Abstract
LeuO is a LysR-type transcriptional regulator (LTTR) that has been described to be a global regulator in Escherichia coli and Salmonella enterica, since it positively and negatively regulates the expression of genes involved in multiple biological processes. LeuO is comprised of an N-terminal DNA-binding domain (DBD) with a winged helix-turn-helix (wHTH) motif and of a long linker helix (LH) involved in dimerization that connects the DBD with the C-terminal effector-binding domain (EBD) or regulatory domain (RD; which comprises subdomains RD-I and RD-II). Here we show that the oligomeric structure of LeuO is a tetramer that binds with high affinity to DNA. A collection of single amino acid substitutions in the LeuO DBD indicated that this region is involved in oligomerization, in positive and negative regulation, as well as in DNA binding. Mutants with point mutations in the central and C-terminal regions of RD-I were affected in transcriptional activation. Deletion of the RD-II and RD-I C-terminal subdomains affected not only oligomerization but also DNA interaction, showing that they are involved in positive and negative regulation. Together, these data demonstrate that not only the C terminus but also the DBD of LeuO is involved in oligomer formation; therefore, each LeuO domain appears to act synergistically to maintain its regulatory functions in Salmonella enterica serovar Typhi.
INTRODUCTION
The leuO gene is located between the leuOABCD and ilvIH operons (1), and its product is involved in the regulation of leuOABCD by a complex cis-acting promoter relay mechanism (2–4). The LeuO regulator belongs to the LysR-type transcriptional regulators (LTTRs) (5) and represses the expression of DsrA RNA and cadAB (6, 7); it is also involved in the activation of bgl and of the yjjQ-bglJ operons in Escherichia coli (8–10) and rovA in Yersinia (11). In Salmonella enterica serovar Typhi, LeuO activates the expression of ompS1 and ompS2 (12, 13). Additionally, LeuO was reported to be a global regulator in S. Typhi, since it positively regulates the expression of assT and STY3070 (casC) and downregulates ompX, tpx, and STY1978 (14–16). LeuO is involved in the virulence of S. Typhimurium both in a mouse model and in Caenorhabditis elegans, and its role in Vibrio cholerae biofilm formation has been reported (17–19).
The role of LeuO as an antagonist of the histone-like nucleoid structuring protein that can act as a transcription silencer (the H-NS protein) has been described in detail for ompS1, where it binds to two sites contained in the F+1 fragment described below, and for the CRISPR-Cas system (13–15, 20). It has also been reported to be a global H-NS protein antagonist in E. coli, S. Typhimurium, and S. Typhi (21, 22). To date, this role as an antagonist of the H-NS protein has not been reported for other LTTRs. Interestingly, LeuO is quiescent in a wild-type (wt) genomic background, due to the negative regulatory effect of the H-NS nucleoid-associated protein and transcriptional silencer, but its expression can be detected in stationary phase and when the levels of phosphate and amino acids are restricted (7, 23–25). More recently, it was shown that leuO expression in E. coli can be activated by the RcsB and BglJ regulators (26).
The LTTRs share features, such as their sequence length (300 to 350 residues) and the high sequence identity of the first 60 residues of the N terminus. These 60 residues are comprised of the DNA-binding domain (DBD) with a winged helix-turn-helix (wHTH) motif and of a long linker helix (LH) that connects the DBD with the C-terminal effector-binding domain (EBD) or regulatory domain (RD). According to the standard nomenclature for the LTTR structure (27–30), RD comprises subdomain RD-I, which is located in the central part of the protein and at the C terminus, and subdomain RD-II, which separates the two parts of RD-I in the central part of the protein and at the C terminus.
The active form of most characterized LTTRs (NahR, CysB, OxyR, CbnR, DntR, ArgP, TsaR, and AphB) is a tetramer (28, 30–35). Nevertheless, there are some LTTRs, such as MetR, CatR, IlvY, NodD3, Nac, and IciA, that form dimers in solution, and this seems to be their active form (36–41). In addition, CrgA forms octamers, and hexamer formation was reported for HsdR (29, 42). Several studies suggest that the C-terminal domain of LTTRs is involved in multimerization, since repression and DNA binding are affected in NahR, CysB, or OxyR with deletions in the C terminus. These effects are thought to be due to defective oligomerization. Moreover, mutants with single point mutations in the C termini of OxyR and CysB were dimeric, indicating that this region is involved in tetramer formation. Interestingly, as dimers these mutants retained the capacity to repress and bind DNA (9, 31, 43–45). In contrast, mutants with deletions in the C-terminal regions of MetR and Nac were not affected in transcriptional regulation and in solution formed dimers that interacted with DNA. These truncated proteins formed dimers, perhaps because they contain an intact LH that is involved in dimerization (28, 30, 36, 40).
In this work, the tetrameric form of the global regulatory protein LeuO is reported. By site-directed and deletion mutagenesis, protein-DNA interaction, transcriptional regulation activity, and oligomerization studies, we observed that each LeuO domain is important to maintenance of its properties.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. S. Typhi IMSS-1 was grown in MA medium (7 g nutrient broth, 1 g yeast extract, 2 g glycerol, 3.75 g K2HPO4, 1.3 g KH2PO4 per liter). The E. coli strains were grown in LB medium (10 g tryptone, 5 g yeast extract, 10 g NaCl per liter). When necessary, the following antibiotics were added to the medium: ampicillin (300 μg/ml), kanamycin (30 μg/ml), tetracycline (10 μg/ml), and chloramphenicol (30 μg/ml). The E. coli strains and S. Typhi were grown aerobically at 37°C.
DNA manipulation and plasmid construction.
The oligonucleotides used in this study are listed in Table S2 in the supplemental material and were provided by the Oligonucleotide Synthesis Unit of the Institute of Biotechnology, Universidad Nacional Autónoma de México (UNAM). Plasmid isolation, restriction enzyme digestions, ligase reactions, transformations, and 5′ end labeling of PCR fragments were performed using standard procedures. PCRs were performed with Taq DNA polymerase (Invitrogen), and restriction and DNA-modifying enzymes were obtained from Roche, Fermentas, or New England BioLabs and used according to the manufacturers' instructions. The construction of deletions was carried out with oligonucleotides to attach NcoI and BamHI sites in frame to amplify the appropriate fragments out of full-length leuO using the pFMTrcleuO-50 plasmid as the template, and the digested fragments were cloned into the pFMTrc12 vector (see Table S1 in the supplemental material). The PCRs used for site-directed mutagenesis of leuO to introduce alanine substitutions at residues L27, L46, S54, L60, T79, P139, S310, V311, C312, K313, and R314 were carried out with the appropriate pairs of primers (see Table S2 in the supplemental material) and the common upper primer 99ACG5′ and lower primer 99CGA3′ using the pFMTrcleuO-50 plasmid as the template; the PCR products were digested with NcoI and BamHI and ligated into pFMTrc12. The plasmids used in the LexA-based two-hybrid system were constructed by PCR using the leuO wild type as the template. Deletions and point mutations in the coding sequence were obtained with the LeuO-FBgl and LeuO-RKpn primers. The fragments were cleaved with BglII-KpnI and cloned into plasmid pSR658-A (see Table S1 in the supplemental material). Plasmids pFMTrcleuO-50, pLeuO-5C, pLeuO-30C, pLeuOS54A, pLeuOT79A, and pLeuOP139A were cleaved with NcoI and HindIII, and fragments were cloned into plasmid pMPM-T6Ω; all fragments had a C-terminal 6×His tag (see Table S1 in the supplemental material). The plasmids derived from pFMTrcleuO-50, pSR658-A, and pMPM-T6Ω were completely sequenced to verify the presence of the inserts.
Purification of LeuO wt and mutant proteins.
For in vitro assays, the S. Typhi LeuO wt and different mutant proteins were overexpressed from plasmids derived from pMPM-T6Ω in E. coli BL21(DE3) (see Table S1 in the supplemental material). The LeuO wt used for size exclusion chromatography was purified from a liter of LB medium and grown to mid-logarithmic phase, and 0.1% l-(+)-arabinose (Sigma-Aldrich) was added to activate expression. The culture was then incubated at 200 rpm at 37°C for 4 h. To purify the LeuO1-309, LeuO1-284, S54A, and P139A mutants, as well as the LeuO wt, the following conditions were used: cultures were grown in 200 ml of LB medium for 2 h at 37°C, and then 0.1% l-(+)-arabinose was added; the cultures were incubated at 200 rpm at 16°C for 20 h, and cells were pelleted by centrifugation. LeuO-6×His and the mutant His-tagged proteins were purified under native (nondenaturing) conditions, and the pellet was resuspended in purification buffer (50 mM NaH2PO4, 500 mM NaCl, 20 mM imidazole, 1 mg/ml lysozyme, 0.1% Triton X-100) and disrupted by sonication. The supernatant was subjected to chromatography on nickel-agarose (Ni-nitrilotriacetic acid [NTA]; Qiagen) according to the manufacturer's instructions. The column was washed with 10 volumes of resuspension buffer with 20 mM imidazole and then with a linear gradient of 20 to 300 mM imidazole. Fractions containing purified LeuO-6×His were selected for SDS-PAGE, loaded into a Slyder-A-Lyzer 10K cassette (Pierce), dialyzed at 4°C against storage buffer (50 mM NaH2PO4, 500 mM NaCl, 0.1% Tween, 5 mM β-mercaptoethanol, 20% glycerol, pH 7.4), and stored at −20°C. The LeuO-6×His wt was purified under denaturing conditions as previously described (13), with some modifications. Briefly, the cells were suspended in lysis buffer (8 M urea, 100 mM NaH2PO4, 10 mM Tris-HCl [pH 8.0]) and disrupted by sonication; the supernatant was subjected to chromatography on nickel-agarose (Ni-NTA; Qiagen); and fractions containing purified LeuO-6×His were selected for SDS-PAGE, loaded onto the polyacrylamide gel, gradually dialyzed at 4°C against decreasing concentrations of urea (4, 1, and 0.2 M), and finally, dialyzed against storage buffer, as described above.
