Abstract
Acidithiobacillus ferrooxidans is a major participant in consortia of microorganisms used for bioleaching. It can obtain energy from the oxidation of Fe2+, H2, S0, and various reduced inorganic sulfur compounds (RISCs). Tetrathionate is a key intermediate during RISC oxidation, hydrolyzed by tetrathionate hydrolase (TetH), and used as sole energy source. In this study, a tetH knockout (ΔtetH) mutant and a tetH overexpression strain were constructed and characterized. The tetH overexpression strain grew better on sulfur and tetrathionate and possessed a higher rate of tetrathionate utilization and TetH activity than the wild type. However, its cell yields on tetrathionate were much lower than those on sulfur. The ΔtetH mutant could not grow on tetrathionate but could proliferate on sulfur with a lower cell yield than the wild type's, which indicated that tetrathionate hydrolysis is mediated only by TetH, encoded by tetH. The ΔtetH mutant could survive in ferrous medium with an Fe2+ oxidation rate similar to that of the wild type. For the tetH overexpression strain, the rate was relatively higher than that of the wild type. The reverse transcription-quantitative PCR (qRT-PCR) results showed that tetH and doxD2 acted synergistically, and doxD2 was considered important in thiosulfate metabolism. Of the two sqr genes, AFE_0267 seemed to play as important a role in sulfide oxidation as AFE_1792. This study not only provides a substantial basis for studying the function of the tetH gene but also may serve as a model to clarify other candidate genes involved in sulfur oxidation in this organism.
INTRODUCTION
Acidithiobacillus ferrooxidans is an extremely acidophilic (pH 2), chemolithoautotrophic bioleaching bacterium of great importance in the biomining industry. It derives energy from the oxidation of hydrogen, ferrous iron, sulfur, or reduced inorganic sulfur compounds (RISCs) for fixation of the carbon dioxide and nitrogen used for synthesizing sugar, protein, nucleic acid, and other important compounds necessary for its growth. In addition, it can grow in the absence of oxygen with ferric iron as the electron acceptor and sulfur as the electron donor. The unusual metabolism and physiological characteristics of A. ferrooxidans make it not only widely applied in the biomining industry but also very attractive to study theoretically from a fundamental biological point of view. Until now, research on its ferrous iron oxidation mechanism have achieved profound results (1–3), and many metabolic models have been posted, among which Quatrini's model, reconstructed by microarray transcript profiling, reverse transcription-quantitative PCR (qRT-PCR) analysis, molecular biology, and biochemistry, was widely recognized (4, 5). However, the study of RISC metabolism in A. ferrooxidans is more complex and complicated due to the involvement of multiple oxidation states of sulfur (from −2 to +6) and some nonenzymatic reactions. The few predicted models were based mainly on transcriptomic data and bioinformatic analysis and lack sufficient support from experimental data (4–6).
Tetrathionate is a key intermediate during RISC oxidation, and it can also be used as the sole energy source for the growth of A. ferrooxidans. Tetrathionate can be hydrolyzed to thiosulfate, sulfate, and sulfur by tetrathionate hydrolase (TetH) (7–9). Compared with TetH from the sulfur-oxidizing strain Acidithiobacillus caldus (10, 11), TetH from A. ferrooxidans was speculated to be phylogenetically distinct (11, 12) and even diverse in its catalytic reaction on account of different cofactor requirements (10, 13). In addition, the subcellular location of TetH in A. ferrooxidans has not been certain until now because it has been reported as a periplasmic (8), outer membrane-associated (12, 14), and extracellular (15) protein. In any case, much progress has been made with regard to the tetH gene and TetH of A. ferrooxidans, such as the identification of the tetH gene, successful refolding of recombinant TetH, and crystallization of TetH (12, 13, 16). However, the detailed function of tetH and its association with other related genes in sulfur metabolism are still not completely understood.
The gene knockout technique has long been a convincing tool for research on gene functions and other studies in a variety of organisms (17–19). On account of the difficulties of genetic manipulation in acidophilic chemolithoautotrophic microorganisms, there have been only three reports on the construction of gene knockout mutants in the strains of Acidithiobacillus (20–22). In 2000, Liu et al. constructed a recA mutant of A. ferrooxidans ATCC 33020 by marker exchange mutagenesis, which first provided a method of investigating the functions of target genes by null mutation in A. ferrooxidans (20). In 2008, van Zyl et al. constructed A. caldus arsB and tetH mutants by the same method. The inhibition of the growth of an A. caldus tetH mutant on tetrathionate was detected; however, no further analyses on the characteristics of the mutant were carried out (21). Moreover, the strategy in the construction of the two above-described knockout mutants was based on marker exchange in that the kanamycin resistance marker was eventually introduced and integrated into the host genomes. This marker exchange mutagenesis not only is a potential threat to biosafety but also cannot be applied to the construction of multiple mutations in one strain, as the selectable markers available for Acidithiobacillus in an acidic environment are quite limited. In 2012, a markerless gene replacement system was constructed based on the homing endonuclease I-SceI in A. ferrooxidans, and the function of the pfkB gene was characterized by comparing the properties of the pfkB knockout mutant with those of the wild type (22). It provided us with a powerful tool to investigate the physiological functions of candidate genes involved in the complex sulfur metabolic pathways in A. ferrooxidans.
In this study, we describe a tetH knockout (ΔtetH) mutant constructed with the markerless gene replacement system and a tetH overexpression strain of A. ferrooxidans. The function of the tetH gene in A. ferrooxidans was characterized by comparing the properties of the ΔtetH mutant with those of the wild type and the tetH overexpression strain grown on different substrates; we examined the growth, enzymatic activities, and transcriptional analysis of related genes involved in sulfur oxidation by qRT-PCR. This study can not only provide a substantial base for investigating the function of the tetH gene in A. ferrooxidans but also serve as a model for future research on other related genes involved in the cascade of sulfur oxidation in this organism.
MATERIALS AND METHODS
Media, bacterial strains, and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were cultured at 37°C in Luria-Bertani (LB) broth with shaking at 180 rpm or on LB agar plates with kanamycin (50 μg/ml) or streptomycin (50 μg/ml) added as required. The strains of A. ferrooxidans were cultured at 30°C with shaking at 180 rpm in 9K basal salts solution (23) adjusted to pH 2.0 with H2SO4 supplemented with 3.3% (wt/vol) FeSO4·7H2O, 1% (wt/vol) S0 or 0.5% (wt/vol) K2S4O6 as an energy source. The concentrated solution of FeSO4·7H2O and K2S4O6 was filtered and added aseptically at the time of inoculation, and S0 was sterilized by intermittent boiling for at least 3 h and added after inoculation.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| A. ferrooxidans | ||
| ATCC 23270 | Type strain | ATCC |
| ΔAF0029 | ATCC 23270 ΔtetH | This study |
| E. coli | ||
| JM109 | recA1 endA1 gryA96 thi-1 hsdR17 (rK− mK+) supE44 relA1λ− lac (F′ proAB lacIq ZΔM15) | 39 |
| DH5αλpir | F− hsdR17 thi-1 gyrA Δ(lacZYA-argF) supE44 recA1ϕ80dΔ(lacZ)M15 relA λpir | 40 |
| S17-1λpir | Tpr Smr recA thi pro hsdR hsdM+ RP4::2-Tc::Mu::Km Tn7λpir lysogen | 41 |
| SM10 | Kmr thi-1 thr leu tonA lacY supE recA::RP4-2–Tc::Mu | 42 |
| Plasmids | ||
| pUC19 | Apr, ColE1 replicon, cloning vector | TaKaRa |
| pKIT | Kmr, R6K replicon, oriTRP4, I-SceI site | 22 |
| pKIT-HA1 | pKIT containing HA1 | This study |
| pKIT-KtetH | pKIT containing homologous sequences flanking the tetH gene from A. ferrooxidans ATCC 23270 | This study |
| pMSD1 | Smr, pBBR1 replicon, mob positive | 43 |
| pMSD1-I-SceI | pMSD1 containing the I-SceI gene | 22 |
| pUC19-probe | pUC19 containing the probe sequence | This study |
| pUC18 | Apr, ColE1 replicon, cloning vector | TaKaRa |
| pEXT20 | Apr, ColE1 replicon, tac promoter, lacIq | 44 |
| pEXT20-tetH | pEXT20 containing the tetH gene from A. ferrooxidans ATCC 23270 | This study |
| pUC18-tac-tetH | pUC18 containing the tetH gene under the control of the tac promoter | This study |
| pMSD1-tac-tetH | pMSD1 containing the tetH gene under the control of the tac promoter | This study |
The compositions of mating medium and solid 2:2 medium for culturing A. ferrooxidans were described previously (24). Kanamycin (100 μg/ml) and streptomycin (100 μg/ml) were used for selection in A. ferrooxidans on solid 2:2 medium plates, and kanamycin (300 μg/ml) and streptomycin (300 μg/ml) were used in 9K liquid medium as required.
