Abstract
The quorum-sensing (QS) system present in the emerging nosocomial pathogen Stenotrophomonas maltophilia is based on the signaling molecule diffusible signal factor (DSF). Production and detection of DSF are governed by the rpf cluster, which encodes the synthase RpfF and the sensor RpfC, among other components. Despite a well-studied system, little is known about its implication in virulence regulation in S. maltophilia. Here, we have analyzed the rpfF gene from 82 S. maltophilia clinical isolates. Although rpfF was found to be present in all of the strains, it showed substantial variation, with two populations (rpfF-1 and rpfF-2) clearly distinguishable by the N-terminal region of the protein. Analysis of rpfC in seven complete genome sequences revealed a corresponding variability in the N-terminal transmembrane domain of its product, suggesting that each RpfF variant has an associated RpfC variant. We show that only RpfC–RpfF-1 variant strains display detectable DSF production. Heterologous rpfF complementation of ΔrpfF mutants of a representative strain of each variant suggests that RpfF-2 is, however, functional and that the observed DSF-deficient phenotype of RpfC–RpfF-2 variant strains is due to permanent repression of RpfF-2 by RpfC-2. This is corroborated by the ΔrpfC mutant of the RpfC–RpfF-2 representative strain. In line with this observations, deletion of rpfF from the RpfC–RpfF-1 strain leads to an increase in biofilm formation, a decrease in swarming motility, and relative attenuation in the Caenorhabditis elegans and zebrafish infection models, whereas deletion of the same gene from the representative RpfC–RpfF-2 strain has no significant effect on these virulence-related phenotypes.
INTRODUCTION
Quorum sensing (QS) is a bacterial cell-cell communication process that allows bacteria to synchronize particular behaviors on a population-wide scale. Within current knowledge, QS in Stenotrophomonas maltophilia depends on the diffusible signal factor QS (DSF-QS) system, which is based mainly on the fatty acid DSF (cis-11-methyl-2-dodecenoic acid) (1, 2). DSF synthesis is fully dependent on RpfF, an enoyl coenzyme A hydratase encoded by the rpf (regulation of pathogenicity factors) cluster, a set of genes that includes all of the components necessary for the synthesis and detection of DSF molecules. In addition to RpfF, rpf encodes the aconitase RpfA, the fatty acid ligase RpfB, the two-component sensor-effector hybrid system RpfC, and the cytoplasmic regulator element RpfG (1, 2). The DSF-QS system was first described in the phytopathogen Xanthomonas campestris pv. campestris, where it plays an important role in virulence regulation (3). Since then, this system has been described in several members of the order Xanthomonadales, including the genera Xanthomonas, Xylella, and Stenotrophomonas, as well as in members of the order Burkholderiales (1, 3–5). The specific functions regulated by the DSF-QS system are dependent on the species, but it has been suggested that it controls several virulence-related phenotypes (6). In the case of S. maltophilia, little is known about the mechanisms implicated in DSF-QS regulation. It has been demonstrated that disruption of DSF signaling has a drastic effect on S. maltophilia K279a, since the rpfF mutant shows reduced swimming motility, reduced exoprotease production, altered lipopolysaccharide, reduced tolerance to a range of antibiotics and to heavy metals, and reduced virulence in a Caenorhabditis elegans infection model (1). In addition, FecA, a ferric citrate receptor, has been shown to be positively regulated by the DSF-QS system. This receptor contributes to the internalization of iron, an essential element for the expression of virulence-related genes (7). In the S. maltophilia WR-C wild-type (WT) strain and a flagellum-defective xanB mutant, flagellum-independent translocation was stimulated not only by the main DSF but also by its derivative 11-methyl-dodecanoic acid (2). Regarding the interaction of S. maltophilia with plants, DSF seems to be involved in oilseed germination, plant colonization, and biofilm architecture in the environmental strain R551-3 (8). Recently, the BDSF system (a DSF variant in Burkholderia species) has also been shown to contribute to the swarming motility phenotype of Burkholderia cenocepacia (9).
In a recent S. maltophilia population study, the authors detected rpfF+ genotypes in 61% of the 89 strains tested, suggesting that an important population of S. maltophilia lacks the rpfF gene (10). With the rapid increase in the number of S. maltophilia sequenced genomes, it is now possible to compare the rpf clusters of different strains. A preliminary analysis showed that all of the genomes sequenced contain the rpfF gene. In addition, at least two rpf cluster variants can be detected on the basis of sequence and genomic organization, with main differences found in the rpfF and rpfC genes. The genetic variation observed in the rpfF gene translates into two distinct protein variants, here named RpfF-1 and RpfF-2. Furthermore, we can associate each of these RpfF variants with a corresponding RpfC variant, i.e., RpfC-1 and RpfC-2, respectively. We have also investigated the DSF production of representative strains from each variant group, revealing that only the strains carrying the RpfF–RpfC-1 variants show detectable DSF production under the conditions assayed. Moreover, characterization of the ΔrpfF mutant of a strain from each RpfF variant group indicates that the virulence-related phenotypes are differently regulated in the two populations.
MATERIALS AND METHODS
Strains and growth conditions.
A panel of 78 S. maltophilia clinical isolates were collected from point prevalence studies in the intensive care units of different European hospitals. For the name, geographic origin, hospital, and isolation source of each strain, see Table S1 in the supplemental material. From this collection, E77 (RpfF-1 variant group) and M30 (RpfF-2 variant group) (11) were used as model strains to characterize ΔrpfF mutants (see Table S2). Escherichia coli OP50 was provided by the Caenorhabditis Genetics Center (CGC). X. campestris pv. campestris 8523/pL6engGUS was obtained from the authors of reference 12.
Bacteria were routinely grown at 37°C in Luria-Bertani (LB) medium on a rotary shaker. When needed, LB was supplemented with tetracycline (Tc) at 17 μg/ml, chloramphenicol (Cm) at 3.2 μg/ml, erythromycin (Erm) at 500 μg/ml, and ampicillin (Ap) at 20 μg/ml. For phenotypic analysis in minimal medium, strains were grown in BM2 medium (62 mM potassium phosphate buffer, pH 7, 2 mM MgSO4, 10 μM FeSO4, supplemented with glucose 0.4%) or a modified M9-salts medium without NH4Cl (0.5% Casamino Acids, 2 mM MgSO4, 0.1 mM CaCl2) and supplemented with 0.2% glucose.
Sequence determination and analysis.
PCR products of 682 to 721 bp containing the rpfF promoter and the region encoding the N-terminal fragment were amplified from all 78 S. maltophilia strains with primers PrpfFtypeUp and PrpfFTypeDw (see Table S3 in the supplemental material) and directly sequenced (Macrogen Inc.). Translation of partial open reading frames (ORFs) to amino acids and sequence alignments were done with MEGA V5.2 (13) and BioEdit, respectively. A phylogenetic tree was constructed with MEGA V5.2 on the basis of a trimmed alignment with the 108 N-terminal residues of RpfF from strain K279a. In parallel, the genomes of strains E77, M30, and UV74 were sequenced and a first draft was constructed (to be reported upon completion). RpfC variant determination was then based on the RpfC sequences from the publicly available sequenced genomes (strains K279a, R551-3, D457, and JV3, with GenBank accession numbers AM743169.1, CP001111.1, HE798556.1, and CP002986.1, respectively) and our draft genome sequences (strains E77, M30, and UV74), by using SMART (14) for the identification and annotation of protein domains.
Generation and complementation of ΔrpfF and ΔrpfC mutants.