Size exclusion chromatography.
Size exclusion chromatography was performed on an Akta system using a Sephacryl S-200 HR column (Amersham Biosciences). The column (volume, 320 ml) was loaded with 2 mg of LeuO wt protein contained in 3 ml of equilibration buffer. The column had previously been washed and equilibrated with 2 column volumes of 50 mM NaH2PO4, 500 mM NaCl, pH 7.4, and protein was eluted at a flow rate of 1.2 ml/min. Elution was monitored at 280 nm. The column was calibrated using blue dextran (≥2,000 kDa) to determine the void volume (Vo), as well as the following molecular mass standards (Sigma): cytochrome c (12.4 kDa), carbonic anhydrase (29 kDa), albumin (66 kDa), alcohol dehydrogenase (150 kDa), and β-amylase 145 (200 kDa). A calibration curve was obtained by plotting the logarithms of the molecular masses of the standards versus their respective Ve/Vo values, where Ve corresponds to the elution volume of each standard. A linear regression with the values was performed to obtain the equation y = 6,922.1e−3.0248x, which was used to calculate the relative molecular mass of LeuO, which was equal to 6,922.1e(−3.0248 × VeLeuO), by overlapping onto the standard curve. The LeuO wt purified under native conditions was loaded onto the column in two independent experiments, and its Ve was monitored. Only one major peak was observed.
β-Galactosidase assay.
The E. coli SU101 strain harboring pSR658-A and LeuO derivative plasmids was grown overnight in LB medium with tetracycline at 37°C under aerobic conditions. It was then subcultured in LB medium with tetracycline (10 μg/ml), kanamycin (30 μg/ml), and IPTG (isopropyl-β-d-thiogalactopyranoside; 1.0 mM). Cells were collected at an A600 of 1.0, and the pellets were stored for β-galactosidase and Western blot assays. The β-galactosidase and microplate protein concentrations were determined as previously described (46).
CAT assays.
S. Typhi strain IMSS-1 harboring plasmid pKK232-9 (ompS1) or pKK232-9 (tpx), together with either plasmid pFMTrcleuO-50, vector pFMTrc12, or derivative plasmids (see Table S1 in the supplemental material), was grown overnight in LB medium at 37°C under aerobic conditions with ampicillin (300 μg/ml) and kanamycin (30 μg/ml) and on the next day was subcultured in MA medium (100 ml) supplemented with ampicillin and kanamycin. IPTG was added to a final concentration of 100 μM at 37°C under aerobic conditions, and bacteria were collected at early log phase (A600 = 1.0) for determination of chloramphenicol acetyltransferase (CAT) activity and Western blotting. CAT activity and protein concentrations were determined as previously described (14, 47).
Western blotting and PAGE.
For Western blot assays, samples were subjected to SDS-PAGE (15% polyacrylamide) and transferred to 0.45-μm-pore-size polyvinylidene difluoride membranes (Immobilon; Millipore) in a semidry electrophoresis unit (Bio-Rad). Membranes were blocked with 5% nonfat milk and incubated with anti-LexA antibody (Abcam), anti-LeuO polyclonal antibodies, and an anti-6×His (Roche) or anti-DnaK (MBL International Corp.) monoclonal antibody. They were then washed with 1× phosphate-buffered saline, 0.05% Tween 20. Immunodetection was performed with a 1:10,000 dilution of horseradish peroxidase-conjugated antirabbit or antimouse antibody (Pierce) for polyclonal or monoclonal antibodies, respectively, and a Western Lightning Plus-ECL chemiluminescence reagent kit was used according to the instructions of the manufacturer (PerkinElmer) in order to visualize the bands.
Protein electrophoresis was performed under native conditions in a 10% nondenaturing polyacrylamide gel with Tris-HCl, pH 8.8, but without SDS and with a Tris-glycine, pH 8.3, running buffer. LeuO-6×His (1.5 μg per lane) was loaded, and after the separation, the proteins were visualized following the first step of the enhanced-background (two-stage) rapid silver-staining protocol (48).
EMSAs.
The DNA fragment from positions −310 to +1 of the regulatory region of the ompS1 gene (referred to here as the DNA F+1 fragment) and the ler structural fragment used as a negative control were generated by PCR with primers 310b-1 and 310-(+1) and primers ler-Kpn and ler-15F, respectively, using plasmids pRO310 and pler-T3 as the templates, respectively (see Tables S1 and S2 in the supplemental material). The conditions used for the binding reactions and electrophoretic mobility shift assays (EMSAs) were those described previously, and 100 ng of each fragment was used (13). The F+1 fragment was 5′ end labeled with [γ-32P]dATP at 3,000 Ci mmol−1 (Amersham Corporation) using polynucleotide kinase (Invitrogen) and purified using a gel extraction kit (Qiagen). Mixtures for EMSA (20 μl) containing 15,000 cpm (see Fig. 2) and 20,000 cpm (see Fig. 5) of the end-labeled F+1 DNA fragment, poly(dI-dC) as a nonspecific competitor, and different concentrations of purified LeuO wt and S54A and P139A mutants were incubated for 20 min at 4°C and then subjected to electrophoresis on 6% polyacrylamide nondenaturing gels in 1× Tris borate-EDTA buffer (pH 8.0) for 3 h at 100 V. The dried gels were subjected to autoradiography to visualize the radiolabeled bands.
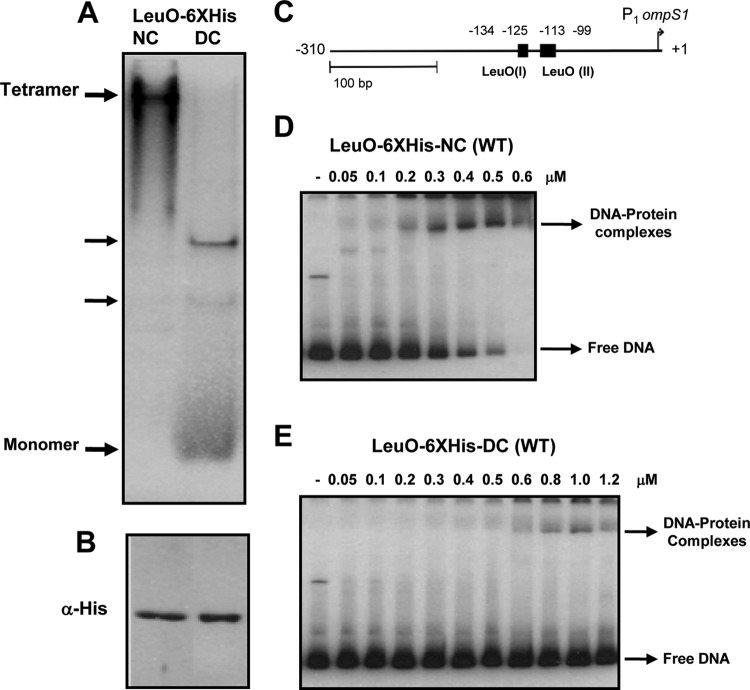
FIG 2.
Tetrameric LeuO wt binds to DNA with a high affinity. The LeuO wt (1.5 μg per lane) purified under native (LeuO6×His-NC) and denaturing (LeuO-6×His-DC) conditions was analyzed in a native 10% polyacrylamide gel (A), in a Western blot after SDS-PAGE with anti-His antibody (B), and by EMSA with different concentrations of LeuO6×His-NC (tetramer) (D) and LeuO-6×His-DC (monomer > dimer > trimer > tetramer) (E). (C) Schematic representation of the F+1 DNA fragment from the ompS1 5′ regulatory region (13) used as a probe for EMSAs.
FIG 5.
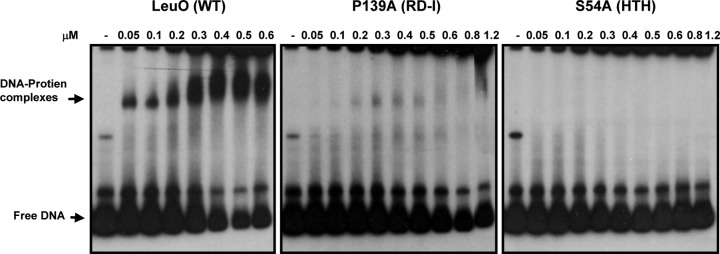
The P139A mutant binds to DNA with a low affinity. The results of EMSAs with the LeuO wt and the P139A and S54A mutants, which were purified under native conditions, bound to the 32P-end-labeled F+1 DNA ompS1 fragment are shown. Different concentrations of LeuO wt and mutant proteins were used, as indicated above each lane.
LexA-based genetic system.
The LexA-based genetic system used to analyze the LeuO wt and the dimerization of the different mutants has been described previously (49, 50). The coding sequences were obtained by PCR with primers LeuO-FBgl and LeuO-RKpn (see Table S2 in the supplemental material) with leuO wt and mutant templates. The resulting fragments were cloned in frame as a BglII-KpnI fragment into the sequence encoding the LexA DNA-binding domain (LexADBD) in pSR658-A, generating plasmids coding for chimeric proteins constituted by LexADBD fused to either the LeuO wt or mutant derivatives (see Table S1 in the supplemental material). The resulting plasmids were verified by DNA sequencing and introduced into E. coli SU101, which contains a chromosomal sulA::lacZ fusion, to monitor the ability of the hybrid proteins to form functional LexADBD dimers, which in turn repress the expression of the sulA::lacZ fusion. The E. coli SU101 strain harboring pSR658-A or LexADBD-LeuO derivative plasmids was grown overnight in LB medium with tetracycline at 37°C under aerobic conditions. It was then subcultured in LB medium with tetracycline (10 μg/ml), kanamycin (30 μg/ml), and IPTG (1 mM) to induce expression of LexADBD-LeuO hybrid proteins. Cells were collected when the optical density at 600 nm (OD600) was 1.0, and the pellets were stored for β-galactosidase and Western blot assays.