Genetic manipulations.
General molecular biological techniques, including the restriction enzyme digestion, ligation, gel electrophoresis, and transformation of a plasmid, were performed as described elsewhere (25). The isolation of genomic DNA from cells of A. ferrooxidans ATCC 23270, the isolation of plasmids, and the recovery of DNA fragments from agarose gels were performed by using, respectively, a bacterial DNA kit, a plasmid minikit I, and a gel extraction kit of Omega Bio-Tek according to the manufacturer's instructions. Restriction enzymes and T4 DNA ligase were purchased from TaKaRa and used as recommended. Primer synthesis and DNA sequencing reactions were performed by the Shanghai BioSune Biotechnology Company. The Southern blot analysis was performed with Amersham Hybond-N+ positively charged nylon transfer membranes (GE Healthcare) and a digoxigenin (DIG)-High Prime DNA labeling and detection starter kit I (Roche) and accomplished according to the manufacturer's instructions.
Construction of the tetH gene knockout mutant of A. ferrooxidans ATCC 23270.
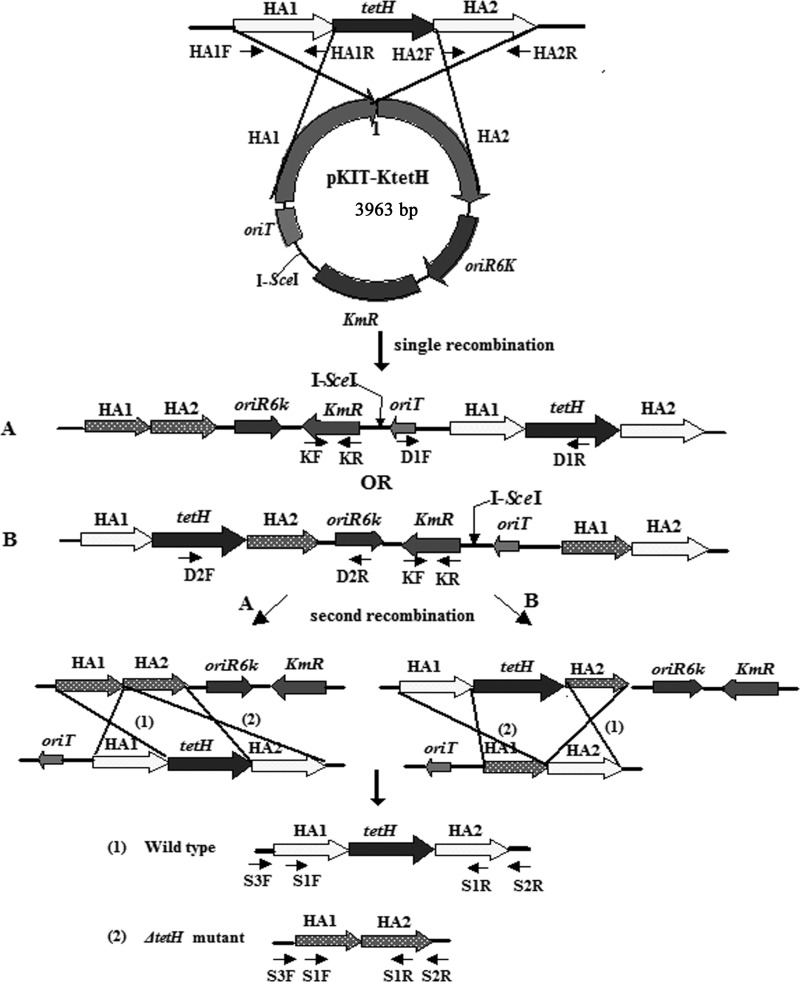
The plasmids involved in the construction of the tetH gene knockout mutant are listed in Table 1, and the designed primers are listed in Table S1 in the supplemental material. A schematic chart for the construction of the ΔtetH mutant of A. ferrooxidans ATCC 23270 is shown in Fig. 1.
FIG 1.
Schematic chart for the construction of the tetH knockout mutant. The suicide plasmid pKIT-KtetH was introduced into A. ferrooxidans ATCC 23270 and integrated into the genome, producing either one of the two types of single recombinants (A and B). Then plasmid pMSD1-I-SceI, carrying the I-SceI gene, was introduced into the single recombinant, resulting in a double-stranded break at the I-SceI site and stimulating the occurrence of the second recombination. As a result, either a tetH gene knockout mutant or a restoration of the wild type was generated, and the kanamycin-resistant gene (KmR) was simultaneously eliminated from the chromosome. The positions of the primers used to identify various recombinants are indicated.
(i) Construction of suicide plasmid pKIT-KtetH.
Plasmid pKIT was used as the backbone for constructing the suicide vector pKIT-KtetH. Two pairs of primers (HA1F/HA1R and HA2F/HA2R) were synthesized to amplify homologous arms 1 and 2 (HA1 and HA2) flanking the tetH gene from the A. ferrooxidans ATCC 23270 genome (GenBank accession number NC_011761). The amplified HA1 sequence (positions 30709 to 31710) and HA2 sequence (positions 32974 to 33982) were first cloned into plasmid pUC19 for sequencing after digestion with EcoRI and HindIII, respectively. Then, HA1 and HA2 were obtained from their sequenced recombinant plasmids by digestion with EcoRI/XbaI and XbaI/SacI, respectively, and inserted into the corresponding sites in pKIT to produce pKIT-KtetH, which was used to inactivate the tetH gene of A. ferrooxidans ATCC 23270.
(ii) Selection of single crossovers.
The suicide plasmid pKIT-KtetH was transformed into E. coli S17-1λpir under the selection of kanamycin resistance, and the resulting E. coli S17-1λpir cells containing pKIT-KtetH were used as donors. The cells of A. ferrooxidans ATCC 23270 were applied as recipients. Conjugation conditions were modified with Peng et al.'s method (24). The donor cells were cultivated in liquid LB medium with kanamycin until exponential phase, and the recipient cells were grown in 9K-S0 medium until stationary phase. Both the donor and recipient cells were collected by centrifugation (6,000 × g, 5 min), washed three times with 2:2 basal salt buffer, and then mixed at a ratio of approximately 1:1 to 1:∼2. Then 100 μl of the cell suspension (approximately 108 cells) was spotted onto the sterilized filter membrane (0.45-μm pore size, 25 mm in diameter) placed on the mating medium. After incubation at 30°C for 5 to 7 days, the cells on the filter membrane were resuspended and diluted with 2:2 basal salt buffer and then plated on 2:2 solid plates with or without kanamycin. After incubation at 30°C for 15 days, colonies grown on 2:2 plates were potential single-crossover mutants. As a control for spontaneous mutation, the recipient cells were plated on the same plates with kanamycin.
(iii) Verification of single-recombination events.
Colonies grown on 2:2 plates with kanamycin were picked randomly, inoculated in 10 ml of 9K-Fe2+ medium, and cultivated at 30°C for 5 days for recovery of growth. Then they were transferred to 9K-S0 medium with kanamycin and cultivated at 30°C for 5 to 7 days. Cells at late exponential phase were collected for extraction of genomic DNA, which was applied as the template for PCR verification of single-recombination events. Primer pair KF/KR was used to amplify a partial sequence of the aph gene for kanamycin resistance, and primer pairs D1F/D1R and D2F/D2R were used to verify the two types (A and B in Fig. 1) of single crossovers.
(iv) Screening for the tetH gene knockout mutant.
