Abstract
More than 50 Helicobacter pylori genes are predicted to encode outer membrane proteins (OMPs), but there has been relatively little experimental investigation of the H. pylori cell surface proteome. In this study, we used selective biotinylation to label proteins localized to the surface of H. pylori, along with differential detergent extraction procedures to isolate proteins localized to the outer membrane. Proteins that met multiple criteria for surface-exposed outer membrane localization included known adhesins, as well as Cag proteins required for activity of the cag type IV secretion system, putative lipoproteins, and other proteins not previously recognized as cell surface components. We identified sites of nontryptic cleavage consistent with signal sequence cleavage, as well as C-terminal motifs that may be important for protein localization. A subset of surface-exposed proteins were highly susceptible to proteolysis when intact bacteria were treated with proteinase K. Most Hop and Hom OMPs were susceptible to proteolysis, whereas Hor and Hof proteins were relatively resistant. Most of the protease-susceptible OMPs contain a large protease-susceptible extracellular domain exported beyond the outer membrane and a protease-resistant domain at the C terminus with a predicted β-barrel structure. These features suggest that, similar to the secretion of the VacA passenger domain, the N-terminal domains of protease-susceptible OMPs are exported through an autotransporter pathway. Collectively, these results provide new insights into the repertoire of surface-exposed H. pylori proteins that may mediate bacterium-host interactions, as well as the cell surface topology of these proteins.
INTRODUCTION
Helicobacter pylori is a Gram-negative bacterium that persistently colonizes the human stomach (1, 2). H. pylori colonization results in gastric inflammation and is a risk factor for the development of gastric adenocarcinoma, gastric lymphoma, and peptic ulcer disease (3, 4). Many H. pylori-induced alterations in host cells are mediated by secreted proteins such as VacA and CagA, which are secreted by type V (autotransporter) and type IV secretion systems, respectively (5, 6). Interactions between H. pylori and the host are also dependent on specialized proteins localized on the surface of H. pylori (7). For example, H. pylori outer membrane protein adhesins mediate H. pylori adherence to gastric epithelial cells (8–10), and surface-exposed components of the cag type IV secretion system (T4SS) have important roles in engaging receptors on host cells (11, 12). Some H. pylori outer membrane proteins (OMPs) can stimulate or inhibit inflammatory responses (13), and others can modulate the activity of the cag type IV secretion system (14–16).
Several approaches have been used to experimentally identify proteins localized on the surface of H. pylori or proteins associated with the H. pylori outer membrane. These approaches have included analyses of susceptibility to protease digestion (17), immunolabeling (18), accessibility to chemical modification (19), and differential detergent solubility (20). These experimental studies have provided useful insights, but most have been limited by the use of only a single method (thus lacking a means for validating results) or the use of methods (such as two-dimensional [2D] gel electrophoresis) that are suboptimal for detecting membrane proteins that are present in low abundance. As an additional complication, it is difficult to separate H. pylori inner and outer membrane proteins using methods optimized for studies of members of the family Enterobacteriaceae (21, 22). Furthermore, H. pylori is prone to undergo autolysis (23), which can potentially lead to artifactual surface exposure of proteins that have an intracellular localization in intact bacteria.
Much of our current knowledge about the outer membrane composition of H. pylori has been deduced from analyses of H. pylori genome sequences. The first analysis of an H. pylori genome sequence (from strain 26695) identified a family of 32 genes that were predicted to encode integral outer membrane proteins (24). These genes are subsequently denoted throughout the manuscript as “annotated OMPs.” A subsequent genomic analysis identified 63 H. pylori genes that were predicted to encode outer membrane proteins and classified these proteins into several different families (25). The largest family, designated the “major OMP family” or Hop family, corresponded to the genes identified in the earlier genomic analysis and was divided into two subfamilies (21 Hop proteins and 12 Hop-related or Hor proteins) (25). Among the 21 Hop proteins, at least five are reported to function as adhesins (BabA, SabA, HopZ, AlpA, and AlpB) (8–10, 26), and several are reported to have porin-like properties (27). In addition to the Hop family of OMPs, several smaller families of putative H. pylori OMPs have been designated, such as Hof proteins, Hom proteins, FecA-like and FrpB-like iron-regulated proteins, and Hef efflux pump proteins (25). The subcellular localization of many of the 63 putative OMPs has not been validated experimentally.
Most of the in silico efforts to identify H. pylori OMPs have been designed to identify OMPs with distinctive C-terminal motifs or β-barrel structures (24). Proteins exported to the surface of H. pylori through pathways such as the flagellar or type IV secretion systems might not possess these features and therefore might not be successfully identified using these approaches. Thus, although the in silico analyses provide a valuable resource for identifying candidate proteins that are likely to be localized to the outer membrane, these analyses offer an incomplete view of the proteins that are potentially exported to the surface of H. pylori. Experimental studies are required in order to gain a more comprehensive understanding of the cell surface proteome and to understand how the production of cell surface proteins might vary under different environmental conditions.
The goals of the current study were to identify and analyze proteins that are localized on the surface of the prototype H. pylori reference strain 26695, to test the hypothesis that the cell surface proteome of H. pylori includes additional proteins besides those that have been predicted based on in silico analyses, and to elucidate the cell surface topology of surface-exposed proteins. To identify surface-exposed proteins, we used multiple complementary biochemical and biophysical methods for protein separation, coupled with robust mass spectrometric methods for protein detection. We report here the identification of proteins that meet multiple criteria for surface-exposed outer membrane localization. These proteins include numerous proteins previously known or predicted to be OMPs, as well as cag pathogenicity island-encoded proteins that are required for activity of the cag type IV secretion system, putative lipoproteins, and additional proteins that were not previously considered to be cell surface components. We show that the majority of Hop and Hom OMPs on the surfaces of intact bacteria are highly susceptible to proteolytic cleavage by an exogenous protease, whereas Hor and Hof proteins are relatively resistant. We present evidence that most of the protease-susceptible OMPs contain a large protease-susceptible extracellular domain exported beyond the outer membrane and a protease-resistant domain at the C terminus with a predicted β-barrel structure. These features suggest that, similar to the secretion of the VacA passenger domain, the N-terminal domains of protease-susceptible OMPs are exported through an autotransporter pathway. Collectively, these results provide new insights into the repertoire of surface-exposed H. pylori proteins that may mediate bacterium-host interactions, as well as the cell surface topology of these proteins.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The variant of Helicobacter pylori strain 26695 used in this study contains an intact cag pathogenicity island (PAI) and a functional cag type IV secretion system (T4SS), but it does not produce flagella (28). A Δcag PAI mutant strain has been described previously (29). Bacteria were grown on either Trypticase soy agar plates supplemented with 5% sheep blood or in sulfite-free brucella broth supplemented with 10% fetal bovine serum (BB-FBS) at 37°C in room air containing 5% CO2.
Generation of rabbit polyclonal antisera.
Anti-UreA serum was obtained from Santa Cruz. CagA, Cag3, CagM, CagT, and CagX, derived from H. pylori 26695, were produced as recombinant proteins in Escherichia coli. Glutathione S-transferase (GST) fusion proteins, maltose-binding protein (MBP) fusion proteins, and 6His-tagged proteins were purified using gluthathione-, maltose-, or nickel-coated beads, respectively. Rabbits were then immunized with the purified proteins. Anti-CagT serum was adsorbed with boiled ΔcagT mutant bacteria prior to use in immunogold electron microscopy (EM) experiments.
Immunoblot analysis.
Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. Proteins were detected by incubating the membrane with primary antisera (diluted 1:5,000 to 1:10,000), followed by horseradish peroxidase-conjugated anti-rabbit IgG as the secondary antibody (Santa Cruz). Labeled proteins were detected by enhanced chemiluminescence (ECL) methodology and visualized using X-ray film.
Immunogold transmission electron microscopy.
H. pylori was grown on Trypticase soy agar plates for 24 h. Broth cultures were inoculated at an initial density of optical density at 600 nm (OD600) of approximately 0.05 and grown for 20 h. Bacteria were harvested in late log phase, corresponding to an OD600 of approximately 0.8, by centrifugation at 3,500 × g for 20 min at 4°C. Surface proteins were labeled using procedures similar to those described previously (28). Briefly, cells were washed once in phosphate-buffered saline (PBS) containing 1 mM CaCl2 and 0.5 mM MgCl2. Bacteria were then fixed with 2% paraformaldehyde (Electron Microscopy Sciences) and 2.5% glutaraldehyde (Electron Microscopy Sciences) in 50 mM sodium cacodylate (Electron Microscopy Sciences) for 30 min at room temperature. Bacteria were then pelleted at 5,000 × g for 10 min. The fixation was quenched with three washes with 0.1% glycine in 50 mM sodium cacodylate. Bacteria were then blocked with 0.1% cold water fish skin gelatin (Electron Microscopy Sciences) for 1 h at 4°C. Bacteria were labeled with CagT antiserum for 1 h at 4°C. Bacteria were washed twice with blocking buffer. Goat anti-rabbit antibody conjugated to 10-nm gold particles (Electron Microscopy Sciences) was applied in blocking buffer for 1 h at room temperature. Samples were washed twice with 50 mM sodium cacodylate. Cells were then placed on Formvar carbon-coated 300-mesh copper grids (Electron Microscopy Sciences) and negatively stained with 0.1% ammonium molybdate (Electron Microscopy Sciences). Samples were imaged with a FEI T-12 transmission electron microscope.
Biotinylation of surface-exposed H. pylori proteins using an amine-reactive biotinylation reagent.
H. pylori was grown on Trypticase soy agar plates for 24 h. Broth cultures were inoculated at an initial density of OD600 of approximately 0.05 and grown for 20 h. Bacteria were harvested in late log phase, corresponding to an OD600 of approximately 0.8, by centrifugation at 3,500 × g for 20 min at 4°C. Surface proteins were labeled using procedures similar to those in a previous publication (19), with several modifications. These included culturing the bacteria in broth instead of plates, use of magnetic beads coated with monomeric avidin (BcMag beads; Bioclone) for purification instead of avidin-agarose, and trichloroacetic acid (TCA) precipitation instead of acetone precipitation. Briefly, the bacterial pellet was washed once, pelleted, and resuspended in labeling buffer (PBS [pH 7.4] containing 1 mM CaCl2, 0.5 mM MgCl2, and 1.6 mM d-biotin [Sigma-Aldrich]). Sulfosuccinimidyl-6-[biotin-amido]hexanoate (sulfo-NHS-LC-biotin; Pierce) was added (final concentration of 200 μM) to the bacterial suspension and incubated for 30 min on ice. The reaction was quenched by the addition of two volumes of TNKCM (50 mM Tris [pH 7.4], 100 mM NaCl, 27 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2) and incubated at room temperature for 10 min. Bacteria were pelleted, washed in TNKCM three times, and resuspended in detergent-free lysis buffer (50 mM Tris [pH 7.4] and 1 mM MgCl2 with EDTA-free protease inhibitor cocktail [Roche]). Bacteria were lysed by sonication, and intact unbroken bacteria were pelleted at 7,000 × g for 10 min. The supernatant was centrifuged at 40,000 × g for 30 min at 4°C. The supernatant was discarded, and the pellet (corresponding to a membrane fraction) was washed three times in TKE (50 mM Tris [pH 7.4], 150 mM KCl, 10 mM EDTA). The pellet was solubilized in TKEZ (50 mM Tris [pH 7.4], 150 mM KCl, 10 mM EDTA, and 2% Zwittergent 3-14 [Fluka] with EDTA-free protease inhibitor cocktail) for 1 h at 4°C. This preparation was centrifuged at 100,000 × g for 1 h at 4°C, and the supernatant (predicted to be enriched in outer membrane proteins) was processed further as described below, to allow purification of biotinylated proteins. A previous study demonstrated that these methods result in minimal labeling of a periplasmic protein (19). A control sample was processed in parallel using exactly the same procedures, except that sulfo-NHS-LC-biotin was omitted from the labeling reaction.