RESULTS
S. Typhi LeuO forms tetramers that bind DNA with high affinity.
To determine the oligomeric state of LeuO from Salmonella enterica serovar Typhi IMSS-1 (see Table S1 in the supplemental material), the leuO gene was cloned in pMPM-T6Ω, and LeuO was overexpressed and purified under native (nondenaturing) conditions (NCs), resulting in LeuO-6×His-NC. LeuO-6×His-NC was subjected to size exclusion chromatography, and a single peak giving an average relative mass of 140.75 kDa was detected. This was calculated using a previously calibrated column with molecular mass standards (Fig. 1A). This indicates that LeuO-6×His-NC is composed of four identical subunits (Fig. 1B) and that LeuO forms a tetramer in solution. Additionally, LeuO-6×His-NC showed only one intense band when analyzed in a native polyacrylamide gel, and that band corresponded to a tetramer (Fig. 2A). Western blot analysis of purified LeuO-6×His-NC with anti-His antibody revealed a single monomeric band of 35.7 kDa (Fig. 2B).
FIG 1.
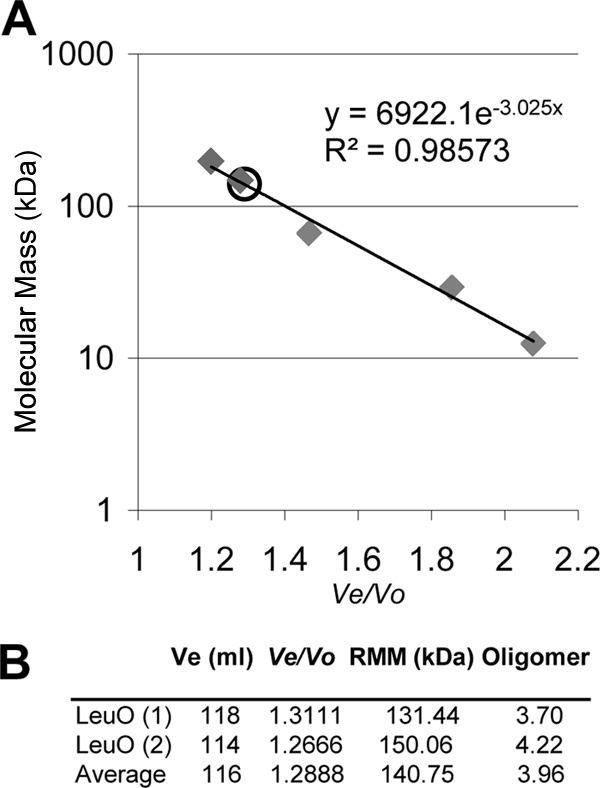
Size exclusion chromatography analysis of LeuO wt. The LeuO wt was purified under native conditions (2 mg/3 ml) and was loaded onto a Sephacryl S-200 column. (A) Calibration curve obtained by plotting the logarithms of the known molecular mass standards versus their respective Ve/Vo values. Blue dextran was used to determine the void volume (Vo) of the column (90 ml). The following protein standards are presented in the standard curve (gray diamonds) (their molecular masses, elution volumes [Ves], and Ve/Vo values are given in parentheses): β-amylase (200 kDa, 108 ml, 1.2), alcohol dehydrogenase (150 kDa, 117 ml, 1.3), albumin (66 kDa, 129 ml, 1.43), carbonic anhydrase (29 kDa, 167 ml, 1.86), and cytochrome c (12.4 kDa, 187 ml, 2.08). (B) LeuO Ve and Ve/Vo values obtained from two independent experiments and the average LeuO Ve and Ve/Vo values; the Ve value was incorporated into the standard curve (open circle in panel A). The relative molecular mass (RMM) of the LeuO wt was calculated using the equation described in Materials and Methods, and the oligomer size of LeuO was calculated as the relative molecular mass/35.7 kDa, where the latter value is the molecular mass of the LeuO monomer.
To analyze the ability of tetrameric LeuO-6×His-NC to bind DNA, a 350-bp DNA fragment (F+1) from the ompS1 promoter region containing two LeuO binding sites (13) was used in an EMSA (Fig. 2C). The results depicted in Fig. 2D show that LeuO-6×His-NC binds at low concentrations, starting at 50 nM.
Previously, LeuO purified under denaturing conditions (LeuO-6×His-DC) did bind DNA, albeit at higher concentrations (13). The analysis of LeuO-6×His-DC in a native polyacrylamide gel showed other bands that migrated faster than the tetramer. The tetramer comigrated with the octameric form of the Ler regulator (a 16-kDa monomer), and the monomer comigrated with the Ler dimer (data not shown). The nature of the bands observed between the tetramer and the monomer is unknown and will be the subject of further studies regarding the structure of LeuO (Fig. 2A). LeuO-6×His-DC was purified to homogeneity according to the findings obtained with Coomassie blue-stained gels (not shown) and analysis by Western blotting (Fig. 2B). When analyzed for DNA binding, it required nearly 10-fold higher concentrations than LeuO-6×His-NC to interact with DNA (Fig. 2E). These results suggest that the tetrameric LeuO in the LeuO-6×His-NC samples binds to DNA with a higher affinity than the other LeuO forms found in LeuO-6×His-DC. In any event, care must be taken to purify LeuO under the appropriate native conditions in order to obtain the best activity.
Residues in LeuO DBD and LH are involved in oligomerization, while an RD-I central subdomain mutant has no effect on oligomer formation.
The DBD is the most conserved region between LTTRs, and mutations in this domain affect DNA binding, activation, and repression. Furthermore, mutations in the DBDs of CysB, OxyR, and GcvA were not transdominant, and it has been suggested that they are affected in oligomer formation (43–45). The LH that is also conserved among LTTRs has a role in dimer formation, and the less conserved RD-I domain is involved in binding and a coinducer response (27–30).
The crystal structure of the characterized LTTRs has shown a similar fold for each subunit (monomer), and in several instances a stable active homotetramer formed as a dimer of dimers has been proposed to be necessary for the biological function of LTTRs (28–35, 51, 52). On the basis these findings, it seems that dimer formation is a process needed to obtain an active LeuO tetrameric form; therefore, we utilized the LexA-based genetic system of homodimerization to evaluate LeuO dimerization. This genetic system was also used in order to be able to assess the oligomeric state of LTTRs since different mutated versions of LeuO constructed in this study could not be overexpressed and purified to homogeneity in the required amounts. This was the main limitation of this study, in spite of the fact that the LeuO wt protein was obtained in sufficient amounts (4 mg per liter of culture).
Hence, in order to determine the involvement of the DBD, the LH, and the RD-I central subdomain of LeuO in oligomerization, fragments with single alanine substitutions at six conserved residues were constructed (Fig. 3) and fused to LexA for dimerization analysis. The LexA-based genetic system detects in vivo protein dimerization in E. coli SU101, in which the reporter fusion sulA::lacZ is controlled by the LexA dimer (dimerization is essential for operator recognition) (50).
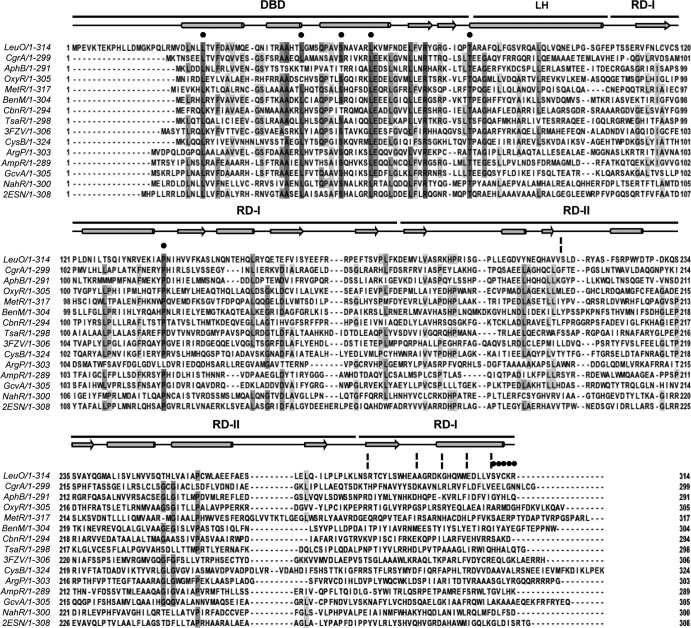
FIG 3.