Plasmid pMSD1-I-SceI, which contained a streptomycin resistance gene and the I-SceI gene, was constructed previously (22). It was introduced into E. coli SM10 by transformation, and the transformed cells containing pMSD1-I-SceI were used as donors for conjugation. Single recombinant cells of A. ferrooxidans ATCC 23270 were used as recipients. The same conjugation procedure as mentioned above was performed, except that incubation at 30°C lasted for 48 to 72 h for the mobilization of pMSD1-I-SceI. To select transconjugants, the cells were plated onto 2:2 solid plates with or without streptomycin. After 15 days of incubation at 30°C, single colonies from 2:2 solid plates with streptomycin were picked and cultured in 10 ml of 9K-Fe2+ medium at 30°C for 5 days for recovery of growth, followed by subculture in 9K-S0 medium containing streptomycin for 5 to 7 days at 30°C. The cells were collected by centrifugation (6,000 × g, 5 min) for extraction of genomic DNA, which was applied as the template for PCR verification. Primer pairs S1, S2, and S3 correspond to primers S1F and S1R, S1F and S2R, and S3F and S2R, respectively, which were employed in the PCR verification to screen for the double-recombination mutants of A. ferrooxidans ATCC 23270.
(v) Elimination of plasmid pMSD1-I-SceI from double-recombination mutants.
The target mutants were consecutively subcultured in 9K-S0 medium without streptomycin at least 3 times and then plated on 2:2 medium without streptomycin. Single colonies were picked and grown again in 9K-S0 medium, and then the cells were harvested for PCR verification to screen for the spontaneous loss of plasmid pMSD1-I-SceI. Primers SMF and SMR were used to amplify the streptomycin resistance gene fragment in pMSD1-I-SceI, so as to verify the elimination of plasmid pMSD1-I-SceI.
(vi) Southern blot analysis.
To confirm the genotype of the ΔtetH mutant, Southern blot analysis was carried out with genomic DNA digested with EcoRV. The probe was designed by using a partial fragment of the downstream homologous arm, HA2, flanking the tetH gene from the A. ferrooxidans ATCC 23270 genome and was amplified with primers ProbeF and ProbeR. After digestion with EcoRI, the amplified 662-bp fragment was inserted into the corresponding site in pUC19, to give pUC19-probe for sequencing. The 650-bp EcoRI fragment digested from pUC19-probe was isolated and used as the probe for Southern blot analysis.
Construction of the tetH gene overexpression strain of A. ferrooxidans ATCC 23270.
The strains and plasmids used to construct the tetH gene overexpression strain of A. ferrooxidans ATCC 23270 are listed in Table 1. The 1,717-bp DNA fragment containing the whole tetH gene (positions 31528 to 33232) with its signal peptide sequence was amplified with primers TF and TR by PCR using A. ferrooxidans ATCC 23270 genomic DNA as the template. After digestion with KpnI and BamHI, the amplified DNA fragment was inserted into the corresponding cloning sites (downstream of the tac promoter) in the vector pEXT20, producing pEXT20-tetH. The expression of the tetH gene under the control of the tac promoter was verified with SDS-PAGE. Then the 1,872-bp tac-tetH fragment was amplified with primers TTF and TR by PCR with pEXT20-tetH as the template. After digestion with PstI and BamHI, the amplified fragment was inserted into the corresponding cloning sites of vector pUC18 to produce pUC18-tac-tetH for DNA sequencing and then subcloned from pUC18-tac-tetH into the corresponding cloning sites of the broad-range-host vector pMSD1, to produce pMSD1-tac-tetH. The transfer of plasmid pMSD1-tac-tetH from E. coli SM10 into A. ferrooxidans ATCC 23270 was performed by using the same conjugation method as described above for plasmid pMSD1-I-SceI. The expression of the tetH gene in pMSD1-tac-tetH in E. coli was detected by SDS-PAGE.
Characterization of the ΔtetH mutant and tetH overexpression strain. (i) Growth properties.
The A. ferrooxidans ATCC 23270 wild type, ΔtetH mutant, and tetH overexpression strain were cultivated in 9K basal salts solution with ferrous iron (Fe2+), sulfur (S0), and tetrathionate (S4O62−) as energy resources. Similar amounts of initial inoculum (approximately 106 cells) of each strain were employed for comparison. Growth in 9K-S0 and 9K-S4O62− was monitored by measuring the optical density at 600 nm after the removal of elemental sulfur by low-speed centrifugation (400 × g, 5 min). Growth in 9K-Fe2+ was monitored by measuring the decrease in the ferrous iron concentration by the o-phenanthroline method (26). The decrease in the S4O62− concentration in 9K-S4O62− medium was measured by the method of cyanolysis (27, 28), which can also indicate the growth of the bacteria.
(ii) TetH activity.
Cells at mid-log phase grown in 9K-S4O62− medium were collected by centrifugation at 13,800 × g for 10 min, washed twice with 0.1 M potassium acetate buffer (pH 3.0), and resuspended in the same buffer. The cells were sonicated intermittently on ice for 20 min. After centrifugation at 13,800 × g for 10 min, the cell debris was discarded and the supernatant was applied as TetH crude extract. Then, a 1-ml reaction mixture was prepared in 1.5-ml Eppendorf tubes, which contained 1 mM K2S4O6, 200 mM K2SO4, 0.1 M acetate buffer (pH 3.0), and 100 μl TetH crude extract. The reaction was initiated by adding the TetH crude extract at 30°C and lasted for 60 min. Then, the reaction tubes were immediately placed in cold ethanol (−20°C), with vibration for 2 min, followed by boiling for 90 s to stop the reaction. The samples were centrifuged at 13,800 × g for 1 min to precipitate the elemental sulfur produced in the reaction. The concentration of tetrathionate remaining in the supernatant was measured by the method of cyanolysis (27, 28). One unit of crude TetH activity (U) was defined as the amount of protein required for the hydrolysis of 1 μmol S4O62− per min.
RNA extraction and qRT-PCR.
Cells at mid-log phase grown in 9K basal salts media with different energy sources were collected by centrifugation, and each independent culture was performed with at least three replicates. Total RNA was extracted by using the RNeasy minikit from Qiagen according to the manufacturer's instructions. DNA was eliminated from the samples by incubating them with RNase-free DNase I (Fermentas) for an appropriate time, and the absence of contaminating DNA was determined by PCR. After we guaranteed the RNA quality and concentration by agarose gel electrophoresis and with a NanoDrop-1000 spectrophotometer (NanoDrop Technologies), cDNA was generated by using a RevertAid first-strand cDNA synthesis kit (Fermentas) for qRT-PCR analysis, which was performed with a LightCycler 480 (Roche) by using SYBR Premix Ex Taq from TaKaRa in triplicate. The thermal cycling conditions were 95°C for 2 min and 40 cycles of 95°C for 20 s, 54°C for 20 s, and 72°C for 15 s, followed by a final extension at 72°C for 2 min and subsequent melting curve analysis. Primers designed for qRT-PCR are listed in Table S1 in the supplemental material (from 29F to 2558R). Two genes, alaS and map, were applied for normalization (29). Relative gene expression values were calculated by using the method of Pfaffl (30).
RESULTS
Construction of the tetH knockout mutant of A. ferrooxidans ATCC 23270.
A markerless gene knockout system was successfully developed in A. ferrooxidans as described by Wang et al. (22). In order to verify the biological functions of the tetH gene in A. ferrooxidans, the construction of a ΔtetH mutant was conceived based on this system. As shown in Fig. 1, the suicide plasmid pKIT-KtetH was first generated by inserting two homologous fragments (HA1 and HA2) flanking the tetH gene into the multiple cloning sites of plasmid pKIT and then transferred into A. ferrooxidans ATCC 23270 by conjugation. Plasmid pKIT-KtetH cannot replicate in A. ferrooxidans ATCC 23270 cells but can be integrated into the genome by homologous recombination via either of plasmid-containing HA1 or HA2 to generate the single-recombination event (two types), which can be selected under the pressure of kanamycin.