For the primers and plasmids used for cloning, see Tables S3 and S4 in the supplemental material, respectively. S. maltophilia E77 ΔrpfF and M30 ΔrpfF and ΔrpfC mutants were obtained by allelic-exchange recombination with an Erm resistance cassette. Briefly, rpfF upstream and downstream flanking regions were amplified by PCR (see Table S3 in the supplemental material) and inserted, flanking the Erm resistance cassette, into the pEX18Tc vector (15), thus generating plasmids pEXE77rpfF and pEXM30rpfF for E77 and M30, respectively. Both strains were electroporated (16) with the respective suicide vectors, and transformants were selected on LB plates containing 500 μg/ml Erm and subsequently streaked onto LB plates containing 17 μg/ml Tc to discard single-crossover events. rpfF deletion was also verified by PCR and DNA sequencing. To generate a ΔrpfC mutant of the M30 strain, the same strategy was used. For the primers used to amplify upstream and downstream regions of rpfC from M30, see Table S3 in the supplemental material. Both fragments were inserted, flanking the Erm resistance cassette, into pEX18Tc, generating pEXM30rpfC. Strain M30 was electroporated, and the mutant candidates were screened and verified with the corresponding primers (see Table S3) as described above.
A fragment of ca. 1,100 bp containing either the E77 or the M30 rpfF ORF and the predicted promoter was amplified by PCR, ligated to pBBR1MCS-Cm (17), and introduced into E77 and/or M30 for either homologous or heterologous trans complementation of ΔrpfF. On the other hand, a fragment of ca. 3,000 bp was amplified from M30 and E77 to generate complementation vectors prpfGCM30 and prpfGCE77 (see Table S4), respectively. These fragments contained the rpfG and rpfC operon with its own promoters. Both fragments were digested with the respective restriction enzymes and ligated into pBBR1MCS1-Cm. Finally, prpfGCM30 and prpfGCE77 were introduced into E77, M30, and the M30 ΔrpfC mutant for either homologous or heterologous trans complementation.
Supernatant DSF extraction.
DSF extraction from culture supernatants was carried out by the ethyl acetate method (3). Briefly, overnight bacterial cultures grown on LB medium were harvested by centrifugation and the supernatant was extracted with the same volume of ethyl acetate. The organic phase was evaporated to dryness with a rotary evaporator, and the residues were dissolved in an appropriate volume of methanol (for supernatant DSF bioassay and analysis by thin-layer chromatography [TLC]) or dichloromethane (for analysis by gas chromatography-mass spectrometry [GC-MS]).
DSF bioassay and TLC analysis.
DSF determination was performed with X. campestris pv. campestris 8523/pL6engGUS (DSF reporter strain) as previously described (12), with a few modifications. Briefly, the DSF reporter strain was grown in 10 ml of NYG medium (0.3% yeast extract, 0.5% peptone, 2% glycerol) supplemented with Tc (10 μg/ml) to an optical density at 600 nm (OD600) of 0.7. Cells were harvested, reconstituted with 1 ml of fresh NYG, added to 100 ml of cold NYG medium containing 1% BD Difco Noble agar (NYGA) supplemented with 80 μg/ml X-Glu (5-bromo-4-chloro-3-indolyl β-d-glucuronide sodium salt; Sigma), and plated into petri plates upon solidification.
For colony-based DSF bioassays, candidate strains were pin inoculated onto plates of NYGA containing X-Glu (80 μg/ml) seeded with the DSF reporter strain and incubated for 24 h at 28°C. The presence of a blue halo around the colony indicates DSF activity.
For supernatant-based DSF bioassays, bacterial cultures were grown in 250 ml of LB for 48 h at 30°C (OD600 of about 4). Supernatants were extracted by the ethyl acetate method, and residues were dissolved in 200 μl of methanol. A 3-μl volume of each sample was deposited into a hand-generated well in a 5.5-cm plate containing NYGA supplemented with 80 μg/ml X-Glu and seeded with the DSF reporter strain to a final OD600 of 0.07. Plates were incubated for 24 h at 30°C. DSF activity was determined by the presence of a blue halo around the well.
For supernatant TLC analysis, 3-μl aliquots of dissolved methanol residues were spotted onto a silica gel 60 TLC plate (20 by 20 cm; Merck) and separated with ethyl acetate-hexane (20:80, vol/vol) as running solvents. TLC plates were subsequently air dried for at least 1 h and overlaid with 100 ml of unsolidified NYGA containing 80 μg/ml X-Glu and the DSF reporter strain at an OD600 of 0.07. TLC plates were incubated overnight at 28°C, and DSF activity was identified by the presence of blue spots.
Identification of DSF molecules from culture supernatants by GC-MS.
Bacterial cultures were grown in 2 liters of LB for 48 h at 30°C with vigorous shaking (250 rpm). Cultures were centrifuged, and supernatants were extracted by the ethyl acetate method. Dry residues were dissolved in 3 ml of dichloromethane. DSF molecules were identified by GC (Agilent Technologies 6890) with an Agilent 19091S-433 column coupled to an MS detector (Hewlett-Packard 5973).
Determination of virulence in a C. elegans model.
C. elegans CF512 [fer-15(b26)II; fem-1(hc17)IV], a strain showing temperature-dependent sterility, was provided by CGC. Nematodes were routinely maintained on NGM plates (1.7% agar, 50 mM NaCl, 0.25% peptone, 1 mM CaCl2, 5 μg/ml cholesterol, 25 mM KH2PO4, 1 mM MgSO4) seeded with E. coli OP50 at 16°C.
Determination of the virulence of S. maltophilia strains in the C. elegans CF512 infection model was based on the “slow killing” method (18). Strains were grown in brain heart infusion broth overnight at 30°C, and 100 μl of each strain culture was spread onto a 5.5-cm-diameter NGM agar plate and incubated at 30°C for 24 h. Each plate was then seeded with 15 to 20 adult hermaphrodite CF512 worms, incubated at 25°C (sterility conditions), and scored for live worms every 24 h. E. coli OP50 was used as a negative control. A worm was considered dead when it no longer responded to touch. Three replicates per strain were prepared.
Determination of virulence in a zebrafish model.
Adult (9- to 12-month-old) WT zebrafish (Danio rerio) were subjected to a 12-h light-dark cycle at 28°C and fed twice daily with dry food. All of the fish used in infection experiments were transferred to an isolated system and acclimated for 3 days before infection. Adult zebrafish (n = 12 per condition) were infected by intraperitoneal injection (19) with 20 μl of a 5 × 108-CFU/ml suspension of each S. maltophilia strain. The strains were previously grown at 28°C on blood agar plates (bioMérieux) for 20 h and collected directly from the plates with sterile phosphate-buffered saline (PBS). Two control groups were injected with PBS, and there were no deaths. Fish were observed daily for signs of disease and death.
One fish from each tank was sacrificed at 72 h postinfection and divided into three sections (anterior, abdominal, and posterior regions) with a sterile surgical blade. All weights were annotated, and every section was homogenized in 3 ml of PBS. After serial dilution, bacteria were plated onto LB medium containing 20 μg/ml Ap (for WT E77), LB containing 500 μg/ml Erm (for the E77 ΔrpfF mutant), or LB supplemented with Cm (for the complemented E77 ΔrpfF mutant). Finally, CFU were counted and divided per gram of tissue. All of the isolates obtained postmortem from infected zebrafish were identified as S. maltophilia on the basis of cell and colony morphology, the analytical profile index, and the 16S rRNA gene sequence (data not shown).
Biofilm formation.
To analyze biofilm formation on a polystyrene surface, 200-μl volumes of bacterial cultures grown to an OD600 of 0.1 in modified M9 or BM2 medium were inoculated into the wells of untreated 96-well microtiter plates (BrandTech 781662) and incubated for 24 h at 30°C. The plates were then washed three times with water, fixed at 60°C for 1 h, and stained for 15 min with 200 μl of 0.1% crystal violet. The dye was discarded, and the plates were rinsed in standing water and allowed to dry for 30 min at 37°C. Crystal violet was dissolved in 250 μl of 95% ethanol for 15 min, and the OD550 of the extracted dye was measured.
Biofilm formation on a glass surface was assayed by inoculating 2 ml of the same medium and adjusted OD as described above into glass tubes and incubating them for 24 h at 30°C with agitation (250 rpm). Biofilm formation was measured by crystal violet staining as described above.
Swarming assay.
