mutant and its parental strain, UA159, were evaluated (Fig. 7A and B). Taken together, these results further suggest that SrtA in S. mutans does not have a significant effect on the production of MVs but does influence their composition.
FIG 7.
SDS-PAGE (A) and Western blot (B) analysis. S. mutans UA159, its AtlA-deficient mutant (630NP), NG8, and its SrtA-deficient mutant (PC339) were grown in FMC broth overnight, and membrane vesicles were prepared from cell-free supernatants by ultracentrifugation. For controls, a set of overnight cultures was killed by incubating at 60°C for 45 min, washed in sterile PBS, resuspended in FMC, and then allowed to incubate at 37°C overnight. For SDS-PAGE analysis, a 10% gel was used. For Western blotting, antibodies against P1, glucan-binding proteins GbpB and GbpC, and pooled glucosyltransferases (Gtf) were used as probes. Relative to the wild type, NG8, the SrtA-deficient mutant had significantly less P1, GbpB, GbpC, and Gtf, which are known to be surface-associated proteins. Equal volumes of membrane vesicle preparations were loaded on the gel.
Deficiency in protein secretion pathway components affects the amount of eDNA.
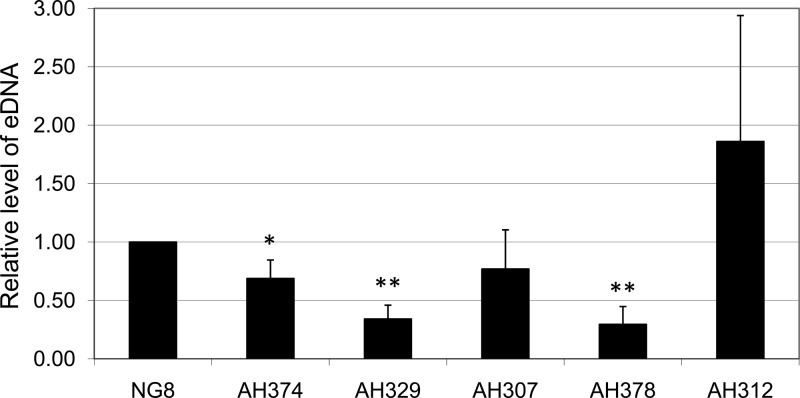
Given that a deficiency in SrtA, a membrane-localized protein, influenced the detection of S. mutans eDNA, especially during growth in biofilms, we further investigated whether known elements of the protein secretion/translocation or membrane protein insertion machinery affected eDNA production. Mutants deficient in ffh (54-kDa homolog of the bacterial signal recognition particle [SRP]), ftsY (the bacterial SRP receptor), scRNA (the small cytoplasmic RNA component of the SRP), and yidC1 and yidC2 (paralogs of the YidC/OxaI/Alb3 family of membrane protein insertases/chaperones) (69–71) were analyzed. Compared to levels for the wild type, significantly less eDNA was measured for mutants lacking yidC1 and, in particular, yidC2 and ffh (Fig. 8). In contrast, the mutant deficient in scRNA demonstrated little difference in the amount of eDNA measured compared to that of the wild type.
FIG 8.
eDNA production by S. mutans mutants. S. mutans wild-type NG8 and its derivatives, AH374 (yidC1), AH329 (ffh), AH307 (ftsY), AH378 (yidC2), and AH312 (scRNA), which are deficient in genes (as specified) that encode components of the protein translocation system, during growth in regular FMC with glucose for 5 h. All data presented here are averages (± standard deviations) from three independent experiments and were further normalized to the wild type, with * and ** indicating differences at significance levels of P < 0.05 and P < 0.01, respectively.
DISCUSSION
This study has demonstrated that eDNA is a functional component of S. mutans biofilms, contributes to S. mutans biofilm formation, and influences structural integrity and stability of biofilms, consistent with previous studies conducted with this bacterium (20, 37), as well as multiple other species (19, 21, 30, 72, 73). Like E. faecalis, S. mutans demonstrated increased eDNA during growth in biofilms compared to that in planktonic cultures, albeit to a lesser degree than E. faecalis. Like S. gordonii, whose hydrogen peroxide-induced eDNA release is most pronounced in actively growing cells (21), S. mutans demonstrated the most significant increases in eDNA during growth in early biofilms when cell lysis is not yet considered significant. However, what triggers eDNA release in S. mutans and why growth in biofilm enhances eDNA production await further investigation.