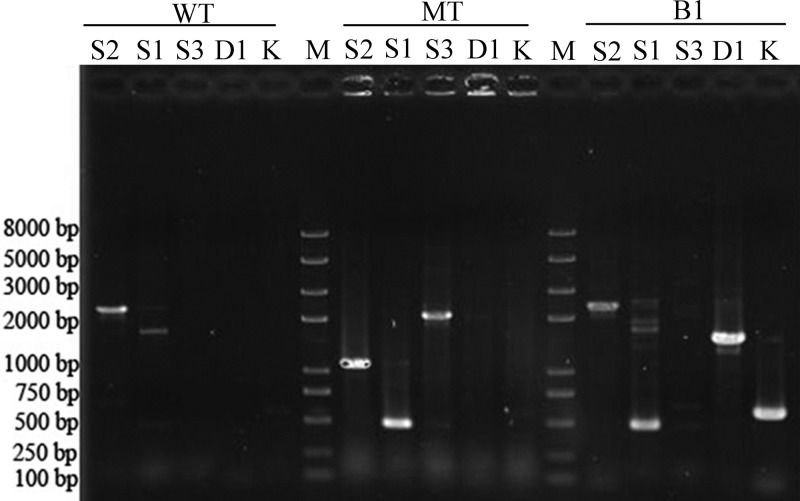
Ten colonies that grew on selective kanamycin plates were randomly picked and cultivated for the identification of a single recombination. A 571-bp fragment could be amplified from the kanamycin resistance aph gene by PCR using primer pair KF/KR in all tested colonies, which proved the occurrence of a single-recombination event in these colonies. Among the 10 colonies, one named A. ferrooxidans B1 was chosen for further identification by PCR using primer pairs D1F/D1R and D2F/D2R. As shown in Fig. 2, a 1,533-bp fragment could be amplified by using primer pair D1F/D1R, which indicated that A. ferrooxidans B1 was the type A single recombinant (Fig. 1).
FIG 2.
PCR analyses of the wild type, single recombinant B1, and ΔtetH mutant of A. ferrooxidans ATCC 23270. WT, MT, and B1 represent the wild type, ΔtetH mutant, and single recombinant B1 of A. ferrooxidans ATCC 23270. S1, S2, S3, D1, and K represent the primer pairs S1F/S1R, S1F/S2R, S3F/S2R, D1F/D1R, and KF/KR, respectively. The numbers on the left indicate the sizes of the fragments based on the molecular size marker (lane M).
According to the strategy of the markerless gene knockout technique, a second recombination event could be stimulated by I-SceI. So, the broad-host-range plasmid pMSD1-I-SceI containing the I-SceI gene, which was constructed previously (22), was transferred from E. coli SM10 to A. ferrooxidans B1 by conjugation. Once transferred to cells of A. ferrooxidans B1, plasmid pMSD1-I-SceI could replicate and stably exist in the transconjugant under the selection pressure of streptomycin. During the growth of A. ferrooxidans B1 harboring pMSD1-I-SceI, the I-SceI expressed by the I-SceI gene from pMSD1-I-SceI could stimulate the event of double recombination in host cells. Since the second recombination event could occur at any time during cell growth and no selectable marker could be employed to concentrate the double-crossover strains, the screening of the ΔtetH mutant had to be performed from a large number of colonies that grew on selective streptomycin plates by PCR. By using the specific primer pairs, different fragments would be amplified with the genomic DNA from the ΔtetH mutant, the wild type, or the single recombinant B1 (see Table S2 in the supplemental material).
One potential mutant was finally screened out among 277 colonies, which was subsequently purified by multiple subculturings alternately in liquid and solid media without streptomycin. During this period, plasmid pMSD1-I-SceI could be lost spontaneously, and the potential double-crossover ΔtetH mutant lacking pMSD1-I-SceI was obtained as a result.
Identification and genetic analysis of the ΔtetH mutant of A. ferrooxidans ATCC 23270.
The identification of the potential ΔtetH mutant of A. ferrooxidans ATCC 23270 was first carried out by PCR. By using primer pairs described in Table S2 and the genomic DNA of the ΔtetH mutant as the template, the fragments amplified by PCR were analyzed and compared with the results obtained by using the genomic DNA of the wild type and of the single recombinant B1 as the template (Fig. 2). As shown in Fig. 2, an 1,122-bp fragment was amplified from the ΔtetH mutant and a larger fragment (2,379 bp) was amplified from the wild type or the single recombinant B1 by using primer pair S2. While using primer pair S1, a 466-bp fragment from the ΔtetH mutant and a 1,723-bp fragment from the wild type were obtained, and the expected 466- and 1,723-bp fragments were detected from the single recombinant B1. When primer pair S3 was employed, a 2,106-bp fragment could be amplified only by using the genomic DNA from the ΔtetH mutant as the template (Fig. 2). The theoretically predicted long fragments (>3,000 bp) in Table S2 for each primer pair in the wild type and single recombinant B1 were too big to be amplified under the conditions used and could not be detected in Fig. 2. All the amplified fragments were in accordance with the expected sizes described in Table S2. The corresponding amplified fragments were also verified by sequencing.
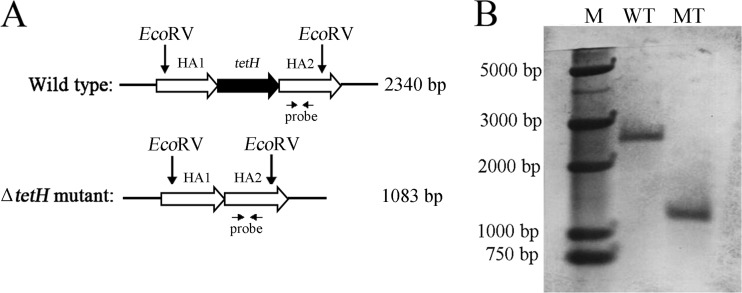
Next, the identification of the potential ΔtetH mutant of A. ferrooxidans ATCC 23270 was performed by Southern blot hybridization (Fig. 3). As shown in Fig. 3A, the partial fragment of HA2 was designed as probe. After being labeled with digoxigenin, the 650-bp EcoRI fragment digested from pUC19-probe was used to probe the EcoRV-digested chromosomal DNA from the wild type and ΔtetH mutant. An expected 2,340-bp band was obtained with the wild type, and a 1,083-bp band was obtained with the mutant (Fig. 3B), which suggested that the tetH gene was successfully knocked out of the genome in the ΔtetH mutant of A. ferrooxidans ATCC 23270.
FIG 3.
Identification of the ΔtetH mutant of A. ferrooxidans ATCC 23270 by Southern blotting. (A) Schematic chart of the genomic region bearing the tetH gene and showing the position of the probe and the sites of EcoRV. The sizes between the two EcoRV sites are indicated on the right. (B) Southern blot analyses of EcoRV-digested genomic DNA from the wild type (WT) and the ΔtetH mutant (MT) of A. ferrooxidans ATCC 23270. Lane M represents the DNA molecular markers, and the sizes of the fragments are indicated on the left.
Construction of the tetH gene overexpression strain of A. ferrooxidans ATCC 23270.
Since the tetH promoter in A. ferrooxidans has not been characterized, the expression of the tetH gene from A. ferrooxidans ATCC 23270 in E. coli cells was first tried by amplifying a fragment containing a longer upstream sequence (∼300 bp) of the tetH gene. By inserting the amplified tetH fragment from the genome of A. ferrooxidans ATCC 23270 into pUC19, the recombinant plasmid pUC19-tetH was constructed. However, no obvious target band in the cell extract of E. coli JM109 containing pUC19-tetH could be detected with SDS-PAGE analysis. The result indicated that the promoter might be absent in the amplified fragment or that some regulators might be absent or not active in E. coli. Thus, the powerful tac promoter was substituted for its own unconfirmed promoter and the construction of plasmid pMSD1-tac-tetH was carried out as described in Materials and Methods. Expression of the tetH gene under the control of the tac promoter could easily be detected by SDS-PAGE in the cell extract of E. coli JM109 containing pUC18-tac-tetH (data not shown). Then, plasmid pMSD1-tac-tetH was transferred from E. coli SM10 into A. ferrooxidans ATCC 23270 by conjugation, and the tetH gene overexpression strain, A. ferrooxidans ATCC 23270 containing pMSD1-tac-tetH, was obtained from the solid 2:2 medium with streptomycin. The existence of plasmid pMSD1-tac-tetH in the overexpression strain was identified by the successful amplification of part of the mob gene, which exists only in plasmid pMSD1-tac-tetH (data not shown).