Swarm agar was made on the basis of modified M9 salts medium without NH4Cl (0.5% Casamino Acids, 2 mM MgSO4, 0.1 mM CaCl2) supplemented with 0.4% glucose and solidified with 0.5% BD Difco Noble agar. Plates containing 20 ml of fresh swarm medium were dried under a laminar-flow hood for 20 min before inoculation. Inoculation was performed with a sterile Drigalski spatula containing biomass from a fresh LB plate by softly depositing it on top of a semisolid modified M9 plate. Inoculated swarm plates were sealed to maintain the humidity and incubated at 28°C for 3 to 5 days.
Quantitative reverse transcription (qRT)-PCR.
Gene expression analysis was performed to determine the ratios of rpfF to rpfC mRNAs in S. maltophilia E77 and M30. Total RNA was isolated from cultures grown under the same conditions as for DSF extraction with a GeneJet RNA purification kit (Thermo Scientific), and DNA was eliminated with TURBO DNase (Ambion, Life Technologies). One microgram of RNA was used to synthesize cDNA with an iScript cDNA synthesis kit (Bio-Rad). Quantitative real-time PCR was performed with the CFX96 real-time PCR system (Bio-Rad), and PCR amplification was detected with SsoAdvanced SYBR green Supermix (Bio-Rad). PCR products of 80 to 110 bp were amplified for rpfC, rpfF, and gyrA; the latter was used as an endogenous gene to normalize gene expression (20). For the primers used, see Table S3 in the supplemental material. Differences in the relative amounts of mRNA for the rpfF-1, rpfC-1, rpfF-2, and rpfC-2 genes were determined by the 2−ΔΔCT method (21). RNA samples were extracted in three different experiments, and results are given as mean values.
Ethics statement.
Zebrafish were handled in compliance with Directive 2010/63/EU of the European Parliament and of the Council on the Protection of Animals Used for Scientific Purposes and with decree 214/1997 of the Government of Catalonia, which regulates the use of animals for experimental and other scientific purposes. Experimental protocols have been reviewed and approved by the Animal and Human Experimentation Ethics Committee of the Universitat Autònoma de Barcelona, Spain (reference number CEEAH-1968).
Nucleotide sequence accession numbers.
All of the amplified rpfF sequences from this S. maltophilia strain collection have been deposited in the GenBank database and assigned accession numbers KJ149475 to KJ149552.
RESULTS
S. maltophilia harbors two RpfF variants that apparently differ in DSF production.
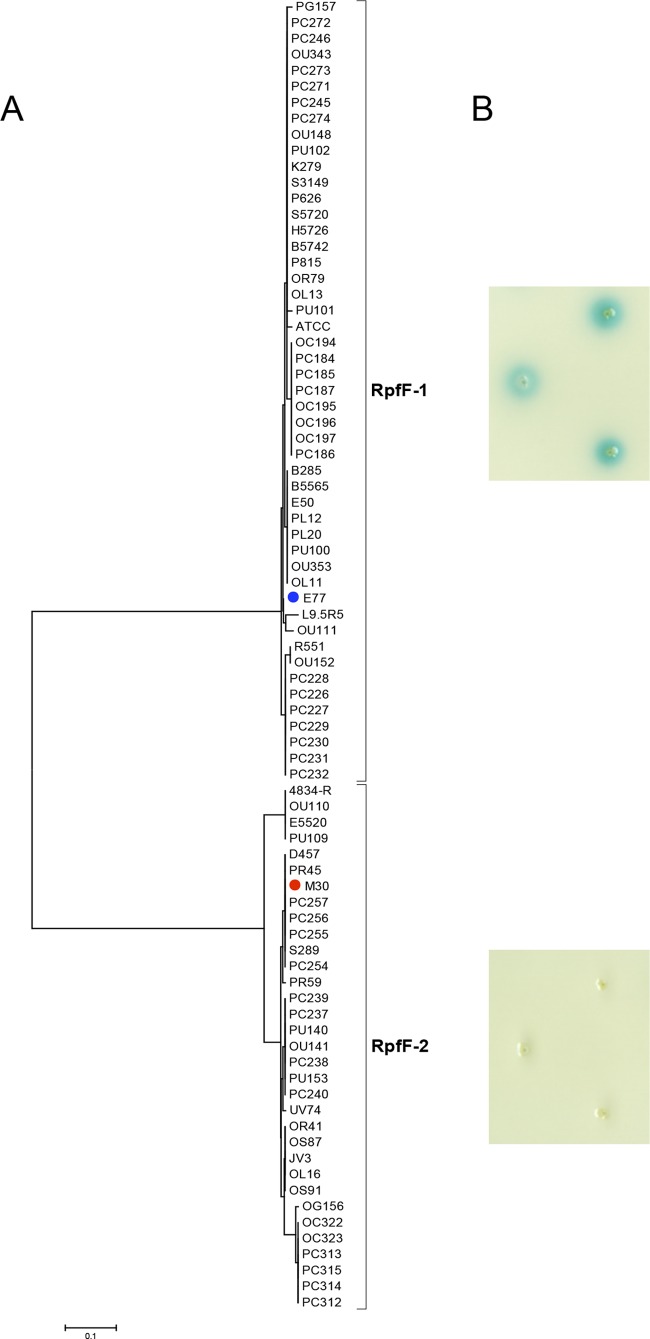
Amplification and sequencing of the corresponding DNA region demonstrated that all of the S. maltophilia strains in this study (see Table S1 in the supplemental material) contain the rpfF gene. However, slightly different rpfF fragment lengths were obtained because of the region's variability. In addition, alignment of the translated N-terminal regions and subsequent phylogenetic analysis revealed that RpfF of S. maltophilia may be distributed into two distinct variants, which we have named RpfF-1 and RpfF-2 (Fig. 1 and 2A and B). The RpfF-1 variant is present in 47 (60.26%) of the 78 strains, whereas RpfF-2 is present in the remaining 31 strains (39.74%) (Fig. 1). Of the additional four complete genome sequences available, K279a and R551-3 contain the RpfF-1 variant and D457 and JV3 contain the RpfF-2 variant.
FIG 1.
(A) Phylogenetic analysis of 82 S. maltophilia strains based on the first 108 amino acids of RpfF. (B) Colony DSF bioassay of three representative strains of each RpfF variant group. Top: E77, ATCC 13637, and K279a (RpfF-1). Bottom: M30, D457, and UV74 (RpfF-2).
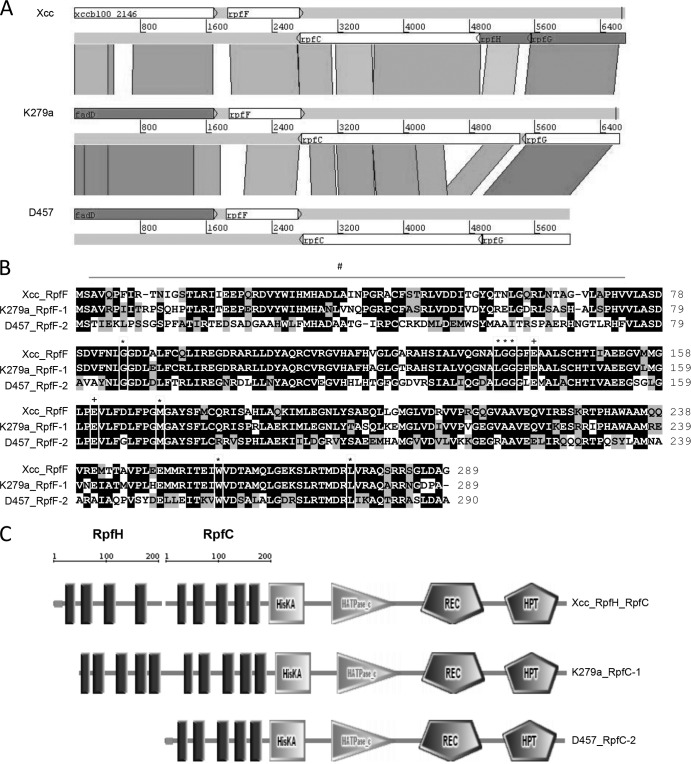
FIG 2.
