Abstract
Allelic replacement mutants were constructed within arginine deiminase (arcA1 and arcA2) to assess the function of the arginine deiminase (ADI) pathway in organic acid resistance and biofilm formation of Staphylococcus epidermidis 1457. A growth-dependent acidification assay (pH ∼5.0 to ∼5.2) determined that strain 1457 devoid of arginine deiminase activity (1457 ΔADI) was significantly less viable than the wild type following depletion of glucose and in the presence of arginine. However, no difference in viability was noted for individual 1457 ΔarcA1 (native) or ΔarcA2 (arginine catabolic mobile element [ACME]-derived) mutants, suggesting that the native and ACME-derived ADIs are compensatory in S. epidermidis. Furthermore, flow cytometry and electron paramagnetic resonance spectroscopy results suggested that organic acid stress resulted in oxidative stress that could be partially rescued by the iron chelator dipyridyl. Collectively, these results suggest that formation of hydroxyl radicals is partially responsible for cell death via organic acid stress and that ADI-derived ammonia functions to counteract this acid stress. Finally, static biofilm assays determined that viability, ammonia synthesis, and pH were reduced in strain 1457 ΔADI following 120 h of growth in comparison to strain 1457 and the arcA1 and arcA2 single mutants. It is hypothesized that ammonia synthesis via the ADI pathway is important to reduce pH stress in specific microniches that contain high concentrations of organic acids.
INTRODUCTION
Staphylococcus epidermidis is the most significant cause of biomaterial-related infections in the nosocomial environment. In contrast to S. aureus, S. epidermidis does not produce a plethora of virulence factors and typically requires a foreign body to act as a nidus of infection where biofilm can form (1). S. epidermidis biofilm is a complex mixture of extracellular DNA (eDNA), protein (Aap, Embp), and polysaccharide (PIA) accumulation molecules (2) and functions to augment antibacterial resistance (3) and increase resistance to the innate immune response (4). In addition, staphylococcal biofilm is fundamentally heterogeneous, displaying multiple oxygen and nutritional gradients and thus creating unique metabolic microenvironments (5, 6). Therefore, based on the defined oxygen and nutritional gradients in addition to other ill-defined factors, differential gene expression occurs throughout staphylococcal biofilms (7).
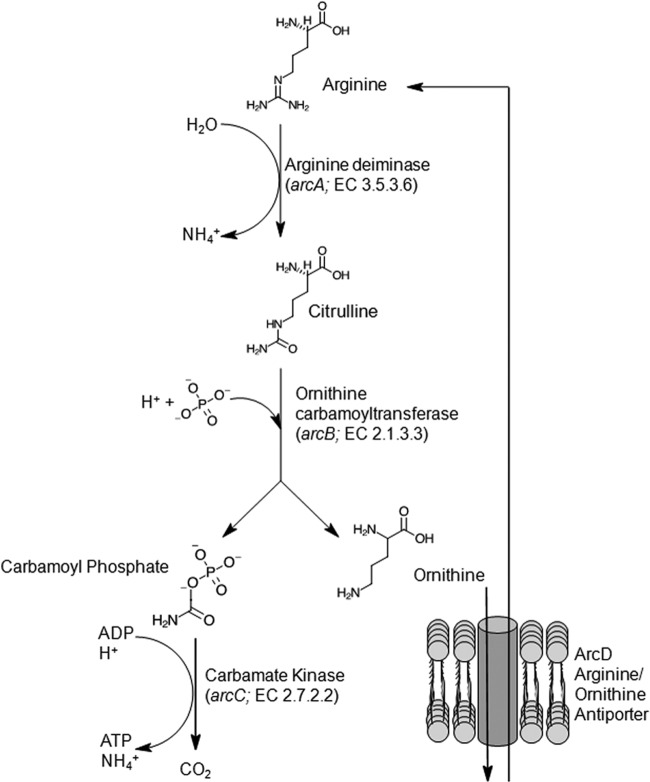
The arginine deiminase (ADI) pathway is highly conserved in bacteria and is composed of three enzymes (ArcA, ArcB, and ArcC), the genes of which are arranged in an operon (arc operon), that catalyze the conversion of arginine to ornithine, ammonia, and carbon dioxide, with the concomitant production of ATP (8) (Fig. 1). The complete conversion of 1 mole of arginine generates 1 mole of ATP and 2 moles of NH4+. The ADI pathway is found across many genera of bacteria, including lactic acid bacteria (9–13), Streptococcus spp. (14, 15), Listeria monocytogenes (16), Staphylococcus spp. (17–19), and Pseudomonas spp. (20); arginine can serve as the sole energy source for growth of a variety of bacteria, including the streptococci (21) and Pseudomonas aeruginosa (22).
FIG 1.
Schematic of ATP and NH4+ generation through the arginine deiminase (ADI) pathway. Arginine is transported into the cell through an arginine/ornithine antiporter (ArcD). Arginine deiminase (ArcA) and ornithine carbamoyltransferase (ArcB) subsequently catalyze the formation of citrulline and carbamoyl phosphate/ornithine, respectively. Carbamate kinase (ArcC) catalyzes the formation of CO2, ATP, and NH4+ from carbamoyl phosphate, whereas ornithine is exported from the cell, facilitating the import of arginine.
Transcription of the arc operon is often arginine dependent via the ArgR family of regulators, including both ArgR and AhrC (23–25), and is also subject to carbon catabolite repression (CCR) via CcpA (9, 26). arc operon expression is also typically induced under anoxic growth conditions (11, 16, 26, 27) and is ArcR dependent in S. aureus (28). One demonstrated function of the ADI pathway in lactic acid bacteria is pH homeostasis and survival under low-pH conditions, mediated by ammonia synthesis, which can increase both cytoplasmic pH and environmental pH (25, 29). Several studies have demonstrated that the ADI pathway is required for survival during acid stress and is also linked to the acid tolerance response in lactic acid bacteria (11, 14, 16, 27, 30–32).
Increased attention was drawn to the ADI pathway within S. aureus due to the identification of a second copy of the arc operon in the community-associated methicillin-resistant USA300 clone (33). This second copy of the arc operon is contained within the arginine catabolic mobile element (ACME), a 30.9-kb pathogenicity island also carrying genes encoding several oligopeptide permeases, a spermine/spermidine N-acetyltransferase (speG), and other putative open reading frames. Genomic analyses have revealed diverse ACMEs present throughout S. epidermidis (34) suggesting that S. epidermidis may be the source for the ACME of USA300 (18, 33). Thurlow and colleagues recently demonstrated that the ACME-encoded ADI pathway and SpeG in S. aureus USA300 coordinate to confer an increased ability to survive and proliferate under acidic growth conditions similar to those found on skin (35). Thus, due to the high prevalence of the ACME-encoded ADI pathway in S. epidermidis, experiments were designed to assess the function of ADI-derived ammonia in pH homeostasis of S. epidermidis to determine its potential function in facilitating growth in acidic microniches such as biofilm.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains, plasmids, and primers used in this study are listed inTables 1 and 2.
TABLE 1.
Bacterial strains and plasmids used in the study
| Bacterial strain, bacteriophage, or plasmid | Notable characteristic(s)a | Source or reference |
|---|---|---|
| Bacterial strains | ||
| E. coli DH5α | Plasmid cloning host and Gram-negative replicon | Invitrogen |
| S. epidermidis 1457 | Wild-type strain used in study | 45 |
| S. epidermidis 1457 ΔarcA1 | arcA1 allelic replacement mutant of 1457, Tetr | This study |
| S. epidermidis 1457 ΔarcA2 | arcA2 allelic replacement mutant of 1457, Tmpr | This study |
| S. epidermidis 1457 ΔADI | arcA1 and arcA2 allelic replacement mutant of 1457, Tetr, Tmpr | This study |
| S. epidermidis 1457 ΔADI/pNF232 | S. epidermidis 1457 arcA1::tetM arcA2::dhfr harboring pNF232 containing arcA1B1D1R1 | This study |
| S. epidermidis 1457 ΔADI/pNF233 | S. epidermidis 1457 arcA1::tetM arcA2::dhfr harboring pNF233 containing arcA2D2R2B2C2 | This study |
| S. aureus RN4220 | Restriction deficient, modification positive | 44 |
| Bacteriophage | ||
| Phage 71 | S. epidermidis transducing phage | 46 |
| Plasmids | ||
| pUC19 | Gram-negative origin of replication, Ampr | Invitrogen |
| pROJ6448 | pE194 containing pC221 nick site functioning in conjugative mobilization, temp sensitive gram-positive origin of replication, Ermr | 42 |
| pNF137 | arcA1 allelic replacement vector, Ampr, Ermr, Tetr | This study |
| pNF142 | arcA2 allelic replacement vector, Ampr, Ermr, Tmpr | This study |
| pCN51 | Staphylococcal shuttle vector containing cadmium-inducible promoter (pCAD) | 82 |
| pNF232 | pCN51 containing arcA1B1D1R1 in SphI-BamHI restriction sites (deleting pCAD) | This study |
| pNF233 | pCN51 containing arcA2D2R2B2C2 in SphI-BamHI restriction sites (deleting pCAD) | This study |
| pJF12 | 3-kb tetM fragment cloned into pCR2.1 (Invitrogen); Ampr, Tetr | 41 |
| pGO558 | 1.7-kb dhfr cloned into the SalI site of pUC19; Ampr, Tmpr | 41 |
Amp, ampicillin; Erm, erythromycin; Tet, tetracycline; Tmp, trimethoprim.
TABLE 2.