Characterization of the tetH knockout mutant and tetH overexpression strain.
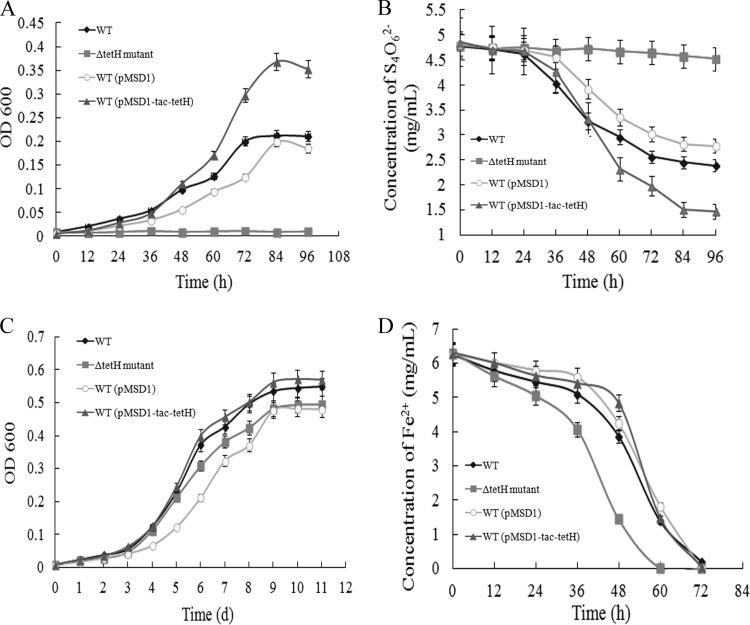
The growth properties of the wild type, ΔtetH mutant, and tetH overexpression strain of A. ferrooxidans ATCC 23270 were investigated and compared when grown in 9K-S4O62−, 9K-S0, and 9K-Fe2+ media (Fig. 4). As streptomycin (300 μg/ml) was added to the medium for cultivation of the tetH overexpression strain, A. ferrooxidans ATCC 23270 containing plasmid pMSD1 was used as its control in the experiments. As shown in Fig. 4A, no growth was detected with the ΔtetH mutant when it was cultivated in S4O62− medium, whereas a significant growth increment was observed with the tetH overexpression strain, compared with the growth not only of the wild type carrying plasmid pMSD1 but also of the wild type without the plasmid (Fig. 4A). The complete inhibition of the cell growth of the ΔtetH mutant with tetrathionate as the sole energy source suggested that TetH is the unique enzyme that decomposes tetrathionate in A. ferrooxidans. The remarkable stimulation of cell growth with the tetH overexpression strain may also prove the effect of TetH on the utilization of tetrathionate. However, the cell yields of all strains were obviously lowered by using S4O62− rather than S0 as the energy source under the conditions used (Fig. 4).
FIG 4.
Characteristics of the wild type, ΔtetH mutant, and tetH overexpression strain of A. ferrooxidans ATCC 23270 grown in 9K basal salts medium to which were added different energy sources. (A) Growth in 9K-S4O62− medium; (B) concentration of S4O62− in 9K-S4O62− medium; (C) growth in 9K-S0 medium; (D) ferrous oxidation in 9K-Fe2+ medium. WT, ΔtetH mutant, WT (pMSD1), and WT (pMSD1-tac-tetH) represent the wild type, ΔtetH mutant, wild type carrying plasmid pMSD1, and tetH overexpression strain of A. ferrooxidans ATCC 23270, respectively. OD 600 indicates the optical density at 600 nm. All the measurements were performed in triplicate. Each data point is given as an arithmetic mean. d, days.
As the decrease of S4O62− in the medium may reflect the growth capacity of the strains, the concentrations of S4O62− during cell growth were monitored and are shown in Fig. 4B. Almost no consumption of S4O62− was detected with the ΔtetH mutant during the whole cultivation process. As expected, the highest hydrolysis of S4O62− occurred in the tetH overexpression strain, whose cell yield was the highest as well. The decreases in the S4O62− concentration (Fig. 4B) were in accordance with the increased cell yields (Fig. 4A) for all strains.
When grown in S0 medium, the ΔtetH mutant showed a relatively reduced growth capacity compared with that of the wild type (Fig. 4C). The addition of streptomycin greatly decreased cell growth, and the wild type containing plasmid pMSD1 grew poorest; however, the tetH overexpression strain grew best among all strains, though streptomycin was also added to its medium (Fig. 4C). The results indicated that the deletion of the tetH gene, which was predicted to play a key role in tetrathionate metabolism, could not completely impede cell growth with S0 as the energy source but could result in a reduced growth capacity. The overexpression of the tetH gene obviously stimulated cell growth of the tetH overexpression strain compared with that of the wild type harboring pMSD1, but only a slight increase in cell yield could be detected compared with that of the wild type without the plasmid. It indicated that the stimulation of cell growth by the overexpression of one gene involved in sulfur metabolism could not greatly enhance the efficiency of sulfur utilization.
The oxidation of Fe2+ by all strains grown in 9K-Fe2+ medium was evaluated, and levels were compared (Fig. 4D). The Fe2+ oxidation rates of the ΔtetH mutant and the wild type were similar, while the lag time to the initiation of Fe2+ oxidation was shorter for the mutant. For the tetH overexpression strain, the Fe2+ oxidation rate was relatively higher than that of the wild type, but the lag time was slightly longer. TetH is a key enzyme involved in sulfur metabolism, and its influence on ferrous oxidation needs further study and discussion.
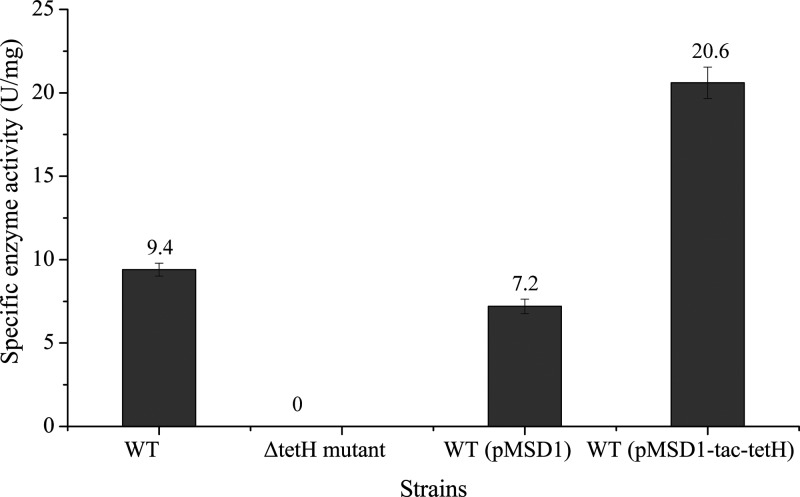
The TetH enzyme activity of the cell extract from all strains grown in 9K-S4O62− medium was measured, and the highest activity was detected with the tetH overexpression strain (Fig. 5). The result for each strain was in accordance with its growth and tetrathionate consumption (Fig. 4). Due to the nondetectable growth of the ΔtetH mutant, its enzyme activity was presumed to be zero. However, by introducing plasmid pMSD1-tac-tetH into the ΔtetH mutant to complement the tetH gene by the same conjugation method used with wild-type A. ferrooxidans, the ΔtetH knockout mutant harboring pMSD1-tac-tetH could grow in 9K-S4O62− and possessed a TetH activity of 17.6 ± 1.1 U/mg in the cell extract. All the results indicated that tetH is the very gene that encodes the TetH responsible for the decomposition of tetrathionate. The TetH activity of the cell extract from all strains grown in 9K-S0 medium was also determined, and no positive result could be obtained. The absent activity might be due to the too-small amount of TetH in cells when elemental sulfur was used as the energy source.
FIG 5.
Specific activities of TetH in the cell extracts of different strains grown in 9K-S4O62− medium. WT, ΔtetH mutant, WT (pMSD1), and WT (pMSD1-tac-tetH) represent the wild type, ΔtetH mutant, wild type carrying plasmid pMSD1, and tetH overexpression strain of A. ferrooxidans ATCC 23270, respectively.
Transcriptional analysis of the tetH knockout mutant and tetH overexpression strain.