Biotinylation of surface-exposed H. pylori proteins using a carboxyl-reactive biotinylation reagent.
H. pylori was cultured as described above for the biotinylation experiments. The bacterial pellet was washed in morpholineethanesulfonic acid (MES) buffer (100 mM MES [pH 6.0], 500 mM NaCl, 5 mM biotin), pelleted at 3,500 × g for 10 min, and resuspended in MES buffer containing 2 mM 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) (Pierce) and 5 mM N-hydroxysulfosuccinimide (S-NHS) (Pierce). The reaction mixture was incubated for 15 min at 4°C. The pH was raised from 6.0 to 7.3 with 1 M sodium carbonate. Amine-PEG3-biotin (PEG3 stands for three polyethylene glycol groups) was added to the reaction mixture (final concentration of 24 mM) and incubated for 2 h at 4°C. Two volumes of TNKCM (50 mM Tris [pH 7.4], 100 mM NaCl, 27 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2) were added to the reaction mixture and incubated at room temperature for 10 min. Bacteria were pelleted, washed in TNKCM three times, and resuspended in detergent-free lysis buffer. Bacteria were lysed by sonication, and intact unbroken bacteria were pelleted at 7,000 × g for 10 min. The supernatant was centrifuged at 40,000 × g for 30 min at 4°C. The supernatant was removed, and the total membrane pellet was washed three times in TKE. Outer membrane proteins were solubilized in TKEZ for 1 h at 4°C (30). Insoluble proteins were pelleted by centrifugation at 100,000 × g for 1 h at 4°C, and the supernatant (containing outer membrane proteins) was processed further as described below. A control sample was processed in parallel using exactly the same procedures, except that EDC, S-NHS, and amine-PEG3-biotin were omitted from the labeling reaction.
Purification of biotinylated proteins.
To purify biotinylated proteins from the preparations described above, the samples were diluted 10 times in phosphate buffer (100 mM NaPO4 [pH 7.4], 150 mM NaCl). Monomeric avidin magnetic beads (Bioclone) were prepared according to the manufacturer's instructions and equilibrated in washing buffer (100 mM NaPO4 [pH 7.4], 150 mM NaCl, 0.2% Zwittergent 3-14). The sample was incubated with equilibrated beads at room temperature, and bound proteins were eluted from the beads with 100 mM NaPO4 (pH 7.4), 150 mM NaCl, 0.2% Zwittergent 3-14, and 5 mM d-biotin (Sigma) for 30 min at room temperature. Eluted proteins were precipitated with a final concentration of 25% trichloroacetic acid.
Bacterial subcellular fractionation.
H. pylori was grown on Trypticase soy agar plates for 24 h and then grown for 20 h in brucella broth to late log phase, corresponding to an OD600 of approximately 0.8. Bacteria were pelleted by centrifugation at 3,500 × g for 20 min at 4°C. The bacterial cells were then fractionated as described previously (31). In brief, the bacterial pellet was washed in 10 mM Tris (pH 8.0) and resuspended in 10 mM Tris (pH 8.0) with EDTA-free protease inhibitor cocktail. Bacteria were lysed by sonication. Intact bacteria were pelleted at 7,000 × g for 10 min. A total membrane fraction was collected by centrifuging the supernatant at 230,000 × g for 1 h at 4°C. Membrane proteins were solubilized with Triton X-100 buffer (10 mM Tris [pH 8.0], 1% Triton X-100, EDTA-free protease inhibitor cocktail) for 30 min at 4°C. This preparation was centrifuged at 230,000 × g for 45 min at 4°C, yielding a soluble fraction (which is predicted to be enriched in inner membrane proteins) and an insoluble fraction (which is predicted to be enriched in outer membrane proteins).
Proteinase K treatment of intact bacteria.
H. pylori was grown on Trypticase soy agar plates for 24 h and then grown for 20 h in BB-FBS broth to late log phase, corresponding to an OD600 of approximately 0.8. Bacteria were pelleted by centrifugation at 3,500 × g for 20 min at 4°C. Bacteria were then washed with PBS and resuspended in Tris buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 1 mM KCl). Proteinase K was added to bacterial suspensions (final concentration of either 100 μg ml−1 at 22°C or 1 mg ml−1 at 4°C) and incubated for 30 min (18, 28, 31). Phenylmethylsulfonyl fluoride (PMSF) was added to a final concentration of 2 mM. Bacteria were pelleted and resuspended in Tris buffer with protease inhibitor cocktail (Roche). Bacteria were then lysed by sonication, and intact cells were removed by centrifugation at 5,000 × g for 10 min. Total membranes were collected by centrifugation at 200,000 × g for 30 min at 4°C. Membrane pellets were washed in Tris buffer and solubilized in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% sodium deoxycholate) containing protease inhibitor cocktail (18, 28, 31).
Mass spectrometric analysis of samples.
To provide a comprehensive analysis of the protein content in samples, the samples were analyzed by multidimensional protein identification technology (MudPIT). Protein preparations were either run about 2 cm into a 10% Bis-Tris NuPAGE gel, stained with colloidal Coomassie blue, and then subjected to in-gel trypsin digestion, or TCA-precipitated proteins were digested with trypsin in solution. MudPIT was performed essentially as described previously (32), using a ThermoFisher LTQ mass spectrometer equipped with a nano-electrospray source and attached to an Eksigent 1D+ or Nanoacuity (Waters) high-pressure liquid chromatograph (HPLC) unit with an autosampler. Peptide tandem mass spectra (MS/MS) were acquired data dependently with one full-scan MS followed by 5 MS/MS scans. Acidified peptides were loaded onto a 150-μm-inner-diameter (ID) biphasic trapping column comprised of 4-cm strong cation exchange resin (Luna [5-μm particle size]; Phenomenex) followed by 4-cm reverse-phase resin (Jupiter [5-μm particle size, 300-Å pore size]). The trapping column was then attached to a 20-cm-long (Jupiter [3-μm particle size, 300-Å pore size]) 100-μm-ID fused silica analytical column packed into a pulled nanospray tip. Preparations of biotinylated proteins and respective unlabeled controls were analyzed by 8-step MudPIT. Preparations from the proteinase K experiment and cell fractions were analyzed by 11-step MudPIT. Salt pulses were performed by using the autosampler to inject 5 μl of ammonium acetate at 0, 100, 150, 200, 300, 500, 750, and 1,000 mM concentrations for the 8-step analyses used in analyzing the biotinylated preparations and at 0, 50, 75, 100, 150, 200, 250, 300, 500, 750, and 1,000 mM for the 11-step analyses. After each salt injection, peptides were separated using a 105-min aqueous-to-organic gradient (2% to 35% acetonitrile [ACN] for all but the last step, which went to 98% ACN). Peptide MS/MS spectra were queried using SEQUEST (full tryptic specificity) and Myrimatch (semitryptic specificity) against an H. pylori strain 26695 database, to which both common contaminants and reversed versions of the proteins had been appended. For analyses of mass spectrometry data from protease susceptibility experiments, we restricted the analysis to peptides that could be uniquely matched to a single protein. For analyses of mass spectrometry results from all other experiments, we analyzed all peptides detected, including those that could be matched to more than one H. pylori protein. Resulting identifications were filtered to an estimated peptide false discovery rate (FDR) of less than 5% and collated by protein using IDPicker 3.0. All subsequent analyses were performed in Excel and R. All reported proteins were identified based on detecting a minimum of two distinct peptides.
Analysis of mass spectrometry data.
All experiments were done in triplicate, and the resulting spectral data were merged prior to analysis (see Table S1 in the supplemental material). Proteins were identified as enriched in one set of preparations compared to another, based on both analysis of fold enrichment (comparing normalized ratios of numbers of assigned spectra, calculated using either QuasiTel model-generated rates or Rsc values, which both account for abundance of spectral counts when analyzing normalized data) and the statistical significance of differences in abundance of spectral counts assigned to each protein (using Fisher's exact test with Bonferroni's correction) (33, 34). Criteria for identifying proteins that were enriched in one set of samples compared to another were chosen based on comparative analysis of data for spectral proteins previously annotated as outer membrane proteins (likely to be surface exposed) and data for proteins annotated either as ribosomal proteins or inner membrane proteins or other non-outer membrane proteins based on annotation (not likely to be surface exposed) (see Fig. S1 and Table S2 in the supplemental material). The application of the selected cutoffs resulted in a protein false discovery rate of <10% (Fig. S1). We then applied these same cutoff criteria in analysis of spectral data corresponding to all proteins identified in the respective preparations (Tables S3, S4, and S5).
Mapping of protein segments that are susceptible to proteinase K digestion.
To systematically identify sites within individual proteins that were susceptible to proteinase K digestion, we analyzed the spectral data from the proteinase K experiment described above (conducted at 4°C). Spectral coverage data from IDPicker 3.0 files were uploaded into R. For each protein to which peptides had been matched, we then quantified the spectral coverage at individual amino acid positions. We analyzed all amino acid sites for which ≥5 assigned spectra were detected in the untreated control sample (designated “residues available for analysis”) and restricted the analysis to proteins with >100 residues available for analysis. Residues with a ≥5-fold-greater number of assigned spectra in the untreated control samples compared to the protease-treated samples were designated protease-susceptible sites. Residues with a <2-fold-greater number of assigned spectra in the untreated control samples compared to the protease-treated samples were designated protease-nonsusceptible sites. Criteria for the designation of protease-susceptible proteins were chosen based on a comparative analysis of annotated OMPs and proteins predicted to have an inner membrane, cytoplasmic, or periplasmic localization (indicated as “non-OMPs”) (see Fig. S1 and Table S2 in the supplemental material). Proteins with a normalized ratio of susceptible residues to nonsusceptible residues of ≥5 and exhibiting a statistically significant difference in relative abundance of susceptible residues compared to nonsusceptible residues (determined by Fisher's exact test with Bonferroni's correction) were considered susceptible to protease treatment (Fig. S1 and Table S2).
RESULTS
Identification of surface-exposed proteins.
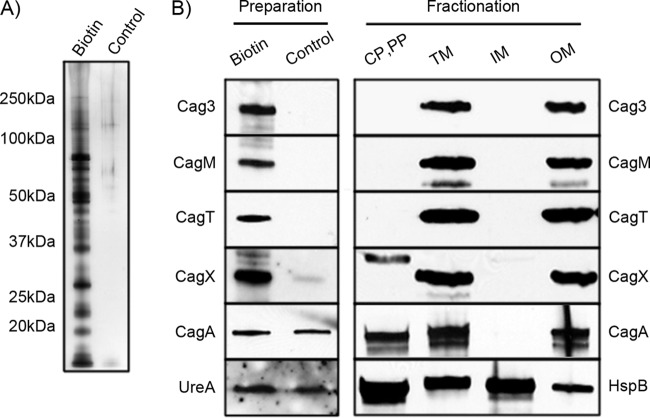
To identify proteins localized on the surface of H. pylori, we used a cell-impermeable amine-reactive biotin conjugation reagent to label surface-exposed proteins on intact bacterial cells. SDS-PAGE and silver staining of proteins purified from the biotinylated bacteria revealed the presence of numerous proteins (Fig. 1A). In parallel, we generated a mock preparation of proteins purified from nonbiotinylated bacteria. This unlabeled control preparation contained a relatively low abundance of proteins compared to the sample of purified biotinylated proteins (Fig. 1A). As a first step in analyzing and comparing these preparations, we immunoblotted the two samples with several available antisera. CagA and UreA were equally abundant in both preparations, whereas Cag3, CagM, CagT, and CagX were more abundant in the biotinylated preparations (Fig. 1B). These results are consistent with an enrichment of specific proteins in the preparation of biotinylated proteins compared to the unlabeled control sample.
FIG 1.