Multiple-sequence alignment of LeuO and several characterized LTTRs. LeuO has a 20-residue extension at the N terminus when its sequence is aligned with the sequences of the other LTTRs. The DNA-binding domain of LeuO comprises residues 21 to 78, the linker helix comprises residues 79 to 105, and the C-terminal regulatory domain comprises residues 106 to 314. Black circles and broken vertical lines highlight the replaced residues and deletions, respectively. The alignments and prediction of the secondary structure (in which the schematic representation above the sequences shows helices as cylinders, β sheets as arrows, and turn segments as lines) of LeuO were performed with the ClustalW and JalView programs (59). Dark gray and pale gray boxes show residues with the highest and the least identity, respectively. LysR family domains DBD, LH, and the C-terminal regulatory domain with two subdomains, RD-I (central, C terminal) and RD-II, are shown at the top. The amino acid sequences used in the alignment are as follows: LeuO (S. enterica serovar Typhi), CgrA (Neisseria meningitidis, GI 7188597), AphB (V. cholerae, GI 5565924), OxyR (E. coli, GI 388479299), MetR (E. coli, GI 388479422), BenM (Acinetobacter baylyi ADP1, GI 2996626), CbnR (Ralstonia eutropha, GI 4210464), TsaR (Comamonas testosteroni T-2, GI 75499536), CysB (E. coli, GI 1787530), ArgP (Mycobacterium tuberculosis H37v, GI 15609122), AmpR (Citrobacter freundii, GI 736669), GcvA (E. coli, GI 388478824), and NahR (Pseudomonas putida, GI 37220701). The PDB accession number for P. aeruginosa Q9I6S0 is 3FZV, and that for P. aeruginosa Q9I641 is 2ESN.
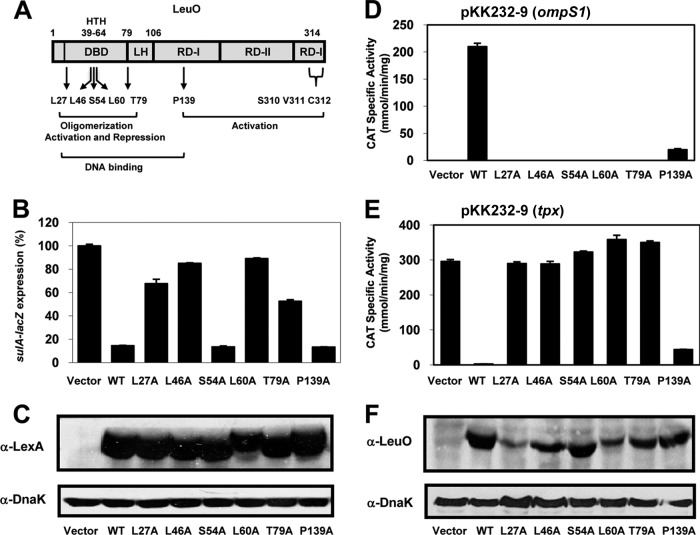
The DBD of LexA (residues 1 to 128) was fused to full-length S. Typhi LeuO (residues 1 to 314) and used as a positive control. This construct is referred to as LexA–LeuO1-314. In the reporter strain with the vector pSR658-A, the level of β-galactosidase activity reflects the level of constitutive expression of the sulA::lacZ fusion, and this level is presented as 100% sulA expression (Fig. 4B). The results showed that LexA–LeuO1-314 (wt) efficiently repressed sulA::lacZ expression (down to 5 to 10%) (Fig. 4B); thus, LeuO can replace the dimerization domain of LexA, and this system was used to probe LeuO dimerization in vivo. Six conserved residues, L27, L46, S54, and L60 (located in the DBD), T79 (located one residue before the predicted LH), and P139A located in the RD-I central subdomain of LeuO (Fig. 3 and 4A), were replaced by alanine. The results showed sulA::lacZ expression of 65% for LexA-L27A, 85% for LexA-L46A and LexA-L60A, and 50% for LexA-T79A. Thus, these mutant proteins had a defective ability to repress the expression of the sulA::lacZ reporter in strain SU101 compared with that of LexA-LeuO (wt) (Fig. 4B).
FIG 4.
Analysis of the dimerization and regulatory functions of LeuO wt and different mutants with alanine-substituted residues in DBD and RD-I. (A) Schematic representation of the domains of the LysR family: DBD with the HTH motif, LH, and C-terminal regulatory domains RD-I (central and C terminal) and RD-II. The locations of the six alanine substitutions are also shown. (B) Dimerization analysis using the LexA-based two-hybrid system. sulA::lacZ expression was analyzed in strain SU101 carrying pSR658-A (vector), which reflects constitutive expression of the fusion and which was set equal to 100%, or pLexA-LeuOL27A, pLexA-LeuOL46A, pLexA-LeuOS54A, pLexA-LeuOL60A, pLexA-LeuOT79A, and pLexA-LeuOP139A. (C) Western blot assays of extracts prepared from cells carrying LexA-LeuO wt or LexA–alanine-substituted DBD and RD-I were carried out using anti-LexA antibody and anti-DnaK monoclonal antibody as a control after SDS-PAGE. The cultures were grown in LB to an OD600 of 1.0, and LexA-fused protein expression was induced with IPTG (1 mM). (D and E) The transcriptional profiles of fusions to the cat reporter gene for the ompS1 (D) and tpx (E) genes in S. Typhi were evaluated in the presence of either the LeuO wt or mutants with six different alanine substitutions in DBD, LH, or RD-I: L27A, L46A, S54A, L60A, T79A, and P139A. (F) Expression of the LeuO wt and alanine-substituted mutants using an anti-LeuO polyclonal antibody and anti-DnaK monoclonal antibody as a control. The cultures were grown in MA medium to an OD595 of 1.0, and expression of the proteins was induced with IPTG (100 μM).
In contrast, LexA-S54A, where S54A is located in the HTH motif, and LexA-P139A, where P139A is located in the RD-I central subdomain, repressed sulA::lacZ reporter expression as effectively as the LexA-LeuO wt (Fig. 4B). Western blot experiments showed similar levels of expression of LexA-LeuO wt and the LexA-fused mutants (Fig. 4C). These results showed that residues L27, L46, L60, and T79 are important in LeuO dimerization, while residues S54 and P139 do not affect dimerization, supporting the involvement of the DBD and LH regions in LeuO oligomerization.
LeuO DBD, LH, and RD-I central subdomain mutants are impaired in transcriptional activation and repression.
To evaluate the effects of mutations in DBD, LH, and the RD-I central domain (Fig. 4A) on transcriptional activation or repression, the genes encoding the LeuO wt and the six alanine-substituted variants were cloned into pFMTrc12 (see Materials and Methods). The resulting plasmids, pFMTrcleuO-50 (wt), pLeuOL27A, pLeuOL46A, pLeuOS54A, pLeuOL60A, pLeuOT79A, and pLeuOP139A, were introduced into strain IMSS-1 harboring the reporter plasmid pKK232-9 (ompS1) or pKK232-9 (tpx). The expression of ompS1 and tpx (as regulatory regions fused to the cat gene) was activated or repressed in the presence of the LeuO wt, respectively (14). The results presented in Fig. 4D and E show that the L27A, L46A, S54A, L60A, and T79A mutations rendered the proteins totally defective in activating or repressing the ompS1 or tpx cat fusion, respectively.
A distinct phenotype was observed for the P139A mutant, whose ability to activate ompS1 transcription was reduced by 10-fold compared to that of the wt (Fig. 4D), yet it was able to substantially repress tpx activity (Fig. 4E). This observation could mean that LeuO binds to DNA differently when it displaces the H-NS protein to turn on a gene than when it binds to impede transcription initiation. The elucidation of this hypothesis is certainly a matter for further research.
Western blot assays of cell extracts (Fig. 4F) showed that the L46A, S54A, T79A, and P139A mutant proteins were expressed at levels similar to the levels of expression of the LeuO wt, whereas the L27A and L60A mutant proteins were detected in smaller amounts, suggesting that they are more unstable. Nevertheless, their expression levels appeared to be sufficient to activate and repress transcription. These results indicate that the L27A, L46A, L60A, and T79A mutants lost their transcriptional function, aside from the fact that they were affected in dimer formation (Fig. 4B). In contrast, the ability of the S54A mutant, which dimerized like the wt (Fig. 4B), to activate and repress transcription was abolished (Fig. 4D and E). This mutant is possibly defective in DNA binding since residue Ser54 is located in the HTH motif (Fig. 3 and 4A). The P139A mutant presented an interesting phenotype, since it dimerized (Fig. 4B), and although it retained only a low level of transcriptional activation, it maintained the capacity to almost fully repress transcription (Fig. 4D and E) and was expressed in amounts similar to those for the wt (Fig. 4F). This mutant protein probably maintains its ability to bind efficiently to tpx DNA but has less affinity for ompS1 DNA. It should be pointed out that for the Western blot assays, the antibody against LeuO that was used was polyclonal; thus, a major reactive band was obtained over a background of other bands.
LeuO DBD, LH, and RD-I central subdomain mutants are defective in DNA binding.
To ascertain the DNA-binding abilities of mutants with mutations in DBD, LH, and the RD-I central subdomain, alanine-substituted variants with six histidine residues in the C terminus were cloned into the pMPM-T6Ω vector, in order to be overexpressed and purified under nondenatured conditions (see Materials and Methods). EMSAs were performed with the 350-bp DNA fragment (F+1) containing the S. Typhi ompS1 promoter region and the LeuO-binding sites (13) and a ler DNA fragment of enteropathogenic E. coli (EPEC) as a negative control (13, 46). By EMSA, the P139A mutant showed no detectable band with retarded migration (data not shown), even though it retained some activating and almost full repressing activity (Fig. 4D and E). However, in a more sensitive gel retardation experiment using a radioactive F+1 fragment, the P139A mutant did show DNA binding to the fragment, although at a much lower affinity than the LeuO wt (Fig. 5). Even with this more sensitive assay, no binding to DNA was observed with the S54A mutant. All these results showed that some residues in the DBD are involved in dimer formation and, thus, are most likely involved in tetramer formation, and these oligomeric defects alter the transcriptional function and DNA binding in vitro.
Deletions in the LeuO C-terminal RD-II and RD-I subdomains affect oligomerization.