(A) Comparison of the rpf cluster in X. campestris pv. campestris and S. maltophilia K279a and D457. The alignment was performed with tblastx (percent identity cutoff, 45%) from the BLAST suite and visualized with the Artemis Comparison Tool. Conserved protein regions are paired by shaded blocks where color intensity is proportional to sequence identity. The scales are relative positions in base pairs. (B) Alignment of RpfF proteins from X. campestris pv. campestris and S. maltophilia K279a (RpfF-1) and D457 (RpfF-2). Symbols: #, hypervariable region; *, binding pocket residues; +, glutamate catalytic residues. (C) SMART software analysis of RpfC and RpfH from X. campestris pv. campestris and RpfC from S. maltophilia K279a and D457, where HisKA is a histidine kinase domain, HATPase_c is a histidine ATPase domain, REC is a CheY-like receiver domain, and HPT is a histidine phosphotransferase domain.
Interestingly, no strain carrying the RpfF-2 variant showed DSF activity when tested with the X. campestris pv. campestris 8523/pL6engGUS bioassay (Fig. 1B; see Fig. S1 in the supplemental material). To corroborate the absence of DSF production in these strains, culture supernatants were analyzed with the DSF reporter bioassay, as well as by TLC and GC-MS (see Materials and Methods) with M30 as a representative strain. DSF production was never detected in M30 supernatants by any of these three techniques, indicating that RpfF-2 does not produce DSF under the conditions assayed (Fig. 3B and 4; see Fig. S2 in the supplemental material).
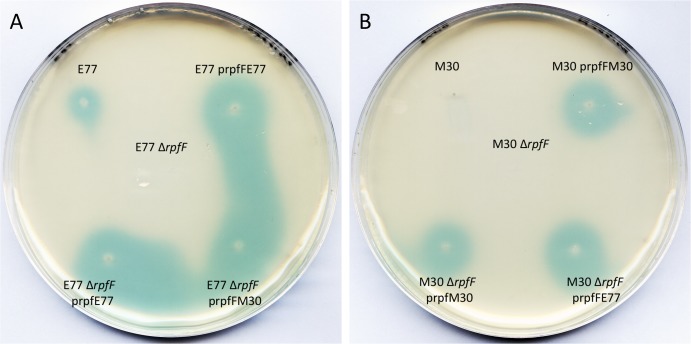
FIG 3.
DSF bioassay of E77 (A) and M30 (B) with their respective ΔrpfF mutants and homologously and heterologously complemented strains and the X. campestris pv. campestris 8523/pL6engGUS reporter strain.
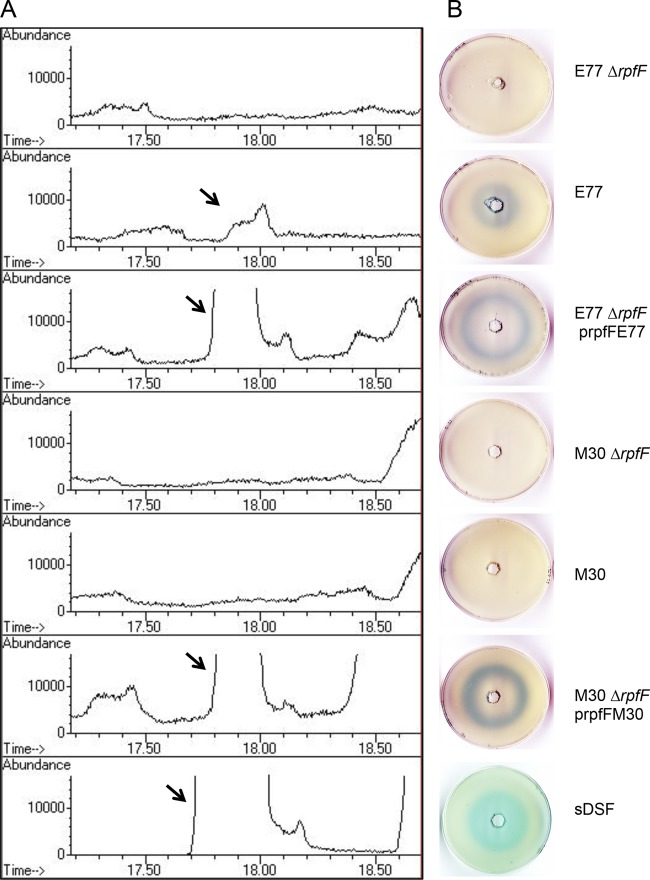
FIG 4.
(A) GC analysis of culture supernatants of E77, M30, and their respective ΔrpfF mutants and complemented strains. (B) DSF bioassay of concentrated supernatants of the same strains from independent extractions.
Initially, the significant differences between the N-terminal regions of the two RpfF variants made us hypothesize that this region could play a direct role in DSF synthesis. However, the residues that form the substrate binding pocket (Leu136, Gly137, Gly138, Gly85, Leu276, Met170, and Trp258), as well as those involved in catalysis (Glu141, Glu161), in X. campestris pv. campestris (22) are conserved in the two variants (Fig. 2B). In order to test the intrinsic capacity of the RpfF-2 variant to produce DSF, we inserted the rpfF gene from M30 (RpfF-2 variant) into an E77 ΔrpfF mutant (RpfF-1 variant) by heterologous complementation. The results obtained demonstrate that RpfF-2 is functional in DSF synthesis, since the E77 ΔrpfF mutant complemented with M30 rpfF showed a big blue halo of DSF diffusion (Fig. 3B). Additionally, insertion of extra copies of its own rpfF gene into WT M30 and the M30 ΔrpfF mutant resulted in DSF production (Fig. 3B), suggesting that RpfF-2 is able to produce DSF but it is repressed in the WT strain under the conditions assayed.
The experiments with deletion mutants and the corresponding complemented strains proved that the blue halo observed in the bioassays is due to the fatty acid produced by the rpfF product (Fig. 3). In addition, MS analysis demonstrated that the signaling factor is DSF (see Fig. S3 in the supplemental material). Moreover, DSF bioassays and TLC analyses of culture supernatants of E77 and M30, their ΔrpfF mutants, and the complemented strains suggested that DSF is the only fatty acid with signaling activity that depends on the RpfF synthase function, since no other differential blue spot was observed when M30 and E77 were compared with each other and with the respective ΔrpfF mutant and complemented strains (see Fig. S2).
Each RpfF variant has an associated RpfC variant, and RpfC-1 contains a TM sensor input domain highly related to the X. campestris pv. campestris RpfH-RpfC complex.
Analyzing the complete rpf cluster in the four S. maltophilia complete genome sequences and in our three draft genome sequences (E77, UV74, and M30), we observed that rpfC also differed significantly between the two S. maltophilia variant groups defined by the rpfF gene (Fig. 2A). Thus, each RpfF variant group appears to have an associated RpfC variant. RpfC-1 (belonging to the RpfF-1 variant strains) and RpfC-2 (belonging to the RpfF-2 variant strains) differ in their N-terminal regions, corresponding to the transmembrane (TM) domain or sensor input domain (Fig. 2C) (23).
It has been postulated that in X. campestris pv. campestris, an additional integral membrane protein, RpfH, participates in DSF sensing (12). In S. maltophilia, the RpfH protein appears to be fused to RpfC-1, generating a sensor input domain with 10 TM regions, as would happen in a putative X. campestris pv. campestris RpfH-RpfC complex. However, the TM domain of the RpfC-2 variant contains only five TM regions. Interestingly, tblastx analysis revealed that the five TM regions present in the RpfC-2 variant are highly related to X. campestris pv. campestris RpfH, while the absent five regions would correspond to the X. campestris pv. campestris RpfC TM domain (Fig. 2A). This indicates that both RpfC variant groups produce a putative RpfH protein but only the RpfC-1 variant contains its own five TM regions in the sensor input domain. The loss of these regions in RpfC-2 could have an implication for DSF detection.