Analysis of purified biotinylated proteins. (A) Biotinylated proteins were purified from biotinylated bacteria, and a control preparation was generated from nonbiotinylated bacteria. Equal volumes of each preparation were analyzed by silver staining. Specific bands were visible in the preparation of biotinylated proteins but not in the control preparation. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel. (B) The preparations from panel A, as well as subcellular fractions, were immunoblotted with antisera to five proteins encoded by the cag PAI, UreA, or HspB. Equal volumes of biotinylated and unlabeled control preparations were loaded into each lane, and standardized amounts of subcellular fractions (25 μg of total protein) were loaded into each lane. The lanes contain soluble proteins predicted to have a cytoplasmic or periplasmic localization (CP,PP), insoluble proteins corresponding to a total membrane preparation (TM), Triton X-100-soluble membrane proteins predicted to have an inner membrane localization (IM), and Triton X-100-insoluble membrane proteins predicted to have an outer membrane localization (OM).
We generated three independent preparations of the biotinylated and unlabeled control samples and analyzed the protein contents of these samples by multidimensional protein identification technology (MudPIT). Consistent with the results of SDS-PAGE and silver staining, the total number of spectra confidently matched to H. pylori proteins (spectral counts) was about 5 times higher in analyses of the biotinylated preparations than in analyses of the control preparations (Table 1; see Table S1 in the supplemental material). In total, we identified >500 H. pylori proteins in these samples (Table S1). The detection of numerous proteins in the control preparations despite very few bands being visible on the silver-stained SDS-polyacrylamide gel is attributed to the high sensitivity of the mass spectrometric detection methods. We employed a statistical approach (based on analysis of spectral counts) to compare the biotinylated preparations with the unlabeled control preparations and identified 85 proteins that were significantly enriched in the biotinylated preparations compared to the control preparations (Table 1; Tables S2 and S3 and Fig. S1).
TABLE 1.
Summary of mass spectrometry data
| Sample | No. of assigned spectral | No. of proteins detectedm |
|---|---|---|
| Biotina | 82,844 | 1,055 |
| Controlb | 16,009 | 941 |
| Biotin>Controlc | NA | 85 |
| TMd | 90,604 | 1,120 |
| CP,PPe | 78,773 | 1,066 |
| TM>CP,PPf | NA | 192 |
| OMg | 64,478 | 1,095 |
| IMh | 74,320 | 1,114 |
| OM>IMi | NA | 156 |
| Untreated 4°Cj | 92,644 | 1,063 |
| Untreated RTj | 75,482 | 1,056 |
| PK 4°Cj | 100,521 | 1,064 |
| PK RTj | 103,021 | 1,063 |
| Untreated>PK 4°Ck | NA | 7 |
| Untreated>PK RTk | NA | 6 |
Biotinylated preparations.
Unlabeled control preparations.
Subset of proteins that were significantly enriched in biotinylated preparations compared to unlabeled control preparations.
Insoluble proteins (in the absence of detergent), predicted to localize to the total membrane (TM) compartment.
Soluble proteins (in the absence of detergent), predicted to localize to the cytoplasmic and periplasmic compartments (CP,PP).
Subset of proteins that were significantly enriched in the insoluble preparations (TM) compared to soluble preparations (CP,PP).
Triton X-100-insoluble proteins, predicted to localize to the outer membrane (OM) compartment.
Triton X-100-soluble proteins, predicted to localize to the inner membrane (IM) compartment.
Subset of proteins that were significantly enriched in the Triton X-100-insoluble preparations compared to the Triton X-100-soluble preparations.
Intact bacteria were incubated in buffer or treated with proteinase K (PK) at 4°C or room temperature (RT).
Subset of proteins for which assigned peptides were detected at significantly higher levels in untreated preparations than in proteinase K-treated preparations.
Results represent the sum of assigned spectra, based on analysis of three preparations. NA, not applicable.
Results represent the total number of identified proteins, based on analysis of three preparations.
We also used biophysical methods independent of biotin labeling to identify proteins associated with the H. pylori outer membrane. This approach was based on the reasoning that most surface-exposed proteins are likely to be associated with the bacterial outer membrane. As a first step, we used detergent-free conditions to generate preparations of soluble H. pylori proteins (predicted to contain cytoplasmic and periplasmic compartments) and insoluble H. pylori proteins (predicted to be derived from both inner and outer membranes, subsequently referred to as the total membrane fraction). Then we fractionated the total membrane preparation based on differential protein solubility in Triton X-100, which allows separation of inner membrane proteins and outer membrane proteins (31). Immunoblotting of these fractions with the same antisera used in Fig. 1A revealed that four Cag proteins (Cag3, CagM, CagT, and CagX) were more abundant in the total membrane fraction than in the cytoplasmic/periplasmic fraction and also more abundant in the outer membrane fraction than in the inner membrane fraction. We generated three independent preparations of each fraction, and the protein composition of these samples was analyzed by MudPIT. We identified 191 proteins that were significantly enriched in the total membrane preparations compared to the soluble protein preparations (Table 1; see Tables S2 and S4 and Fig. S1 in the supplemental material), and we identified 155 proteins that were significantly enriched in the Triton X-100-insoluble outer membrane fractions compared to the Triton X-100-soluble inner membrane fractions (Table 1; Tables S2 and S5 and Fig. S1).
To evaluate the efficacy of the procedures described above for enrichment of putative outer membrane proteins, we analyzed the presence of proteins in each preparation that were previously predicted by Tomb et al. to be outer membrane proteins (“annotated OMPs”) (24) and subsequently classified as Hop or Hop-related proteins (25). There was a significant enrichment of proteins with an outer membrane protein annotation in preparations of biotinylated proteins compared to preparations of control (nonbiotinylated) proteins (P < 0.0001) (see Fig. S2 in the supplemental material). Similarly, there was an enrichment in annotated OMPs when comparing the total membrane preparations with the soluble protein preparations (cytoplasmic/periplasmic proteins) (P < 0.0001) and when comparing the outer membrane preparations with the inner membrane preparations (P < 0.0001) (Fig. S2). These analyses indicated that, as expected, each of the experimental approaches resulted in an enrichment of annotated outer membrane proteins.
Proteins meeting multiple criteria for surface-exposed outer membrane localization.
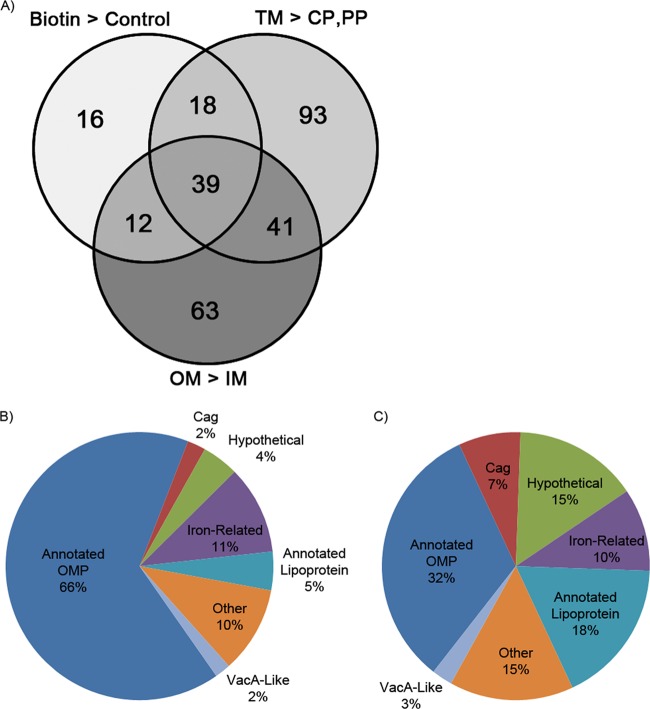
We next sought to identify proteins that met multiple criteria for surface-exposed outer membrane localization, thereby providing stronger evidence compared to the use of only a single method. Specifically, we sought to identify proteins that met three criteria: (i) significantly enriched in the biotinylated preparations compared to the control (nonbiotinylated) preparations, (ii) significantly enriched in the total membrane preparation compared to the soluble fraction, and (iii) significantly enriched in the Triton X-100-insoluble outer membrane fraction compared to the Triton X-100-soluble inner membrane fraction. Thirty-nine proteins (localized in the center of the Venn diagram shown in Fig. 2A) met all three of these criteria (Table 2). In an analysis of the detected peptides corresponding to this set of 39 proteins, 66% of the spectra were assigned to proteins previously annotated by Tomb et al. (24) as outer membrane proteins (Fig. 2B), which is significantly higher than the corresponding values for any of the individual preparations (see Fig. S2 in the supplemental material). Thirteen of these 39 proteins were annotated by Tomb et al. as predicted OMPs (corresponding to 12 Hop proteins and 1 Hop-related or Hor protein) (24), and 20 were annotated by Alm et al. (25) as predicted OMPs [including three annotated as iron(III) dicitrate transport (FecA) proteins, an iron-regulated FrpB protein, and the VacA-like protein FaaA (HP0609/0610)] (18). The set of 39 proteins also includes numerous proteins not previously annotated as OMPs. These include three Cag proteins (Cag3, CagM, and CagT), each of which is required for the function of the cag T4SS that translocates the effector protein CagA into gastric epithelial cells (cag T4SS) (35), several annotated as lipoproteins (including Lpp20 and HP1571) (36), two homologs of the flagellar sheath adhesin HpaA (HP0410 and HP0492) (19, 37), a protein annotated as “protective surface antigen D15” which corresponds to an outer membrane assembly protein BamA ortholog (HP0655), a secreted serine protease (HtrA [HP1019]) (38, 39), a protein annotated as a conserved secreted protein or peptidyl prolyl cis-trans isomerase (HP0977), a plasminogen-binding protein (PgbB [HP0863]) (40), and several proteins identified annotated as “hypothetical proteins” (Table 2 and Fig. 2C).
FIG 2.
Use of multiple criteria to identify surface-exposed outer membrane proteins. (A) Venn diagram of proteins identified as enriched in the biotinylated preparation compared to the control preparation (Biotin > Control), enriched in the total membrane preparation compared to the soluble fraction (TM > CP,PP), and enriched in a Triton X-100-insoluble preparation compared to a Triton X-100-soluble preparation (OM > IM). Thirty-nine proteins met all three criteria. (B and C) Characteristics of the 39 proteins in the center segment of the Venn diagram, based on proportion of spectral counts (B) or proportion of annotated proteins in each class (C). Annotated OMPs represent Hop and Hor OMPs.
TABLE 2.