The C-terminal domains of LTTRs are involved in oligomerization, although the specific amino acid regions involved in tetramerization have not been determined due to the poor conservation of this domain (53, 54). Accordingly, LeuO shares a high degree of sequence similarity with other LTTRs at the N-terminal DNA-binding domain and less conservation at the C-terminal inducer-binding domain (Fig. 3).
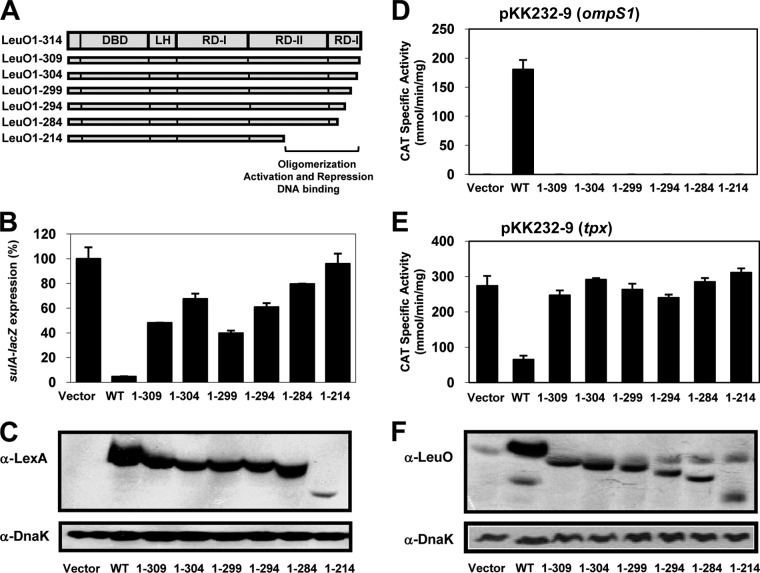
To determine the role of the C-terminal domain of LeuO in oligomer formation, a collection of sequentially truncated forms of LeuO fused to LexA was constructed. The selection of deletions was based on some that have been analyzed in other LTTRs (9, 31, 36, 40, 43–45). Thus, fragments of leuO encoding amino acid residues 1 to 309, 1 to 304, 1 to 299, 1 to 294, 1 to 284, and 1 to 214 (Fig. 6A), lacking 5, 10, 15, 20, 30, and 100 amino acids, respectively, were cloned into plasmid pSR658-A (see Materials and Methods), and strain SU101 was transformed with the resulting plasmids. The results presented in Fig. 6B show that all the truncated proteins fused to LexA were defective in repressing sulA::lacZ expression to various extents. The reporter gene expressions were 50% for pLexA-LeuO1-309, 70% for pLexA-LeuO1-304, 40% for pLexA-LeuO1-299, 60% for pLexA-LeuO-1-294, 80% for pLexA-LeuO1-284, and 90% for pLexA-LeuO1-214. Thus, all the mutants had dimerization defects compared with the dimerization ability of LexA-LeuO wt (Fig. 6B).
FIG 6.
Analysis of dimerization and regulatory functions of LeuO wt and C-terminal deletion mutants. (A) Schematic representation of the domains of the LysR family: DBD, LH, and the C-terminal regulatory domain with two subdomains, RD-I (central, C terminal) and RD-II. The localization of the deletions in the C-terminal domain is also shown. (B) Dimerization analysis. sulA::lacZ expression in strain SU101 carrying pSR658-A (vector), the expression of which was set equal to 100%, or carrying pLexA-LeuO (wt), pLexA-LeuO1-309, pLexA-LeuO1-304, pLexA-LeuO1-299, pLexA-LeuO1-294, pLexA-LeuO-1-284, or pLexA-LeuO1-214 is shown. (C) Expression of the LeuO wt and deletion mutants fused to LexA was evaluated by Western blotting using an anti-LexA polyclonal antibody and an anti-DnaK monoclonal antibody as a control after SDS-PAGE. The cultures were grown in LB medium to an OD600 of 1.0, and LexA-fused protein expression was induced with IPTG (1.0 mM). (D and E) Transcriptional profiles of fusions to the cat reporter gene for the ompS1 and tpx genes in S. Typhi in the presence of either the LeuO wt or mutants with different deletions in the C terminus: LeuO1-309, LeuO1-304, LeuO1-299, LeuO1-294, LeuO1-284, and LeuO1-214. (F) Expression of the LeuO wt and the deletion mutants was evaluated by Western blotting using an anti-LeuO polyclonal antibody and an anti-DnaK monoclonal antibody as a control. The cultures were grown in MA medium to an OD595 of 1.0, and expression of the proteins was induced with IPTG (100 μM).
Western blot assays of extracts prepared from cells carrying the LexA-LeuO wt and LexA-LeuO truncated proteins using anti-LexA antibody showed that all the proteins were expressed at levels similar to those of the wt, with the exception of LexA-LeuO1-214, which was expressed at very low levels (Fig. 6C). It was unexpected that all the LeuO truncated variants had dimerization defects, as they have an intact LH (which is involved in dimer formation) (28–30, 34, 35). The effect observed in the truncated protein lacking only 5 residues has not been observed for other LTTRs (40, 44, 45). This is clear evidence that the C-terminal RD-II and RD-I subdomains of LeuO are involved not only in tetramer formation but also in dimerization.
The transcriptional function of LeuO C-terminal variants with truncations in the RD-II and RD-I subdomains is completely abolished.
To determine whether the proteins with truncations in the C terminus that are affected in dimer formation (Fig. 6B) are impaired in their transcriptional function in vivo, their capacity to activate or repress expression was explored. The various versions with deletions of the leuO gene were cloned into plasmid pFMTrc12 (see Materials and Methods), and the resulting plasmids were named pLeuO1-309, pLeuO1-304, pLeuO1-299, pLeuO1-294, pLeuO1-284, and pLeuO1-214 (Fig. 6A). These were introduced into strain IMSS-1 harboring either plasmid pKK232-9 (ompS1) or plasmid pKK232-9 (tpx). As expected, in accordance with their dimerization defects, all truncated proteins were unable to activate or repress gene transcription (Fig. 6D and E).
To verify that the phenotypes observed for these truncated proteins were not due to very low or no expression, Western blot analysis with a polyclonal anti-LeuO antibody was performed, and it showed that the truncated proteins were expressed in amounts similar to those of the LeuO wt, with the exception of LeuO1-214, which was expressed at lower but still detectable levels that would appear to be sufficient for functionality (Fig. 6F). These results show that all the truncated proteins that are affected in dimerization are also completely defective in their regulatory function, and this suggests that even though they have an intact DBD, they are also affected in DNA binding.
LeuO variants with C-terminal truncations fail to bind DNA.
The DNA-binding capacity of two truncated proteins, LeuO1-309 and LeuO1-284, affected in dimerization (Fig. 6B) and in their regulatory function (Fig. 6D and E) was determined. The fragments encoding the LeuO1-309 and LeuO1-284 truncated proteins with six histidine residues in the C terminus were cloned into the pMPM-T6Ω vector in order to be overexpressed and purified (see Materials and Methods). These two LeuO truncated variants were the ones that could be obtained at relatively higher yields for use in EMSAs using the 350-bp DNA fragment (F+1). The truncated proteins LeuO1-309 and LeuO1-284 did not form any detectable shifted bands; hence, they could not bind DNA even at the higher protein concentrations compared with the concentrations of LeuO wt used (data not shown).
All these observations show that the C- and N-terminal domains have a role in dimer formation and thus suggest that they have a role in tetramer formation and that the oligomeric state of LeuO is essential to exert transcriptional regulation and DNA binding.
LeuO mutants with point mutations in the last 5 residues of the C terminus are able to dimerize but are differentially affected in activation and repression.
Given the dimerization defects of LexA–LeuO1-309C (Fig. 6B), it was of interest to know if proteins with single alanine substitutions in the last 5 residues showed the same phenotype. Using the two-hybrid system, we observed that all LexA-fused mutants with point mutations repressed expression of the lacZ reporter as efficiently as the LexA-LeuO wt; therefore, they maintained the capacity to dimerize (Fig. 7A). To verify the expression of the LexA-fused point mutants, a Western blot assay was performed using extracts of cells carrying LexA-LeuO wt and LexA-S310A, LexA-V311A, LexA-C312A, LexA-K313A, and LexA-R314A. As observed in Fig. 7B, all fused proteins were expressed at levels similar to the level for the LexA-LeuO wt, showing that they were stable and functional.
FIG 7.
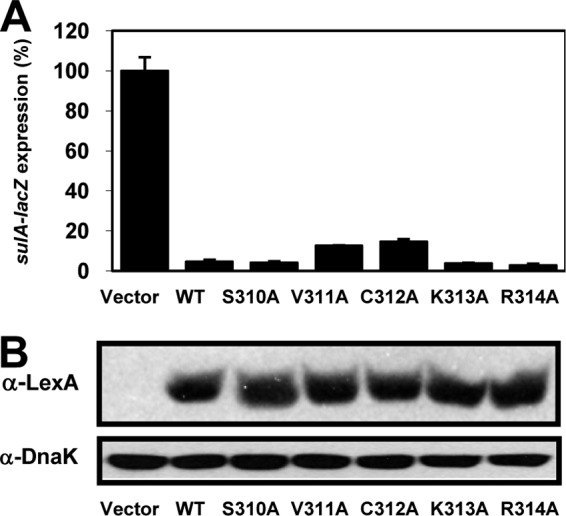
Dimerization analysis of mutants with single alanine substitutions of the last 5 residues of the RD-I subdomain at the C terminus by the LexA-based two-hybrid system. sulA::lacZ expression in strain SU101 carrying the pSR658-A (vector) was set equal to 100%. (A) Analyses of pLexA-LeuO (wt), pLexA-LeuOS310A, pLexA-LeuOV311A, pLexA-LeuOC312A, pLexA-LeuOK313A, and pLexA-LeuOR314A. (B) Western blot assay of extracts from cells carrying the LexA-LeuO wt and all the LexA-point mutants after SDS-PAGE. The gels were probed with anti-LexA antibody (top) and, for DnaK detection, with an anti-DnaK antibody as a control (bottom).