RpfF-2 is permanently repressed by RpfC-2.
In order to study the implication of each RpfC variant in DSF synthesis repression, DSF producer strain E77 was provided with both RpfC variants in trans. Since rpfC is expected to be cotranscribed jointly with rpfG in the rpfGC operon in both the rpf-1 and rpf-2 clusters, we generated vectors prpfGC-E77 and prpfGC-M30. The in trans repression vectors resulted in a reduction of E77 DSF synthesis in both cases. However, while E77 harboring the prpfGC-1 vector showed only a small decrease in DSF synthesis, provision of prpfGC-2 resulted in strong inhibition of DSF production (Fig. 5A), suggesting that RpfC-2 is a stronger repressor of RpfF activity. In order to corroborate this hypothesis, we generated a ΔrpfC-2 mutant of strain M30. Consistent with the previous result, the M30 ΔrpfC mutant became a DSF producer strain (Fig. 5B). Complementation of the M30 ΔrpfC mutant with vectors prpfGC-E77 and prpfGC-M30 in trans led to a scenario similar to that obtained with the E77 strain (Fig. 5B). To further characterize the relationship between RpfF and RpfC in the two variants, the expression of the gene pairs rpfF-1–rpfC-1 and rpfF-2–rpfC-2 was quantified by qRT-PCR using the 2−ΔΔCT method with gyrA as an endogenous control. Thus, expression in WT E77 was 5.16-fold ± 0.59-fold for rpfF-1 and 2.69-fold ± 0.29-fold for rpfC-1 (rpfF-1/rpfC-1 ratio of 1.92), while in WT M30 it was 1.57-fold ± 0.23-fold for rpfF-2 and 1.65-fold ± 0.25-fold for rpfC-2 (rpfF-2/rpfC-2 ratio of 0.95) (see Fig. S4 in the supplemental material).
FIG 5.
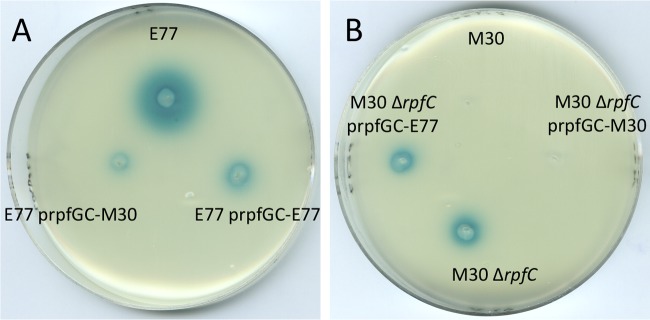
(A) DSF bioassay of WT E77 and E77 complemented with vectors prpfGCE77 and prpfGCM30. (B) DSF bioassay of WT M30, the M30 ΔrpfC mutant, and the M30 ΔrpfC mutant complemented with vectors prpfGCE77 and prpfGCM30.
ΔrpfF mutants display different virulence-associated phenotypes as a function of the native RpfF variant.
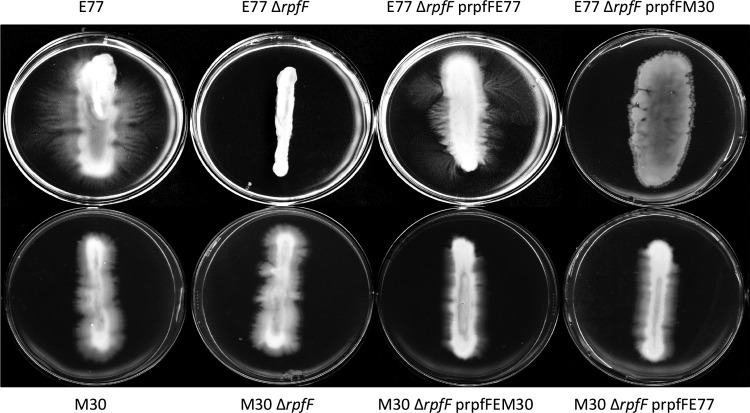
We have investigated the implication of the two RpfF variants for virulence-associated phenotypes such as biofilm formation and swarming motility. We had previously observed that swarming activation of S. maltophilia is faster with streak inoculation than with pin inoculation, suggesting that a high-density population facilitates the initiation of this type of motion (unpublished results). This supports the idea that QS could be involved in swarming activation in S. maltophilia. To corroborate this hypothesis, we tested the ability of E77 and M30 ΔrpfF mutants to swarm on modified M9 medium with a 0.5% agar concentration, relative to that of the WT strains. WT E77 displays tendril-like motility, whereas WT M30 hardly swarms, likely because of its DSF deficiency (Fig. 6). The E77 ΔrpfF mutant shows a clear motility loss and phenotype restoration when rpfF is complemented in trans. On the contrary, the swarming motility of the M30 ΔrpfF mutant is not significantly different from that of the WT M30 strain, suggesting that RpfF does not intervene in swarming control in M30. However, this behavior does not seem to be strictly linked to the RpfF variant. Thus, on the one hand the E77 ΔrpfF mutant displayed an atypical nontendril swarming morphology when heterologously complemented with the M30 rpfF gene. On the other, heterologous complementation of the M30 ΔrpfF mutant with the E77 rpfF gene resulted in motility similar to that of the M30 ΔrpfF mutant strain.
FIG 6.
Swarming motility assay of E77, M30, and their ΔrpfF mutants and homologously and heterologously complemented strains on modified M9 medium solidified with 0.5% Noble agar and incubated at 30°C for 4 days.
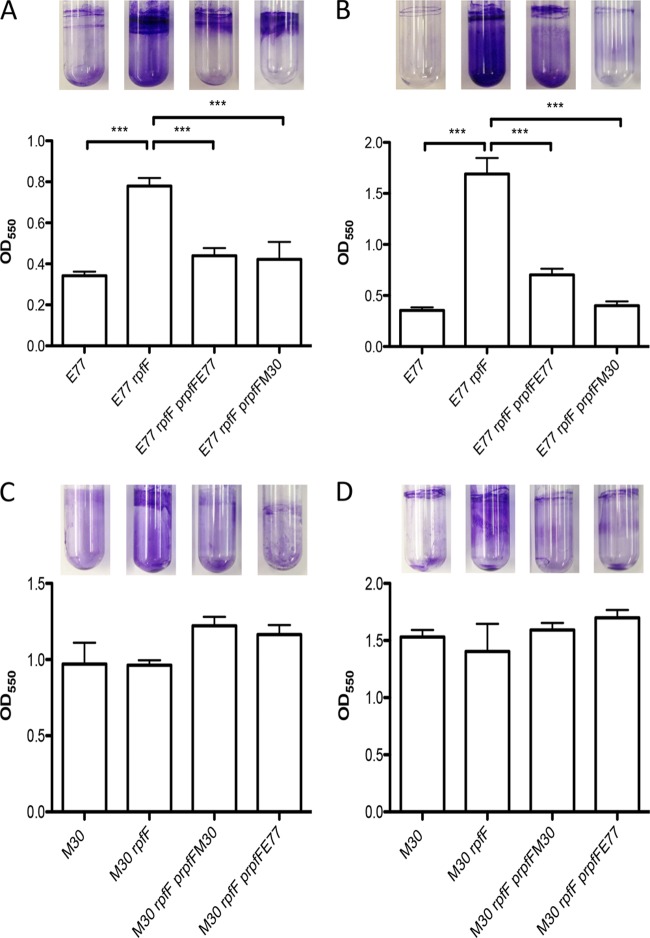
Biofilm formation by E77, M30, and the respective ΔrpfF mutants and complemented variants on a polystyrene or glass surface was also evaluated under two different medium conditions (M9 and BM2). The results show that M30 has a higher capacity than E77 to form biofilm under both growth conditions (Fig. 7), contrary to the ability to swarm, suggesting that DSF production may inversely regulate these two behaviors in S. maltophilia. Additionally, the results also indicate that biofilm formation is altered only in the E77 ΔrpfF mutant, showing a significant increase relative to that of WT E77. Homologous and heterologous complementation with the respective RpfF variants restores almost WT E77 levels of biofilm formation (P < 0.0005) on both glass and plastic surfaces (Fig. 7A and B). On the other hand, the M30 strain, the M30 ΔrpfF mutant, and the homologously and heterologously complemented strains show similar levels of biofilm formation (Fig. 7C and D). As for the regulation of swarming motility or the ability to produce DSF, it therefore appears that the regulation of biofilm formation is not strictly dependent on the RpfF variant but on one or more components associated with this variant, in particular, RpfC. Specifically, the results suggest that RpfF is involved in the regulation of biofilm formation and swarming motility only in strains that natively carry RpfF-1 (even when this is replaced with RpfF-2). This is likely connected to the ability of these strains to produce DSF.