Proteins meeting multiple criteria for surface-exposed outer membrane proteinsa
| Original annotation | Alternate protein name(s) | Gene no.b |
|---|---|---|
| Outer membrane protein (Omp2) | HopD | HP0025 |
| Hypothetical protein | HP0036 | |
| Hypothetical protein | HP0120 | |
| Outer membrane protein (Omp4) | HorB | HP0127 |
| Outer membrane protein (Omp7) | HopF/HopX | HP0252 |
| Hypothetical protein | HP0358 | |
| Putative neuraminyllactose-binding hemagglutinin homolog (HpaA) | HP0410 | |
| Outer membrane protein (Omp12) | HopJ | HP0477 |
| Lipoprotein, putative | HpaA | HP0492 |
| cag pathogenicity island protein (Cag3) | Cag3 | HP0522 |
| cag pathogenicity island protein (Cag12) | CagT | HP0532 |
| cag pathogenicity island protein (Cag16) | CagM | HP0537 |
| Penicillin-binding protein 1A (PBP-1A) | HP0597 | |
| Membrane fusion protein (MtrC) | HefB | HP0606 |
| Hypothetical proteinc | FaaA | HP0609c |
| Toxin-like outer membrane proteinc | FaaA | HP0610c |
| Outer membrane protein (Omp13) | HopH/OipA | HP0638 |
| Protective surface antigen D15 | BamA | HP0655 |
| Iron(III) dicitrate transport protein (FecA) | FecA-1 | HP0686 |
| Lipoprotein, putative | HP0746 | |
| Iron(III) dicitrate transport protein (FecA) | FecA-2 | HP0807 |
| Lipoprotein, putative | HP0838 | |
| Lipoprotein, putative | PgbB | HP0863 |
| Outer membrane protein (Omp19) | BabB/HopT | HP0896 |
| Outer membrane protein (Omp20) | HopC/AlpA | HP0912 |
| Outer membrane protein (Omp21) | HopB/AlpB | HP0913 |
| Outer membrane protein (Omp22) | HopK | HP0923 |
| Conserved hypothetical secreted protein | HP0977 | |
| Lipoprotein, putative | HP1002 | |
| Serine protease (HtrA) | HP1019 | |
| Outer membrane protein (Omp25) | HopI | HP1156 |
| Outer membrane protein (Omp26) | HopL | HP1157 |
| Hypothetical protein | HP1173 | |
| Outer membrane protein (Omp27) | HopQ | HP1177 |
| Outer membrane protein (Omp28) | BabA/HopS | HP1243 |
| Paralyzed flagellar protein (PflA) | HP1274 | |
| Iron(III) dicitrate transport protein (FecA) | FecA-3 | HP1400 |
| Membrane-associated lipoprotein (Lpp20) | HP1456 | |
| Iron-regulated outer membrane protein (FrpB) | FrpB-3 | HP1512 |
| Rare lipoprotein A (RlpA) | HP1571 |
These proteins correspond to the 39 proteins that are depicted in the center of the Venn diagram shown in Fig. 2.
Gene number or locus tag.
HP0609 and HP0610 were originally annotated as two separate proteins, but they correspond to a single protein (VacA-like protein FaaA).
We also analyzed the proteins that met only two criteria for outer membrane localization, corresponding to relevant segments of the Venn diagram in Fig. 2A. One subset of 18 proteins (enriched in the biotinylation preparations and the total membrane preparations, but not in the Triton X-100-insoluble membrane preparations) included six annotated OMPs (BabC or HopU [HP0317], HomA [HP0710], HopM [HP0227], HopA [HP0229], HorF [HP0671], and HopN [HP1342]) and HpaA (a flagellar sheath adhesin; HP0797) (41). A high proportion of the proteins in the other segments of the Venn diagram (Fig. 2A) corresponded to proteins predicted to have an intracellular localization, and these probably represent false-positive results.
Collectively, these experiments detected numerous proteins on the surface of H. pylori that had previously been predicted to have an outer membrane localization (“annotated OMPs”), as well as additional proteins that were not previously predicted to be localized on the H. pylori surface. The evidence for surface localization is strongest for the set of proteins meeting multiple criteria for surface-exposed outer membrane localization, depicted in the center of the Venn diagram in Fig. 2A and in Table 2.
Immunogold EM analysis of CagT.
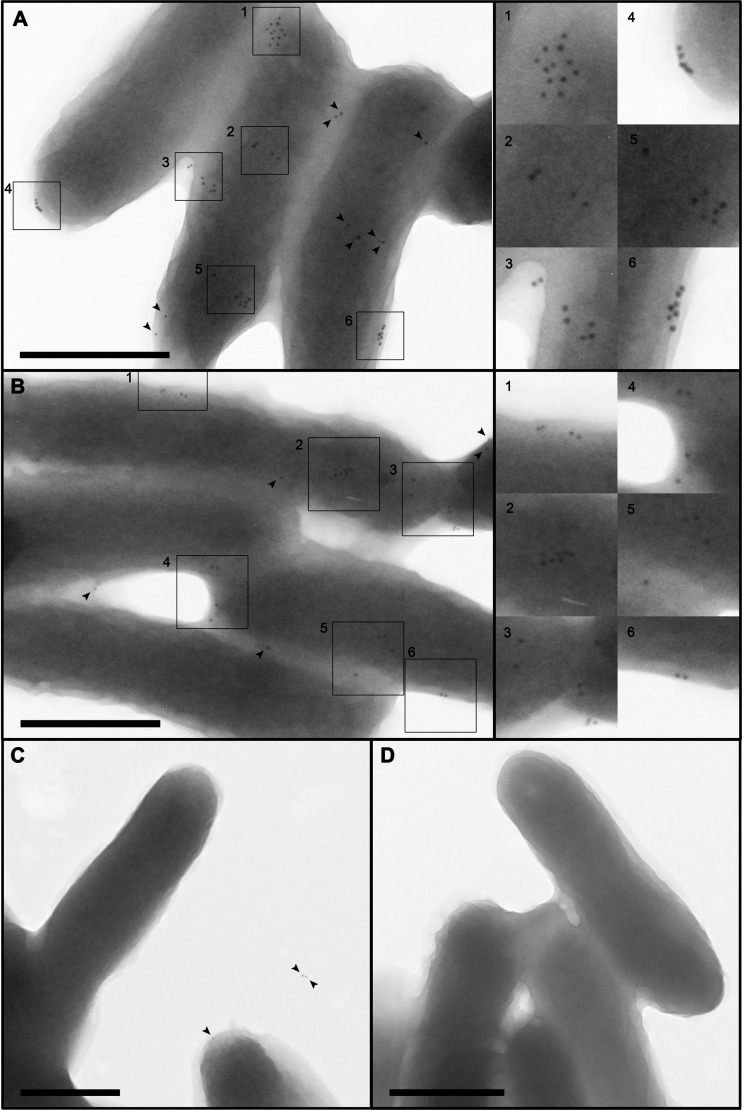
To provide further evidence for the surface-exposed localization of proteins not previously annotated as OMPs, we selected one of the proteins that met multiple criteria for outer membrane localization (CagT) and undertook further analysis of this protein by immunogold electron microscopy. In immunogold labeling experiments with an anti-CagT antiserum, we detected a significantly greater number of gold particles on the surfaces of wild-type bacteria than on the surfaces of Δcag PAI mutant bacteria, which fail to produce CagT (P < 0.0001) (Fig. 3). In images of the wild-type bacteria, multiple gold particles were often clustered together, which suggests that CagT is nonrandomly distributed on the bacterial surface. These immunogold labeling studies corroborate the surface localization of CagT and provide further validation of the experimental approaches used in this study for the identification of surface-exposed proteins.
FIG 3.
Localization of CagT by immunogold EM analysis. (A to C) H. pylori strains were immunolabeled with rabbit antiserum to CagT, followed by secondary antibodies conjugated to 10-nm gold particles. (A and B) Wild-type (WT) strain. (C) Δcag PAI mutant strain. Higher-magnification images of the regions in boxes, containing a high density of gold particles, are shown to the right of panels A and B. Arrowheads designate additional gold particles. (D) Immunogold labeling of the WT strain using secondary antibodies conjugated to gold particles, without primary antiserum. The number of gold particles detected on WT bacteria was significantly higher than the number detected on the cag ΔPAI mutant strain (mean 3.6 gold particles per WT cell and 1.8 gold particles per cag ΔPAI mutant cell, based on analysis of >200 bacteria of each type; P value < 0.0001, Welch two-sample t test). Bars, 500 nm.
Experimental evidence of amino-terminal signal sequence cleavage.
Many surface-exposed proteins are predicted to undergo cleavage of an N-terminal signal sequence during Sec-dependent export across the inner membrane. To experimentally identify surface-exposed proteins that underwent cleavage of amino-terminal signal sequences, we analyzed the spectral data from the biotinylation experiments and sought to identify peptides that had been generated by non-tryptic-cleavage events occurring near the amino termini of proteins. Fourteen of the 39 surface-exposed proteins shown in Table 2 showed evidence of amino-terminal non-tryptic-cleavage events consistent with Sec-dependent signal sequence processing (see Table S6 in the supplemental material). An alignment of these sequences, relative to the sites of nontryptic cleavage, is shown in Fig. 4. The consensus sequence includes a stretch of approximately 10 uncharged or hydrophobic residues upstream of the cleavage site, a semiconserved leucine at position −3, and an alanine at position −1. An acidic patch of 1 to 3 residues was detected immediately downstream from the cleavage site (Table S6). Interestingly, the signal peptide prediction software SignalP 4.1 (42) predicted the existence of only 7 of these 14 cleavage sites, and for one of the proteins (HopI [HP1156]), the cleavage site predicted by SignalP differed by one amino acid position compared to what was experimentally detected. This discrepancy suggests that the currently available signal peptide prediction software is not optimally designed for analysis of signal peptides in epsilonproteobacteria such as H. pylori.
FIG 4.
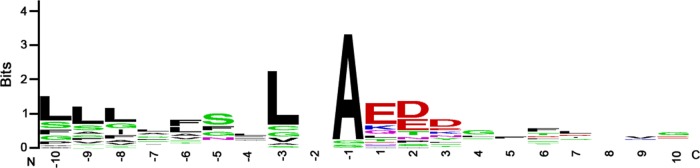
Experimentally detected sites of signal peptide cleavage. We identified peptides in the preparations of biotinylated proteins that corresponded to non-tryptic-cleavage events and that could be matched to sites near the N termini of proteins, suggesting the occurrence of signal peptide cleavage. This analysis was restricted to peptides that could be matched to a single protein. Amino-terminal nontryptic cleavage was detected for 14 of the 39 surface-exposed proteins shown in Table 2 (see Table S6 in the supplemental material). These sequences were then aligned using WebLogo (weblogo.berkeley.edu). In the alignment, the numbering of positions is relative to the detected non-tryptic-cleavage site. The N terminus (N) and C terminus (C) are indicated.
Multiple classes of C-terminal motifs in surface-exposed proteins.
Targeting of proteins to the outer membrane is often dependent on the presence of a C-terminal motif required for interaction with the Bam complex (43, 44). Therefore, we analyzed the C-terminal sequences of the 39 surface-exposed proteins shown in Table 2. The most commonly identified amino acids at the C-terminal position were phenylalanine (n = 16), lysine (n = 9), and tyrosine (n = 5) (see Table S7 in the supplemental material).
There were several similarities in the C-terminal sequences of the proteins terminating in phenylalanine or tyrosine. Specifically, 13 of the 39 surface-exposed proteins shown in Table 2 contain closely related C-terminal sequences characterized by a conserved tyrosine at the −5 position, a conserved arginine at the −13 position, either a valine (7 proteins) or phenylalanine (6 proteins) at position −9, and semiconserved tyrosines at positions −8 (11 proteins) and −15 (10 proteins) (Fig. 5). Thus, the consensus C-terminal motif for these proteins is (Y/F)XR XXX (V/F)Y XXY XXX (F/Y), and three of these proteins (HopQ, BabA, and BabB) contain an identical C-terminal motif (VYLNYVFAY). All 13 of these C-terminal sequences contain hydrophobic residues at the −1, −3, −5, −7, −8, −9, and −15 positions, which is consistent with a motif that allows insertion of proteins into the outer membrane through a BamA (or Omp85)-dependent process (43, 45). Twelve of the 13 proteins containing this motif are classified as Hop proteins, and one is classified as a Hop-related protein (HorB [HP0127]). Eight of the 21 proteins with C-terminal phenylalanines or tyrosines, including the VacA-like protein FaaA, contain C-terminal sequences that are somewhat different from the consensus sequence described above, but still contain hydrophobic residues at the −3, −5, −7, and −9 positions.
FIG 5.
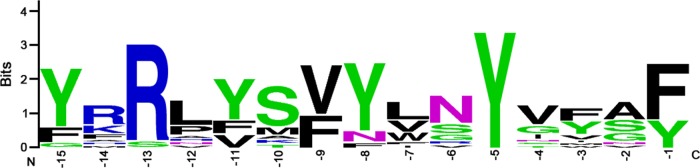
Analysis of a conserved C-terminal motif. Among the 39 surface-exposed proteins shown in Table 2, 15 contain a distinctive C-terminal motif, characterized by conserved residues at positions −15, −13, −9, −5, and −1 (see Table S7 in the supplemental material). The numbers correspond to the distance from the carboxyl terminus. Sequences were aligned using WebLogo (weblogo.berkeley.edu).