Membrane vesicles are nanovesicular structures originally found to bleb from the outer membrane of Gram-negative bacteria (55, 74). More recently, studies have shown that Gram-positive bacteria also naturally produce MVs that share some common features with MVs of Gram-negative organisms (52, 53, 62, 75). Our results suggest that S. mutans produces MVs during growth. This is the third Gram-positive bacterium thus far, in addition to B. subtilis and S. aureus, known to produce MVs (52, 61, 62). S. mutans MVs can serve as a means for protein as well as DNA release, properties demonstrated in Gram-negative bacteria (18, 74) but as yet undescribed in Gram-positive bacteria. To determine whether a specific region(s) of the chromosomal DNA was preferentially released, quantitative PCR was used to assess amplification of genes located in defined regions of the chromosome. MVs were analyzed using chromosomal DNA as a control and basis for comparison. Results showed no major differences between the eDNA samples and the chromosomal DNA, suggesting that eDNA release is nonselective in S. mutans, consistent with findings in some other bacteria (76, 77). Our results also demonstrate that the formation of S. mutans MVs is an active process, since none were observed when overnight cultures were heat killed and prepared in a manner similar to that of live cells. In a recent study by the Whitchurch group using high-resolution phase-contrast microscope and fluorescence microscope imaging, P. aeruginosa was shown to release DNA in bursts that were concomitant with cellular explosion (60). This “explosive lysis” was credited with providing large amounts of MVs and with eDNA production. It is currently unknown whether such explosive lysis exists in S. mutans or other bacterial species and, if so, whether it contributes to MV or eDNA production. We also evaluated whether the AtlA autolysin contributes to protein release by MVs in S. mutans but did not observe any difference between an atlA-deficient mutant and the parent strain under the conditions studied.
S. mutans is known for its copious production of exopolysaccharides during growth in the presence of sucrose (16). We have also recently shown that like several other bacteria, S. mutans forms formic acid-sensitive/protease-resistant amyloid-like materials in biofilms (3). The results presented here indicate that the nanofibers observed in S. mutans biofilms are comprised of DNA based on their sensitivity to DNase I. However, the reduction of the fibrous matrices following treatment with dextranase and formic acid also suggests that the eDNA nanofibers interact with the glucans and possibly with amyloid fibrils, which are sensitive to formic acid dissolution. Extracellular DNA nanofibers consistently appeared to be interwoven/integrated with glucans under FE-SEM when S. mutans biofilms were grown in the presence of sucrose. In support of this interpretation, an in vitro adherence assay also demonstrated that eDNA significantly enhanced glucan-mediated adherence to saliva-coated hydroxylapatite, while no such effect was observed in the absence of glucans. A recent report on Myxococcus xanthus also showed that eDNA was recruited by extracellular polysaccharides and integrated into the extracellular matrix (57). However, an eDNA-glucan interaction and its role in S. mutans adherence has not been reported thus far. The lack of significant effects of eDNA alone on S. mutans adherence in the current study differs from a recent report by Das et al. (36). Unlike the glass surface used by the Das group, we used an apatitic surface that was coated with saliva to mimic pellicle-covered teeth in the mouth. The presence of salivary constituents is known to drastically influence S. mutans binding and biofilm formation (5). Therefore, the presence of a salivary pellicle on the apatitic surface could have masked or diminished the effects of eDNA and likely contributed to such a discrepancy in binding efficiency. Nevertheless, our results clearly show that eDNA-glucan interactions play a crucial role in sucrose-dependent S. mutans adherence.