FIG 7.
Biofilm formation by E77, M30, and their respective ΔrpfF mutants and homologously and heterologously complemented strains on polystyrene (plots) and glass (tubes) surfaces in M9 (A and C) and BM2 (B and D) minimal media. ***, P < 0.0005.
The ΔrpfF-1 mutant, but not the ΔrpfF-2 mutant, shows attenuation in C. elegans.
To elucidate the direct implication of each RpfF variant in S. maltophilia virulence in vivo, the killing ability of E77, M30, and the respective ΔrpfF mutants and complemented strains was tested in C. elegans. Although the WT E77 and M30 strains showed similar virulence capacities in the C. elegans model (with times required to kill 50% of the nematodes, 6.04 and 4.99 days, respectively), significant attenuation was observed here for the E77 ΔrpfF mutant (Fig. 8). In line with the observations made for the phenotypes analyzed previously, the virulence of the E77 ΔrpfF mutant is restored after complementation with either its own rpfF gene (RpfF-1 variant) or the M30 rpfF gene (RpfF-2 variant), indicating once more that E77 is able to respond to heterologous DSF production in an RpfC-1 variant background (Fig. 8). Infection with the M30 ΔrpfF mutant shows no significant differences from WT M30. These results suggest again that the RpfF-RpfC pair may regulate virulence only in those strains carrying the variant 1 combination.
FIG 8.
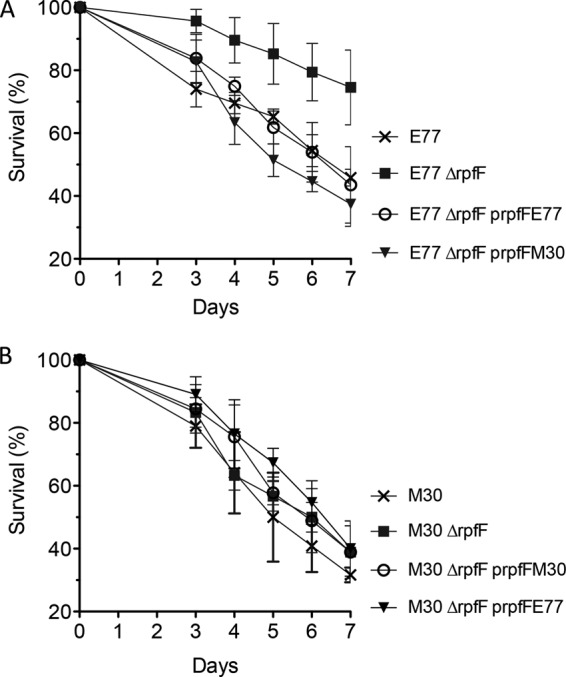
Determination of virulence of E77 (A), M30 (B), and their respective ΔrpfF mutants and homologously and heterologously complemented strains in a C. elegans CF512 model of infection.
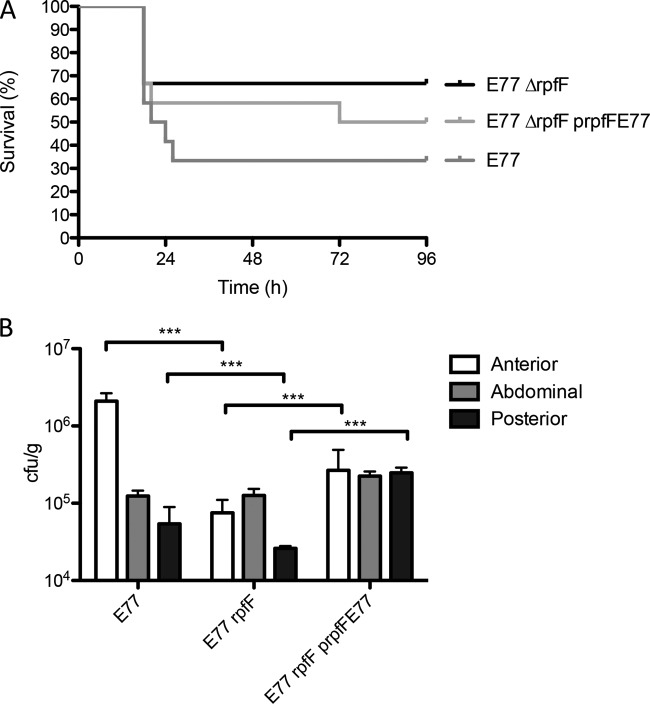
The ΔrpfF-1 mutant shows attenuation in zebrafish due to its inability to disseminate through fish tissues.
E77, its ΔrpfF mutant, and the complemented strain were evaluated in zebrafish as a vertebrate model. Similar results were obtained, corroborating that RpfF-1 is involved in virulence regulation (Fig. 9A). Interestingly, recovery of bacteria from sacrificed fish from each tank at 72 h postinjection showed the ability of WT E77 to disseminate through the fish body from the abdominal region to the anterior and posterior regions. On the contrary, the E77 ΔrpfF mutant does not seem to be able to colonize those regions effectively. Complementation of rpfF partially restores its body dissemination capacity (Fig. 9B).
FIG 9.
(A) Virulence of E77, the ΔrpfF mutant, and the complemented strain in 9-month-old zebrafish. (B) Bacterial recovery from different regions of the bodies of sacrificed fish at 72 h postinfection with E77, the ΔrpfF mutant, or the complemented strain. ***, P < 0.0005.
DISCUSSION
We have characterized 78 S. maltophilia clinical strains isolated from diverse sources in different European hospitals for the rpfF gene. We have first demonstrated that the 78 strains contain the rpfF gene but the RpfF product is distributed into two different variants that we have named RpfF-1 and RpfF-2 (Fig. 1 and 2A). We also show that the isolates produce two RpfC variants, each associated with one of the RpfF variants (Fig. 2A). The two RpfC variants are different in the N-terminal region, which corresponds to a TM domain (Fig. 2C) thought to participate in DSF sensing in several Xanthomonas species (12). In S. maltophilia, the RpfC-1 variant contains 10 TM regions that display high similarity to the putative X. campestris pv. campestris RpfH-RpfC TM complex (Fig. 2A and C). On the other hand, the RpfC-2 variant has only five TM regions, which appear to be related to the X. campestris pv. campestris RpfH TM domain rather than that of X. campestris pv. campestris RpfC (Fig. 2A). This phenomenon is also observed in Xylella fastidiosa, Xanthomonas oryzae, and Pseudoxanthomonas species, suggesting that the RpfC-2 variant is widely distributed among the members of the order Xanthomonadales that share the DSF-QS system. Nevertheless, protein sequence comparison shows a high similarity between the RpfC and RpfH TM domains, suggesting that a duplication event (for X. campestris pv. campestris rpfC to rpfH and S. maltophilia rpfC-1) or a deletion (for S. maltophilia rpfC-2) may have occurred.