Primers used in the studya
| Product | Forward primer | Reverse primer |
|---|---|---|
| arcA1B1D1R1 | 2009 CCGGGCATGCAAGACGACACCAGGATTTGC | 2010 CCGGGGATCCATTATGTATGAGTGATACGC |
| arcA2D2R2B2C2 | 2011 CCGGGCATGCACATCTCCTGGCTTTGCTCA | 2012 CCGGGGATCCAATAGCCTAGAACATGGTG |
| Upstream and 5′ region of arcA1 | 1068 ATCCTAGGATCCGCAGATATTATTGATTAC | 1069 ATCCTATCTAGACGTCATCTATAAATTGC |
| Downstream and 3′ region of arcA1 | 1070 ATCCTAGTCGACCGTCCCGGTGTGGTTGT | 1071 ATCCTACTGCAGCGGCAGTATGAATGTTATC |
| Upstream and 5′ region of arcA2 | 1074 ATCCTAGGATCCGGCAAGACATTTTGGAG | 1075 ATCCTATCTAGACTCGCGTACTTCTGGCTC |
| Downstream and 3′ region of arcA2 | 1076 ATCCTAGTCGACGCTATTGGTATATCAGAACG | 1077 ATCCTACTGCAGCTTGGAAGTGTGTTACCTCC |
| tetM tetracycline resistance cassette | 1638 ATCCTAGGATCCGGTACATGATTACAGATAC | 1639 ATCCTAGGATCCCGATCTCCTCCTTTCCAC |
| dhfr trimethoprim resistance cassette | 887 GGCATAGGATCCCGATTGTCAGGCTTAATGG | 888 GGCATAGGATCCGCAACTTAGGGAATGTTTATGG |
| arcA1 DNA probe | 1130 GTACGCAAAGAAGAAATACAACTTG | 1129 TGCTGAATGTGTAGTGAATTTGTCG |
| arcA2 DNA probe | 1682 CTTAGATCCAATGCCCAACC | 1683 CATACATCTTGGGCCTCCGC |
Underlined portions of the primers correspond to restriction enzyme recognition sites. GCATGC, SphI; GGATCC, BamHI; TCTAGA, XbaI; GTCGAC, SalI; CTGCAG, PstI; GCTAGC, NheI.
Culture media and growth conditions.
Staphylococcal strains were grown in tryptic soy broth (TSB; Becton, Dickinson [BD], Franklin Lakes, NJ) or tryptic soy agar (TSA; BD). Escherichia coli DH5α was grown in lysogeny broth (LB) or agar (Becton, Dickinson). Antibiotics were purchased from Sigma (St. Louis, MO.) and used at the following concentrations: ampicillin at 50 μg/ml, tetracycline at 10 μg/ml, trimethoprim at 10 μg/ml, and erythromycin at 10 μg/ml for S. epidermidis and S. aureus and 500 μg/ml for E. coli. Anaerobic growth studies were performed using a 10:1 flask-to-volume ratio in media supplemented with 1.0 g/liter l-cysteine hydrochloride monohydrate (Sigma), with shaking at 250 rpm at 37°C (to ensure that cells did not settle), in an anaerobic growth chamber (Coy Laboratory Products, Inc., Grass Lakes, MI).
Transcriptional profiling.
S. epidermidis 1457 grown under microaerobic growth conditions (5:3 flask-to-volume ratio, 125 rpm, 37°C) in TSB was used to inoculate Stovall flow cells (Stovall, Greensboro, NC). Flow cell biofilms were grown in TSB at 37°C using plastic convertible flow cells (24 mm by 40 mm by 8 mm) and a flow rate of 0.5 ml/min. RNA was isolated from the biofilm within the flow cells at 24 and 72 h of growth, and the transcriptional profiles were compared. Briefly, RNA was converted to cDNA, biotinylated, and subsequently hybridized to S. epidermidis PMDsepi1a GeneChips (Affymetrix, Santa Clara, CA). PMDsepi1a contains the complete genomes of S. epidermidis ATCC 1228 (36) and RP62a (37) and publicly available S. epidermidis sequences. cDNA samples were isolated from three independent experiments to ensure biological reproducibility. Signal intensity values for each predicted open reading frame and intergenic region were normalized to the median signal intensity value for each GeneChip. Comparisons of signal intensity values of 1457 RNA isolated at 24 h and 1457 RNA isolated at 72 h showing at least a 2-fold difference and the appropriate t test value (P ≤ 0.05) were considered significant. Data were normalized and analyzed using GeneSpring 6.2 software (Silicon Genetics, Redwood City, CA). KOBAS 2 was utilized to identify statistically enriched pathways using the S. epidermidis ATCC 12228 genome annotation (38).
Construction of allelic replacement mutants and complementation vectors.
PCR primers for amplification of S. epidermidis 1457 arginine deiminase loci were designed using the genome sequence of S. epidermidis ATCC 12228 (GenBank accession no. AE015929) and are listed in Table 2. pNF137, the 1457 ΔarcA1 allelic replacement construct, was first created by insertion of a 1,005-bp arcA1 upstream PCR product (using primers 1068 and 1069; Table 2) into the BamHI and XbaI sites of pUC19 (39). Second, a 942-bp arcA1 downstream PCR product was amplified (using primers 1070 and 1071; Table 2) and ligated into the SalI and PstI sites of the pUC19 polylinker. tetM, the minocycline resistance cassette (40), was inserted into the BamHI site following amplification from pJF12 (41) using primers 1638 and 1639. Finally, the temperature-sensitive pE194 derivative pROJ6448 was ligated to the plasmid via the PstI site (42). pNF142, the 1457 ΔarcA2 allelic replacement construct, was created by first inserting an 884-bp arcA2 upstream PCR product (using primers 1074 and 1075; Table 2) into the BamHI and XbaI sites of pUC19. Second, a 1,017-bp arcA2 downstream PCR product was amplified (using primers 1076 and 1077; Table 2) and inserted into the SalI and PstI sites of the pUC19 polylinker. Next, dhfr, the trimethoprim resistance cassette, was amplified from pGO558 (41) using primers 887 and 888 (Table 2) and inserted into the BamHI site. Finally, pROJ6448 was ligated to the plasmid via the PstI site. Throughout, ligation products were transformed into chemically competent E. coli DH5α (Invitrogen, Carlsbad, CA). Completed constructs were electroporated into the RN4220 restriction-negative S. aureus strain using previously described protocols (43, 44). Modified plasmid DNA was isolated using Wizard Plus MidiPrep kits (Promega, Madison, WI) and electroporated into S. epidermidis 1457 (45). S. epidermidis 1457 isolates containing allelic replacement constructs were grown to mid-exponential phase at 30°C with erythromycin, diluted 1:100 in fresh TSB, and incubated at 45°C (nonpermissive temperature) overnight. The culture was diluted 1:100 again the following day and incubated again at 45°C. The culture was then diluted and plated on TSA, TSA containing minocycline (1457 ΔarcA1), or TSA containing trimethoprim (1457 ΔarcA2) and incubated at 45°C. Colonies of 1457 ΔarcA1 or 1457 ΔarcA2 that were resistant to minocycline or trimethoprim, respectively, and erythromycin susceptible were selected for further study. Following PCR and Southern blot confirmation of gene disruption, mutant alleles were backcrossed to wild-type (WT) S. epidermidis 1457 using transducing phage 71 as previously described (46).
For complementation analysis, the native arc operon (arcA1B1D1R1) and the acquired arc operon (arcA2D2R2B2C2) were amplified (including their putative native promoters) using primer pair 2009 and 2010 and primer pair 2011 and 2012, respectively (Table 2). Each was cloned into the SphI and BamHI sites of pCN51 (removing the cadmium-inducible promoter) to produce pNF232 (native arc operon) and pNF233 (ACME-carried, acquired arc operon). Plasmids were electroporated into and purified from RN4220 and subsequently electroporated into 1457 ΔADI as described above.
RNA isolation, Northern analysis, and metabolite quantification.
For Northern analysis of biofilm-grown cells, 100 μl of S. epidermidis 1457, grown under microaerobic growth conditions overnight, was used to inoculate Stovall flow cells. Flow cell biofilms were grown in TSB at 37°C using plastic convertible flow cells (24 mm by 40 mm by 8 mm) and a flow rate of 0.5 ml/min. RNA was isolated from the biofilm within the flow cells at 24, 48, and 72 h of growth and subjected to Northern analysis. DNA probes for Northern analysis were PCR labeled with digoxigenin-labeled dUTP (Roche, Indianapolis, IN) using primer pair 1129 and 1130 and primer pair 1682 and 1683 for arcA1 and arcA2, respectively (Table 2). Flow cell effluent was collected, and glucose, lactic acid, acetic acid, and ammonia were assayed using kits from R-Biopharm, Inc. (Washington, MO). Free-amino-acid analysis was performed by the Protein Structure Core Facility, University of Nebraska Medical Center (UNMC), using a Hitachi L-8800 amino acid analyzer.
Static biofilm assay.
Assays measuring the production of biofilm were performed using the methods of Christensen et al. (47). Briefly, overnight cultures were grown aerobically and used to inoculate 200 μl of TSB to a final optical density at 600 nm (OD600) of 0.05 in 96-well flat-bottom polystyrene plates. Plates were incubated statically at 37°C for 8, 12, 16, or 32 h, at which time the cells were washed vigorously, stained with crystal violet, and analyzed using a spectrophotometer at A595.
Survival during growth-dependent acidification.
Aerobically grown overnight cultures were diluted to an OD600 of 0.05 in 100 ml of TSB media containing 35 mM glucose and additional 5 mM arginine (final concentration of ∼7.5 mM, as TSB typically contains ∼2.5 mM free arginine) in 1-liter baffled flasks. For the duration of the experiment, flasks were shaken at 250 rpm at 37°C. Aliquots were removed at 0, 24, 48, 72, 96, and 120 h after inoculation. At each time point, cultures were measured for cell density at OD600, serially diluted and plated onto TSA for enumeration, and centrifuged at 6,000 × g and the supernatant was removed for pH, glucose, acetic acid, ammonia, and amino acid analysis as described above. Experiments using the iron chelator dipyridyl (2, 2′-dipyridyl; Sigma) were performed by adding dipyridyl at a concentration of 200 μM at 24 h postinoculation.
Determination of pHi.
Intracellular pH (pHi) was determined by measurement of a pH-sensitive fluorescent compound, carboxyfluorescein diacetate, succinimidyl ester (CFDA SE) (Life Technologies, Grand Island, NY), as described previously (48). Briefly, 5 ml of culture was removed at the 24-h time point of the growth-dependent acidification assay described above. Cells were collected by centrifugation, and the culture medium was removed. Cells were washed twice in 10 mM phosphate buffer (pH 7.0) and then labeled by incubation with 10 μM CFDA SE in phosphate buffer for 15 min at 37°C (48). Cells were washed once with phosphate buffer, and excess compound was removed during one 15-min incubation at 37°C in phosphate buffer–1 mM glucose. Cells were washed twice in phosphate buffer and resuspended in McIlvaine's citric acid buffer solution at pH 5, 6, and 7. Fluorescence (excitation, 490 nm; emission, 525 nm) was measured using a Tecan Infinite 200 Pro plate reader. Standard curves for both wild-type and mutant strains were determined by permeabilization of labeled cells with 1 μg/ml valinomycin and 1 μg/ml nigericin.