In P. aeruginosa as well as several enteric bacteria, peptidoglycan-associated outer membrane proteins that tether the outer membrane to the underlying peptidoglycan have been shown to significantly affect the formation of outer membrane vesicles (53). In evaluation of the nature of the nanofibers, we employed mutant strains lacking srtA, which encodes sortase A, the membrane-localized transpeptidase known for its role in anchoring of a number of surface-associated proteins to the peptidoglycan (6, 64, 65). Our data indicate that a deficiency of SrtA in S. mutans significantly reduced eDNA detection, especially during growth in biofilms. To the best of our knowledge, a role for SrtA in eDNA production has not yet been reported in other Gram-positive bacteria. Interestingly, no major differences were observed between the mutant deficient in SrtA and the parent strain in MV production, but many proteins observed by SDS-PAGE, including P1, GbpB, GbpC, and Gtfs, were notably less abundant in the MVs derived from the SrtA-deficient mutant than from the parent strain. In S. mutans, P1, GbpC, and several other surface-associated proteins are known to be SrtA substrates (7, 68). Therefore, it is not a surprise that SrtA deficiency in S. mutans caused a pronounced alteration in multiple surface-associated proteins. In addition, SrtA has been reported to affect gene expression (7, 68); therefore, both direct and indirect effects may contribute to the altered protein profiles of SrtA-deficient mutant cells. We speculate that surface-associated proteins contribute to the structure and function of eDNA in biofilms and to biofilm formation. As a result of SrtA deficiency, a protein(s) involved in eDNA-cell interactions would not be properly anchored on the surface and would result in the loss of eDNA nanofibers observed by FE-SEM, as well as a reduction in biofilm formation. Studies are ongoing to examine whether surface-associated proteins interact directly with S. mutans eDNA. Besides its role in anchoring surface-associated proteins, S. mutans and S. gordonii SrtA proteins influence the expression of genes, including those encoding adhesin WapA and fructanase FruA in S. mutans and adhesins SspA/B, CshA/B, AbpA/B, and Has in S. gordonii (7, 68). The reduced eDNA observed in SrtA-deficient S. mutans, especially within biofilms as visualized by FE-SEM, suggests that SrtA's contribution to S. mutans biology includes effects downstream of protein anchoring and cell surface biogenesis that influence eDNA and nanofiber formation.
Active export of chromosomal DNA by the type IV secretion system has recently been described in Neisseria gonorrhoeae (31). Mutations constructed in seven putative type IV secretion genes greatly reduced or completely eliminated DNA secretion. Although S. mutans does not have a type IV secretion system, we analyzed S. mutans strains deficient in highly conserved elements of protein secretion and translocation pathways for their respective impacts on eDNA production. Ffh (for SRP54) and scRNA are core elements of the bacterial signal recognition particle (SRP) cotranslational protein translocation pathway, and FtsY is the membrane-bound SRP receptor (70). YidC1 and YidC2 are paralogues of the ubiquitous YidC/OxaI/Alb3 family of membrane-integral chaperones and insertases (71, 78). Loss of any of the S. mutans SRP pathway components Ffh, scRNA, and FtsY, as well as YidC2, results in severe acid, osmotic, and oxidative stress sensitivity with decreased membrane-associated F-ATPase activity by the mutant strains (70). In contrast, loss of YidC1 is less severe and does not result in obvious stress sensitivity. Losses of Ffh, YidC2, and YidC1 all impede biofilm formation, although with different manifestations (79). The respective substrates of the SRP pathway and YidC2 and YidC1 are largely unknown in S. mutans, although these systems likely overlap and appear to work in concert (72). The integral membrane proteins YidC1 and YidC2 would be expected to mediate membrane protein insertion, but they have also been found to influence the amount and functional maturation of cell surface-localized proteins, including adhesin P1 (71). In the current study, S. mutans mutants lacking Ffh, YidC2, and, to a lesser extent, YidC1 all demonstrated less eDNA than the parental strain. This may partially explain their previously observed defects in biofilm formation (77) and again points to potential downstream effects and a level of complexity in the machinery responsible for membrane biogenesis, since the same proteins also influence cell surface-localized molecules. Our results further suggest that generation of a cell envelope with the correct protein composition is required for eDNA release and subsequent eDNA-cell interactions.
As an organism with an obligate biofilm lifestyle, S. mutans possesses multiple mechanisms to colonize the tooth surface (2, 16, 80, 81). Our results provide evidence that eDNA is a functional component within S. mutans biofilms, and that it too plays a substantial role in bacterial adherence and biofilm initiation. While the underlying mechanisms await further investigation, active release of eDNA is triggered in response to cell-surface interaction. The highly structured eDNA network interacts with the substratum, as well as with close and distant cells, contributing to surface and intercellular adhesion. During growth in sucrose, eDNA and the adhesive glucan products of the Gtf enzymes also interact, further strengthening the extracellular matrix scaffold. Such a polymeric scaffold would facilitate inter- and intracellular interactions and biofilm accumulation, as well as provide greater stability and resistance to mechanical stress (18, 19, 57).