A previous S. maltophilia population study suggested that an important group of S. maltophilia isolates lack rpfF (10). PCR-based typing of 89 strains showed an rpfF+ prevalence of 61.8%, while the remaining 38.2% were considered to be rpfF mutants. On the basis of our sequence analysis, we can conclude that the work of Pompilio and collaborators (10) failed to detect rpfF because the primers they used were designed to hybridize within the most variable region of this gene; more specifically, those primers do not amplify rpfF in strains carrying what we have defined as variant 2. Accordingly, we hypothesize that all of the S. maltophilia strains analyzed in the study by Pompilio et al. and showing an rpfF+ genotype belong to the RpfF-1 variant group, whereas the rpfF mutant strains would belong to the RpfF-2 variant group. Interestingly, our analysis of rpfF from a collection of 82 S. maltophilia strains shows similar RpfF variant frequencies in the population. RpfF-1 is present in 59.75% of the strains (including K279a and R551), whereas RpfF-2 is present in 40.25% (including D457 and JV3). Taking the two studies together (171 strains), strains carrying the RpfF-1 variant appear to be more commonly isolated than those carrying the RpfF-2 variant, with relative prevalences of ca. 60 and 40%, respectively.
Surprisingly, we have observed that only strains carrying the RpfC–RpfF-1 pair produce DSF under WT conditions, while strains belonging to the RpfC–RpfF-2 variant group require extra copies of their own rpfF gene (Fig. 3 and 4) or the absence of the repressor component RpfC-2 (Fig. 5) to achieve detectable DSF production levels. These results indicate that RpfF-2 is able to synthesize DSF but the production of this signaling molecule is permanently repressed by RpfC-2 under the conditions assayed. It has been shown that the stoichiometric balance between RpfF and RpfC is crucial for DSF production in many members of the order Xanthomonadales. In X. campestris pv. campestris, RpfC physically interacts with the RpfF active site, inhibiting DSF synthesis activity (12, 22, 24). RpfC has also been shown to repress the RpfF activity of X. fastidiosa (25). Analysis of mRNA levels in E77 and M30 by qRT-PCR shows that the rpfF/rpfC expression ratio in the DSF producer strain (variant 1) is double that found in the nonproducer one (variant 2), suggesting, together with the observation that variant 2 strains complemented with extra rpfF copies produce DSF, that the different phenotypes of the two variants may be partly due to the different regulation of the stoichiometry of these two components. It has also been suggested that in X. campestris pv. campestris, RpfC could play a positive-feedback role in DSF synthesis, liberating active RpfF upon the detection of DSF molecules (22). Assuming similar mechanisms in S. maltophilia, we hypothesize that DSF production in RpfC–RpfF-1 strains is due to the presence of a competent sensor input domain, i.e., composed of 10 TM regions, in RpfC-1, which would enable the liberation of active RpfF-1 upon DSF detection and the subsequent synthesis of DSF. On the other hand, the missing TM regions in RpfC-2 would render this factor incompetent for DSF sensing, leading to permanent inhibition of RpfF-2 by RpfC-2 in a situation of equal numbers of copies. Demonstrating that RpfC-1 liberates free active RpfF after DSF detection and understanding its mechanism or unveiling why the S. maltophilia population produces two RpfC variants and what implications it may have for DSF-mediated regulation are questions that require further studies. The possibility that RpfC–RpfF-2 variant strains may produce DSF under specific environmental conditions or that RpfF-2 may produce a different yet undetected DSF derivative cannot be ruled out. Comparison of GC-MS spectra from M30, its ΔrpfF mutant, and the complemented strain did not, however, reveal any peak compatible with the mass of a DSF derivative.
It is well known that the DSF-QS system regulates certain virulence traits in many bacteria (1, 3, 9, 12, 26–29). To determine the possible implication of each RpfF variant for virulence regulation, we generated an rpfF deletion mutant for a strain representative of each variant group, i.e., E77 for the RpfF-1 variant group and M30 for the RpfF-2 group. All of the phenotypes evaluated in M30 were unaltered in the ΔrpfF mutant and in the corresponding complemented strain, suggesting that RpfC–RpfF-2 variant strains may not use the DSF-QS system to regulate these virulence factors, likely because of their inability to produce and sense DSF molecules under the conditions assayed. On the contrary, the E77 ΔrpfF mutant showed attenuation in both the C. elegans (Fig. 8A) and zebrafish (Fig. 9A) infection models, proving that DSF-mediated regulation affects the virulence of RpfC–RpfF-1 strains. Moreover, the recovery of bacteria from sacrificed fishes at 72 h postinjection showed that E77 is able to disseminate to the anterior and posterior regions through the fish body, while the E77 ΔrpfF mutant had serious problems in crossing intraperitoneal barriers (Fig. 9B). This is in concordance with the results showing a loss of swarming motility (Fig. 6) and a drastic increase in biofilm formation capacity (Fig. 7A and B) by the E77 ΔrpfF mutant, two important virulence-related traits that would explain attenuation in the animal models and especially in the zebrafish experiments. Much evidence of the implication of RpfF and DSF-like fatty acids in bacterial motility has indeed been reported (1, 2, 9, 30). Our results thus reinforce previous evidence that one of the main functions of DSF-QS is to regulate bacterial motility. Many studies have also demonstrated the implication of DSF-like molecules in biofilm regulation. There is, however, some controversy about whether DSF-like molecules may act by stimulating or inhibiting the sessile or motile bacterial lifestyle. Thus, DSF molecules have been shown to positively regulate biofilm formation in X. oryzae pv. oryzae (31), B. cenocepacia (9, 28), and X. fastidiosa (4, 26). On the contrary, in X. campestris pv. campestris, the DSF-mediated QS acts as a negative regulator of biofilm development (32–34). Additionally, fatty acid-mediated biofilm dispersion is not restricted to species with the DSF-QS system. For example, the fatty acid cis-2-decenoic acid produced by Pseudomonas aeruginosa PAO1 stimulates biofilm dispersion in several Gram-positive and Gram-negative bacteria (35, 36). Our findings indicate that the DSF-QS system in S. maltophilia E77 has a regulatory function similar to that described for X. campestris pv. campestris, where DSF also plays an important role in preventing biofilm formation and stimulating bacterial motility.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funding under the Seventh Research Framework Programme of the European Union (HEALTH-F3-2009-223101) and the Spanish MICINN (BFU2010-17199). I.G. acknowledges support from the Catalan AGAUR (2009SGR-00108). I.R. and J.V. acknowledge support from the SATURN project (European Community FP7, HEALTH-F3-2009-241796).
We thank C. Prat and J. Domínguez (Servei de Microbiologia, Hospital Universitari Germans Trias i Pujol, Institut d'Investigació Germans Trias i Pujol, Universitat Autònoma de Barcelona, Badalona, Spain) and J. L. Martínez (Centro Nacional de Biotecnología, CSIC, Cantoblanco, Madrid, Spain) for providing some of the clinical strains used in this study.
Footnotes
Published ahead of print 25 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01540-14.