Long-term static-biofilm survival.
Aerobically grown overnight cultures were diluted to an OD600 of 0.1 in TSB media supplemented with 35 mM glucose and 5 mM arginine, and 2-ml aliquots were transferred into a 24-well plate. Plates were incubated statically at 37°C. Every 24 h, representative wells were scraped and the cells resuspended into the culture media. Cells were dilution plated for enumeration and then centrifuged at 6,000 × g, and the supernatant was analyzed for pH and ammonia content.
Electron paramagnetic resonance (EPR) spectroscopy.
Aliquots from the growth-dependent acidification assay were removed at 72 h, centrifuged, and resuspended to an OD600 of 10 U in 1 ml of KDD buffer (Krebs-HEPES buffer [pH 7.4]; 99 mM NaCl, 4.69 mM KCl, 2.5 mM CaCl2, 1.2 mM MgSO4, 25 mM NaHCO3, 1.03 mM KH2PO4, 5.6 mM d-glucose, 20 mM HEPES, 5 μM diethyldithiocarbamic acid sodium salt, 25 μM deferoxamine). Samples were then incubated for 15 min at room temperature with a 200 μM concentration of a cell-permeable reactive oxygen species (ROS)-sensitive spin probe, 1-hydroxy-3-methoxycarbonyl-2,2,5,5-tetramethylpyrrolidone (CMH; Noxygen Science Transfer and Diagnostics, Elzach, Germany). Analysis was performed using a Bruker e-scan EPR spectrometer set to a field sweep width of 60.0 G, microwave frequency of 9.75 kHz, microwave power of 21.90 mW, modulation amplitude of 2.37 G, conversion time of 10.24 ms, and time constant of 40.96 ms.
Flow cytometry.
Aliquots from the growth-dependent acidification assay were removed at 24 and 72 h and resuspended in phosphate-buffered saline (PBS) buffer to a final concentration of 107 cells per ml and stained for 30 min with 5 mM 5-cyano-2,3-ditoyl tetrazolium chloride (CTC) and 15 μM 3-(p-hydroxyphenyl)fluorescein (HPF). Stained cells were analyzed by fluorescence-activated cell sorter (FACS) analysis at a flow rate of approximately 1,000 cells/s, and a total of 10,000 events were collected for each sample. Samples were excited at 488 nm, and emission was detected at 530 ± 30 nm (HPF) and 695 ± 40 nm (CTC). FlowJo software was used for raw data analysis.
Nucleotide sequence accession numbers.
The sequences of the native arc operon arcA1B1D1R1 determined by draft genome analysis in this study have been deposited in GenBank under accession no. KF707929 and KF707930.
RESULTS
S. epidermidis 1457 contains two complete copies of the arc operon encoding the ADI pathway.
Draft genome sequence analysis demonstrated that strain 1457 contained two copies of the arc operon, one native (arcA1B1D1R1; GenBank accession no. KF707929 and KF707930) and one acquired on the ACME (arcA2D2R2B2C2) (Fig. 2). Based on the presence of opp3 and arcA2D2R2B2C2, the ACME of 1457 was considered type I (18) and the argR2 and arcC2 regions had 99% nucleotide identity with the S. aureus USA300 argR2 and arcC2 region (GenBank accession no. CP000255). The native argR1 and arc operon and arcC1 in S. epidermidis 1457 had 99% and 100% nucleotide identity, respectively, to the native argR1 arc operon in S. epidermidis ATCC 12228 (36). Note that the native arc operon (S. epidermidis ATCC 12228 SE_2218-SE_2214) and arcC1 (S. epidermidis ATCC 12228 SE_0228) are not chromosomally linked as is found with the ACME-derived arc operon.
FIG 2.
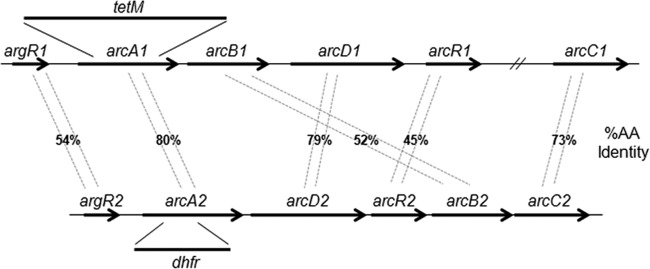
Comparison of native and acquired arc operons in S. epidermidis 1457. Percentages of amino acid (AA) identity between genes of the native (arcA1B1D1R1) and acquired (arcA2D2R2B2C2) arc operons are shown. arcA1 and arcA2 allelic replacement is also indicated. Note that the arginine-dependent transcriptional regulator, consisting of argR1 and argR2, is an independent transcriptional unit encoded upstream of arcA1 and arcA2.
Arginine deiminase contributes to long-term survival under acid conditions.
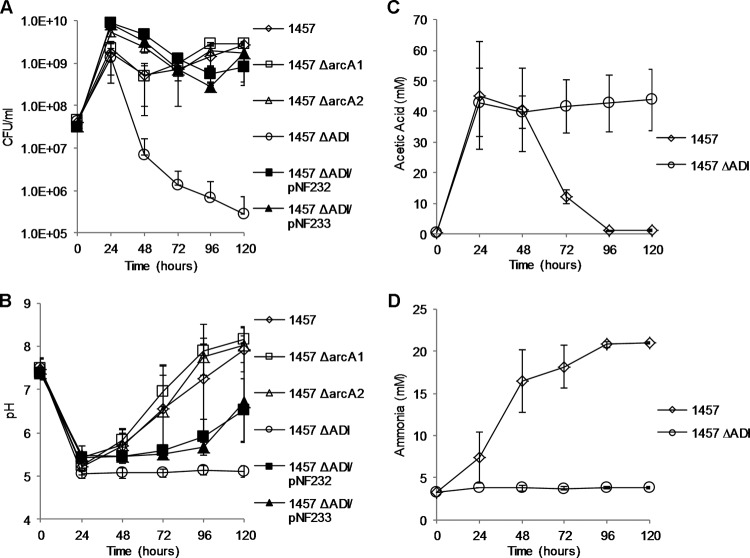
To assess the function of arginine deiminase in acid survival, arcA1 and arcA2 allelic replacement mutants were constructed in S. epidermidis strain 1457 using tetM and dhfr markers, respectively (Fig. 2). An arcA1 arcA2 double mutant was subsequently constructed through Φ71 transduction of the ΔarcA1 locus to 1457 ΔarcA2. In contrast to S. aureus (35), Listeria monocytogenes (16), and Streptococcus suis (27), a growth defect of 1457 ΔarcA1, 1457 ΔarcA2, or 1457 ΔarcA1 ΔarcA2 (here referred to as 1457 ΔADI) was not detected in comparison to 1457 in TSB adjusted to pH 4.0, 4.5, 5.0, 5.5, 6.0, or 6.5 with HCl, lactic acid, pyruvic acid, or acetic acid (data not shown). Therefore, based on the previous observations that arginine deiminase transcription (arcA1) is glucose repressed in S. aureus (28), we hypothesized that the function of arginine deiminase with relation to pH stress would be observed only in acidified media where glucose had been depleted. To this end, strains 1457, 1457 ΔarcA1, 1457 ΔarcA2, and 1457 ΔADI were grown in TSB supplemented with 35 mM glucose and 5 mM arginine for 5 days under aerobic conditions and sampled daily for viable count, pH, and acetic acid, glucose, and ammonia concentration. Under aerobic growth conditions in TSB containing 14 mM glucose, glucose is catabolized to acetate and excreted into the medium. When glucose is depleted, however, acetate is rapidly extracted from the medium and further catabolized via the tricarboxylic acid (TCA) cycle, generating reducing power and metabolic intermediates (49). Furthermore, ammonia is subsequently generated via amino acid catabolism that results in a sharp rise in extracellular pH. In contrast, aerobic growth in TSB containing 35 mM glucose results in the accumulation of acetate in the culture medium; however, the acetate is not extracted from the extracellular milieu until approximately 48 h of growth, and the pH of the medium remains low (∼4.8 to ∼5.0) within this time frame (50). In addition, it was reasoned that TSB supplemented with additional arginine (5 mM) would result in ample substrate for ADI pathway-dependent ammonia production. Therefore, this growth-dependent acidification assay using TSB containing 35 mM glucose and 5 mM arginine was utilized to determine if 1457 ΔADI could survive the organic acid stress conditions that are known to develop in this growth medium. It was determined that, by 24 h of growth, the glucose in the medium was depleted (data not shown) and that there was no significant difference in growth yield between the arcA mutants and strain 1457 (Fig. 3A). However, the growth yield of 1457 ΔADI had dropped 2 log10 by 48 h of growth and ∼4 log10 by 120 h (Fig. 3A). No significant growth differences between strains 1457, 1457 ΔarcA1, and 1457 ΔarcA2 were noted after 120 h of growth, suggesting that both ADI operons were active in this assay and were able to compensate for each other (Fig. 3A). In addition, 1457 ΔADI could be complemented by both the native (pNF232) and acquired (pNF233) ADI operon (Fig. 3A). Furthermore, after 24 h of growth, the pH of the medium dropped from 7.4 to ∼5.2 in strain 1457 and in the arcA single mutants; however, the pH of the growth medium of 1457 ΔADI was ∼5.0 and remained at that value for 120 h (Fig. 3B). In contrast, presumably due to extraction of acetate from the growth medium and subsequent ammonia production via amino acid metabolism and arginine deiminase activity, the extracellular pH of 1457 and the arcA single mutants gradually rose from 5.2 to ∼8.0 after 120 h of growth (Fig. 3B). Acetate accumulation and extraction from the growth medium were confirmed using 1457 and 1457 ΔADI; acetate was gradually depleted from the medium of 1457, whereas the acetate concentration remained at ∼40 mM in that of 1457 ΔADI (Fig. 3C). Lastly, the ammonia concentration was determined in the growth media of 1457 and 1457 ΔADI (Fig. 3D). By 48 h of growth, the growth medium of 1457 contained ∼17 mM ammonia whereas the level of ammonia in that of 1457 ΔADI was negligible. Note that, by 48 h of growth, acetate had not yet been extracted from the culture medium of 1457 and yet the pH rose from 5.2 to 5.8. Acetate consumption stimulates TCA cycle activity and subsequent amino acid catabolism via NH3 production. Collectively, these data suggest that ADI-dependent ammonia production before 48 h of growth in this assay protected the cell from the damaging effects of organic acid stress, allowing further extraction and use of available carbon sources, including acetate, in the growth medium.