In summary, our results demonstrate that previously unrecognized factors contribute to the presence and quantity of S. mutans eDNA. These include MVs, components of protein secretion and translocation pathways, and the SrtA enzyme essential for the localization of multiple cell surface-localized proteins. The amount of eDNA measured also appeared to be responsive to changes in environmental conditions and growth mode in particular. As a functional component of the extracellular matrix, eDNA likely interacts with exopolysaccharides and possibly amyloid, enhancing cell-surface and cell-cell interactions, facilitating biofilm formation, and contributing to structural integrity and stability of biofilms. Current effort is directed to further investigate the mechanisms that underlie eDNA production, transport, and eDNA-cell interactions.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by NIH/NIDCR grants DE19452 to Z.T.W., DE08007 and DE21789 to L.J.B., and DE22529 to M.I.K. and an NSF grant (EFRI-1137186) to H.K.
We thank Wandy L. Beatty at Washington University Department of Microbiology, St. Louis, MO, for her assistance in EM analysis; Jibao He at Tulane University Electron Microscope Laboratory for cryo-EM analysis; Kar Man (Edmond) Leung at the University of Southern California Departments of Biological Sciences and Physics and Astronomy for AFM analysis; and James B. McLachlan at the Tulane University School of Medicine Department of Microbiology and Immunology for assistance with flow cytometry.
Footnotes
Published ahead of print 18 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01493-14.
REFERENCES
- 1.Lee SF, Progulske-Fox A, Bleiweis AS. 1988. Molecular cloning and expression of a Streptococcus mutans major surface protein antigen, P1 (I/II), in Escherichia coli. Infect. Immun. 56:2114–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crowley PJ, Brady LJ, Michalek SM, Bleiweis AS. 1999. Virulence of a spaP mutant of Streptococcus mutans in a gnotobiotic rat model. Infect. Immun. 67:1201–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oli MW, Otoo HN, Crowley PJ, Heim KP, Nascimento MM, Ramsook CB, Lipke PN, Brady LJ. 2012. Functional amyloid formation by Streptococcus mutans. Microbiology 158:2903–2916. 10.1099/mic.0.060855-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nobbs AH, Lamont RJ, Jenkinson HF. 2009. Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73:407–450. 10.1128/MMBR.00014-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahn SJ, Wen ZT, Brady LJ, Burne RA. 2008. Characteristics of biofilm formation by Streptococcus mutans in the presence of saliva. Infect. Immun. 76:4259–4268. 10.1128/IAI.00422-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SF, Boran TL. 2003. Roles of sortase in surface expression of the major protein adhesin P1, saliva-induced aggregation and adherence, and cariogenicity of Streptococcus mutans. Infect. Immun. 71:676–681. 10.1128/IAI.71.2.676-681.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levesque CM, Voronejskaia E, Huang YC, Mair RW, Ellen RP, Cvitkovitch DG. 2005. Involvement of sortase anchoring of cell wall proteins in biofilm formation by Streptococcus mutans. Infect. Immun. 73:3773–3777. 10.1128/IAI.73.6.3773-3777.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowen WH, Schilling K, Giertsen E, Pearson S, Lee SF, Bleiweis A, Beeman D. 1991. Role of a cell surface-associated protein in adherence and dental caries. Infect. Immun. 59:4604–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schilling KM, Bowen WH. 1992. Glucans synthesized in situ in experimental salivary pellicle function as specific binding sites for Streptococcus mutans. Infect. Immun. 60:284–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vacca-Smith AM, Bowen WH. 1998. Binding properties of streptococcal glucosyltransferases for hydroxyapatite, saliva-coated hydroxyapatite, and bacterial surfaces. Arch. Oral Biol. 43:103–110. 10.1016/S0003-9969(97)00111-8 [DOI] [PubMed] [Google Scholar]