REFERENCES
- 1.Fouhy Y, Scanlon K, Schouest K, Spillane C, Crossman L, Avison MB, Ryan RP, Dow JM. 2007. Diffusible signal factor-dependent cell-cell signaling and virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J. Bacteriol. 189:4964–4968. 10.1128/JB.00310-07 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Huang T-P, Lee Wong AC. 2007. Extracellular fatty acids facilitate flagella-independent translocation by Stenotrophomonas maltophilia. Res. Microbiol. 158:702–711. 10.1016/j.resmic.2007.09.002 [DOI] [PubMed] [Google Scholar]
- 3.Barber CE, Tang JL, Feng JX, Pan MQ, Wilson TJ, Slater H, Dow JM, Williams P, Daniels MJ. 1997. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24:555–566. 10.1046/j.1365-2958.1997.3721736.x [DOI] [PubMed] [Google Scholar]
- 4.Beaulieu ED, Ionescu M, Chatterjee S, Yokota K, Trauner D, Lindow S. 2013. Characterization of a diffusible signaling factor from Xylella fastidiosa. mBio 4:e00539–00512. 10.1128/mBio.00539-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boon C, Deng Y, Wang L-H, He Y, Xu J-L, Fan Y, Pan SQ, Zhang L-H. 2008. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J. 2:27–36. 10.1038/ismej.2007.76 [DOI] [PubMed] [Google Scholar]
- 6.Deng Y, Wu J, Tao F, Zhang L-H. 2011. Listening to a new language: DSF-based quorum sensing in Gram-negative bacteria. Chem. Rev. 111:160–173. 10.1021/cr100354f [DOI] [PubMed] [Google Scholar]
- 7.Huang T-P, Wong ACL. 2007. A cyclic AMP receptor protein-regulated cell-cell communication system mediates expression of a FecA homologue in Stenotrophomonas maltophilia. Appl. Environ. Microbiol. 73:5034–5040. 10.1128/AEM.00366-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alavi P, Müller H, Cardinale M, Zachow C, Sánchez MB, Martínez JL, Berg G. 2013. The DSF quorum sensing system controls the positive influence of Stenotrophomonas maltophilia on plants. PLoS One 8:e67103. 10.1371/journal.pone.0067103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng Y, Schmid N, Wang C, Wang J, Pessi G, Wu D, Lee J, Aguilar C, Ahrens CH, Chang C, Song H, Eberl L, Zhang L-H. 2012. Cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc. Natl. Acad. Sci. U. S. A. 109:15479–15484. 10.1073/pnas.1205037109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pompilio A, Pomponio S, Crocetta V, Gherardi G, Verginelli F, Fiscarelli E, Dicuonzo G, Savini V, D'Antonio D, Di Bonaventura G. 2011. Phenotypic and genotypic characterization of Stenotrophomonas maltophilia isolates from patients with cystic fibrosis: genome diversity, biofilm formation, and virulence. BMC Microbiol. 11:159. 10.1186/1471-2180-11-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrer-Navarro M, Planell R, Yero D, Mongiardini E, Torrent G, Huedo P, Martínez P, Roher N, Mackenzie S, Gibert I, Daura X. 2013. Abundance of the quorum-sensing factor Ax21 in four strains of Stenotrophomonas maltophilia correlates with mortality rate in a new zebrafish model of infection. PLoS One 8:e67207. 10.1371/journal.pone.0067207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slater H, Alvarez-Morales A, Barber CE, Daniels MJ, Dow JM. 2000. A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol. Microbiol. 38:986–1003. 10.1046/j.1365-2958.2000.02196.x [DOI] [PubMed] [Google Scholar]
- 13.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultz J, Milpetz F, Bork P, Ponting CP. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95:5857–5864. 10.1073/pnas.95.11.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- 16.Choi K-H, Kumar A, Schweizer HP. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391–397. 10.1016/j.mimet.2005.06.001 [DOI] [PubMed] [Google Scholar]
- 17.Kovach ME, Phillips RW, Elzer PH, Roop RM, II, Peterson KM. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800–802 [PubMed] [Google Scholar]
- 18.Tan MW, Mahajan-Miklos S, Ausubel FM. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 96:715–720. 10.1073/pnas.96.2.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinkel MD, Eames SC, Philipson LH, Prince VE. 2010. Intraperitoneal injection into adult zebrafish. J. Vis. Exp. 42:2126. 10.3791/2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gould VC, Avison MB. 2006. SmeDEF-mediated antimicrobial drug resistance in Stenotrophomonas maltophilia clinical isolates having defined phylogenetic relationships. J. Antimicrob. Chemother. 57:1070–1076. 10.1093/jac/dkl106 [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 22.Cheng Z, He Y-W, Lim SC, Qamra R, Walsh MA, Zhang L-H, Song H. 2010. Structural basis of the sensor-synthase interaction in autoinduction of the quorum sensing signal DSF biosynthesis. Structure 18:1199–1209. 10.1016/j.str.2010.06.011 [DOI] [PubMed] [Google Scholar]
- 23.Ryan RP, Fouhy Y, Garcia BF, Watt SA, Niehaus K, Yang L, Tolker-Nielsen T, Dow JM. 2008. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol. Microbiol. 68:75–86. 10.1111/j.1365-2958.2008.06132.x [DOI] [PubMed] [Google Scholar]
- 24.Wang L-H, He Y, Gao Y, Wu JE, Dong Y-H, He C, Wang SX, Weng L-X, Xu J-L, Tay L, Fang RX, Zhang L-H. 2004. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol. Microbiol. 51:903–912. 10.1046/j.1365-2958.2003.03883.x [DOI] [PubMed] [Google Scholar]
- 25.Ionescu M, Baccari C, Da Silva AM, Garcia A, Yokota K, Lindow SE. 2013. Diffusible signal factor (DSF) synthase RpfF of Xylella fastidiosa is a multifunction protein also required for response to DSF. J. Bacteriol. 195:5273–5284. 10.1128/JB.00713-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chatterjee S, Wistrom C, Lindow SE. 2008. A cell-cell signaling sensor is required for virulence and insect transmission of Xylella fastidiosa. Proc. Natl. Acad. Sci. U. S. A. 105:2670–2675. 10.1073/pnas.0712236105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deng Y, Boon C, Eberl L, Zhang L-H. 2009. Differential modulation of Burkholderia cenocepacia virulence and energy metabolism by the quorum-sensing signal BDSF and its synthase. J. Bacteriol. 191:7270–7278. 10.1128/JB.00681-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy Y, Yang L, Twomey KB, Sass A, Tolker-Nielsen T, Mahenthiralingam E, Dow JM, Ryan RP. 2010. A sensor kinase recognizing the cell-cell signal BDSF (cis-2-dodecenoic acid) regulates virulence in Burkholderia cenocepacia. Mol. Microbiol. 77:1220–1236. 10.1111/j.1365-2958.2010.07285.x [DOI] [PubMed] [Google Scholar]
- 29.Twomey KB, O'Connell OJ, McCarthy Y, Dow JM, O'Toole GA, Plant BJ, Ryan RP. 2012. Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa. ISME J. 6:939–950. 10.1038/ismej.2011.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryan RP, McCarthy Y, Kiely PA, O'Connor R, Farah CS, Armitage JP, Dow JM. 2012. Dynamic complex formation between HD-GYP, GGDEF and PilZ domain proteins regulates motility in Xanthomonas campestris. Mol. Microbiol. 86:557–567. 10.1111/mmi.12000 [DOI] [PubMed] [Google Scholar]
- 31.Rai R, Ranjan M, Pradhan BB, Chatterjee S. 2012. Atypical regulation of virulence-associated functions by a diffusible signal factor in Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 25:789–801. 10.1094/MPMI-11-11-0285-R [DOI] [PubMed] [Google Scholar]
- 32.Dow JM, Crossman L, Findlay K, He Y-Q, Feng J-X, Tang J-L. 2003. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc. Natl. Acad. Sci. U. S. A. 100:10995–11000. 10.1073/pnas.1833360100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He Y-W, Xu M, Lin K, Ng Y-JA, Wen C-M, Wang L-H, Liu Z-D, Zhang H-B, Dong Y-H, Dow JM, Zhang L-H. 2006. Genome scale analysis of diffusible signal factor regulon in Xanthomonas campestris pv. campestris: identification of novel cell-cell communication-dependent genes and functions. Mol. Microbiol. 59:610–622. 10.1111/j.1365-2958.2005.04961.x [DOI] [PubMed] [Google Scholar]
- 34.Tao F, Swarup S, Zhang L-H. 2010. Quorum sensing modulation of a putative glycosyltransferase gene cluster essential for Xanthomonas campestris biofilm formation. Environ. Microbiol. 12:3159–3170. 10.1111/j.1462-2920.2010.02288.x [DOI] [PubMed] [Google Scholar]
- 35.Davies DG, Marques CNH. 2009. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191:1393–1403. 10.1128/JB.01214-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amari DT, Marques CNH, Davies DG. 2013. The putative enoyl-coenzyme A hydratase DspI is required for production of the Pseudomonas aeruginosa biofilm dispersion autoinducer cis-2-decenoic acid. J. Bacteriol. 195:4600–4610. 10.1128/JB.00707-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.