The transcriptional responses to the knockout or overexpression of the tetH gene were further investigated when sulfur (S0), tetrathionate (S4O62−), or ferrous iron (Fe2+) was used as the energy source. A subset of genes involved in sulfur metabolism in A. ferrooxidans ATCC 23270 was selected (listed in Table 2), and the relative mRNA levels of these genes were determined by qRT-PCR analysis against the wild type (Table 2). As shown in Table 2, when cultivated in 9K-S0 and 9K-Fe2+ media, the mRNA of tetH was not detectable in the ΔtetH mutant, which is in accordance with the previous genetic determination. However, in the tetH overexpression strain, the higher level of tetH expression than in the wild type, whatever the growth conditions, was expected due to a higher copy number of the tetH gene and the tetH gene under the control of Ptac in the plasmid.
TABLE 2.
Relative mRNA levels of a subset of selected genes involved in sulfur metabolism in response to the knockout of tetH and tetH overexpressiona
| Gene | Locus | Gene description | Log2 ratio medianb |
|||||
|---|---|---|---|---|---|---|---|---|
| 9K-S0 |
9K-K2S4O6 |
9K-FeSO4 |
||||||
| A | B | A | B | A | B | |||
| tetH | AFE_0029 | Tetrathionate hydrolase | —* | 3* | — | 12.7* | — | 2.3* |
| doxD2 | AFE_0044 | DoxD-like family protein | −1.8* | 2.6* | — | 10.4* | −1.6* | 1.4 |
| doxD1 | AFE_0048 | DoxD-like family protein | −0.6 | 0.9 | — | 1.5* | −0.9 | 0.3 |
| sqr | AFE_0267 | Sulfide quinone reductase, putative | −1.4 | 2.3* | — | 1.3 | −0.6 | −1.2 |
| sdo | AFE_0269 | Metallo-beta-lactamase family protein | 1.2 | −0.2 | — | −0.5 | −1.1 | −0.4 |
| sat | AFE_0539 | Sulfate adenylyltransferase, putative adenylylsulfate kinase | 1.3 | −0.6 | — | 1.6* | −0.2 | −0.6 |
| sqr | AFE_1792 | Sulfide quinone reductase, putative | −1.0 | 1.9* | — | 0.9 | −0.5 | −0.7 |
| hdrB | AFE_2554 | Heterodisulfide reductase subunit B, homolog | −1.1 | 1.7* | — | 1.1 | −1.1 | −0.9 |
| rhd | AFE_2558 | Rhodanase-like domain protein | −1.4 | 1.5* | — | 1.5* | −0.9 | −0.5 |
Determined by qRT-PCR analysis against the wild-type A. ferrooxidans ATCC 23270 cultivated in different media.
A, the ΔtetH mutant of A. ferrooxidans; B, the tetH overexpression strain of A. ferrooxidans; —, no mRNA of the relevant genes was detected in the ΔtetH mutant. Genes for which the log2 ratio of the median was larger than |1.5| (corresponding to genes induced more than 2.8-fold) are considered differentially expressed (indicated with *). The locus of a relevant gene and its description are those of the GenBank genome annotation of NC_011761. The negative numbers represent downregulation, while positive numbers represent upregulation. The qRT-PCRs were performed in triplicate, with at least three independent biological replicates.
When cultured in 9K-K2S4O6 medium, the mRNAs of all selected genes, including tetH, were not detected in the ΔtetH mutant due to no growth (Table 2). In the tetH overexpression strain cultivated in 9K-K2S4O6 medium, the genes tetH, doxD2, doxD1, sat, and rhd were upregulated, and among them, we detected much higher mRNA levels of tetH and doxD2. The relative mRNA levels of the two genes were also much higher than those in 9K-S0 medium. Tetrathionate can be hydrolyzed by TetH to thiosulfate, sulfate, and sulfur. The doxD genes, encoding thiosulfate:quinone oxidoreductase, were predicted to be involved in thiosulfate decomposition (5). So, the results indicated that the transcription of tetH and doxD2 could be induced by tetrathionate and thiosulfate. The transcription levels of the genes predicted to function in elemental sulfur metabolism, such as sqr and hdrB, did not show obvious changes in 9K-K2S4O6 medium compared with that in 9K-S0 medium, which might suggest that these genes do not play crucial roles in tetrathionate metabolism.
For the tetH overexpression strain cultured in 9K-S0 medium (Table 2), the transcription of tetH, doxD2, the two sqr genes, hdrB, and rhd were upregulated, among which tetH and doxD2 might be driven by the overexpression of the tetH gene from plasmid pMSD1-tac-tetH, resulting in the speedup of the hydrolysis of tetrathionate and the production of thiosulfate and sulfur. The lesser extent of increased expression of tetH and doxD2 compared with that in 9K-K2S4O6 medium could be explained by the lower concentration of thiosulfate produced during the metabolism of sulfur. Accordingly, transcription of doxD2 was downregulated in the ΔtetH mutant. Of the two doxD genes, doxD2 showed positive signals, consistently with the knockout or overexpression of the tetH gene, while no apparent changes were detected in doxD1 expression. This indicated that the doxD2 gene might play a main part in thiosulfate decomposition. In addition, the upregulation of the transcription of the two sqr genes, hdrB, and rhd in the tetH overexpression strain cultured in 9K-S0 medium demonstrated that TetH might play an indirect role in the transcriptional regulation of the genes involved in RISC oxidation. As for the two sqr genes, they behaved the same way but with slightly larger changes for AFE_0267 than for AFE_1792. However, most of the research on sulfide metabolism has been focused on AFE_1792 (31–34). In our study, the qRT-PCR data indicated that AFE_0267 might also play a key role in H2S metabolism after elemental sulfur enters the cells.
The relative mRNA levels of the selected genes of the tested strains cultivated in 9K-Fe2+ medium were also investigated (Table 2). As expected, no mRNA of tetH was detected in the ΔtetH mutant, and upregulated transcription of tetH was detected in the tetH overexpression strain. Most of the genes did not show obvious changes in transcriptional level, suggesting that the two energy source-metabolic systems of sulfur and ferrous iron in A. ferrooxidans ATCC 23270 are relatively independent.
DISCUSSION
It is well established that A. ferrooxidans can obtain energy by the oxidation of ferrous iron or RISCs for their chemolithoautotrophic growth in acidic environments. The study on oxidation of RISCs by A. ferrooxidans has proved particularly challenging, and the proposed models of sulfur metabolism were built based merely on the bioinformatic predictions of its genome sequence, coupled with analyses of its transcriptomic or proteomic experimental data (4, 5). The functions of most genes involved in the oxidation of RISCs remain uncertain, though some genes have been characterized by cloning and expressing them in Escherichia coli (12, 13, 32, 36). Considering the marked differences in the growth requirements between the genera Acidithiobacillus and Escherichia, the periplasmic or outer membrane proteins of A. ferrooxidans, such as some enzymes and electron carriers related to sulfur oxidation, might find it hard to function in E. coli. So, the most effective method relies mainly on direct proof of functional identification by the knockout and overexpression of a certain gene in A. ferrooxidans itself. Unfortunately, to the best of our knowledge, no progress on this problem has been reported until now due to the difficulties of genetic manipulation in A. ferrooxidans.
The markerless gene deletion method which was newly established for the extremely acidophilic organism A. ferrooxidans (22) was used in the construction of the tetH knockout mutant. The potential single recombinants of A. ferrooxidans were screened with little effort after the first transfer of a suicide plasmid due to the existence of a selectable marker of kanamycin integrated into the chromosome. However, the double-recombination events may occur randomly during cell growth, which makes it really laborious to obtain potential double-recombination mutants among the transconjugants without any selectable markers. Considering the difficulties of making replicate plates of A. ferrooxidans colonies, the negative-selection method was incapable of convenient employment in A. ferrooxidans. An improved method for screening double-recombination mutants based on colony color has been established lately in our lab and will be applied in the knockout of other genes of A. ferrooxidans.