- 11.Xiao J, Klein MI, Falsetta ML, Lu B, Delahunty CM, Yates JR, III, Heydorn A, Koo H. 2012. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 8:e1002623. 10.1371/journal.ppat.1002623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banas JA, Vickerman MM. 2003. Glucan-binding proteins of the oral streptococci. Crit. Rev. Oral Biol. Med. 14:89–99. 10.1177/154411130301400203 [DOI] [PubMed] [Google Scholar]
- 13.Burne RA. 1998. Oral streptococci. Products of their environment. J. Dent. Res. 77:445–452 [DOI] [PubMed] [Google Scholar]
- 14.Koo H, Xiao J, Klein MI, Jeon JG. 2010. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J. Bacteriol. 192:3024–3032. 10.1128/JB.01649-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregoire S, Xiao J, Silva BB, Gonzalez I, Agidi PS, Klein MI, Ambatipudi KS, Rosalen PL, Bauserman R, Waugh RE, Koo H. 2011. Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle on hydroxyapatite surfaces. Appl. Environ. Microbiol. 77:6357–6367. 10.1128/AEM.05203-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowen WH, Koo H. 2011. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 45:69–86. 10.1159/000324598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch DJ, Fountain TL, Mazurkiewicz JE, Banas JA. 2007. Glucan-binding proteins are essential for shaping Streptococcus mutans biofilm architecture. FEMS Microbiol. Lett. 268:158–165. 10.1111/j.1574-6968.2006.00576.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. 2002. Extracellular DNA required for bacterial biofilm formation. Science 295:1487. 10.1126/science.295.5559.1487 [DOI] [PubMed] [Google Scholar]
- 19.Lappann M, Claus H, van Alen T, Harmsen M, Elias J, Molin S, Vogel U. 2010. A dual role of extracellular DNA during biofilm formation of Neisseria meningitidis. Mol. Microbiol. 75:1355–1371. 10.1111/j.1365-2958.2010.07054.x [DOI] [PubMed] [Google Scholar]
- 20.Petersen FC, Tao L, Scheie AA. 2005. DNA binding-uptake system: a link between cell-to-cell communication and biofilm formation. J. Bacteriol. 187:4392–4400. 10.1128/JB.187.13.4392-4400.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itzek A, Zheng L, Chen Z, Merritt J, Kreth J. 2011. Hydrogen peroxide-dependent DNA release and transfer of antibiotic resistance genes in Streptococcus gordonii. J. Bacteriol. 193:6912–6922. 10.1128/JB.05791-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kreth J, Vu H, Zhang Y, Herzberg MC. 2009. Characterization of hydrogen peroxide-induced DNA release by Streptococcus sanguinis and Streptococcus gordonii. J. Bacteriol. 191:6281–6291. 10.1128/JB.00906-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beenken KE, Spencer H, Griffin LM, Smeltzer MS. 2012. Impact of extracellular nuclease production on the biofilm phenotype of Staphylococcus aureus under in vitro and in vivo conditions. Infect. Immun. 80:1634–1638. 10.1128/IAI.06134-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodman SD, Obergfell KP, Jurcisek JA, Novotny LA, Downey JS, Ayala EA, Tjokro N, Li B, Justice SS, Bakaletz LO. 2011. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 4:625–637. 10.1038/mi.2011.27 [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Burne RA. 2011. The major autolysin of Streptococcus gordonii is subject to complex regulation and modulates stress tolerance, biofilm formation, and extracellular-DNA release. J. Bacteriol. 193:2826–2837. 10.1128/JB.00056-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guiton PS, Hung CS, Kline KA, Roth R, Kau AL, Hayes E, Heuser J, Dodson KW, Caparon MG, Hultgren SJ. 2009. Contribution of autolysin and sortase a during Enterococcus faecalis DNA-dependent biofilm development. Infect. Immun. 77:3626–3638. 10.1128/IAI.00219-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rice KC, Bayles KW. 2003. Death's toolbox: examining the molecular components of bacterial programmed cell death. Mol. Microbiol. 50:729–738. 10.1046/j.1365-2958.2003.t01-1-03720.x [DOI] [PubMed] [Google Scholar]