FIG 3.
Five-day growth-dependent acidification survival assay. S. epidermidis 1457, 1457 ΔarcA1, 1457 ΔarcA2, 1457 ΔADI, 1457 ΔADI/pNF232, and 1457 ΔADI/pNF233 were grown aerobically in TSB supplemented with 35 mM glucose and 5 mM arginine and sampled daily for determinations of cell density (A), extracellular pH (B), acetic acid (C), and ammonia concentration (D). Plasmids pNF232 and pNF233 contain the native (arcA1B1D1R1) and acquired (arcA2D2R2B2C2) ADI operons, respectively. Error bars represent standard deviations of the means for the results of three independent experiments.
To directly measure the intracellular pH (pHi) following organic acid stress, we utilized the pH-dependent, intracellular fluorescent compound CFDA SE (Fig. 4). Strain 1457 and 1457 ΔADI cells were washed after 24 h of growth in TSB containing 35 mM glucose and 5 mM arginine and resuspended into McIlvaine's citric acid buffer solution at pH 5, 6, and 7. Growth was assayed at 24 h instead of 48 h because (i) the glucose concentration had been depleted at 24 h of growth, suggesting that glucose repression of the ADI pathway was not active at this time point, and (ii) the viability of 1457 ΔADI had dropped ∼2 log10 by 48 h of growth and we did not want to measure the pHi during cell death processes. At a neutral pH of 7.0, there was no difference between the pHi values determined for 1457 and 1457 ΔADI; however, a significant difference was noted between the mean pHi values when 1457 and 1457 ΔADI were resuspended in buffer at pH 5 and 6 (the mean values for 1457 and 1457 ΔADI at pH 5 and 6 were pHi 6.2 and 5.8 and pHi 6.7 and 6.3, respectively) (Fig. 4). Collectively, these results suggest that 1457 ΔADI is unable to maintain a more neutral pHi in the presence of a weak organic acid in the absence of ADI-derived ammonia.
FIG 4.
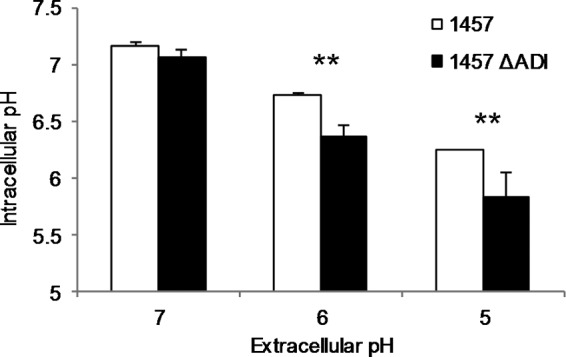
1457 ΔADI has a reduced ability to maintain an neutral intracellular pH in an acidic extracellular environment. Strain 1457 or 1457 ΔADI was grown in TSB supplemented with 35 mM glucose and 5 mM arginine. After 24 h, aliquots were removed, washed, and labeled with CFDase for measurement of intracellular pH. Labeled cells were incubated in McIlvaine's sodium citrate buffer at pH 5, 6, and 7, and the fluorescence was measured for determination of intracellular pH. Data were analyzed using the Student t test (n = 3; **, significant difference from strain 1457 with P < 0.05).
ADI-dependent pH stress mediates oxidative stress.
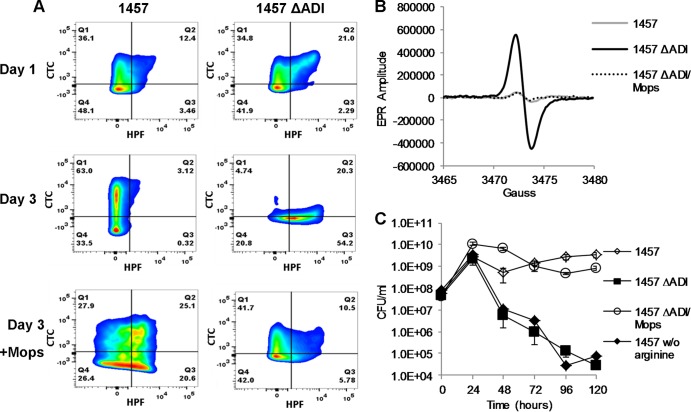
Recent studies of Streptococcus thermophilus found that manganese-containing superoxide dismutase (MnSOD) was required for growth and survival in media acidified to pH 3.5 by lactic acid (51). Furthermore, chelation of free intracellular soluble iron partially rescued pH-mediated death in an MnSOD mutant, suggesting that lactic acid dissociates protein-iron complexes, facilitating hydroxyl radical formation via Fenton chemistry (51–54). Thus, the following experiments were designed to determine if ADI-dependent pH stress, via acetic acid, mediated oxidative stress in S. epidermidis 1457. Both strains 1457 and 1457 ΔADI were grown aerobically in TSB containing 35 mM glucose and 5 mM arginine, and the cells were stained with both CTC and HPF following 24 and 72 h of growth and subjected to FACS analysis. Reduction of CTC into red fluorescent formazan results in the intracellular accumulation of the dye and is an indicator of respiration (55), whereas HPF is a fluorescent reporter used to detect oxygen radicals (hydroxyl, OH) (56). Consistent with our growth analyses (Fig. 3A), following 24 h of growth, no significant difference between 1457 and 1457 ΔADI was detected using FACS analysis (Fig. 5A and Table 3); however, by 72 h of growth, a significant shift occurred where 55% of 1457 ΔADI was stained with HPF but only 14.8% of 1457, indicating significant oxidative stress in the former. In addition, consistent with the hypothesis that respiration would be most active in cells following activation of the TCA cycle and acetate catabolism, CTC reduction was primarily detected in 1457 following 72 h of growth. As predicted from the growth analysis (Fig. 3A), no CTC reduction was detected in 1457 ΔADI at 72 h (Fig. 5A). To confirm that the production of oxygen radicals was dependent upon acidic pH, both 1457 and 1457 ΔADI were grown in TSB containing 35 mM glucose and 5 mM arginine that was buffered with 100 mM MOPS [3-(N-morpholino) propanesulfonic acid] to a pH of 7.3. As predicted, the addition of MOPS buffer rescued the oxidative-stress phenotype, further demonstrating that the ammonia produced by ADI functions in pH homeostasis, preventing subsequent intracellular acidification and oxidative stress (Fig. 5A). Electron paramagnetic resonance spectroscopy was utilized to confirm the oxidative stress observed in 1457 ΔADI following growth for 72 h in TSB containing 35 mM glucose and 5 mM arginine (Fig. 5B); we detected an approximately 12-fold increase in oxygen radicals from 1457 ΔADI compared to 1457. Again as expected, the addition of MOPS buffer to the growth medium of 1457 ΔADI resulted in a significant decrease of oxygen radicals similar to that seen with 1457 (Fig. 5B).
FIG 5.
Long-term growth of 1457 ΔADI results in oxidative stress conditions and subsequent cell death. (A) Strain 1457 and 1457 ΔADI cells growing in TSB supplemented with 35 mM glucose and 5 mM arginine with or without buffering by MOPS were removed at 24 and 72 h postinoculation, doubly labeled with CTC and HPF, and measured by flow cytometry. (B) Cells were also removed at 72 h for analysis by electron paramagnetic resonance spectroscopy (EPR). (C) Survival of cultures was monitored daily for 5 days by cell enumeration.
TABLE 3.
Flow cytometry
| Time (h) | Sample | % of cells ± SDa |
|||
|---|---|---|---|---|---|
| CTC+ (Quad 1) | CTC+ HPF+ (Quad 2) | HPF+ (Quad 3) | Unstained (Quad 4) | ||
| 24 | 1457 | 38.5 ± 4.2 | 12.1 ± 9.0 | 4.7 ± 2.3 | 44.8 ± 14.3 |
| 24 | 1457 ΔADI | 33.6 ± 7.2 | 16.2 ± 13.9 | 3.9 ± 2.9 | 46.4 ± 20.9 |
| 72 | 1457 | 62.7 ± 11.6 | 11.3 ± 10.7 | 3.5 ± 4.1 | 22.4 ± 9.2 |
| 72 | 1457/MOPS | 34.8 ± 20.8 | 23.4 ± 26.2 | 17.6 ± 17.8 | 24.2 ± 23.2 |
| 72 | 1457 ΔADI | 21.0 ± 12.2 | 24.4 ± 11.3 | 31.4 ± 17.5 | 23.2 ± 9.2 |
| 72 | 1457 ΔADI/MOPS | 38.3 ± 4.9 | 20.2 ± 13.7 | 13.9 ± 11.5 | 27.6 ± 2.4 |
Data represent percentages of cells stained with HPF and/or CTC or left unstained. Quad, quadrant.
Additionally, strains 1457 and 1457 ΔADI were grown aerobically for 5 days in TSB containing 35 mM glucose and 5 mM arginine as shown in Fig. 3A. As suggested by the flow cytometry data in Fig. 5A, the addition of MOPS buffer rescued survival of 1457 ΔADI, demonstrating that extracellular acidification, resulting in oxidative stress, was partially responsible for cell death of S. epidermidis in this assay (Fig. 5C). Furthermore, to determine the arginine dependence of this phenotype, 1457 was grown in TSB with no additional arginine supplementation (final free arginine concentration, ∼2.5 mM). Under these growth conditions, it was determined that without excess arginine for use as a substrate by ADI generating NH4+, the growth yield of 1457 dropped ∼4 log10, phenocopying 1457 ΔADI (Fig. 5C).