Eighteen of the 39 proteins did not contain a C-terminal phenylalanine or tyrosine; in nine cases, the C-terminal amino acid was lysine, and in two cases, the C-terminal amino acid was arginine. There was no apparent conservation of C-terminal sequences among the 9 proteins with C-terminal lysines or among the larger set of 18 proteins terminating in amino acids other than phenylalanine or tyrosine. However, many of these proteins had C-terminal segments that were characterized by an abundance of basic residues (lysine and arginine) (see Table S7 in the supplemental material). Interestingly, several of these proteins were previously annotated as lipoproteins. We hypothesized that the amine-reactive biotinylation reagent might preferentially label these C-terminal basic residues, leading to an increased representation of these proteins in the preparation of purified biotinylated proteins. To address this possibility, we biotinylated surface-exposed proteins with a cell-impermeable carboxyl-reactive biotinylation reagent and then purified and analyzed the biotinylated proteins. By using this approach, we identified 32 proteins that were significantly enriched in the biotinylated preparation compared to the unlabeled control (Table S8). Twenty-seven of these 32 proteins were also identified as surface exposed in experiments using the amine-linked biotinylation method (Fig. S3). Thus, the surface-exposed proteins identified when using a carboxyl-reactive biotinylation reagent were similar to those identified when using the amine-reactive biotinylation reagent. Eight of the 27 surface-exposed proteins identified using both methods terminated in either a lysine or arginine, and the proportion of surface-exposed proteins terminating in either lysine or arginine was similar in experiments using the two methods (3 of 32 proteins with the carboxyl-reactive method and 9 of 85 proteins with the amine-reactive method). This provides evidence that the proteins identified using the amine-linked biotinylation approach were not identified simply as a consequence of preferentially labeling C-terminal basic residues.
Susceptibility of surface-exposed proteins to proteinase K digestion and cell surface topology of surface-exposed proteins.
We next undertook experiments designed to assess the susceptibility of surface-exposed proteins on intact bacteria to digestion by an exogenous protease. These experiments provided an additional approach for localizing specific proteins to the surface of H. pylori and also allowed us to test the hypothesis that specific groups of surface-exposed OMPs might differ in susceptibility to proteolytic cleavage. We treated intact bacteria with proteinase K under conditions previously shown to preferentially digest surface-exposed proteins but not intracellular proteins (28, 31). We then used MudPIT to identify proteins for which there were significantly reduced numbers of assigned spectra detected in protease-treated bacteria compared to control bacteria. We identified seven such proteins that were highly susceptible to proteolysis: VacA (HP0887), BabA (HP1243), BabB (HP0896), BabC (HopU [HP0317]), HopQ (HP1177), HomA (HP0710), and HomD (HP1453) (Table 3; see Table S9 in the supplemental material). Proteinase K treatment did not cause any significant reduction in the abundance of peptides originating from inner membrane proteins, such as FixO (HP0145) and FtsH (HP1069) (Table S9). The results were similar in experiments conducted at 4°C and at 22°C (Table 3).
TABLE 3.
Surface-exposed proteins highly susceptible to proteinase K digestion, based on total assigned spectraa
| Original annotation | Alternative name(s) | Gene no. | Spectral countb |
P valuec |
Fold changed |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| 4°C |
22°C |
4°C | 22°C | 4°C | 22°C | |||||
| Untreated | Prot K | Untreated | Prot K | |||||||
| Vacuolating cytotoxin | VacA | HP0887 | 986 | 185 | 793 | 183 | <0.0001 | <0.0001 | 5.85 | 5.98 |
| Outer membrane protein (Omp9) | HopU | HP0317 | 653 | 92 | 567 | 24 | <0.0001 | <0.0001 | 7.75 | 32 |
| Outer membrane protein (Omp27) | HopQ | HP1177 | 1,446 | 773 | 1,229 | 760 | <0.0001 | <0.0001 | 2.06 | 2.24 |
| Outer membrane protein (Omp19) | BabB/HopT | HP0896 | 577 | 95 | 510 | 33 | <0.0001 | <0.0001 | 6.63 | 21 |
| Outer membrane protein (Omp28) | BabA/HopS | HP1243 | 341 | 20 | 273 | 10 | <0.0001 | <0.0001 | 18.23 | 35.8 |
| Conserved hypothetical protein | HomA | HP0710 | 264 | 114 | 184 | 107 | <0.0001 | <0.0001 | 2.52 | 2.36 |
| Conserved hypothetical protein | HomD | HP1453 | 45 | 13 | 37 | 17 | <0.005 | 0.1463 | 3.68 | 2.94 |
| Total assigned spectra | 89,222 | 97,275 | 89,222 | 99,781 | ||||||
Intact H. pylori bacteria were incubated with proteinase K at 4°C or 22°C, and membrane fractions from the treated bacteria (as well as untreated controls) were then analyzed by MudPIT.
The combined results of three independent experiments for each condition (untreated or proteinase K treated [Prot K]) are shown.
Results for proteinase K-treated bacteria were compared with results for untreated bacteria, using Fisher's exact test with Bonferonni's multiple test correction.
The magnitude of changes in protein abundance in response to proteinase K digestion was calculated based on analysis of 2Rsc.
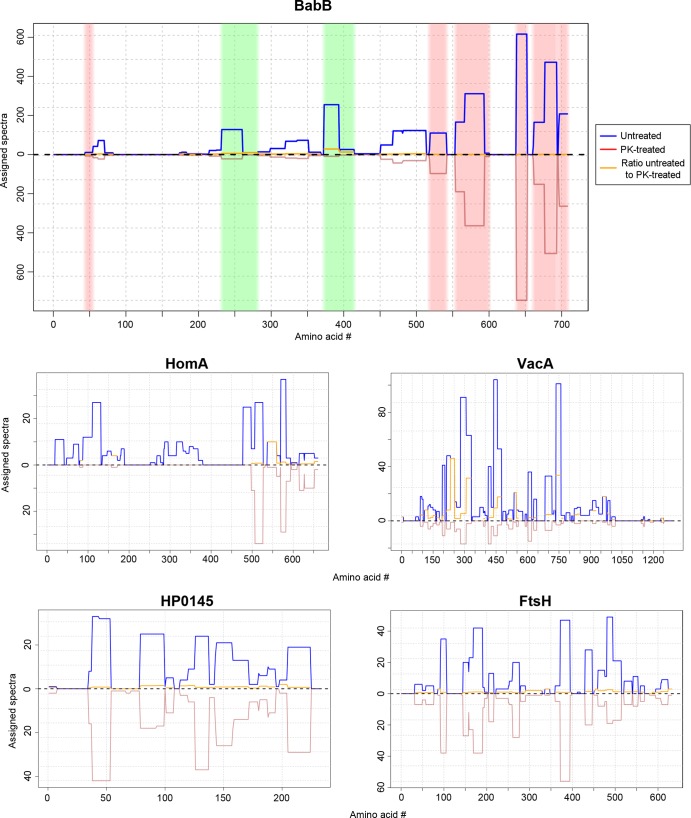
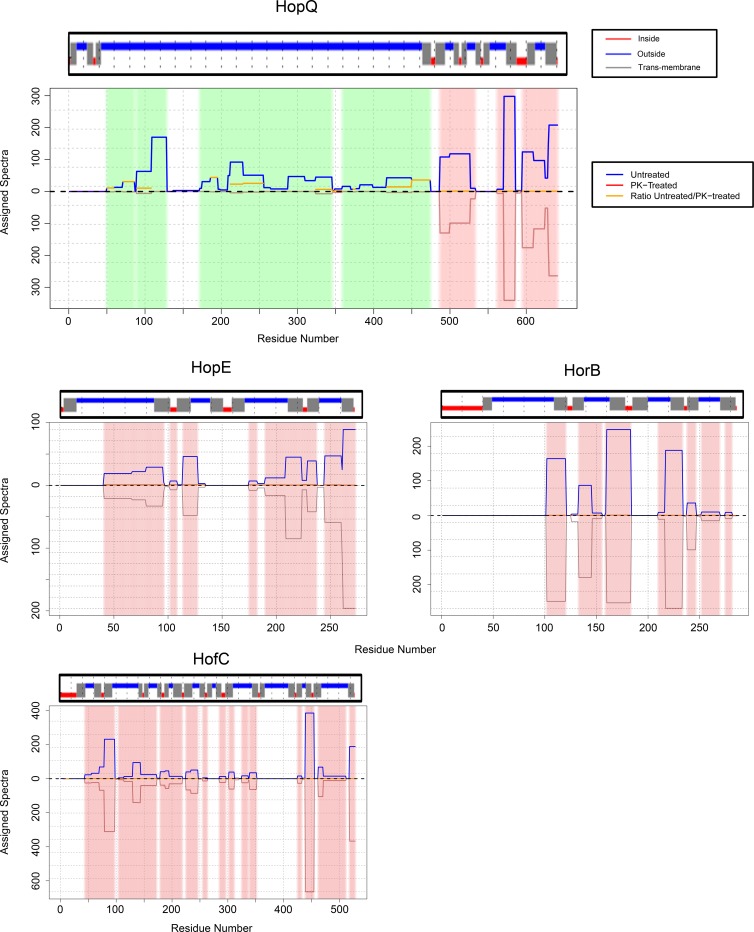
The methodology described above, based on analysis of all spectra assigned to individual proteins, allowed us to identify proteins that were highly susceptible to proteolytic digestion by an exogenous protease, but this is probably not a sensitive approach for identifying proteins that undergo proteolysis at relatively few sites. Therefore, we systematically analyzed the assigned spectra for each detected protein at the level of individual amino acids, which permitted a more in-depth assessment of sites where proteins had undergone proteolysis. The identification of protease-susceptible sites using this approach is illustrated in the top panel of Fig. 6. Using this approach, six of the seven proteins described above were identified as susceptible to proteolytic digestion, and nine additional protease-susceptible proteins were identified (Fig. 6 and Table 4). HomD, identified as protease susceptible in the earlier analysis (Table 3), was excluded from the current analysis because the mass spectral data did not meet the inclusion criteria. Thirteen of the 15 protease-susceptible proteins met multiple criteria for surface-exposed localization in the earlier biotinylation and cell fractionation experiments (see Fig. S4 in the supplemental material). Additionally, VacA was identified as susceptible to proteinase K treatment. The other protein exhibiting evidence of protease susceptibility (cell division inhibitor MinD) is presumed to represent a false-positive result. A systematic comparison of the protease susceptibility of annotated OMPs detected in the proteomic analysis revealed that the majority of Hop proteins were susceptible to proteolytic digestion, but several Hop proteins (HopJ, HopK, HopE, and HopG) were resistant (Table 5). In comparison to Hop proteins, the Hof and Hor families of OMPs were relatively resistant to extracellular protease digestion (Table 5).
FIG 6.
Susceptibility of surface-exposed proteins to proteinase K treatment. Intact H. pylori cells were treated with proteinase K (1 mg ml−1) or buffer control at 4°C and then analyzed by MudPIT. Three experiments were performed, and the results were merged. We then analyzed the assigned spectra for each detected protein at the level of individual amino acids, and sites susceptible to proteolytic cleavage or resistant to proteolytic cleavage were identified, based on criteria defined in Materials and Methods. The blue lines (top half of graphs) depict the numbers of assigned spectra from experiments with control (untreated) bacteria, the red lines (bottom half of graphs) depict the numbers of assigned spectra from experiments with proteinase K (PK)-treated bacteria, and the orange lines indicate the ratio of assigned spectra in untreated compared to protease-treated bacteria. Data for 5 representative proteins are shown. In the top panel, the green shading illustrates protease-susceptible regions in BabB (exhibiting a ≥5-fold difference in the spectral counts when comparing untreated bacteria and protease-treated bacteria), and the pink shading illustrates protease-resistant regions (exhibiting a <2-fold difference in the spectral counts). The designation of protease-susceptible or protease-resistant regions was restricted to residues for which ≥5 assigned spectra were detected in the untreated samples. The N-terminal portions of BabB, HomA, and VacA were highly susceptible to protease digestion, whereas intracellular proteins such as the cytochrome c oxidase subunit FixO (HP0145) and the cell division protein FtsH were resistant to protease digestion.