- 28.Ahn SJ, Rice KC, Oleas J, Bayles KW, Burne RA. 2010. The Streptococcus mutans Cid and Lrg systems modulate virulence traits in response to multiple environmental signals. Microbiology 156:3136–3147. 10.1099/mic.0.039586-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas VC, Hiromasa Y, Harms N, Thurlow L, Tomich J, Hancock LE. 2009. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol. Microbiol. 72:1022–1036. 10.1111/j.1365-2958.2009.06703.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnes AM, Ballering KS, Leibman RS, Wells CL, Dunny GM. 2012. Enterococcus faecalis produces abundant extracellular structures containing DNA in the absence of cell lysis during early biofilm formation. mBio 3:e00193–00112. 10.1128/mBio.00193-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamilton HL, Dominguez NM, Schwartz KJ, Hackett KT, Dillard JP. 2005. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol. Microbiol. 55:1704–1721. 10.1111/j.1365-2958.2005.04521.x [DOI] [PubMed] [Google Scholar]
- 32.Perry JA, Cvitkovitch DG, Levesque CM. 2009. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol. Lett. 299:261–266. 10.1111/j.1574-6968.2009.01758.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wenderska IB, Lukenda N, Cordova M, Magarvey N, Cvitkovitch DG, Senadheera DB. 2012. A novel function for the competence inducing peptide, XIP, as a cell death effector of Streptococcus mutans. FEMS Microbiol. Lett. 336:104–112. 10.1111/j.1574-6968.2012.02660.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahn SJ, Qu MD, Roberts E, Burne RA, Rice KC. 2012. Identification of the Streptococcus mutans LytST two-component regulon reveals its contribution to oxidative stress tolerance. BMC Microbiol. 12:187. 10.1186/1471-2180-12-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahn SJ, Wen ZT, Burne RA. 2007. Effects of oxygen on virulence traits of Streptococcus mutans. J. Bacteriol. 189:8519–8527. 10.1128/JB.01180-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das T, Sharma PK, Krom BP, van der Mei HC, Busscher HJ. 2011. Role of eDNA on the adhesion forces between Streptococcus mutans and substratum surfaces: influence of ionic strength and substratum hydrophobicity. Langmuir 27:10113–10118. 10.1021/la202013m [DOI] [PubMed] [Google Scholar]
- 37.Klein MI, DeBaz L, Agidi S, Lee H, Xie G, Lin AH, Hamaker BR, Lemos JA, Koo H. 2010. Dynamics of Streptococcus mutans transcriptome in response to starch and sucrose during biofilm development. PLoS One 5:e13478. 10.1371/journal.pone.0013478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434–14439. 10.1073/pnas.172501299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ayakawa GY, Boushell LW, Crowley PJ, Erdos GW, McArthur WP, Bleiweis AS. 1987. Isolation and characterization of monoclonal antibodies specific for antigen P1, a major surface protein of mutans streptococci. Infect. Immun. 55:2759–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terleckyj B, Willett NP, Shockman GD. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect. Immun. 11:649–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loo CY, Corliss DA, Ganeshkumar N. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374–1382. 10.1128/JB.182.5.1374-1382.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen ZT, Burne RA. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196–1203. 10.1128/AEM.68.3.1196-1203.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bitoun JP, Liao S, Yao X, Ahn S-J, Isoda R, Nguyen AH, Brady LJ, Burne RA, Abranches A, Wen ZT. 2012. BrpA is involved in regulation of cell envelope stress responses in Streptococcus mutans. Appl. Environ. Microbiol. 78:2914–2922. 10.1128/AEM.07823-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wen ZT, Burne RA. 2004. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 186:2682–2691. 10.1128/JB.186.9.2682-2691.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koo H, Seils J, Abranches J, Burne RA, Bowen WH, Quivey RG., Jr 2006. Influence of apigenin on gtf gene expression in Streptococcus mutans UA159. Antimicrob. Agents Chemother. 50:542–546. 10.1128/AAC.50.2.542-546.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen ZT, Baker HV, Burne RA. 2006. Influence of BrpA on critical virulence attributes of Streptococcus mutans. J. Bacteriol. 188:2983–2992. 10.1128/JB.188.8.2983-2992.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wen ZT, Nguyen AH, Bitoun JP, Abranches J, Baker HV, Burne RA. 2011. Transcriptome analysis of LuxS-deficient Streptococcus mutans grown in biofilms. Mol. Oral Microbiol. 26:2–18. 10.1111/j.2041-1014.2010.00581.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1992. Current protocols in molecular biology, p 17.19.11–17.19.13, vol V John Wiley and Sons, Inc., New York, NY [Google Scholar]