Finally, the deleterious effects of oxygen radicals revealed via Fenton chemistry (57) are known to be mediated via free intracellular iron (Fe2+) (52, 53). Therefore, we reasoned that the addition of iron chelators should partially protect 1457 ΔADI against pH-mediated oxidative stress. Thus, 1457 and 1457 ΔADI were grown aerobically in TSB containing 35 mM glucose and 5 mM arginine as shown in Fig. 3A. Following 24 h of growth, the iron chelator dipyridyl (200 μm) was added to the culture and cell viability was assessed at 24, 48, 72, 96, and 120 h (Fig. 6). Growth analysis comparing the log10-transformed cell populations of treated and untreated cultures indicated that treatment of 1457 ΔADI with dipyridyl offered a partial restoration of viability after 120 h. Taken together, these data indicate that arginine catabolism via arginine deiminase protects against cell death by generating NH4+, which reduces intracellular pH stress and the subsequent iron-dependent oxidative stress that is partially responsible for cell death.
FIG 6.
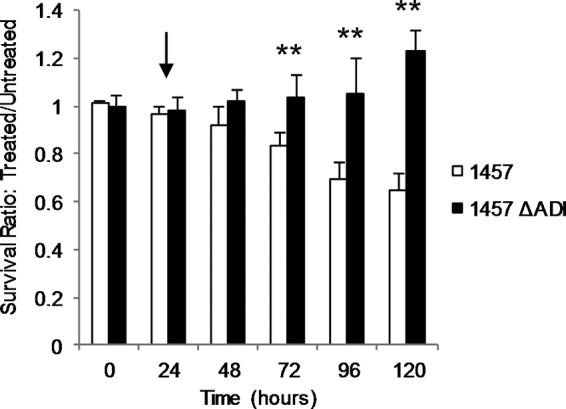
Iron chelation increases survival of 1457 ΔADI. Strains 1457 and 1457 ΔADI were grown aerobically in TSB supplemented with 35 mM glucose and 5 mM arginine. Following 24 h of growth, 200 μM dipyridyl (DP) was added and culture viability assessed after 24, 48, 72, 96, and 120 h. The graph represents the ratio of log10-transformed cell populations of treated and untreated cultures (n = 3). *, P < 0.05. The arrow indicates the time at which dipyridyl was added.
Arginine deiminase is induced as S. epidermidis biofilm matures.
As arginine deiminase activity functions to protect the cell against pH-dependent stress, we hypothesized that arginine deiminase activity would be important in protection against acidic microniches in a mature biofilm. Indeed, genes within the arginine deiminase operon are among the few genes consistently upregulated during biofilm growth in multiple S. aureus strains and in S. epidermidis (58–63). To identify genes differentially regulated as a S. epidermidis 1457 biofilm matures from 24 to 72 h, total RNAs were isolated from a flow cell biofilm at 24 and 72 h and compared using transcriptional profiling (see Table S1 in the supplemental material). As previously noted in other studies comparing planktonic growth and biofilm growth, genes upregulated as a S. epidermidis biofilm matures include those that function in microaerobic/anaerobic metabolism and respiration, including complex 1 NADH oxidoreductase (snoD) (64), cytochrome d ubiquinol oxidase, fumarate hydratase (fumC), and lactate dehydrogenase (ddh) (65). KOBAS 2 analysis (see Table S2) (38) identified several metabolic pathways that were enriched as a biofilm matured from 24 to 72 h, including pyruvate metabolism, TCA cycle activity, and glycolysis/gluconeogenesis. Also noted were the enrichment of arginine/proline metabolism and, specifically, induction of the arginine deiminase (ADI) operon (S. epidermidis ATCC 12228 SE0103-SE0106) carried within the ACME island (17, 33, 36). Northern analysis confirmed induction of the ACME-derived arcA2 gene during biofilm maturation, with little expression detected at 24 h compared to 48 and 72 h (Fig. 7A). Conversely, arcA1 was constitutively expressed from 24 to 72 h and, as predicted by the transcriptional profiling experiments, was not upregulated as the biofilm matured. To further detect and document arginine deiminase activity during biofilm growth, culture supernatant was collected during flow cell biofilm maturation and arginine, ornithine, citrulline, and ammonia concentrations were measured (Fig. 7B). As predicted from the transcriptional profiling experiments, as arginine was depleted from the media, citrulline and ornithine, both byproducts of arginine deiminase activity, accumulated in the supernatant. In addition, the level of ammonia, which is generated through arginine deiminase activity and is indicative of amino acid catabolism, increased from 24 to 72 h. As previously demonstrated by Zhu and colleagues (66), microaerobic growth conditions within the biofilm led to enhanced organic acid production as glucose was depleted (Fig. 7C). In contrast to the acetate generated during highly aerobic growth (Fig. 3C), biofilm growth led to a predominance of lactate (∼15 mM following 72 h of growth) compared to 1.1 mM and 0.7 mM formate and acetate, respectively. Combined, these data provide insight into the collective metabolic characteristics of a maturing S. epidermidis biofilm; both the native and acquired ADI pathways are active, and the reduced oxygen tension provides an environment where pyruvate is oxidized to lactate and acetate, which presumably results in acidic microniches within the biofilm.
FIG 7.
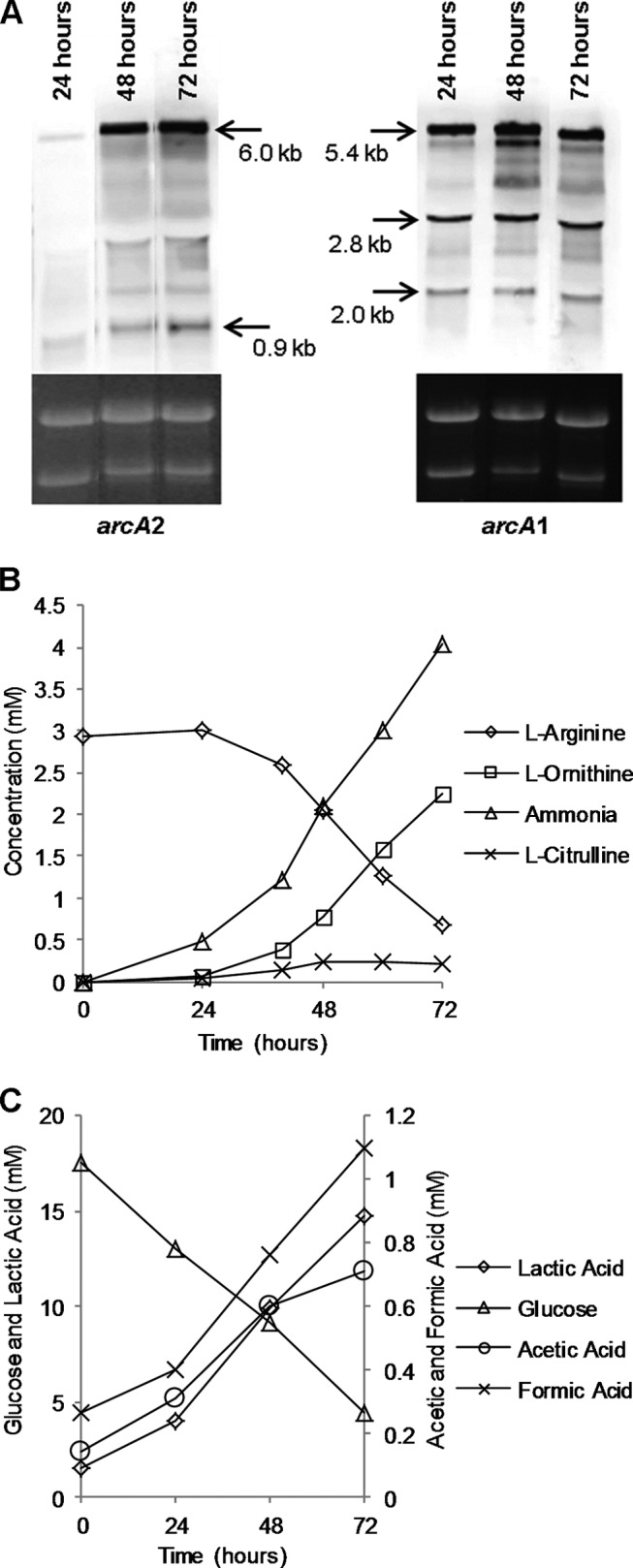
Analysis of maturing flow cell biofilms. (A) Total RNA was extracted from cells growing in maturing biofilms and used for Northern blot analysis of arcA1 and arcA2. (B and C) Spent medium effluent from a maturing flow cell biofilm was collected and subjected to analysis for products of arginine deiminase activity (B) and glucose metabolism (C). (D) Biofilm-grown cells were removed at 72 h postinoculation and subjected to paramagnetic resonance spectroscopy (EPR) analysis.
ADI contributes to long-term biofilm viability.
Upon observing that arginine deiminase genes were upregulated during biofilm maturation, we hypothesized that ADI contributes to S. epidermidis biofilm growth and/or maturation. Therefore, static, 24-h biofilm assays were performed in TSB with S. epidermidis strains 1457, 1457 ΔarcA1, 1457 ΔarcA2, and 1457 ΔADI. However, as previously observed by Zhu and colleagues (66), no biofilm biomass differences between the wild-type and arcA-deficient strains were observed (data not shown). Therefore, based on our previous observations that 1457 ΔADI had decreased viability under conditions of long-term acidic stress, both 1457 and 1457 ΔADI biofilms were grown in TSB containing 35 mM glucose and 5 mM arginine in a 24-well microtiter plate (Fig. 8). It was determined that the pH levels of the medium after 5 days of incubation were approximately 6.0 and 5.35 for 1457 and 1457 ΔADI, respectively (Fig. 8A). In addition, 1457 ΔADI biofilms produced significantly less NH4+ than the WT biofilms (Fig. 8B) and the viability of the biofilm biomass was reduced 0.5 log10 (Fig. 8C). No difference between 1457 and 1457 ΔarcA1 and 1457 ΔarcA2 with respect to biofilm viability, pH, or ammonia was observed (data not shown). Importantly, complementation of 1457 ΔADI with either pNF232 (native arc operon) or pNF233 (ACME-acquired arc operon) was able to rescue extracellular pH, ammonia synthesis, and viability (Fig. 8).
FIG 8.