Next, the effect on the regulatory function of these mutants not affected in dimerization was analyzed. The five variants were cloned into pFMTrc12 (see Materials and Methods), generating plasmids pLeuOS310A, pLeuOV311A, pLeuOC312A, pLeuOK313A, and pLeuOR314A, which were introduced into strain IMSS-1 harboring plasmids pKK232-9 (ompS1) and pKK232-9 (tpx). Only the K313A and R314A mutants were able to activate the ompS1-cat fusion at a level similar to that for the LeuO wt; the S310A, V311A, and C312A mutants were not able to do so (Fig. 8A). Interestingly, all five variants repressed the tpx-cat fusion (Fig. 8B). Western blot analyses were performed to verify the expression of these mutant proteins at a level similar to that of the LeuO wt (Fig. 8C).
FIG 8.
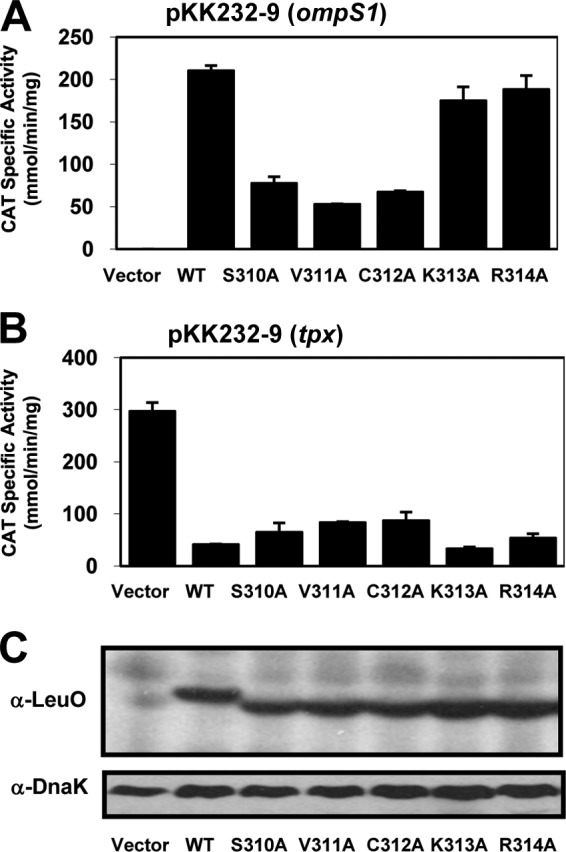
Effect of single alanine substitutions in the last 5 residues of the RD-I subdomain at the C terminus on LeuO regulatory functions. (A and B) Transcriptional profiles of ompS1 and tpx genes in S. Typhi with the LeuO wt or with five mutants with different alanine-substituted residues: LeuOS310A, LeuOV311A, LeuOC312A, LeuOK313A, and LeuOR314A. (C) Expression of the LeuO wt and mutated proteins was evaluated by Western blotting after SDS-PAGE using an anti-LeuO polyclonal antibody and an anti-DnaK monoclonal antibody as a control. The cultures were grown in MA medium to an OD595 of 1.0, and expression of the proteins was induced with 100 μM IPTG.
These results show that deletion of the last 5 LeuO residues of the RD-I terminal subdomain affects dimerization but that independent substitutions in each residue do not. Interestingly, mutants with two of these mutations (K313A and K314A) located at the end of the protein (Fig. 3) retained the capacity to activate and repress gene expression (Fig. 8A and B). In contrast, mutants with mutation of the remaining three residues that are part of the last predicted helix in LeuO (Fig. 3) still repressed but were affected in activating transcription (Fig. 8A and B). A similar phenotype was observed in the P139A mutant (Fig. 4D and E), in which the mutation was located in the RD-I central subdomain (Fig. 3). This indicates that residues in the RD-I central and terminal subdomains are involved in transcriptional activation.
DISCUSSION
In this work, the oligomeric structure of LeuO from Salmonella enterica serovar Typhi IMSS-1 was determined to be a tetramer (Fig. 1) that binds with high affinity to DNA (Fig. 2D). This oligomeric form was obtained when LeuO was purified under native conditions (LeuO-6×His-NC). In contrast, LeuO purified under denaturing conditions (LeuO-6×His-DC) bound to the same DNA with a lower affinity (Fig. 2E). Interestingly, both LeuO-6×His-NC and LeuO-6×His-DC showed DNA-protein complexes with identical mobilities (Fig. 2D and E). LeuO-6×His-DC did not form complexes that migrated faster than those of LeuO-6×His-NC, even at higher protein concentrations, suggesting that in both cases the tetramer is the form that interacts with DNA (i.e., for LeuO-6×His-DC, the tetramer was present at very low concentration) or else that other oligomeric forms found in the LeuO-6×His-DC preparation bound with a low affinity, to result in complexes similar to those formed by the tetramer (Fig. 2E).
Curiously, DntR forms tetramers in solution and crystallizes as a homodimer (33). ArgP crystallizes as a dimer and also forms dimers in solution, but it has been proposed that it functions as a tetramer due to cooperative binding of dimers to dimers at two binding sites during DNA contact, thus enhancing the recruitment of RNA polymerase (30, 55). Other LTTRs, such as MetR, NodD3, and Nac, form functional dimers that have been shown to interact with DNA and activate or repress transcription. For these regulators, the C terminus appears to be dispensable for DNA-binding and transcriptional activity (36, 38, 40). Given this, it could be that LeuO binds to DNA as a dimer via a dimer-dimer interaction, which would be favored by the presence of DNA. Nevertheless, our findings suggest that the LeuO tetramer is the form that binds DNA in vitro (Fig. 2D and E). Therefore, LeuO is part of a subclass of LTTRs that includes NahR, TrpI, CysB, BenM, CbnR, DntR, TsaR, AphB, and ArgP, which have been shown to form homotetramers in solution. In most cases, the formation of an active homotetramer is necessary for their biological function (28–35, 51, 52).
Since the LeuO tetramer is required for DNA binding, the involvement of DBD, the C-terminal RD-I central and terminal subdomains, and the RD-II subdomain in oligomer formation was evaluated using alanine substitutions and deletions. Single alanine substitutions were constructed in six conserved residues in DBD and the RD-I central subdomain (Fig. 3). The L27A mutant (in which the mutation is located in an extended region of the N terminus), the L46A and L60A mutants (in which the mutation is within the HTH motif), and the T79A mutant (in which the mutation is proximal to the LH) had dimerization defects (Fig. 4B). As expected, these mutants were unable to activate and repress ompS1 and tpx expression, respectively (Fig. 4D and E) and to interact with DNA (data not shown).
In agreement with these results, the K42E, R60E, and L71W mutants, whose mutations were located in the DBD and LH of ArgP, were shown to be involved in the dissociation of dimers into monomers, thus supporting the interactions between the C terminus and the DBD (30).
The S54A mutant (in which the mutation was located in the HTH) and the P139A mutant (in which the mutation was located in the region corresponding to RD-I) dimerized like the wt (Fig. 4B), so these mutants are likely to form tetramers.
Despite these findings, the S54A mutant was affected in transcriptional activation and DNA binding (Fig. 4D and E). Accordingly, residue Ser54, located in the HTH motif, is one of the most highly conserved residues in the recognition helix among LTTRs (Fig. 3), and these residues have been proposed to contact the DNA major groove (29). Ser54 corresponds to Ser35 in AmpR, Ser33 in OxyR, and Ser38 in CysB and GcvA, and these residues are required for DNA binding but not oligomerization (27, 43–45). On the other hand, the P139A mutant retained some ability to activate and almost fully repress transcription (Fig. 4D and E), and accordingly, it was able to bind to DNA in vitro but with less affinity (Fig. 5). The P139 residue is located in the RD-I central subdomain (Fig. 3 and 4A), which is involved in binding and the inducer response in LTTRs (27, 56). However, it is not known whether LeuO requires an inducer in vivo. All these data show that some residues in the DBD of LeuO are involved in dimer formation, since Ala substitutions of these residues affected DNA binding and transcriptional activity. Therefore, this region could be important for dimer stability or in protein folding. Others residues in the DBD are not involved in oligomer formation yet are important for DNA binding and transcriptional function.
The C-terminal regulatory domain comprises the major portion of the LTTR proteins and contains two subdomains, RD-I and RD-II (Fig. 6A). Although the C-terminal regulatory domain is involved in oligomerization, the specific region involved has not been determined due to low levels of sequence conservation (Fig. 3). All truncated proteins in the C-terminal regulatory domain of LeuO were defective in dimerization (Fig. 6B), transcriptional activation and repression (Fig. 6D and E), and DNA binding in vitro (not shown). This held even in a mutant in which the last 5 C-terminal residues were deleted.
It was expected that the mutant with this deletion would retain the capacity to dimerize, since proteins with deletions of 5, 11, and 14 amino acids from the C termini of CysB, OxyR, and GcvA, respectively, function almost like the wild type or become constitutively active and are thus able to bind to DNA (43–45). Nevertheless, deletion of 9 residues from the C terminus of NahR severely affected transcription and DNA binding. Proteins lacking 16 or 22 residues in CysB and OxyR, respectively, were mainly affected in repression but were constitutively active. These truncated proteins formed dimers, suggesting that these regions are involved in tetramerization. A different effect was observed for mutants with a A233V point mutation in OxyR and a A227D point mutation in CysB, which still repressed and activated transcription constitutively and bound DNA as dimers (9, 31, 43–45).