- 49.Burne RA, Wen ZT, Chen YM, Penders JEC. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol. 181:2863–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romero D, Aguilar C, Losick R, Kolter R. 2010. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. U. S. A. 107:2230–2234. 10.1073/pnas.0910560107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wunder D, Bowen WH. 1999. Action of agents on glucosyltransferases from Streptococcus mutans in solution and adsorbed to experimental pellicle. Arch. Oral Biol. 44:203–214. 10.1016/S0003-9969(98)00129-0 [DOI] [PubMed] [Google Scholar]
- 52.Rivera J, Cordero RJ, Nakouzi AS, Frases S, Nicola A, Casadevall A. 2010. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc. Natl. Acad. Sci. U. S. A. 107:19002–19007. 10.1073/pnas.1008843107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wessel AK, Liew J, Kwon T, Marcotte EM, Whiteley M. 2013. Role of Pseudomonas aeruginosa peptidoglycan-associated outer membrane proteins in vesicle formation. J. Bacteriol. 195:213–219. 10.1128/JB.01253-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wen TZ, Suntharaligham P, Cvitkovitch DG, Burne RA. 2005. Trigger factor in Streptococcus mutans is involved in stress tolerance, competence development, and biofilm formation. Infect. Immun. 73:219–225. 10.1128/IAI.73.1.219-225.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Renelli M, Matias V, Lo RY, Beveridge TJ. 2004. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 150:2161–2169. 10.1099/mic.0.26841-0 [DOI] [PubMed] [Google Scholar]
- 56.Lemos JA, Abranches J, Koo H, Marquis RE, Burne RA. 2010. Protocols to study the physiology of oral biofilms. Methods Mol. Biol. 666:87–102. 10.1007/978-1-60761-820-1_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu W, Li L, Sharma S, Wang J, McHardy I, Lux R, Yang Z, He X, Gimzewski JK, Li Y, Shi W. 2012. DNA builds and strengthens the extracellular matrix in Myxococcus xanthus biofilms by interacting with exopolysaccharides. PLoS One 7:e51905. 10.1371/journal.pone.0051905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cazzola M, Bergamaschi G, Dezza L, Arosio P. 1990. Manipulations of cellular iron metabolism for modulating normal and malignant cell proliferation: achievements and prospects. Blood 75:1903–1919 [PubMed] [Google Scholar]
- 59.Spatafora GA, Moore MW. 1998. Growth of Streptococcus mutans in an iron-limiting medium. Methods Cell Sci. 20:217–221. 10.1023/A:1009755424668 [DOI] [Google Scholar]
- 60.Turnbull L, Whitchurch CB. 2012. Explosive cell lysis in bacterial biofilms produces membrane vesicles and extracellular DNA, abstr. S1:4 Abstr. 6th ASM Conf. Biofilms. ASM Press, Washington, DC [Google Scholar]
- 61.Popkin TJ, Theodore TS, Cole RM. 1971. Electron microscopy during release and purification of mesosomal vesicles and protoplast membranes from Staphylococcus aureus. J. Bacteriol. 107:907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee EY, Choi DY, Kim DK, Kim JW, Park JO, Kim S, Kim SH, Desiderio DM, Kim YK, Kim KP, Gho YS. 2009. Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 9:5425–5436. 10.1002/pmic.200900338 [DOI] [PubMed] [Google Scholar]
- 63.Gurung M, Moon DC, Choi CW, Lee JH, Bae YC, Kim J, Lee YC, Seol SY, Cho DT, Kim SI, Lee JC. 2011. Staphylococcus aureus produces membrane-derived vesicles that induce host cell death. PLoS One 6:e27958. 10.1371/journal.pone.0027958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Igarashi T, Asaga E, Goto N. 2003. The sortase of Streptococcus mutans mediates cell wall anchoring of a surface protein antigen. Oral Microbiol. Immunol. 18:266–269. 10.1034/j.1399-302X.2003.00076.x [DOI] [PubMed] [Google Scholar]