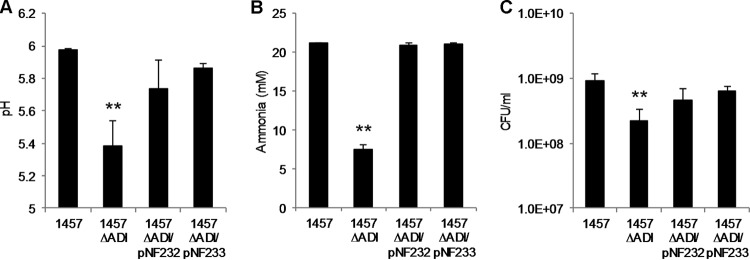
Arginine deiminase contributes to long-term (5-day) static biofilm survival. Overnight cultures were diluted in TSB supplemented with 35 mM glucose and 5 mM arginine to an OD600 of 0.100 and incubated statically in 24-well plates at 37°C. (A and B) Cell-free supernatant was collected for measurement of pH (A) and ammonia (B). (C) Representative wells were scraped, and the cells were resuspended and dilution plated for enumeration. Plasmids pNF232 and pNF233 contain the native (arcA1B1D1R1) and acquired (arcA2D2R2B2C2) ADI operons, respectively. Data were analyzed using the Student t test (n = 3; **, significant difference from strain 1457 with P < 0.05).
DISCUSSION
Levels of resistance to acid stress in Gram-positive bacteria differ (67) and depend on such factors as, among others, the F1-F0-ATPase (68), glutamate decarboxylase (69), electrogenic transport via lactate-malate antiporters (70), and production of alkali, including NH4+ via arginine deiminase (14, 16, 25, 27). However, although the staphylococci encounter various acidic niches in the human host, including the skin (pH 4.2 to 5.9) (71) or an abscess (pH 6.2 to 7.3) (72), the innate ability of these organisms to thrive in these niches is ill defined. Transcriptional profiling experiments have suggested that S. aureus utilizes multiple defense mechanisms to counteract acid stress, including induction of urease and NADH+ dehydrogenases (73, 74); however, the importance of these systems in acid resistance has not been experimentally addressed. Importantly, acid stress has been shown to induce an oxidative stress response in S. aureus, including transcriptional induction of superoxide dismutase (sodA), catalase (katA), alkyl hydroperoxide reductase (ahpC/ahpF), and the thioredoxin genes trxA and trxB (73). In fact, earlier studies showed a direct link between acid stress and oxidative stress, as S. aureus defective in sodA is more susceptible to severe acid stress (75). Similarly, more recent studies in S. thermophilus found that the cytotoxic effects of both acid stress and oxidative stress were very similar, as acid stress induced generation of oxygen radicals, stimulating Fenton chemistry and subsequent OH formation (51). Furthermore, those investigators found that manganese-dependent superoxide dismutase and, predictably, other antioxidant enzymes protected the cell against the deleterious effects of acid stress via reduction of superoxide radicals.
Synthesis of NH3 via ADI or urease and subsequent protonation generating NH4+, thus increasing the intracellular and extracellular pH, are well documented and are critical for growth and survival of many bacterial species that reside in the low-pH niche of the human oral cavity (67). However, we are just now beginning to understand the function of many metabolic pathways, including the ADI pathway, in the context of staphylococcal colonization or infection. A few S. aureus strain backgrounds, most prominently USA300 ST8, carry two copies of the ADI pathway where the second copy is located within the ACME island (17). Unlike other S. aureus strains where the native arc operon is induced under anoxic growth conditions via ArcR (28), native arc activity is not detected in USA300 strains and the ACME-carried arc operon is constitutively transcribed (35). It is hypothesized that NH3 production via the ACME-carried arc operon allows enhanced growth of S. aureus in acidic microenvironments such as the skin. Indeed, S. aureus USA300 containing the ACME-carried arc operon was more resistant to media acidified to pH 5.0 by lactic acid (35). Interestingly, excess ornithine produced via the ADI pathway is toxic to staphylococci due to the subsequent generation of polyamines; S. aureus counteracts this ADI-induced toxicity through an ACME-encoded spermine/spermidine acetyltransferase (SpeG) (35, 76). Therefore, available data suggest that the ACME-carried arc operon functions to enhance skin colonization and thus transfer of S. aureus USA300. Based on this hypothesis, it is not surprising that the majority of USA300 infections are skin and soft tissue based (77). In contrast to S. aureus, the natural niche of S. epidermidis is the skin; accordingly, approximately 50% of S. epidermidis strains contain the ACME-encoded ADI pathway, suggesting a potential beneficial function in skin colonization (18).
Given this background information, we were surprised to find that our initial attempts to determine a function of ADI in S. epidermidis acid resistance failed. These growth experiments were performed in acidified (via lactic, acetic, pyruvic, or hydrochloric acid) media with or without glucose and excess arginine; no differences in growth or survival of wild-type 1457 versus ADI-deficient strains were noted. Therefore, we chose to exploit the acidification of media during growth in excess glucose to test for long-term survival. During aerobic growth of S. epidermidis, glucose is preferentially catabolized to pyruvate via glycolysis. When glucose is in excess, pyruvate is converted to acetate and excreted from the cell. This extracellular pool of acetic acid results in a sharp decline in pH and, thus, growth-dependent acidification of the media. After glucose is depleted, however, the organism consumes the extracellular acetate, converting it to acetyl-coenzyme A (acetyl-CoA) and subsequently oxidizing it via the TCA cycle, and in combination with NH3 production via amino acid catabolism, urease, and/or arginine deiminase activity, the medium is restored to a more neutral pH (78). However, when staphylococci grow in media containing 35 mM glucose, the acetate is typically not oxidized until approximately 48 h of growth (50), allowing adequate exposure to an acidic pH of approximately 5.0 in the absence of glucose and presence of arginine. Under these conditions, survival of 1457 ΔADI was reduced ∼4 log10 by 120 h of growth in comparison to 1457, suggesting a specific function of ADI in organic acid resistance. It was also determined that both 1457 ΔarcA1 and 1457 ΔarcA2 did not have growth defects under these growth conditions, suggesting that each ADI copy is active in S. epidermidis 1457. These data are in contrast to those presented in S. aureus strain SH1000 (28) or USA300 (35), where transcription of the native arc operon was induced only under anoxic growth conditions or was not detected, respectively. Collectively, our data suggest that as the pH of the extracellular media nears the pKa of acetic acid (4.76), the protonated form of acetic acid can permeate the cell membrane and dissociate, thus mediating intracellular acid stress and cell death. However, under these conditions, both the native ADI and acquired ADI can prevent intracellular pH stress through production of alkali via NH3.
We also confirmed through HPF staining and electron paramagnetic resonance analysis that organic acid stress mediated oxidative stress in the absence of ADI-dependent alkali (Fig. 5A and B). Although it is currently unclear how acetic acid functions to initiate reactive oxygen species (ROS) formation, previous studies have suggested that acid stress may decrease the function of the respiratory chain, generating promiscuous transfer of electrons to molecular oxygen (75). Furthermore, excess superoxide radicals have the potential to liberate iron from iron-sulfur clusters, generating Fenton chemistry, subsequent hydroxyl radical formation, and damage to proteins, DNA, and lipids. Therefore, as predicted, quenching of free iron via dipyridyl (Fig. 6) was able to rescue viability of 1457 ΔADI partially during growth-dependent acidification experiments, suggesting that formation of hydroxyl radicals via oxidative stress is partially responsible for cell death in this assay.
Lastly, S. epidermidis primarily causes disease via formation of biofilm on abiotic surfaces. Previous investigators have shown upregulation of the arc operon during biofilm formation (58, 62, 79) compared to planktonic growth, suggesting that staphylococcal biofilms may undergo significant acid stress. Indeed, we propose that the growth-dependent acidification assay (Fig. 3A) may mimic microenvironment pH stress in a maturing biofilm; high rates of glycolytic activity within certain biofilm structures may yield high local concentrations of organic acid due to diffusion limitations within the interior of the biofilm (80). Transcriptional profiling and Northern blot experiments (Fig. 7A; see also Table S1 in the supplemental material) confirmed that both the native and acquired ADI operons were active as a biofilm matured; interestingly, however, the native ADI was active in early biofilm formation whereas the ACME-acquired ADI was induced as a biofilm matured from 24 to 72 h, suggesting differential transcriptional regulation between these two operons. Furthermore, ADI activity could be detected between 24 and 48 h of biofilm growth through depletion of arginine and the accumulation of ornithine, citrulline, and ammonia in the cell effluent (Fig. 7B). As glucose was depleted from the effluent, byproducts of pyruvate catabolism, including lactate (∼15 mM), acetate (∼0.7 mM), and formate (∼1.1 mM), were detected in the medium (Fig. 7C), reflecting the microaerobic and anaerobic environment and potential acidic stress in certain microniches. However, using a variety of biofilm growth environments, including flow cell and static conditions, we were unable to detect a significant, consistent difference between 1457 and 1457 ΔADI in the levels of biofilm mass. We reasoned, therefore, that the major beneficial effect of ADI during biofilm development is NH4+ production to compensate for acidic stress and protect against cellular death in particular niches of the biofilm. We therefore performed static biofilm assays and determined that levels of viability, ammonia, and pH were indeed reduced in 1457 ΔADI following 120 h of growth. Thus, the ADI pathway is beneficial for cell viability during S. epidermidis biofilm growth. We hypothesize that ammonia synthesis via the ADI pathway is important to reduce pH stress in specific microniches that contain high concentrations of organic acids. Subsequent oxidative stress within a biofilm most likely varies in a manner dependent on the growth environment and/or depth within the biofilm structure. Indeed, one reason why viability of 1457 ΔADI is less affected during biofilm growth than during aerobic growth may be the small amount of oxygen associated with biofilm environments. Nevertheless, recent studies found that genes which counteract oxidative stress, such as sodA, were upregulated in S. aureus and S. epidermidis biofilms; antioxidants were able to reduce oxidative stress-mediated mutability, supporting the idea of the presence of oxygen radicals within a biofilm (81).