TABLE 4.
Proteins identified as susceptible to extracellular proteinase K digestion, based on analysis of individual amino acid sites
| Original annotation | Alternative name(s) | Gene no. | Protein lengtha | No. of amino acid residues |
Normalized ratio (PK-susceptible residues to nonsusceptible residues)d | |
|---|---|---|---|---|---|---|
| Available for analysisb | Susceptible to proteasec | |||||
| Outer membrane protein (Omp5) | HopM | HP0227 | 691 | 356 | 143 | 26.9 |
| Outer membrane protein (Omp6) | HopA | HP0229 | 483 | 283 | 197 | 33.7 |
| Outer membrane protein (Omp7) | HopF | HP0252 | 487 | 296 | 85 | 6.0 |
| Outer membrane protein (Omp9) | HopU/BabC | HP0317 | 745 | 480 | 157 | 16.5 |
| Cell division inhibitor | MinD | HP0331 | 268 | 121 | 40 | 8.5 |
| Toxin-like outer membrane protein | FaaA | HP0610e | 1,943 | 199 | 39 | 5.3 |
| Outer membrane protein (Omp13) | HopH/OipA | HP0638 | 305 | 120 | 31 | 5.2 |
| Conserved hypothetical protein | HomA | HP0710 | 660 | 283 | 205 | 38.6 |
| Vacuolating cytotoxin | VacA | HP0887 | 1,290 | 642 | 377 | 57.7 |
| Outer membrane protein (Omp19) | BabB | HP0896 | 708 | 442 | 92 | 9.5 |
| Outer membrane protein (Omp20) | HopC/AlpA | HP0912 | 515 | 360 | 208 | 23.4 |
| Outer membrane protein (Omp21) | HopB/AlpB | HP0913 | 529 | 273 | 135 | 14.4 |
| Outer membrane protein (Omp27) | HopQ | HP1177 | 641 | 495 | 367 | 45.7 |
| Outer membrane protein (Omp28) | BabA | HP1243 | 733 | 444 | 190 | 21.2 |
| Outer membrane protein (Omp29) | HopN | HP1342 | 691 | 356 | 143 | 26.9 |
Total number of amino acids in the protein.
Total number of amino acids with ≥5 assigned spectra in the untreated samples.
Total number of amino acids with ≥5-fold-more assigned spectra in the untreated samples compared to proteinase K-treated samples.
Ratio of proteinase K (PK)-susceptible residues compared to nonsusceptible residues, normalized by total residues for each category.
HP0609 and HP0610 were originally annotated as two separate proteins, but they correspond to a single protein (VacA-like protein FaaA).
TABLE 5.
Protease susceptibility of annotated OMPs
| OMP | Gene no. | Protein length (no. of amino acids) | No. of amino acid residues |
% residues susceptible to proteasec | |
|---|---|---|---|---|---|
| Available for analysisa | Susceptible to proteaseb | ||||
| Hop family | |||||
| HopH | HP0638 | 305 | 120 | 31 | 25.83 |
| HopJ | HP0477 | 367 | 129 | 0 | 0 |
| HopK | HP0923 | 369 | 129 | 0 | 0 |
| HopE | HP0706 | 273 | 162 | 0 | 0 |
| HopL | HP1157 | 1,230 | 221 | 34 | 15.38 |
| HopG | HP0254 | 431 | 233 | 0 | 0 |
| HopI | HP1156 | 696 | 259 | 46 | 17.76 |
| HopB/AlpB | HP0913 | 529 | 273 | 135 | 49.45 |
| HopA | HP0229 | 483 | 283 | 197 | 69.61 |
| HopF | HP0252 | 487 | 296 | 85 | 28.72 |
| HopM | HP0227 | 691 | 356 | 143 | 40.17 |
| HopN | HP1342 | 691 | 356 | 143 | 40.17 |
| HopC/AlpA | HP0912 | 515 | 360 | 208 | 57.78 |
| HopD | HP0025 | 711 | 436 | 52 | 11.93 |
| HopT/BabB | HP0896 | 708 | 442 | 92 | 20.81 |
| HopS/BabA | HP1243 | 733 | 444 | 190 | 42.79 |
| HopU/BabC | HP0317 | 745 | 480 | 157 | 32.71 |
| HopQ | HP1177 | 641 | 495 | 367 | 74.14 |
| Hop-related family | |||||
| HorI | HP1113 | 277 | 24 | 0 | 0 |
| HorC | HP0324 | 254 | 77 | 20 | 25.97 |
| HorE | HP0472 | 186 | 85 | 0 | 0 |
| HorL | HP1395 | 242 | 85 | 20 | 23.53 |
| HorF | HP0671 | 270 | 123 | 0 | 0 |
| HorB | HP0127 | 286 | 125 | 0 | 0 |
| HorJ | HP1469 | 248 | 131 | 0 | 0 |
| HorH | HP1107 | 230 | 132 | 25 | 18.94 |
| HorK | HP1501 | 388 | 221 | 0 | 0 |
| Hom family | |||||
| HomC | HP0373 | 700 | 25 | 25 | 100 |
| HomD | HP1453 | 746 | 26 | 26 | 100 |
| HomA | HP0710 | 660 | 283 | 205 | 72.44 |
| Hof family | |||||
| HofD | HP0487 | 480 | 87 | 0 | 0 |
| HofF | HP0788 | 499 | 94 | 0 | 0 |
| HofG | HP0914 | 514 | 177 | 0 | 0 |
| HofB | HP1083 | 479 | 266 | 10 | 3.76 |
| HofC | HP0486 | 528 | 326 | 0 | 0 |
| HofE | HP0782 | 455 | 0 | 0 | 0 |
Total number of amino acids with ≥5 assigned spectra in the untreated samples.
Total number of amino acids with ≥5-fold-more assigned spectra in the untreated samples compared to proteinase K-treated samples.
Percentage of residues available for analysis (defined in footnote a) that were susceptible to proteolysis (defined in footnote b).
In an analysis of the regions within individual proteins that were susceptible or resistant to proteolysis, we noted that many annotated OMPs (especially those in the Hop family) were highly susceptible to protease digestion within the amino-terminal portion of the protein (comprising roughly two-thirds of the entire length of the protein), whereas the carboxy-terminal regions of these proteins were protease resistant. This result was observed for 10 proteins (BabA, BabB, BabC, HopQ, HomA, HopA, HopM, HopN, AlpA, and AlpB) (Fig. 6 and 7 and data not shown). The protease-resistant C-terminal regions of these proteins ranged from about 150 to 250 amino acids in length. Analysis of these proteins with the β-barrel prediction program BOCTOPUS predicts that the C-terminal 160 to 300 amino acids of these proteins correspond to multiple transmembrane β-strand segments consistent with a β-barrel structure and that predicted transmembrane segments are absent or infrequently present elsewhere within these proteins (Fig. 7). These data suggest that most of the protease-susceptible OMPs contain a large protease-susceptible extracellular domain exported beyond the outer membrane and a protease-resistant β-barrel domain at the C terminus.
FIG 7.
Resistance of predicted β-barrel regions to digestion by proteinase K. This figure shows an analysis of the protease susceptibility of 4 H. pylori OMPs, using the approach shown in Fig. 6. The blue lines (top half of graphs) depict the numbers of assigned spectra from experiments with control (untreated) bacteria, and the red lines (bottom half of graphs) depict the numbers of assigned spectra from experiments with proteinase K-treated bacteria. The green shading illustrates protease-susceptible regions (exhibiting a ≥5-fold difference in the spectral counts when comparing untreated bacteria and protease-treated bacteria), and the pink shading illustrates protease-resistant regions (exhibiting a <2-fold difference in the spectral counts). The designation of protease-susceptible or protease-resistant regions was restricted to residues for which ≥5 assigned spectra were detected in the untreated samples. The locations of transmembrane β-strands were predicted using the program BOCTOPUS and are shown above each graph (red indicates predicted periplasm-facing residues, gray indicates predicted transmembrane β-strands, and blue indicates predicted extracellular facing residues). A large N-terminal portion of HopQ is highly susceptible to proteolytic digestion, whereas the C-terminal portion of HopQ is resistant to proteolytic digestion. The other 3 OMPs (HopE, HorB, and HofC) are relatively resistant to proteolysis. The resistant C-terminal region of HopQ corresponds to a region predicted by BOCTOPUS to have multiple transmembrane β-strands, consistent with a β-barrel structure. Three protease-resistant OMPs (HopE, HorB, and HofC) are each predicted to have predominantly β-barrel structure.
DISCUSSION
In this study, we used multiple complementary methods to identify and analyze proteins localized on the surface of H. pylori. We identified 39 proteins that met three criteria for surface localization or outer membrane localization (i.e., identification by biotinylation, enriched in a membrane fraction compared to a soluble fraction, and enriched in a Triton X-100-insoluble outer membrane fraction compared to a Triton X-100-soluble inner membrane fraction), thereby providing stronger evidence for surface localization compared to the use of only a single method. Among these 39 proteins, many were previously known or predicted to be localized on the bacterial surface, whereas others were not previously known to be surface exposed or associated with the outer membrane.
Our primary method involved biotinylation of surface-exposed proteins with a membrane-impermeable biotinylation reagent. A potential concern with biotinylation-based methods is the possibility that periplasmic or inner membrane proteins might be labeled, but a previous study showed that the approach used in this study results in minimal labeling of the abundant E. coli periplasmic protein MalE (19). The biotinylation experiments allowed us to identify 85 putative surface-exposed H. pylori proteins (see Table S3 in the supplemental material). Eighteen putative surface-exposed proteins were identified in a previous study that used similar biotinylation-based methodology (19). Three surface-exposed proteins were identified in both the current study and the previous study: an HpaA-like protein (HP0410), CagM, and the secreted serine protease HtrA (19, 38, 39). There were several differences when comparing the methodology of the current study with that used in a previous study (19). First, the current study used a MudPIT-based methodology for identifying proteins, whereas the previous study used 2D gel-based methods; limitations in the use of 2D gel methods for detection of membrane proteins may account for the detection of relatively few annotated OMPs in the previous study. Second, the current study compared preparations of biotinylated proteins with control nonbiotinylated preparations, which allowed the exclusion of proteins that interacted nonspecifically with the beads used in the purification process. Third, the current study used monomeric avidin-coated magnetic beads instead of avidin-agarose beads. Finally, the current study analyzed bacteria grown in broth culture, whereas the previous study analyzed bacteria grown on plates. Collectively, these modified approaches were helpful in enhancing the detection of outer membrane proteins and reducing contamination from cytoplasmic proteins. To determine whether the bacterial growth conditions had a major influence on the results, we conducted experiments in which we compared preparations of biotinylated proteins derived from bacteria grown in broth culture with preparations of biotinylated proteins derived from bacteria grown on blood agar plates. In comparison to preparations of biotinylated proteins from bacteria grown in broth, preparations of biotinylated proteins from bacteria grown on solid medium had a lower proportion of assigned spectra for annotated OMPs (relative proportion of total assigned spectra for bacteria grown on plates versus in broth = 0.5) and a higher proportion of assigned spectra for annotated intracellular proteins (relative proportion of total assigned spectra for bacteria grown on plates versus in broth = 9.9) (see Tables S10 and S11 in the supplemental material).