- 65.Igarashi T, Asaga E, Goto N. 2004. Roles of Streptococcus mutans dextranase anchored to the cell wall by sortase. Oral Microbiol. Immunol. 19:102–105. 10.1046/j.0902-0055.2003.00123.x [DOI] [PubMed] [Google Scholar]
- 66.Ahn SJ, Burne RA. 2006. The atlA operon of Streptococcus mutans: role in autolysin maturation and cell surface biogenesis. J. Bacteriol. 188:6877–6888. 10.1128/JB.00536-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown TA, Jr, Ahn SJ, Frank RN, Chen YY, Lemos JA, Burne RA. 2005. A hypothetical protein of Streptococcus mutans is critical for biofilm formation. Infect. Immun. 73:3147–3151. 10.1128/IAI.73.5.3147-3151.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nobbs AH, Vajna RM, Johnson JR, Zhang Y, Erlandsen SL, Oli MW, Kreth J, Brady LJ, Herzberg MC. 2007. Consequences of a sortase A mutation in Streptococcus gordonii. Microbiology 153:4088–4097. 10.1099/mic.0.2007/007252-0 [DOI] [PubMed] [Google Scholar]
- 69.Crowley PJ, Svensater G, Snoep JL, Bleiweis AS, Brady LJ. 2004. An ffh mutant of Streptococcus mutans is viable and able to physiologically adapt to low pH in continuous culture. FEMS Microbiol. Lett. 234:315–324. 10.1111/j.1574-6968.2004.tb09550.x [DOI] [PubMed] [Google Scholar]
- 70.Hasona A, Crowley PJ, Levesque CM, Mair RW, Cvitkovitch DG, Bleiweis AS, Brady LJ. 2005. Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. Proc. Natl. Acad. Sci. U. S. A. 102:17466–17471. 10.1073/pnas.0508778102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palmer SR, Crowley PJ, Oli MW, Ruelf MA, Michalek SM, Brady LJ. 2012. YidC1 and YidC2 are functionally distinct proteins involved in protein secretion, biofilm formation and cariogenicity of Streptococcus mutans. Microbiology 158:1702–1712. 10.1099/mic.0.059139-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vilain S, Pretorius JM, Theron J, Brozel VS. 2009. DNA as an adhesin: Bacillus cereus requires extracellular DNA to form biofilms. Appl. Environ. Microbiol. 75:2861–2868. 10.1128/AEM.01317-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harmsen M, Lappann M, Knochel S, Molin S. 2010. Role of extracellular DNA during biofilm formation by Listeria monocytogenes. Appl. Environ. Microbiol. 76:2271–2279. 10.1128/AEM.02361-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mashburn LM, Whiteley M. 2005. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature 437:422–425. 10.1038/nature03925 [DOI] [PubMed] [Google Scholar]
- 75.Remis JP, Wei D, Gorur A, Zemla M, Haraga J, Allen S, Witkowska HE, Costerton JW, Berleman JE, Auer M. 2014. Bacterial social networks: structure and composition of Myxococcus xanthus outer membrane vesicle chains. Environ. Microbiol. 16:598–610. 10.1111/1462-2920.12187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59:1114–1128. 10.1111/j.1365-2958.2005.05008.x [DOI] [PubMed] [Google Scholar]
- 77.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 104:8113–8118. 10.1073/pnas.0610226104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Funes S, Hasona A, Bauerschmitt H, Grubbauer C, Kauff F, Collins R, Crowley PJ, Palmer SR, Brady LJ, Herrmann JM. 2009. Independent gene duplications of the YidC/Oxa/Alb3 family enabled a specialized cotranslational function. Proc. Natl. Acad. Sci. U. S. A. 106:6656–6661. 10.1073/pnas.0809951106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hasona A, Zuobi-Hasona K, Crowley PJ, Abranches J, Ruelf MA, Bleiweis AS, Brady LJ. 2007. Membrane composition changes and physiological adaptation by Streptococcus mutans signal recognition particle pathway mutants. J. Bacteriol. 189:1219–1230. 10.1128/JB.01146-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lemos JA, Quivey RG, Jr, Koo H, Abranches J. 2013. Streptococcus mutans: a new Gram-positive paradigm? Microbiology 159:436–445. 10.1099/mic.0.066134-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakano K, Matsumura M, Kawaguchi M, Fujiwara T, Sobue S, Nakagawa I, Hamada S, Ooshima T. 2002. Attenuation of glucan-binding protein C reduces the cariogenicity of Streptococcus mutans: analysis of strains isolated from human blood. J. Dent. Res. 81:376–379. 10.1177/154405910208100604 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.