All these results strongly suggest that several LysR regulators, including LeuO, function as tetramers and that the C terminus (the RD-I and RD-II subdomains) is important for proper oligomer formation.
Interestingly, a single alanine substitution in each of the last 5 residues of LeuO did not affect dimerization (Fig. 7A) or repression (Fig. 8B). This suggests that all five mutants retained the ability to bind DNA, since they were able to repress tpx. The S310A, V311A, and C312A mutants, which were affected in the ability to activate expression (Fig. 8A), could be affected in their ability to interact efficiently with the ompS1 regulatory region, like the P139A mutant, which bound DNA with less affinity (Fig. 5), or could be affected in their capacity to displace the H-NS protein to derepress expression (13). Interestingly, these residues, like P139, are located in the RD-I C-terminal and central subdomain and are part of the last LeuO helix (Fig. 3).
The activation and repression mechanisms have been shown to be different depending on the oligomeric form of the Nac regulator: as a dimer it is capable of activating transcription and binding to DNA, and it weakly represses since it binds at one site, yet it needs to form a tetramer to promote strong repressive binding at two sites of the target regulatory region (57). Nevertheless, as discussed above, the LeuO tetramer binds with a high affinity to DNA in vitro and is the oligomeric form obtained under native purification conditions (Fig. 1 and Fig. 2A). The coexistence of dimers and tetramers described for Nac was not found for LeuO-NC, but the possibility that some of the LeuO mutants studied here form dimers in vivo that retain the capacity to repress expression in vivo cannot be discarded. How these mutants are able to repress tpx expression but not efficiently activate ompS1 is a matter for future study, since the mechanisms used by LTTRs for repression of target genes that are not oriented divergently from the regulator are poorly documented (56).
By analysis of mutants with site-directed and deletion mutations, we observed that LeuO shares some features with other characterized LTTRs, such as the functionality of its DBD and the involvement of the C-terminal domain, including the last 5 residues, in oligomerization. In addition, we found that each LeuO domain is also important in oligomerization, transcriptional regulatory functions, and protein-DNA interactions. These findings suggest a possible interaction between the C terminus and the DBD which might be important for the formation and stability of the dimer and, thus, for tetramer formation. Even though the LeuO sequence alignment with other LTTRs showed the conservation of domains and residues (Fig. 3), up to this point, further research on the three-dimensional structure of LeuO should help provide an understanding of whether its domains correspond to those previously defined for other LTTRs (Fig. 3). Therefore, complex structure-function relationships in LeuO may account for its regulatory functions, acting as a repressor, antirepressor, or activator of a wide variety of nonrelated genes, the number of which is increasing (21, 22, 58).
Supplementary Material
ACKNOWLEDGMENTS
E.C. was supported by grants from CONACyT, Mexico (no. 82383 and 179946). C.G. was supported by a predoctoral fellowship from CONACyT, Mexico (no. 184646), and from IHL DGAPA-UNAM (no. 1N-203312).
We thank Dimitris Georgellis and Alejandro Huerta for technical help in protein purification and gel filtration experiments, respectively; Marcos Fernández-Mora for the anti-LeuO polyclonal antibody; Ana Lucía Gallego-Hernández for providing plasmids pKK232-9 (ompS1) and pKK232-9 (tpx); Michael Dunn for revising the manuscript; and the Oligonucleotide Synthesis Facility of the Institute of Biotechnology, UNAM, for providing the oligonucleotides used in this study.
Footnotes
Published ahead of print 21 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01484-14.
REFERENCES
- 1.Hertzberg KM, Gemmill R, Jones J, Calvo JM. 1980. Cloning of an EcoRI-generated fragment of the leucine operon of Salmonella typhimurium. Gene 8:135–152. 10.1016/0378-1119(80)90033-5 [DOI] [PubMed] [Google Scholar]
- 2.Chen D, Bowater R, Dorman CJ, Lilley DM. 1992. Activity of a plasmid-borne leu-500 promoter depends on the transcription and translation of an adjacent gene. Proc. Natl. Acad. Sci. U. S. A. 89:8784–8788. 10.1073/pnas.89.18.8784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu HY, Tan J, Fang M. 1995. Long-range interaction between two promoters: activation of the leu-500 promoter by a distant upstream promoter. Cell 82:445–451. 10.1016/0092-8674(95)90433-6 [DOI] [PubMed] [Google Scholar]
- 4.Wu HY, Fang M. 2003. DNA supercoiling and transcription control: a model from the study of suppression of the leu-500 mutation in Salmonella typhimurium topA- strains. Prog. Nucleic Acids Res. Mol. Biol. 73:43–68. 10.1016/S0079-6603(03)01002-X [DOI] [PubMed] [Google Scholar]
- 5.Henikoff S, Haughn GW, Calvo JM, Wallace JC. 1988. A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. U. S. A. 85:6602–6606. 10.1073/pnas.85.18.6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi X, Bennett GN. 1995. Effects of multicopy LeuO on the expression of the acid-inducible lysine decarboxylase gene in Escherichia coli. J. Bacteriol. 177:810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klauck E, Böhringer J, Hengge-Aronis R. 1997. The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol. Microbiol. 25:559–569. 10.1046/j.1365-2958.1997.4911852.x [DOI] [PubMed] [Google Scholar]
- 8.Ueguchi C, Ohta T, Seto C, Suzuki T, Mizuno T. 1998. The leuO gene product has a latent ability to relieve bgl silencing in Escherichia coli. J. Bacteriol. 180:190–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartowsky E, Normark S. 1991. Purification and mutant analysis of Citrobacter freundii AmpR, the regulator for chromosomal AmpC β-lactamase. Mol. Microbiol. 5:1715–1725. 10.1111/j.1365-2958.1991.tb01920.x [DOI] [PubMed] [Google Scholar]
- 10.Stratmann T, Madhusudan S, Schnetz K. 2008. Regulation of the yjjQ-bglJ operon, encoding LuxR-type transcription factors, and the divergent yjjP gene by H-NS and LeuO. J. Bacteriol. 190:926–935. 10.1128/JB.01447-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrenz MB, Miller VL. 2007. Comparative analysis of the regulation of rovA from the pathogenic yersiniae. J. Bacteriol. 189:5963–5975. 10.1128/JB.00528-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernández-Mora M, Puente JL, Calva E. 2004. OmpR and LeuO positively regulate the Salmonella enterica serovar Typhi ompS2 porin gene. J. Bacteriol. 186:2909–2920. 10.1128/JB.186.10.2909-2920.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De la Cruz MA, Fernández-Mora M, Guadarrama C, Flores-Valdez MA, Bustamante VH, Vázquez A, Calva E. 2007. LeuO antagonizes H-NS and StpA-dependent repression in Salmonella enterica ompS1. Mol. Microbiol. 66:727–743. 10.1111/j.1365-2958.2007.05958.x [DOI] [PubMed] [Google Scholar]
- 14.Hernández-Lucas I, Gallego-Hernández AL, Encarnación S, Fernández-Mora M, Martínez-Batallar AG, Salgado H, Oropeza R, Calva E. 2008. The LysR-type transcriptional regulator LeuO controls expression of several genes in Salmonella enterica serovar Typhi. J. Bacteriol. 190:1658–1670. 10.1128/JB.01649-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Medina-Aparicio L, Rebollar-Flores JE, Gallego-Hernández AL, Vázquez A, Olvera L, Gutiérrez-Ríos RM, Calva E, Hernández-Lucas I. 2011. The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica serovar Typhi. J. Bacteriol. 193:2396–2407. 10.1128/JB.01480-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallego-Hernández AL, Hernández-Lucas I, De la Cruz MA, Olvera L, Morett E, Medina-Aparicio L, Ramírez-Trujillo JA, Vázquez A, Fernández-Mora M, Calva E. 2012. Transcriptional regulation of the assT-dsbL-dsbI gene cluster in Salmonella enterica serovar Typhi IMSS-1 depends on LeuO, H-NS, and specific growth conditions. J. Bacteriol. 194:2254–2264. 10.1128/JB.06164-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11. 10.1371/journal.ppat.0020011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodríguez-Morales O, Fernández-Mora M, Hernández-Lucas I, Vázquez A, Puente JL, Calva E. 2006. Salmonella enterica serovar Typhimurium ompS1 and ompS2 mutants are attenuated for virulence in mice. Infect. Immun. 74:1398–1402. 10.1128/IAI.74.2.1398-1402.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moorthy S, Watnick PI. 2005. Identification of novel stage-specific genetic requirements through whole genome transcription profiling of Vibrio cholerae biofilm development. Mol. Microbiol. 57:1623–1635. 10.1111/j.1365-2958.2005.04797.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westra ER, Pul U, Heidrich N, Jore MM, Lundgren M, Stratmann T, Wurm R, Raine A, Mescher M, Van Heereveld L, Mastop M, Wagner EG, Schnetz K, Van Der Oost J, Wagner R, Brouns SJ. 2010. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 77:1380–1393. 10.1111/j.1365-2958.2010.07315.x [DOI] [PubMed] [Google Scholar]
- 21.Shimada T, Bridier A, Briandet R, Ishihama A. 2011. Novel roles of LeuO in transcription regulation of E. coli genome: antagonistic interplay with the universal silencer H-NS. Mol. Microbiol. 82:378–397. 10.1111/j.1365-2958.2011.07818.x [DOI] [PubMed] [Google Scholar]