The set of proteins meeting three criteria for surface localization (Table 2) included three cag pathogenicity island (PAI)-encoded proteins that are components of the cag T4SS (Cag3, CagM, and CagT) (46). We also detected surface-exposed proteins that were previously annotated as lipoproteins. Among the proteins shown in Table 2, five proteins contain a sequence motif known as a lipobox (required for the recognition of posttranslational lipidation machinery), based on analysis by DOLOP (47) (HpaA paralog HP0492, CagT HP0532, predicted lipoprotein HP1002, Lpp20 [HP1456], and predicted lipoprotein HP1571). In E. coli and many other well-studied Gram-negative species, most lipoproteins are anchored to either the outer membrane or the inner membrane and face the periplasm (48), but some lipoproteins may also be surface exposed and exported to the bacterial surface by an unknown mechanism (for example, JlpA in Campylobacter jejuni and TbpB in Neisseria meningitidis) (43, 49, 50). The findings in the current study suggest that the surface of H. pylori is decorated with multiple lipoproteins. The Lol system, which is required for lipoprotein export, is considerably different in H. pylori compared to many other bacterial species (7), and this could be a factor that influences the localization of H. pylori lipoproteins. Finally, we detected multiple surface-exposed proteins that are annotated as “hypothetical proteins.” Further study will be required to investigate the localization and functions of these proteins in greater detail.
Several proteins previously reported to be localized to the surface of H. pylori are not included on the list of proteins in Table 2, which indicates that this is not a comprehensive list of the proteins exported to the H. pylori surface. Therefore, we also analyzed the proteins that met two criteria consistent with a surface-exposed outer membrane localization (Fig. 2; see Tables S3, S4, and S5 in the supplemental material). This analysis revealed that the set of proteins enriched in the biotinylated preparations and enriched in the total membrane preparations (but not enriched in the Triton X-100-insoluble outer membrane preparation) contained numerous proteins that are known or predicted to be localized to the outer membrane, such as HopA, HopM, HopN, HopU/BabC, and HorF.
Although the experiments reported in this article provide a much more comprehensive understanding of the H. pylori cell surface proteome than has been previously reported, there are several limitations of the current study. First, we analyzed mainly bacteria that were cultured in a single growth condition (late log phase from broth culture). These conditions were selected based on the results of a previous microarray gene expression study, which reported that several virulence-associated genes were maximally expressed in the late log phase of growth (51). Additional surface-exposed proteins might be detectable if alternate conditions were studied. Identification of surface-exposed proteins under well-defined laboratory conditions, as described in this study, provides an important foundation for future studies investigating how the surface proteins of H. pylori change under alternate growth conditions. Second, the mass spectrometry-based approach used in these studies might fail to detect surface proteins that contain a small number of tryptic cleavage sites, surface proteins with extensive posttranslational modifications, or surface proteins expressed in low abundance. Finally, we studied only a single strain, which is known to lack production of at least four outer membrane proteins (HopP/SabA, HopO/SabB, HopZ, and HomB) (24, 25) and lacks flagella (28).
Many of the surface-exposed proteins identified in this study are predicted to be exported through a process that involves Sec-dependent cleavage of an amino-terminal signal sequence. In support of this view, we detected non-tryptic-cleavage events near the amino termini of numerous proteins. The presence in many of these proteins of a conserved C-terminal motif ending in phenylalanine or tyrosine is consistent with insertion of these proteins into the outer membrane through a Bam-dependent process (43–45). Most of the Hop or Hor proteins contained the consensus C-terminal motif (Y/F)XR XXX (V/F)Y XXY XXX (F/Y). Surprisingly, many of the surface-exposed proteins detected in this study, including several putative lipoproteins, did not contain a C-terminal motif terminating in phenylalanine or tyrosine, but instead terminated in lysine. The mechanism by which these and other proteins lacking a typical C-terminal motif are exported to the bacterial surface is not known.
As an additional approach for analyzing the cell surface proteome of H. pylori, we subjected intact bacteria to treatment with proteinase K and sought to identify a subset of surface-exposed proteins that were susceptible to proteolysis. Initial studies, based on analyzing the total numbers of spectral counts assigned to individual proteins, allowed us to identify seven surface-exposed proteins (VacA, BabA, HopQ, BabB, BabC [HopU], HomA, and HomD) that were highly susceptible to protease digestion. VacA is a toxin that is secreted through an autotransporter pathway (5, 52) and subsequently can be released as a soluble protein into the extracellular space or remain attached to the bacterial surface (5, 53). BabA is an H. pylori adhesin that binds to the Lewis b antigen on host cells (8), and HopQ is an outer membrane protein that influences the activity of the cag type IV secretion system (14). In contrast to VacA, BabA, and HopQ, virtually nothing is known about the functions of BabB, BabC, HomA, and HomD. BabA, BabB, BabC and HopQ are all classified as Hop proteins, whereas HomA and HomD are classified within a small family of OMPs known as Hom proteins (25). A previous study used proteinase K treatment coupled with 2D gel electrophoretic methods to identify five protease-susceptible surface-exposed H. pylori proteins (17). Two of the proteinase K-susceptible proteins identified in the previous study (VacA and HopQ) (17) were also shown to be protease susceptible in the current study.
We also analyzed the assigned spectra from the proteinase K experiments at the level of individual amino acids, which provided a more sensitive approach for detecting proteins that were susceptible to proteolytic cleavage. By using this approach, we identified 15 proteins that were protease susceptible. Most of these were identified in the previous biotinylation and cell fractionation experiments (see Fig. S4 in the supplemental material). Conversely, the majority of surface-exposed proteins identified in the biotinylation and cell fractionation experiments were not susceptible to proteolytic cleavage. Thus, the proteinase K experiments identified a specialized subset of protease-susceptible surface-exposed proteins with properties that differ from those of protease-resistant surface-exposed proteins.
The set of protease-susceptible proteins included numerous OMPs classified within the Hop or Hom family, as well as VacA and a VacA-like protein (FaaA) (Table 4). Among the protease-susceptible Hop and Hom proteins, the C-terminal regions were resistant to protease digestion, whereas the N-terminal portions were susceptible. These data suggest that the N-terminal portions of these OMPs are exported beyond the outer membrane. The protease-resistant C-terminal domains of these OMPs are predicted to have multiple transmembrane β-strands consistent with a β-barrel structure (based on the use of the BOCTOPUS program), and all contain the conserved C-terminal motif shown in Fig. 5. On the basis of similarities between the protease susceptibility patterns of VacA and OMPs, we suggest that the N-terminal domains of these OMPs are exported beyond the outer membrane through an autotransporter (type V secretion) pathway, similar to the secretion of the VacA passenger domain (5, 52). In contrast to VacA, which undergoes proteolysis to cleave the passenger domain from the β-barrel domain, there is not yet any evidence that the protease-susceptible OMPs undergo similar proteolytic cleavage.
Although a subset of annotated OMPs were susceptible to proteolytic digestion, many others were relatively resistant (Table 5). All three Hom proteins detected in this study (HomA, HomC, and HomD) and many of the Hop proteins exhibited evidence of susceptibility to proteolytic cleavage, whereas Hof and Hor proteins were relatively resistant (Table 5). There are several possible explanations that may account for the observed differences in susceptibility to proteolytic cleavage, including variation among the proteins in the proportion of β-barrel structure, differences among the proteins in the orientation of large loops (projecting toward the periplasm or the bacterial surface), differences in physical accessibility of extracellular domains to the protease active site, and differences in amino acid composition. In contrast to the protease-susceptible Hop proteins, at least one of the Hop proteins resistant to proteolytic cleavage (HopE) is predicted to have a structure comprised of predominantly transmembrane β-strands, based on analysis by BOCTOPUS (Fig. 7), which likely accounts for its resistance to proteolysis. Similarly, HorB and HofC are predicted to have structures comprised of predominantly transmembrane β-strands, and these proteins were resistant to proteolysis (Fig. 7). Thus, the current data suggest that β-barrel-containing segments of H. pylori OMPs are relatively resistant to proteolysis, whereas large extracellular domains of H. pylori OMPs with non-β-barrel structures are often susceptible to proteolysis.
The passenger domains of most proteins secreted through an autotransporter (type V secretion) pathway, including H. pylori VacA, have β-helical structures (54, 55). Interestingly, analysis of the protease-sensitive putative autotransporter proteins identified in this study using the BetaWrapPro program predicted the existence of β-helical structure within VacA (consistent with the crystal structure of this protein) (54, 55), but not within the other protease-sensitive OMPs. Thus, it seems likely that the N-terminal portions of many protease-susceptible H. pylori OMPs differ in structure compared to most known autotransporter passenger domains. In support of this view, a recent X-ray crystallographic study of the N-terminal region of H. pylori SabA (an OMP that is related to BabA, but not produced by the H. pylori strain analyzed in this study) revealed primarily α-helical motifs instead of the β-helical motifs characteristic of most autotransporter passenger domains (54, 56).
In summary, these experiments provide valuable new insights into the repertoire of proteins present on the surface of H. pylori. In addition, our detailed analysis of the susceptibility of surface proteins to proteinase K digestion provides the first systematic analysis of the topology of H. pylori cell surface proteins and provides experimental evidence in support of an autotransporter mode of export for many of these proteins. Several of the surface-exposed proteins identified in this study are known to have important roles in mediating H. pylori-host interactions, and the functions of others are not yet known. The surface-exposed proteins with large extracellular domains identified in this study are likely to be localized in close proximity to host cells, and therefore, we suggest that these proteins in particular may have important roles in mediating interactions with the host.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (R01 AI068009, AI039657, and CA116087) and the Department of Veterans Affairs. Experiments in the Vanderbilt Proteomics facility and Cell Imaging Shared Resource were supported by the Vanderbilt University Digestive Disease Research Center (NIH grant P30DK058404) and the Vanderbilt University Ingram Cancer Center.
We thank members of the Cover lab for helpful discussions, and we thank David Tabb for providing advice on statistical analysis of mass spectrometry results.
Footnotes
Published ahead of print 25 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01768-14.