Although it was generally considered very difficult to return a certain gene to the extremely acidophilic organism A. ferrooxidans (6, 37, 38), the tetH gene was successfully introduced into A. ferrooxidans ATCC 23270 by conjugation, and an engineering strain overexpressing the tetH gene under the control of the powerful tac promoter from a broad-host-range plasmid, pMSD1-tac-tetH, was constructed. Therefore, except for the elimination in function by being knocked out, the tetH gene could also be elucidated by its increase in function on account of its overexpression. In this study, the significant growth increment of the tetH overexpression strain, the lack of growth of the ΔtetH mutant, and the restored growth of the ΔtetH mutant complemented with the tetH gene were detected by using tetrathionate (S4O62−) as the sole energy source, which provided sufficient proof that tetH acts as the sole gene to encode the tetrathionate hydrolase (TetH) for the decomposition of tetrathionate in A. ferrooxidans ATCC 23270.
Since TetH was thought to be an induced enzyme involved in RISC oxidation (12), little research has focused on its role in Fe2+ oxidation. It was reported that TetH could show a tetrathionate-dependent Fe2+-producing activity due to the enzymatic catalysis of tetrathionate hydrolysis followed by a chemical Fe3+ reduction, to give Fe2+ by the produced thiosulfate (14). Although the evidence that TetH could be purified from Fe2+-grown cells proved the expression of tetH in Fe2+ medium (14), the relation between TetH and Fe2+ oxidation is still not clear. Therefore, the influence on Fe2+ oxidation by knocking out and overexpressing the tetH gene in A. ferrooxidans needs further study.
It is well established that multiple genes are involved in the sulfur-metabolic pathways of A. ferrooxidans, among which the tetH gene takes part just in the dissimilation of tetrathionate, an intermediate during sulfur oxidation (4–6, 38). The corresponding reduced and increased growth obtained, respectively, by the ΔtetH mutant and the tetH overexpression strain when grown in 9K-S0 medium clearly indicated that TetH influenced the whole metabolism of sulfur to some extent. The existence of TetH in sulfur-grown A. ferrooxidans cells has been proved, but the procedures resulted in approximately 93-fold purification (14), indicating a quite small amount of TetH in sulfur-grown cells. This might be the reason why we failed to detect the TetH activities of strains grown on sulfur. A similar negative result was also reported for Acidithiobacillus caldus, in which TetH activity was not detected in the crude enzyme extract when the organism was cultivated in liquid medium with elemental sulfur as the energy source (10). As TetH is thought to be responsible for the hydrolysis of tetrathionate to thiosulfate, sulfur, and sulfate, the upregulation of doxD2 in the tetH overexpression strain and its downregulation in the ΔtetH mutant according to qRT-PCR analysis indicated that the predicted doxD2 operon (5, 36) might play an important role in thiosulfate metabolism in A. ferrooxidans. This could be supported by the identification of the doxD2 cotranscribed gene AFE_0042, which encodes the thiosulfate dehydrogenase responsible for the enzymatic metabolism of thiosulfate (36). With regard to the two sqr genes, AFE_0267 seemed to play as important a role in sulfide oxidation as AFE_1792 according to qRT-PCR analysis, while only AFE_1792 was reported to be upregulated among sulfur-induced genes (5). Moreover, AFE_0267 is much closer to AFE_0269, a newly identified sdo gene encoding sulfur dioxygenase (35) in the genome of A. ferrooxidans ATCC 23270 in our studies (unpublished data). So, the function of AFE_0267 needs further elucidation. In any case, the genes selected by qRT-PCR analysis were quite limited, and the influence on the whole metabolic network by deletion of the tetH gene still needs further transcriptomic analysis.
In conclusion, this is the first report on the characterization of the tetH gene by construction of a tetH overexpression strain and a tetH knockout mutant of A. ferrooxidans. The successful construction of the ΔtetH mutant of A. ferrooxidans demonstrates that the markerless gene deletion technology is no doubt a useful and stable tool for clarifying the functions of certain genes by knocking them out in A. ferrooxidans. This study on the tetH gene could be considered a model for investigating other candidate genes involved in the whole sulfur oxidation network, including genes that might completely block sulfur oxidation through application of ferrous iron as an energy source, and relevant research is under way in our lab.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation (31370084), the National Basic Research Program (2010CB630902), and the Key Scientific and Technological Project (2010GSF10626) of Shandong Province, People's Republic of China.
Footnotes
Published ahead of print 11 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01472-13.
REFERENCES
- 1.Bonnefoy V, Holmes DS. 2012. Genomic insights into microbial iron oxidation and iron uptake strategies in extremely acidic environments. Environ. Microbiol. 14:1597–1611. 10.1111/j.1462-2920.2011.02626.x [DOI] [PubMed] [Google Scholar]
- 2.Bird LJ, Bonnefoy V, Newman DK. 2011. Bioenergetic challenges of microbial iron metabolisms. Trends Microbiol. 19:330–340. 10.1016/j.tim.2011.05.001 [DOI] [PubMed] [Google Scholar]
- 3.Bruscella P, Appia-Ayme C, Levican G, Ratouchniak J, Jedlicki E, Holmes DS, Bonnefoy V. 2007. Differential expression of two bc1 complexes in the strict acidophilic chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans suggests a model for their respective roles in iron or sulfur oxidation. Microbiology 153:102–110. 10.1099/mic.0.2006/000067-0 [DOI] [PubMed] [Google Scholar]
- 4.Quatrini R, Appia-Ayme C, Denis Y, Ratouchniak J, Veloso F, Valdes J, Lefimil C, Silver S, Roberto F, Orellana O, Denizot F, Jedlicki E, Holmes DS, Bonnefoy V. 2006. Insights into the iron and sulfur energetic metabolism of Acidithiobacillus ferrooxidans by microarray transcriptome profiling. Hydrometallurgy 83:263–272. 10.1016/j.hydromet.2006.03.030 [DOI] [Google Scholar]
- 5.Quatrini R, Appia-Ayme C, Denis Y, Jedlicki E, Holmes DS, Bonnefoy V. 2009. Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10:394. 10.1186/1471-2164-10-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valdes J, Pedroso I, Quatrini R, Dodson RJ, Tettelin H, Blake R, II, Eisen JA, Holmes DS. 2008. Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics 9:597. 10.1186/1471-2164-9-597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meulenberg R, Pronk JT, Hazeu W, Bos P, Kuenen JG. 1992. Oxidation of reduced sulfur compounds by intact cells of Thiobacillus acidophilus. Arch. Microbiol. 157:161–168 [Google Scholar]
- 8.de Jong GAH, Hazeu W, Bos P, Kuenen JG. 1997. Polythionate degradation by tetrathionate hydrolase of Thiobacillus ferrooxidans. Microbiology 143:499–504. 10.1099/00221287-143-2-499 [DOI] [PubMed] [Google Scholar]
- 9.de Jong, GAH. Hazeu W, Bos P, Kuenen JG. 1997. Isolation of tetrathionate hydrolase from Thiobacillus acidophilus. Eur. J. Biochem. 243:678–683. 10.1111/j.1432-1033.1997.00678.x [DOI] [PubMed] [Google Scholar]
- 10.Bugaytsova Z, Lindstrom EB. 2004. Localization, purification and properties of a tetrathionate hydrolase from Acidithiobacillus caldus. Eur. J. Biochem. 271:272–280. 10.1046/j.1432-1033.2003.03926.x [DOI] [PubMed] [Google Scholar]
- 11.Rzhepishevska OI, Valdes J, Marcinkeviciene L, Gallardo CA, Meskys R, Bonnefoy V, Holmes DS, Dopson M. 2007. Regulation of a novel Acidithiobacillus caldus gene cluster involved in metabolism of reduced inorganic sulfur compounds. Appl. Environ. Microbiol. 73:7367–7372. 10.1128/AEM.01497-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanao T, Kamimura K, Sugio T. 2007. Identification of a gene encoding a tetrathionate hydrolase in Acidithiobacillus ferrooxidans. J. Biotechnol. 132:16–22. 10.1016/j.jbiotec.2007.08.030 [DOI] [PubMed] [Google Scholar]