In conclusion, we have determined that the ADI pathway is crucial for resistance to acid stress in S. epidermidis 1457 and that each of the two copies of ADI was able to compensate for the loss of the other copy and facilitate acid resistance in our in vitro organic acid resistance assay and during biofilm growth. Based on the prevalence of the ACME island-encoded ADI in both S. aureus and S. epidermidis, the function of this operon must be highly advantageous and may be selectively induced under certain growth conditions such as colonization of the skin. We also found that ADI-dependent alkali protected the cell against pH-induced oxidative stress and that viability could be partially rescued with iron scavengers. Further work is required to ascertain the mechanisms underlying cell death during pH stress; however, our data suggest that oxidative stress is partially responsible. Lastly, a greater understanding of acid resistance mechanisms in S. epidermidis and S. aureus is required first to determine which infection niche(s) requires acid resistance and second to design novel treatment modalities to circumvent these highly conserved systems.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by Public Health Service grant P01AI083211 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print 11 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00051-14.
REFERENCES
- 1.Rogers KL, Fey PD, Rupp ME. 2009. Coagulase-negative staphylococcal infections. Infect. Dis. Clin. North Am. 23:73–98. 10.1016/j.idc.2008.10.001 [DOI] [PubMed] [Google Scholar]
- 2.Fey PD, Olson ME. 2010. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 5:917–933. 10.2217/fmb.10.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olson ME, Slater SR, Rupp ME, Fey PD. 2010. Rifampicin enhances activity of daptomycin and vancomycin against both a polysaccharide intercellular adhesin (PIA)-dependent and -independent Staphylococcus epidermidis biofilm. J. Antimicrob. Chemother. 65:2164–2171. 10.1093/jac/dkq314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, Engebretsen IL, Bayles KW, Horswill AR, Kielian T. 2011. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J. Immunol. 186:6585–6596. 10.4049/jimmunol.1002794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rani SA, Pitts B, Beyenal H, Veluchamy RA, Lewandowski Z, Davison WM, Buckingham-Meyer K, Stewart PS. 2007. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J. Bacteriol. 189:4223–4233. 10.1128/JB.00107-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6:199–210. 10.1038/nrmicro1838 [DOI] [PubMed] [Google Scholar]
- 7.Moormeier DE, Endres JL, Mann EE, Sadykov MR, Horswill AR, Rice KC, Fey PD, Bayles KW. 2013. Use of microfluidic technology to analyze gene expression during Staphylococcus aureus biofilm formation reveals distinct physiological niches. Appl. Environ. Microbiol. 79:3413–3424. 10.1128/AEM.00395-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cunin R, Glansdorff N, Pierard A, Stalon V. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budin-Verneuil A, Maguin E, Auffray Y, Ehrlich DS, Pichereau V. 2006. Genetic structure and transcriptional analysis of the arginine deiminase (ADI) cluster in Lactococcus lactis MG1363. Can. J. Microbiol. 52:617–622. 10.1139/w06-009 [DOI] [PubMed] [Google Scholar]
- 10.De Angelis M, Mariotti L, Rossi J, Servili M, Fox PF, Rollan G, Gobbetti M. 2002. Arginine catabolism by sourdough lactic acid bacteria: purification and characterization of the arginine deiminase pathway enzymes from Lactobacillus sanfranciscensis CB1. Appl. Environ. Microbiol. 68:6193–6201. 10.1128/AEM.68.12.6193-6201.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernández M, Zúñiga M. 2006. Amino acid catabolic pathways of lactic acid bacteria. Crit. Rev. Microbiol. 32:155–183. 10.1080/10408410600880643 [DOI] [PubMed] [Google Scholar]
- 12.Vrancken G, Rimaux T, Weckx S, De Vuyst L, Leroy F. 2009. Environmental pH determines citrulline and ornithine release through the arginine deiminase pathway in Lactobacillus fermentum IMDO 130101. Int. J. Food Microbiol. 135:216–222. 10.1016/j.ijfoodmicro.2009.07.035 [DOI] [PubMed] [Google Scholar]
- 13.Zúñiga M, Champomier-Verges M, Zagorec M, Pérez-Martínez G. 1998. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J. Bacteriol. 180:4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Degnan BA, Fontaine MC, Doebereiner AH, Lee JJ, Mastroeni P, Dougan G, Goodacre JA, Kehoe MA. 2000. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect. Immun. 68:2441–2448. 10.1128/IAI.68.5.2441-2448.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong K. 2006. Characterization of the arginine deiminase of Streptococcus equi subsp. zooepidemicus. Can. J. Microbiol. 52:868–876. 10.1139/w06-041 [DOI] [PubMed] [Google Scholar]
- 16.Ryan S, Begley M, Gahan CG, Hill C. 2009. Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ. Microbiol. 11:432–445. 10.1111/j.1462-2920.2008.01782.x [DOI] [PubMed] [Google Scholar]
- 17.Diep BA, Stone GG, Basuino L, Graber CJ, Miller A, des Etages SA, Jones A, Palazzolo-Ballance AM, Perdreau-Remington F, Sensabaugh GF, DeLeo FR, Chambers HF. 2008. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 197:1523–1530. 10.1086/587907 [DOI] [PubMed] [Google Scholar]
- 18.Miragaia M, de Lencastre H, Perdreau-Remington F, Chambers HF, Higashi J, Sullam PM, Lin J, Wong KI, King KA, Otto M, Sensabaugh GF, Diep BA. 2009. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PLoS One 4:e7722. 10.1371/journal.pone.0007722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pi B, Yu M, Chen Y, Yu Y, Li L. 2009. Distribution of the ACME-arcA gene among meticillin-resistant Staphylococcus haemolyticus and identification of a novel ccr allotype in ACME-arcA-positive isolates. J. Med. Microbiol. 58:731–736. 10.1099/jmm.0.007351-0 [DOI] [PubMed] [Google Scholar]
- 20.Stalon V, Mercenier A. 1984. L-Arginine utilization by Pseudomonas species. J. Gen. Microbiol. 130:69–76 [DOI] [PubMed] [Google Scholar]
- 21.Deibel RH. 1964. Utilization of arginine as an energy source for the growth of Streptococcus faecalis. J. Bacteriol. 87:988–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vander Wauven C, Pierard A, Kley-Raymann M, Haas D. 1984. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160:928–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsen R, Buist G, Kuipers OP, Kok J. 2004. ArgR and AhrC are both required for regulation of arginine metabolism in Lactococcus lactis. J. Bacteriol. 186:1147–1157. 10.1128/JB.186.4.1147-1157.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsen R, Kok J, Kuipers OP. 2005. Interaction between ArgR and AhrC controls regulation of arginine metabolism in Lactococcus lactis. J. Biol. Chem. 280:19319–19330. 10.1074/jbc.M413983200 [DOI] [PubMed] [Google Scholar]
- 25.Fulde M, Willenborg J, de Greeff A, Benga L, Smith HE, Valentin-Weigand P, Goethe R. 2011. ArgR is an essential local transcriptional regulator of the arcABC operon in Streptococcus suis and is crucial for biological fitness in an acidic environment. Microbiology 157:572–582. 10.1099/mic.0.043067-0 [DOI] [PubMed] [Google Scholar]
- 26.Dong Y, Chen YY, Burne RA. 2004. Control of expression of the arginine deiminase operon of Streptococcus gordonii by CcpA and Flp. J. Bacteriol. 186:2511–2514. 10.1128/JB.186.8.2511-2514.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gruening P, Fulde M, Valentin-Weigand P, Goethe R. 2006. Structure, regulation, and putative function of the arginine deiminase system of Streptococcus suis. J. Bacteriol. 188:361–369. 10.1128/JB.188.2.361-369.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Makhlin J, Kofman T, Borovok I, Kohler C, Engelmann S, Cohen G, Aharonowitz Y. 2007. Staphylococcus aureus ArcR controls expression of the arginine deiminase operon. J. Bacteriol. 189:5976–5986. 10.1128/JB.00592-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Burne RA. 2009. Multiple two-component systems modulate alkali generation in Streptococcus gordonii in response to environmental stresses. J. Bacteriol. 191:7353–7362. 10.1128/JB.01053-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez-Ordóñez A, Fernández A, Bernardo A, López M. 2010. Arginine and lysine decarboxylases and the acid tolerance response of Salmonella Typhimurium. Int. J. Food Microbiol. 136:278–282. 10.1016/j.ijfoodmicro.2009.09.024 [DOI] [PubMed] [Google Scholar]
- 31.Bearson BL, Lee IS, Casey TA. 2009. Escherichia coli O157: H7 glutamate- and arginine-dependent acid-resistance systems protect against oxidative stress during extreme acid challenge. Microbiology 155:805–812. 10.1099/mic.0.022905-0 [DOI] [PubMed] [Google Scholar]
- 32.Budin-Verneuil A, Pichereau V, Auffray Y, Ehrlich DS, Maguin E. 2005. Proteomic characterization of the acid tolerance response in Lactococcus lactis MG1363. Proteomics 5:4794–4807. 10.1002/pmic.200401327 [DOI] [PubMed] [Google Scholar]
- 33.Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739. 10.1016/S0140-6736(06)68231-7 [DOI] [PubMed] [Google Scholar]
- 34.Barbier F, Lebeaux D, Hernandez D, Delannoy AS, Caro V, Francois P, Schrenzel J, Ruppe E, Gaillard K, Wolff M, Brisse S, Andremont A, Ruimy R. 2011. High prevalence of the arginine catabolic mobile element in carriage isolates of methicillin-resistant Staphylococcus epidermidis. J. Antimicrob. Chemother. 66:29–36. 10.1093/jac/dkq410 [DOI] [PubMed] [Google Scholar]
- 35.Thurlow LR, Joshi GS, Clark JR, Spontak JS, Neely CJ, Maile R, Richardson AR. 2013. Functional modularity of the arginine catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13:100–107. 10.1016/j.chom.2012.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang YQ, Ren SX, Li HL, Wang YX, Fu G, Yang J, Qin ZQ, Miao YG, Wang WY, Chen RS, Shen Y, Chen Z, Yuan ZH, Zhao GP, Qu D, Danchin A, Wen YM. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49:1577–1593. 10.1046/j.1365-2958.2003.03671.x [DOI] [PubMed] [Google Scholar]
- 37.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426–2438. 10.1128/JB.187.7.2426-2438.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. 2011. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acid Res. 39:W316–W322. 10.1093/nar/gkr483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. 10.1016/0378-1119(85)90120-9 [DOI] [PubMed] [Google Scholar]