REFERENCES
- 1.Atherton JC, Blaser MJ. 2009. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J. Clin. Invest. 119:2475–2487. 10.1172/JCI38605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cover TL, Blaser MJ. 2009. Helicobacter pylori in health and disease. Gastroenterology 136:1863–1873. 10.1053/j.gastro.2009.01.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amieva MR, El-Omar EM. 2008. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology 134:306–323. 10.1053/j.gastro.2007.11.009 [DOI] [PubMed] [Google Scholar]
- 4.Polk DB, Peek RM., Jr 2010. Helicobacter pylori: gastric cancer and beyond. Nat. Rev. Cancer 10:403–414. 10.1038/nrc2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cover TL, Blanke SR. 2005. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat. Rev. Microbiol. 3:320–332. 10.1038/nrmicro1095 [DOI] [PubMed] [Google Scholar]
- 6.Hatakeyama M. 2011. Anthropological and clinical implications for the structural diversity of the Helicobacter pylori CagA oncoprotein. Cancer Sci. 102:36–43. 10.1111/j.1349-7006.2010.01743.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liechti G, Goldberg JB. 2012. Outer membrane biogenesis in Escherichia coli, Neisseria meningitidis, and Helicobacter pylori: paradigm deviations in H. pylori. Front. Cell. Infect. Microbiol. 2:29. 10.3389/fcimb.2012.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L, Boren T. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373–377. 10.1126/science.279.5349.373 [DOI] [PubMed] [Google Scholar]
- 9.Mahdavi J, Sonden B, Hurtig M, Olfat FO, Forsberg L, Roche N, Angstrom J, Larsson T, Teneberg S, Karlsson KA, Altraja S, Wadstrom T, Kersulyte D, Berg DE, Dubois A, Petersson C, Magnusson KE, Norberg T, Lindh F, Lundskog BB, Arnqvist A, Hammarstrom L, Boren T. 2002. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 297:573–578. 10.1126/science.1069076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Odenbreit S, Till M, Hofreuter D, Faller G, Haas R. 1999. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol. Microbiol. 31:1537–1548. 10.1046/j.1365-2958.1999.01300.x [DOI] [PubMed] [Google Scholar]
- 11.Kwok T, Zabler D, Urman S, Rohde M, Hartig R, Wessler S, Misselwitz R, Berger J, Sewald N, Konig W, Backert S. 2007. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature 449:862–866. 10.1038/nature06187 [DOI] [PubMed] [Google Scholar]
- 12.Jimenez-Soto LF, Kutter S, Sewald X, Ertl C, Weiss E, Kapp U, Rohde M, Pirch T, Jung K, Retta SF, Terradot L, Fischer W, Haas R. 2009. Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD-independent manner. PLoS Pathog. 5:e1000684. 10.1371/journal.ppat.1000684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sause WE, Castillo AR, Ottemann KM. 2012. The Helicobacter pylori autotransporter ImaA (HP0289) modulates the immune response and contributes to host colonization. Infect. Immun. 80:2286–2296. 10.1128/IAI.00312-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belogolova E, Bauer B, Pompaiah M, Asakura H, Brinkman V, Ertl C, Bartfeld S, Nechitaylo TY, Haas R, Machuy N, Salama N, Churin Y, Meyer TF. 2013. Helicobacter pylori outer membrane protein HopQ identified as a novel T4SS-associated virulence factor. Cell. Microbiol. 15:1896–1912. 10.1111/cmi.12158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jimenez-Soto LF, Clausen S, Sprenger A, Ertl C, Haas R. 2013. Dynamics of the Cag-type IV secretion system of Helicobacter pylori as studied by bacterial co-infections. Cell. Microbiol. 15:1924–1937. 10.1111/cmi.12166 [DOI] [PubMed] [Google Scholar]
- 16.Ishijima N, Suzuki M, Ashida H, Ichikawa Y, Kanegae Y, Saito I, Boren T, Haas R, Sasakawa C, Mimuro H. 2011. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J. Biol. Chem. 286:25256–25264. 10.1074/jbc.M111.233601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabarth N, Hurvitz R, Schmidt M, Zimny-Arndt U, Jungblut PR, Meyer TF, Bumann D. 2005. Identification of Helicobacter pylori surface proteins by selective proteinase K digestion and antibody phage display. J. Microbiol. Methods 62:345–349. 10.1016/j.mimet.2005.04.030 [DOI] [PubMed] [Google Scholar]
- 18.Radin JN, Gaddy JA, Gonzalez-Rivera C, Loh JT, Algood HM, Cover TL. 2013. Flagellar localization of a Helicobacter pylori autotransporter protein. mBio 4(2):e00613–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabarth N, Lamer S, Zimny-Arndt U, Jungblut PR, Meyer TF, Bumann D. 2002. Identification of surface proteins of Helicobacter pylori by selective biotinylation, affinity purification, and two-dimensional gel electrophoresis. J. Biol. Chem. 277:27896–27902. 10.1074/jbc.M204473200 [DOI] [PubMed] [Google Scholar]
- 20.Baik SC, Kim KM, Song SM, Kim DS, Jun JS, Lee SG, Song JY, Park JU, Kang HL, Lee WK, Cho MJ, Youn HS, Ko GH, Rhee KH. 2004. Proteomic analysis of the sarcosine-insoluble outer membrane fraction of Helicobacter pylori strain 26695. J. Bacteriol. 186:949–955. 10.1128/JB.186.4.949-955.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doig P, Trust TJ. 1994. Identification of surface-exposed outer membrane antigens of Helicobacter pylori. Infect. Immun. 62:4526–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Exner MM, Doig P, Trust TJ, Hancock RE. 1995. Isolation and characterization of a family of porin proteins from Helicobacter pylori. Infect. Immun. 63:1567–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phadnis SH, Parlow MH, Levy M, Ilver D, Caulkins CM, Connors JB, Dunn BE. 1996. Surface localization of Helicobacter pylori urease and a heat shock protein homolog requires bacterial autolysis. Infect. Immun. 64:905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. 10.1038/41483 [DOI] [PubMed] [Google Scholar]
- 25.Alm RA, Bina J, Andrews BM, Doig P, Hancock RE, Trust TJ. 2000. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect. Immun. 68:4155-4168. 10.1128/IAI.68.7.4155-4168.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peck B, Ortkamp M, Diehl KD, Hundt E, Knapp B. 1999. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 27:3325–3333. 10.1093/nar/27.16.3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doig P, Exner MM, Hancock RE, Trust TJ. 1995. Isolation and characterization of a conserved porin protein from Helicobacter pylori. J. Bacteriol. 177:5447–5452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaffer CL, Gaddy JA, Loh JT, Johnson EM, Hill S, Hennig EE, McClain MS, McDonald WH, Cover TL. 2011. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog. 7:e1002237. 10.1371/journal.ppat.1002237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Busler VJ, Torres VJ, McClain MS, Tirado O, Friedman DB, Cover TL. 2006. Protein-protein interactions among Helicobacter pylori cag proteins. J. Bacteriol. 188:4787–4800. 10.1128/JB.00066-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kutter S, Buhrdorf R, Haas J, Schneider-Brachert W, Haas R, Fischer W. 2008. Protein subassemblies of the Helicobacter pylori Cag type IV secretion system revealed by localization and interaction studies. J. Bacteriol. 190:2161–2171. 10.1128/JB.01341-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pham KT, Weiss E, Jimenez Soto LF, Breithaupt U, Haas R, Fischer W. 2012. CagI is an essential component of the Helicobacter pylori Cag type IV secretion system and forms a complex with CagL. PLoS One 7:e35341. 10.1371/journal.pone.0035341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacCoss MJ, McDonald WH, Saraf A, Sadygov R, Clark JM, Tasto JJ, Gould KL, Wolters D, Washburn M, Weiss A, Clark JI, Yates JR., III 2002. Shotgun identification of protein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. U. S. A. 99:7900–7905. 10.1073/pnas.122231399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. 2005. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4:1487–1502. 10.1074/mcp.M500084-MCP200 [DOI] [PubMed] [Google Scholar]
- 34.Li M, Gray W, Zhang H, Chung CH, Billheimer D, Yarbrough WG, Liebler DC, Shyr Y, Slebos RJ. 2010. Comparative shotgun proteomics using spectral count data and quasi-likelihood modeling. J. Proteome Res. 9:4295–4305. 10.1021/pr100527g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer W, Puls J, Buhrdorf R, Gebert B, Odenbreit S, Haas R. 2001. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 42:1337–1348 [DOI] [PubMed] [Google Scholar]
- 36.Keenan J, Oliaro J, Domigan N, Potter H, Aitken G, Allardyce R, Roake J. 2000. Immune response to an 18-kilodalton outer membrane antigen identifies lipoprotein 20 as a Helicobacter pylori vaccine candidate. Infect. Immun. 68:3337–3343. 10.1128/IAI.68.6.3337-3343.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duncan SS, Valk PL, McClain MS, Shaffer CL, Metcalf JA, Bordenstein SR, Cover TL. 2013. Comparative genomic analysis of East Asian and non-Asian Helicobacter pylori strains identifies rapidly evolving genes. PLoS One 8:e55120. 10.1371/journal.pone.0055120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoy B, Lower M, Weydig C, Carra G, Tegtmeyer N, Geppert T, Schroder P, Sewald N, Backert S, Schneider G, Wessler S. 2010. Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep. 11:798–804. 10.1038/embor.2010.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bumann D, Aksu S, Wendland M, Janek K, Zimny-Arndt U, Sabarth N, Meyer TF, Jungblut PR. 2002. Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori. Infect. Immun. 70:3396–3403. 10.1128/IAI.70.7.3396-3403.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jonsson K, Guo BP, Monstein HJ, Mekalanos JJ, Kronvall G. 2004. Molecular cloning and characterization of two Helicobacter pylori genes coding for plasminogen-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 101:1852–1857. 10.1073/pnas.0307329101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones AC, Logan RP, Foynes S, Cockayne A, Wren BW, Penn CW. 1997. A flagellar sheath protein of Helicobacter pylori is identical to HpaA, a putative N-acetylneuraminyllactose-binding hemagglutinin, but is not an adhesin for AGS cells. J. Bacteriol. 179:5643–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 43.Bos MP, Robert V, Tommassen J. 2007. Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol. 61:191–214. 10.1146/annurev.micro.61.080706.093245 [DOI] [PubMed] [Google Scholar]
- 44.Hagan CL, Silhavy TJ, Kahne D. 2011. β-Barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80:189–210. 10.1146/annurev-biochem-061408-144611 [DOI] [PubMed] [Google Scholar]
- 45.Struyvé M, Moons M, Tommassen J. 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218:141–148. 10.1016/0022-2836(91)90880-F [DOI] [PubMed] [Google Scholar]
- 46.Fischer W. 2011. Assembly and molecular mode of action of the Helicobacter pylori Cag type IV secretion apparatus. FEBS J. 278:1203–1212. 10.1111/j.1742-4658.2011.08036.x [DOI] [PubMed] [Google Scholar]
- 47.Babu MM, Priya ML, Selvan AT, Madera M, Gough J, Aravind L, Sankaran K. 2006. A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J. Bacteriol. 188:2761–2773. 10.1128/JB.188.8.2761-2773.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masuda K, Matsuyama S, Tokuda H. 2002. Elucidation of the function of lipoprotein-sorting signals that determine membrane localization. Proc. Natl. Acad. Sci. U. S. A. 99:7390–7395. 10.1073/pnas.112085599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewis LA, Rohde K, Gipson M, Behrens B, Gray E, Toth SI, Roe BA, Dyer DW. 1998. Identification and molecular analysis of lbpBA, which encodes the two-component meningococcal lactoferrin receptor. Infect. Immun. 66:3017–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nothaft H, Szymanski CM. 2010. Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8:765–778. 10.1038/nrmicro2383 [DOI] [PubMed] [Google Scholar]
- 51.Thompson LJ, Merrell DS, Neilan BA, Mitchell H, Lee A, Falkow S. 2003. Gene expression profiling of Helicobacter pylori reveals a growth-phase-dependent switch in virulence gene expression. Infect. Immun. 71:2643–2655. 10.1128/IAI.71.5.2643-2655.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fischer W, Buhrdorf R, Gerland E, Haas R. 2001. Outer membrane targeting of passenger proteins by the vacuolating cytotoxin autotransporter of Helicobacter pylori. Infect. Immun. 69:6769–6775. 10.1128/IAI.69.11.6769-6775.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ilver D, Barone S, Mercati D, Lupetti P, Telford JL. 2004. Helicobacter pylori toxin VacA is transferred to host cells via a novel contact-dependent mechanism. Cell. Microbiol. 6:167–174. 10.1046/j.1462-5822.2003.00349.x [DOI] [PubMed] [Google Scholar]
- 54.Junker M, Schuster CC, McDonnell AV, Sorg KA, Finn MC, Berger B, Clark PL. 2006. Pertactin beta-helix folding mechanism suggests common themes for the secretion and folding of autotransporter proteins. Proc. Natl. Acad. Sci. U. S. A. 103:4918–4923. 10.1073/pnas.0507923103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gangwer KA, Mushrush DJ, Stauff DL, Spiller B, McClain MS, Cover TL, Lacy DB. 2007. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proc. Natl. Acad. Sci. U. S. A. 104:16293–16298. 10.1073/pnas.0707447104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pang SS, Nguyen ST, Perry AJ, Day CJ, Panjikar S, Tiralongo J, Whisstock JC, Kwok T. 27 December 2013. The three-dimensional structure of the extracellular adhesion domain of the sialic acid-binding adhesin SabA from Helicobacter pylori. J. Biol. Chem. 10.1074/jbc.M113.513135 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.