- 13.Kanao T, Matsumoto C, Shiraga K, Yoshida K, Takada J, Kamimura K. 2010. Recombinant tetrathionate hydrolase from Acidithiobacillus ferrooxidans requires exposure to acidic conditions for proper folding. FEMS Microbiol. Lett. 309:43–47. 10.1111/j.1574-6968.2010.02019.x [DOI] [PubMed] [Google Scholar]
- 14.Sugio T, Taha TM, Takeuchi F. 2009. Ferrous iron production mediated by tetrathionate hydrolase in tetrathionate-, sulfur-, and iron-grown Acidithiobacillus ferrooxidans ATCC 23270 cells. Biosci. Biotechnol. Biochem. 73:1381–1386. 10.1271/bbb.90036 [DOI] [PubMed] [Google Scholar]
- 15.Beard S, Paradela A, Albar JP, Jerez CA. 2011. Growth of Acidithiobacillus ferrooxidans ATCC 23270 in thiosulfate under oxygen-limiting conditions generates extracellular sulfur globules by means of a secreted tetrathionate hydrolase. Front. Microbiol. 2:79. 10.3389/fmicb.2011.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanao T, Kosaka M, Yoshida K, Nakayama H, Tamada T, Kuroki R, Yamada H, Takada J, Kamimura K. 2013. Crystallization and preliminary X-ray diffraction analysis of tetrathionate hydrolase from Acidithiobacillus ferrooxidans. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 69:692–694. 10.1107/S1744309113013419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zarschler K, Janesch B, Zayni S, Schaffer C, Messner P. 2009. Construction of a gene knockout system for application in Paenibacillus alvei CCM 2051T, exemplified by the S-layer glycan biosynthesis initiation enzyme Wsfp▽. Appl. Environ. Microbiol. 75:3077–3085. 10.1128/AEM.00087-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camp S, Zhang L, Krejci E, Dobbertin A, Bernard V, Girard E, Duysen EG, Lockridge O, Jaco AD, Taylor P. 2010. Contributions of selective knockout studies to understanding cholinesterase disposition and function. Chem. Biol. Interact. 187:72–77. 10.1016/j.cbi.2010.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar R, Shimizu K. 2011. Transcriptional regulation of main metabolic pathways of cyoA, cydB, fnr, and fur gene knockout Escherichia coli in C-limited and N-limited aerobic continuous cultures. Microb. Cell Fact. 10:3–17. 10.1186/1475-2859-10-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Guiliani N, Appia-Ayme C, Borne F, Ratouchniak J, Bonnefoy V. 2000. Construction and characterization of a recA mutant of Thiobacillus ferrooxidans by marker exchange. J. Bacteriol. 182:2269–2276. 10.1128/JB.182.8.2269-2276.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Zyl LJ, Munster JM, Rawlings DE. 2008. Construction of arsB and tetH mutants of the sulfur-oxidizing bacterium Acidithiobacillus caldus by marker exchange. Appl. Environ. Microbiol. 74:5686–5694. 10.1128/AEM.01235-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Liu X, Liu S, Yu Y, Lin J, Lin J, Pang X, Zhao J. 2012. Development of a markerless gene replacement system for Acidithiobacillus ferrooxidans and construction of a pfkB mutant. Appl. Environ. Microbiol. 78:1826–1835. 10.1128/AEM.07230-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Silverman MP, Lundgren DG. 1959. Studies on the chemoautotrophic iron bacterium Ferrobacillus ferrooxidans. J. Bacteriol. 77:642–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng JB, Yan WM, Bao XZ. 1994. Plasmid and transposon transfer to Thiobacillus ferrooxidans. J. Bacteriol. 176:2892–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26.Herrera L, Ruiz P, Aguillon JC, Fehrmann A. 1989. A new spectrophotometric method for the determination of ferrous iron in the presence of ferric iron. J. Chem. Technol. Biotechnol. 44:171–181 [Google Scholar]
- 27.Kelly DP, Chambers LA, Trudinger PA. 1969. Cyanolysis and spectrophotometric estimation of trithionate in mixture with thiosulfate and tetrathionate. Anal. Chem. 41:898–902. 10.1021/ac60276a029 [DOI] [Google Scholar]
- 28.Nor YM, Tabatabai MA. 1975. Colorimetric determination of microgram quantities of thiosulfate and tetrathionate. Anal. Lett. 8:537–547. 10.1080/00032717508058238 [DOI] [Google Scholar]
- 29.Nieto P, Covarrubias P, Jedlicki E, Holmes D, Quatrini R. 2009. Selection and evaluation of reference genes for improved interrogation of microbial transcriptomes: case study with the extremophile Acidithiobacillus ferrooxidans. BMC Mol. Biol. 10:63–73. 10.1186/1471-2199-10-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfaffl M. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wakai S, Kikumoto M, Kanao T, Kamimura K. 2004. Involvement of sulfide:quinone oxidoreductase in sulfur oxidation of an acidophilic iron-oxidizing bacterium, Acidithiobacillus ferrooxidans NASF-1. Biosci. Biotechnol. Biochem. 68:2519–2528. 10.1271/bbb.68.2519 [DOI] [PubMed] [Google Scholar]
- 32.Wakai S, Tsujita M, Kikumoto M, Manchur MA, Kanao T, Kamimura K. 2007. Purification and characterization of sulfide:quinone oxidoreductase from an acidophilic iron-oxidizing bacterium, Acidithiobacillus ferrooxidans. Biosci. Biotechnol. Biochem. 71:2735–2742. 10.1271/bbb.70332 [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Cherney MM, Solomonson M, Liu J, James MN, Weiner JH. 2009. Preliminary X-ray crystallographic analysis of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65:839–842. 10.1107/S1744309109027535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cherney MM, Zhang Y, Solomonson M, Weiner JH, James MN. 2010. Crystal structure of sulfide:quinone oxidoreductase from Acidithiobacillus ferrooxidans: insights into sulfidotrophic respiration and detoxification. J. Mol. Biol. 398:292–305. 10.1016/j.jmb.2010.03.018 [DOI] [PubMed] [Google Scholar]
- 35.Rohwerder T, Sand W. 2003. The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 149:1699–1709. 10.1099/mic.0.26212-0 [DOI] [PubMed] [Google Scholar]
- 36.Kikumoto M, Nogami S, Kanao T, Takada J, Kamimura K. 2013. Tetrathionate-forming thiosulfate dehydrogenase from the acidophilic, chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 79:113–120. 10.1128/AEM.02251-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmes D, Bonnefoy V. 2007. Genetic and bioinformatic insights into iron and sulfur oxidation mechanisms of bioleaching organisms, p 281–307 In Rawlings D, Johnson B. (ed), Biomining. Springer-Verlag, Berlin, Germany [Google Scholar]
- 38.Valdes J, Pedroso I, Quatrini R, Holmes DS. 2008. Comparative genome analysis of Acidithiobacillus ferrooxidans, A. thiooxidans and A. caldus: insights into their metabolism and ecophysiology. Hydrometallurgy 94:180–184. 10.1016/j.hydromet.2008.05.039 [DOI] [Google Scholar]
- 39.Nomura CT, Taguchi K, Gan Z, Kuwabara K, Tanaka T, Takase K, Doi Y. 2005. Expression of 3-ketoacyl-acyl carrier protein reductase (fabG) genes enhances production of polyhydroxyalkanoate copolymer from glucose in recombinant Escherichia coli JM109. Appl. Environ. Microbiol. 71:4297-4306. 10.1128/AEM.71.8.4297-4306.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin X, Wheatcroft R, Chambers JR, Liu B, Zhu J, Gyles CL. 2009. Contributions of O island 48 to adherence of enterohemorrhagic Escherichia coli O157: H7 to epithelial cells in vitro and in ligated pig ileal loops. Appl. Environ. Microbiol. 75:5779–5786. 10.1128/AEM.00507-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kienesberger S, Gorkiewicz G, Joainig MM, Scheicher SR, Leitner E, Zechner EL. 2007. Development of experimental genetic tools for Campylobacter fetus. Appl. Environ. Microbiol. 73:4619–4630. 10.1128/AEM.02407-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- 43.Meng J, Wang H, Liu X, Lin J, Pang X, Lin J. 2013. Construction of small plasmid vectors for use in genetic improvement of the extremely acidophilic Acidithiobacillus caldus. Microbiol. Res. 168:469–476. 10.1016/j.micres.2013.04.003 [DOI] [PubMed] [Google Scholar]
- 44.Dykxhoorn DM, Pierre RS, Linn T. 1996. A set of compatible tac promoter expression vectors. Gene 177:133–136. 10.1016/0378-1119(96)00289-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.