- 40.Nesin M, Svec P, Lupski JR, Godson GN, Kreiswirth B, Kornblum J, Projan SJ. 1990. Cloning and nucleotide sequence of a chromosomally encoded tetracycline resistance determinant, tetA(M), from a pathogenic, methicillin-resistant strain of Staphylococcus aureus. Antimicrob. Agents Chemother. 34:2273–2276. 10.1128/AAC.34.11.2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Handke LD, Slater SR, Conlon KM, O'Donnell ST, Olson ME, Bryant KA, Rupp ME, O'Gara JP, Fey PD. 2007. σB and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis. Can. J. Microbiol. 53:82–91. 10.1139/w06-108 [DOI] [PubMed] [Google Scholar]
- 42.Projan SJ, Archer GL. 1989. Mobilization of the relaxable Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J. Bacteriol. 171:1841–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Augustin J, Gotz F. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 54:203–207 [DOI] [PubMed] [Google Scholar]
- 44.Kreiswirth BN, Lofdahl S, Betley MJ, O'Reilly M, Schlievert PM, Bergdoll MS, Novick RP. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709–712. 10.1038/305709a0 [DOI] [PubMed] [Google Scholar]
- 45.Mack D, Siemssen N, Laufs R. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nedelmann M, Sabottke A, Laufs R, Mack D. 1998. Generalized transduction for genetic linkage analysis and transfer of transposon insertions in different Staphylococcus epidermidis strains. Zentralbl. Bakteriol. 287:85–92. 10.1016/S0934-8840(98)80151-5 [DOI] [PubMed] [Google Scholar]
- 47.Christensen GD, Simpson WA, Bisno AL, Beachey EH. 1982. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect. Immun. 37:318–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breeuwer P, Drocourt J, Rombouts FM, Abee T. 1996. A novel method for continuous determination of the intracellular pH in bacteria with the internally conjugated fluorescent probe 5 (and 6-)-carboxyfluorescein succinimidyl ester. Appl. Environ. Microbiol. 62:178–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Somerville GA, Said-Salim B, Wickman JM, Raffel SJ, Kreiswirth BN, Musser JM. 2003. Correlation of acetate catabolism and growth yield in Staphylococcus aureus: implications for host-pathogen interactions. Infect. Immun. 71:4724–4732. 10.1128/IAI.71.8.4724-4732.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patton TG, Rice KC, Foster MK, Bayles KW. 2005. The Staphylococcus aureus cidC gene encodes a pyruvate oxidase that affects acetate metabolism and cell death in stationary phase. Mol. Microbiol. 56:1664–1674. 10.1111/j.1365-2958.2005.04653.x [DOI] [PubMed] [Google Scholar]
- 51.Bruno-Bárcena JM, Azcárate-Peril MA, Hassan HM. 2010. Role of antioxidant enzymes in bacterial resistance to organic acids. Appl. Environ. Microbiol. 76:2747–2753. 10.1128/AEM.02718-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flint DH, Tuminello JF, Emptage MH. 1993. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem. 268:22369–22376 [PubMed] [Google Scholar]
- 53.Keyer K, Imlay JA. 1996. Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Nat. Acad. Sci. 93:13635–13640. 10.1073/pnas.93.24.13635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rehncrona S, Hauge HN, Siesjo BK. 1989. Enhancement of iron-catalyzed free radical formation by acidosis in brain homogenates: differences in effect by lactic acid and CO2. J. Cereb. Blood Flow Metab. 9:65–70. 10.1038/jcbfm.1989.9 [DOI] [PubMed] [Google Scholar]
- 55.Smith JJ, McFeters GA. 1996. Effects of substrates and phosphate on INT (2-(4-iodophenyl)-3-(4-nitrophenyl)-5-phenyl tetrazolium chloride) and CTC (5-cyano-2,3-ditolyl tetrazolium chloride) reduction in Escherichia coli. J. Appl. Bacteriol. 80:209–215. 10.1111/j.1365-2672.1996.tb03212.x [DOI] [PubMed] [Google Scholar]
- 56.Setsukinai K, Urano Y, Kakinuma K, Majima HJ, Nagano T. 2003. Development of novel fluorescence probes that can reliably detect reactive oxygen species and distinguish specific species. J. Biol. Chem. 278:3170–3175. 10.1074/jbc.M209264200 [DOI] [PubMed] [Google Scholar]
- 57.Fenton HJH. 1894. Oxidation of tartaric acid in the presence of iron. J. Chem. Soc. Trans. 65:899–910. 10.1039/CT8946500899 [DOI] [Google Scholar]
- 58.Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4665–4684. 10.1128/JB.186.14.4665-4684.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brady RA, Leid JG, Camper AK, Costerton JW, Shirtliff ME. 2006. Identification of Staphylococcus aureus proteins recognized by the antibody-mediated immune response to a biofilm infection. Infect. Immun. 74:3415–3426. 10.1128/IAI.00392-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Castro SL, Nelman-Gonzalez M, Nickerson CA, Ott CM. 2011. Induction of attachment-independent biofilm formation and repression of Hfq expression by low-fluid-shear culture of Staphylococcus aureus. Appl. Environ. Microbiol. 77:6368–6378. 10.1128/AEM.00175-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagarajan V, Smeltzer MS, Elasri MO. 2009. Genome-scale transcriptional profiling in Staphylococcus aureus: bringing order out of chaos. FEMS Microbiol. Lett. 295:204–210. 10.1111/j.1574-6968.2009.01595.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Resch A, Rosenstein R, Nerz C, Gotz F. 2005. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 71:2663–2676. 10.1128/AEM.71.5.2663-2676.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yao Y, Sturdevant DE, Villaruz A, Xu L, Gao Q, Otto M. 2005. Factors characterizing Staphylococcus epidermidis invasiveness determined by comparative genomics. Infect. Immun. 73:1856–1860. 10.1128/IAI.73.3.1856-1860.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bayer AS, McNamara P, Yeaman MR, Lucindo N, Jones T, Cheung AL, Sahl HG, Proctor RA. 2006. Transposon disruption of the complex I NADH oxidoreductase gene (snoD) in Staphylococcus aureus is associated with reduced susceptibility to the microbicidal activity of thrombin-induced platelet microbicidal protein 1. J. Bacteriol. 188:211–222. 10.1128/JB.188.1.211-222.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Richardson AR, Libby SJ, Fang FC. 2008. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science 319:1672–1676. 10.1126/science.1155207 [DOI] [PubMed] [Google Scholar]
- 66.Zhu Y, Weiss EC, Otto M, Fey PD, Smeltzer MS, Somerville GA. 2007. Staphylococcus aureus biofilm metabolism and the influence of arginine on polysaccharide intercellular adhesin synthesis, biofilm formation, and pathogenesis. Infect. Immun. 75:4219–4226. 10.1128/IAI.00509-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cotter PD, Hill C. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67:429–453. 10.1128/MMBR.67.3.429-453.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arikado E, Ishihara H, Ehara T, Shibata C, Saito H, Kakegawa T, Igarashi K, Kobayashi H. 1999. Enzyme level of enterococcal F1Fo-ATPase is regulated by pH at the step of assembly. Eur. J. Biochem. 259:262–268. 10.1046/j.1432-1327.1999.00031.x [DOI] [PubMed] [Google Scholar]
- 69.Feehily C, O'Byrne CP, Karatzas KA. 2013. Functional gamma-aminobutyrate shunt in Listeria monocytogenes: role in acid tolerance and succinate biosynthesis. Appl. Environ. Microbiol. 79:74–80. 10.1128/AEM.02184-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Poolman B, Molenaar D, Smid EJ, Ubbink T, Abee T, Renault PP, Konings WN. 1991. Malolactic fermentation: electrogenic malate uptake and malate/lactate antiport generate metabolic energy. J. Bacteriol. 173:6030–6037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ehlers C, Ivens UI, Moller ML, Senderovitz T, Serup J. 2001. Females have lower skin surface pH than men. A study on the surface of gender, forearm site variation, right/left difference and time of the day on the skin surface pH. Skin Res. Technol. 7:90–94 [DOI] [PubMed] [Google Scholar]
- 72.Bessman AN, Page J, Thomas LJ. 1989. In vivo pH of induced soft-tissue abscesses in diabetic and nondiabetic mice. Diabetes 38:659–662 [DOI] [PubMed] [Google Scholar]
- 73.Bore E, Langsrud S, Langsrud O, Rode TM, Holck A. 2007. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 153:2289–2303. 10.1099/mic.0.2007/005942-0 [DOI] [PubMed] [Google Scholar]
- 74.Weinrick B, Dunman PM, McAleese F, Murphy E, Projan SJ, Fang Y, Novick RP. 2004. Effect of mild acid on gene expression in Staphylococcus aureus. J. Bacteriol. 186:8407–8423. 10.1128/JB.186.24.8407-8423.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clements MO, Watson SP, Foster SJ. 1999. Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J. Bacteriol. 181:3898–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Joshi GS, Spontak JS, Klapper DG, Richardson AR. 2011. Arginine catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol. Microbiol. 82:9–20. 10.1111/j.1365-2958.2011.07809.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.DeLeo FR, Otto M, Kreiswirth BN, Chambers HF. 2010. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375:1557–1568. 10.1016/S0140-6736(09)61999-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Somerville GA, Proctor RA. 2009. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol. Mol. Biol. Rev. 73:233–248. 10.1128/MMBR.00005-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yao Y, Sturdevant DE, Otto M. 2005. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J. Infect. Dis. 191:289–298. 10.1086/426945 [DOI] [PubMed] [Google Scholar]
- 80.Stewart PS. 2003. Diffusion in biofilms. J. Bacteriol. 185:1485–1491. 10.1128/JB.185.5.1485-1491.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ryder VJ, Chopra I, O'Neill AJ. 2012. Increased mutability of Staphylococci in biofilms as a consequence of oxidative stress. PLoS One 7:e47695. 10.1371/journal.pone.0047695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Charpentier E, Anton AI, Barry P, Alfonso B, Fang Y, Novick RP. 2004. Novel cassette-based shuttle vector system for gram-positive bacteria. Appl. Environ. Microbiol. 70:6076–6085. 10.1128/AEM.70.10.6076-6085.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.