- 22.Dillon SC, Espinosa E, Hokamp K, Ussery DW, Casadesús J, Dorman CJ. 2012. LeuO is a global regulator of gene expression in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 85:1072–1089. 10.1111/j.1365-2958.2012.08162.x [DOI] [PubMed] [Google Scholar]
- 23.VanBogelen RA, Olson ER, Wanner BL, Neidhardt FC. 1996. Global analysis of proteins synthesized during phosphorus restriction in Escherichia coli. J. Bacteriol. 178:4344–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang M, Wu HY. 1998. Suppression of leu-500 mutation in topA+ Salmonella typhimurium strains. The promoter relay at work. J. Biol. Chem. 273:29929–29934 [DOI] [PubMed] [Google Scholar]
- 25.Fang M, Majumder A, Tsai KJ, Wu HY. 2000. ppGpp-dependent leuO expression in bacteria under stress. Biochem. Biophys. Res. Commun. 276:64–70. 10.1006/bbrc.2000.3440 [DOI] [PubMed] [Google Scholar]
- 26.Stratmann T, Pul U, Wurm R, Wagner R, Schnetz K. 2012. RcsB-BglJ activates the Escherichia coli leuO gene, encoding an H-NS antagonist and pleiotropic regulator of virulence determinants. Mol. Microbiol. 83:1109–1123. 10.1111/j.1365-2958.2012.07993.x [DOI] [PubMed] [Google Scholar]
- 27.Schell MA. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597–626. 10.1146/annurev.mi.47.100193.003121 [DOI] [PubMed] [Google Scholar]
- 28.Muraoka S, Okumura R, Ogawa N, Nonaka T, Miyashita K, Senda T. 2003. Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J. Mol. Biol. 328:555–566. 10.1016/S0022-2836(03)00312-7 [DOI] [PubMed] [Google Scholar]
- 29.Sainsbury S, Lane LA, Ren J, Gilbert RJ, Saunders NJ, Robinson CV, Stuart DI, Owens RJ. 2009. The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators. Nucleic Acids Res. 37:4545–4558. 10.1093/nar/gkp445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X, Lou Z, Fu S, Yang A, Shen H, Li Z, Feng Y, Bartlam M, Wang H, Rao Z. 2010. Crystal structure of ArgP from Mycobacterium tuberculosis confirms two distinct conformations of full-length LysR transcriptional regulators and reveals its function in DNA binding and transcriptional regulation. J. Mol. Biol. 396:1012–1024. 10.1016/j.jmb.2009.12.033 [DOI] [PubMed] [Google Scholar]
- 31.Schell MA, Brown PH, Raju S. 1990. Use of saturation mutagenesis to localize probable functional domains in the NahR protein, a LysR-type transcription activator. J. Biol. Chem. 265:3844–3850 [PubMed] [Google Scholar]
- 32.Hryniewicz MM, Kredich NM. 1994. Stoichiometry of binding of CysB to the cysJIH, cysK, and cysP promoter regions of Salmonella typhimurium. J. Bacteriol. 176:3673–3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smirnova IA, Dian C, Leonard GA, McSweeney S, Birse D, Brzezinski P. 2004. Development of a bacterial biosensor for nitrotoluenes: the crystal structure of the transcriptional regulator DntR. J. Mol. Biol. 340:405–418. 10.1016/j.jmb.2004.04.071 [DOI] [PubMed] [Google Scholar]
- 34.Monferrer D, Tralau T, Kertesz MA, Dix I, Sola M, Uson I. 2010. Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold. Mol. Microbiol. 75:1199–1214. 10.1111/j.1365-2958.2010.07043.x [DOI] [PubMed] [Google Scholar]
- 35.Taylor JL, De Silva RS, Kovacikova G, Lin W, Taylor RK, Skorupski K, Kull FJ. 2012. The crystal structure of AphB, a virulence gene activator from Vibrio cholerae, reveals residues that influence its response to oxygen and pH. Mol. Microbiol. 83:457–470. 10.1111/j.1365-2958.2011.07919.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maxon ME, Wigboldus J, Brot N, Weissbach H. 1990. Structure-function studies on Escherichia coli MetR protein, a putative prokaryotic leucine zipper protein. Proc. Natl. Acad. Sci. U. S. A. 87:7076–7079. 10.1073/pnas.87.18.7076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsek MR, Shinabarger DL, Rothmel RK, Chakrabarty AM. 1992. Roles of CatR and cis,cis-muconate in activation of the catBC operon, which is involved in benzoate degradation in Pseudomonas putida. J. Bacteriol. 174:7798–7806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher RF, Long SR. 1993. Interactions of NodD at the nod box: NodD binds to two distinct sites on the same face of the helix and induces a bend in the DNA. J. Mol. Biol. 233:336–348. 10.1006/jmbi.1993.1515 [DOI] [PubMed] [Google Scholar]
- 39.Bender RA. 1991. The role of the NAC protein in the nitrogen regulation of Klebsiella aerogenes. Mol. Microbiol. 5:2575–2580. 10.1111/j.1365-2958.1991.tb01965.x [DOI] [PubMed] [Google Scholar]
- 40.Muse WB, Bender RA. 1999. The amino-terminal 100 residues of the nitrogen assimilation control protein (NAC) encode all known properties of NAC from Klebsiella aerogenes and Escherichia coli. J. Bacteriol. 181:934–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thöny B, Hwang DS, Fradkin L, Kornberg A. 1991. iciA, an Escherichia coli gene encoding a specific inhibitor of chromosomal initiation of replication in vitro. Proc. Natl. Acad. Sci. U. S. A. 88:4066–4070. 10.1073/pnas.88.10.4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gong W, Xiong G, Maser E. 2012. Oligomerization and negative autoregulation of the LysR-type transcriptional regulator HsdR from Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 132:203–211. 10.1016/j.jsbmb.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 43.Kullik I, Stevens J, Toledano MB, Storz G. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for DNA binding and multimerization. J. Bacteriol. 177:1285–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lochowska A, Iwanicka-Nowicka R, Plochocka D, Hryniewicz MM. 2001. Functional dissection of the LysR-type CysB transcriptional regulator. Regions important for DNA binding, inducer response, oligomerization, and positive control. J. Biol. Chem. 276:2098–2107. 10.1074/jbc.M007192200 [DOI] [PubMed] [Google Scholar]
- 45.Jourdan AD, Stauffer GV. 1998. Mutational analysis of the transcriptional regulator GcvA: amino acids important for activation, repression, and DNA binding. J. Bacteriol. 180:4865–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flores-Valdez MA, Puente JL, Calva E. 2003. Negative osmoregulation of the Salmonella ompS1 porin gene independently of OmpR in an hns background. J. Bacteriol. 185:6497–6506. 10.1128/JB.185.22.6497-6506.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez-Laguna Y, Calva E, Puente JL. 1999. Autoactivation and environmental regulation of bfpT expression, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 33:153–166. 10.1046/j.1365-2958.1999.01460.x [DOI] [PubMed] [Google Scholar]
- 48.Joo WA, Speicher DW. 2007. Protein detection in gels without fixation. Curr. Protoc. Protein Sci. Chapter 10:Unit 10.6. 10.1002/0471140864.ps1006s48 [DOI] [PubMed] [Google Scholar]
- 49.Daines DA, Silver RP. 2000. Evidence for multimerization of Neu proteins involved in polysialic acid synthesis in Escherichia coli K1 using improved LexA-based vectors. J. Bacteriol. 182:5267–5270. 10.1128/JB.182.18.5267-5270.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dmitrova M, Younes-Cauet G, Oertel-Buchheit P, Porte D, Schnarr M, Granger-Schnarr M. 1998. A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in Escherichia coli. Mol. Gen. Genet. 257:205–212. 10.1007/s004380050640 [DOI] [PubMed] [Google Scholar]
- 51.Bundy BM, Collier LS, Hoover TR, Neidle EL. 2002. Synergistic transcriptional activation by one regulatory protein in response to two metabolites. Proc. Natl. Acad. Sci. U. S. A. 99:7693–7698. 10.1073/pnas.102605799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruangprasert A, Craven SH, Neidle EL, Momany C. 2010. Full-length structures of BenM and two variants reveal different oligomerization schemes for LysR-type transcriptional regulators. J. Mol. Biol. 404:568–586. 10.1016/j.jmb.2010.09.053 [DOI] [PubMed] [Google Scholar]
- 53.Knapp GS, Hu JC. 2009. The oligomerization of CynR in Escherichia coli. Protein Sci. 18:2307–2315. 10.1002/pro.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knapp GS, Tsai JW, Hu JC. 2009. The oligomerization of OxyR in Escherichia coli. Protein Sci. 18:101–107. 10.1002/pro.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee Y, Lee H, Yim J, Hwang D. 1997. The binding of two dimers of IciA protein to the dnaA promoter 1P element enhances the binding of RNA polymerase to the dnaA promoter 1P. Nucleic Acids Res. 25:3486–3489. 10.1093/nar/25.17.3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maddocks SE, Oyston PC. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609–3623. 10.1099/mic.0.2008/022772-0 [DOI] [PubMed] [Google Scholar]
- 57.Rosario CJ, Bender RA. 2005. Importance of tetramer formation by the nitrogen assimilation control protein for strong repression of glutamate dehydrogenase formation in Klebsiella pneumoniae. J. Bacteriol. 187:8291–8299. 10.1128/JB.187.24.8291-8299.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hernández-Lucas I, Calva E. 2012. The coming of age of the LeuO regulator. Mol. Microbiol. 85:1026–1028. 10.1111/j.1365-2958.2012.08175.x [DOI] [PubMed] [Google Scholar]
- 59.Clamp M, Cuff J, Searle SM, Barton GJ. 2004. The Jalview Java alignment editor. Bioinformatics 20:426–427. 10.1093/bioinformatics/btg430 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.